Abstract
Until recently, mass spectrometry (MS) was not often associated with the analysis of protein conformation and dynamics but rather as a method to measure molecular weight and quantify molecules. However, by taking advantage of labeling methods such as hydrogen exchange (HX), many details about protein conformation, dynamics and interactions can be revealed by mass spectrometry. In the current work we provide an update that covers hydrogen exchange theory as it applies to HX MS protocols, explain in detail the practice of HX MS including data analysis and interpretation, and highlight recent advancements in technology which greatly increase the depth of information gained from the technique.
Key terms for indexing: proteins, mass spectrometry, hydrogen exchange
INTRODUCTION
Until recently, mass spectrometry (MS) was not often associated with the analysis of protein conformation and dynamics but rather as a method to measure molecular weight and quantify molecules. However, by taking advantage of labeling methods such as hydrogen exchange (HX), many details about protein conformation, dynamics and interactions can be revealed by mass spectrometry. In the current work we provide an update that covers hydrogen exchange theory as it applies to HX MS protocols, explain in detail the practice of HX MS including data analysis and interpretation, and highlight recent advancements in technology which greatly increase the depth of information gained from the technique.
The hydrogen exchange phenomenon, whereby labile hydrogens in proteins exchange positions with hydrogens in the surrounding solvent, has been known for some time (Hvidt and Linderstrom-Lang, 1954; Hvidt and Nielsen, 1966; Englander and Poulsen, 1969). HX occurs all the time for all proteins but can go unnoticed unless an isotope of hydrogen other than protium (H) is introduced. Tritium oxide (T2O) was employed at one time for labeling, but now deuterium oxide (D2O) is mostly used. Any spectroscopic technique or experimental method capable of distinguishing between the isotopes of hydrogen can be used to measure the exchange, including measurements of density, radioactivity, NMR, infrared spectroscopy or mass spectrometry.
The deuterium atom (D) has a mass of 2.014 amu whereas protium (H) has a mass of 1.008 amu; therefore, if a protein were to be deuterated, its mass would increase slightly. Measuring the mass of a deuterated protein with a mass spectrometer indicates how much deuterium was incorporated. The first demonstrated use of HX MS came shortly after the development of electrospray ionization (Chowdhury et al., 1990; Katta and Chait, 1991). An extension of the method was described by Prof. David Smith and developed throughout the 1990s (Zhang and Smith, 1993; Smith et al., 1997). Hydrogen exchange mass spectrometry has now been the subject of several comprehensive reviews (Hoofnagle et al., 2003; Wales and Engen, 2006b; Tsutsui and Wintrode, 2007; Brier and Engen, 2008). Recent developments in this field offer unparalleled limits of detection, low sample consumption requirements, the promise of single amino acid resolution, potential for automation and the ability to analyze increasingly complex mixtures of proteins.
HYDROGEN EXCHANGE BACKGROUND
The theory of hydrogen exchange itself is fairly well understood and has been reviewed extensively (Woodward et al., 1982; Englander and Kallenbach, 1983). The following section highlights basic hydrogen exchange principles which are important for HX MS experimental design and data analysis.
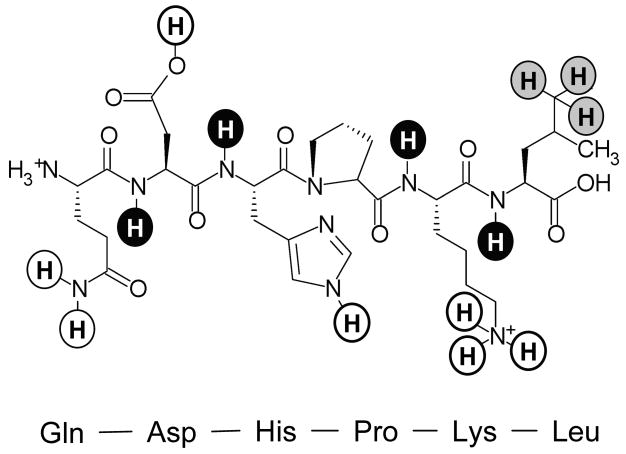
Proteins contain a number of hydrogens (Figure 1). Hydrogens bonded to carbon essentially do not exchange. The exchangeable hydrogens in sidechains can become deuterated but their exchange rates are too rapid to be measured by MS (Englander et al., 1985; Bai et al., 1993) and they are purposefully exchanged back to hydrogen during analysis (Zhang and Smith, 1993) (see also below). The most interesting hydrogens for analysis by MS are the backbone amide hydrogens. The exchange rates of these hydrogens are in a range that can be followed by MS. Conveniently, every amino acid has a backbone amide hydrogen (except for proline) meaning that each amino acid can be used as an exchange sensor. It is also the backbone amide hydrogens that hold secondary structural elements (α-helicies and β-sheets) together.
Figure 1.
Types of hydrogens in proteins. The six residue peptide Gln-Asp-His-Pro-Lys-Leu illustrates backbone amide hydrogens (black), non-exchangeable hydrogens bonded to carbon (grey) and exchangeable hydrogens which cannot be monitored with HX MS because their rates are too fast (white). In this six residue peptide, there are only four backbone amide hydrogens that can be monitored by MS as proline does not have a backbone amide hydrogen and the exchange rates of the N-terminus amide hydrogens are similar to those of hydrogens in sidechains, such that they cannot be measured by MS.
The exchange rate of amide hydrogens is primarily dictated by four factors: pH, temperature, solvent accessibility, and intramolecular hydrogen bonding. Given a particular temperature and pH, the exchange rate for each amide hydrogen in an unstructured peptide can be predicted based on its sequence (Molday et al., 1972; Bai et al., 1993). The primary structure of a peptide may affect amide hydrogen exchange rates by as much as 30-fold due to the sidechains on neighboring residues. Secondary, tertiary, and quaternary structure can decrease the rate of exchange at individual amide positions by a factor of 108. This drastic reduction in exchange rate is what allows HX to be a sensitive probe for conformational change.
Hydrogen exchange is both acid- and base-catalyzed and the rate of exchange for any hydrogen is given by:
| Equation 1 |
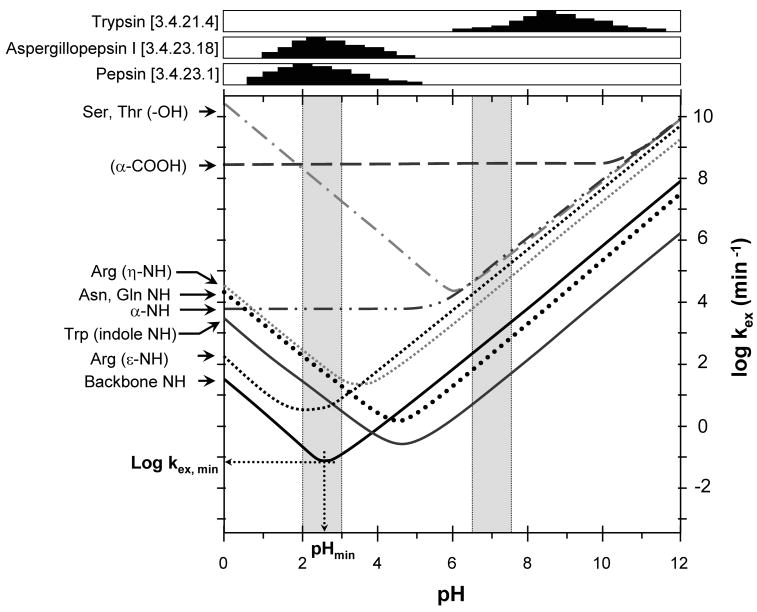
where kint is the intrinsic exchange rate constant and kH and kOH are the acid and base catalyzed exchange rate constants, respectively. At physiological pH, where most labeling experiments are performed (see Figure 2), the base-catalyzed exchange mechanism is dominant. The pH minimum for exchange of the average backbone amide hydrogen is around 2.5, but will vary slightly depending on the neighboring residues. All other types of hydrogens (side chain groups, etc) exchange at least two orders of magnitude faster at this pH. The connection between the rate of exchange and the pH is a key aspect that makes an HX MS experiment possible, as described below.
Figure 2.
Exchange rates for various types of hydrogens as a function of pH. The data shown are for hydrogens in unstructured peptides. Labeling (pH ~7.0) and quench (pH ~2.5) regions are highlighted in grey. Activity (arbitrary units) of trypsin and the acid proteases aspergillopepsin I and porcine pepsin (EC numbers are noted) as a function of pH are shown at the top [activity values derived from: aspergillopepsin I – (Tello-Solis and Hernandez-Arana, 1995), pepsin – (Bohak, 1969; Brier et al., 2007), trypsin – (Woodard et al., 2003)]. A full bar represents 100% activity. Note that pepsin activity is in the pH range where the HX rate for backbone amide hydrogens is at its minimum. This figure was adapted from (Brier and Engen, 2008) and is used with permission.
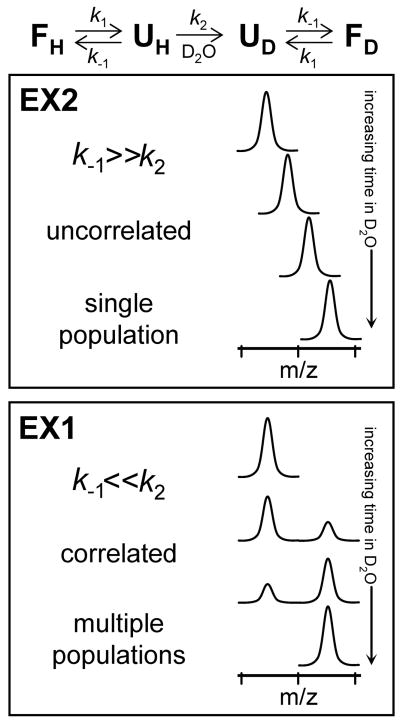
Hydrogen exchange into folded proteins can be described by a two-process model (Kim and Woodward, 1993; Bai et al., 1994; Li and Woodward, 1999) which will not be described in great detail here [see (Smith et al., 1997; Wales and Engen, 2006b) for more details]. The first process is exchange from the folded form, expressed as:
| Equation 2 |
where F is the folded form of the protein and the subscripts H and D refer to hydrogen and deuterium respectively. Exchange from the second process occurs from partially unfolded forms and is described by:
| Equation 3 |
where F is the folded form, U is the unfolded form, k2 is the intrinsic rate of exchange, and k1 and k-1 are the unfolding and refolding rate constants, respectively. Localized unfolding events (Equation 3) can break intramolecular hydrogen bonds and expose hydrogens to solvent; such an unfolded region can now exchange as if it were an unstructured peptide.
For most proteins under physiological conditions k-1 ≫ k2 and the protein must undergo multiple unfolding events until the region becomes deuterated. This type of exchange is referred to as EX2 kinetics and results in peaks in mass spectra that gradually increase in mass as the protein is exposed to D2O for longer periods of time (see Figure 3). In the case where k-1 ≪ k2, called EX1 kinetics, multiple amide hydrogens in a region can become simultaneously (i.e., cooperatively) deuterated during a single unfolding event. The mass spectra of a protein in the EX1 regime are very different from those found in the EX2 regime (Figure 3) (Miranker et al., 1993; Deng et al., 1999b; Weis et al., 2006b). In EX1, two distinct populations are revealed, the less massive being the population of molecules which have not unfolded and the more massive being the population of molecules which have unfolded and become labeled. Observation of EX1 kinetics is much more infrequent than EX2 kinetics but provides important clues about protein dynamics in solution.
Figure 3.
Protein unfolding and deuterium labeling kinetics in hydrogen exchange mass spectrometry. The folding/unfolding and labeling scheme (Equation 3) is shown at the top and resulting spectra for both EX2 and EX1 kinetics are shown below. This figure is reproduced from (Weis et al., 2006b) with permission.
PROTOCOLS
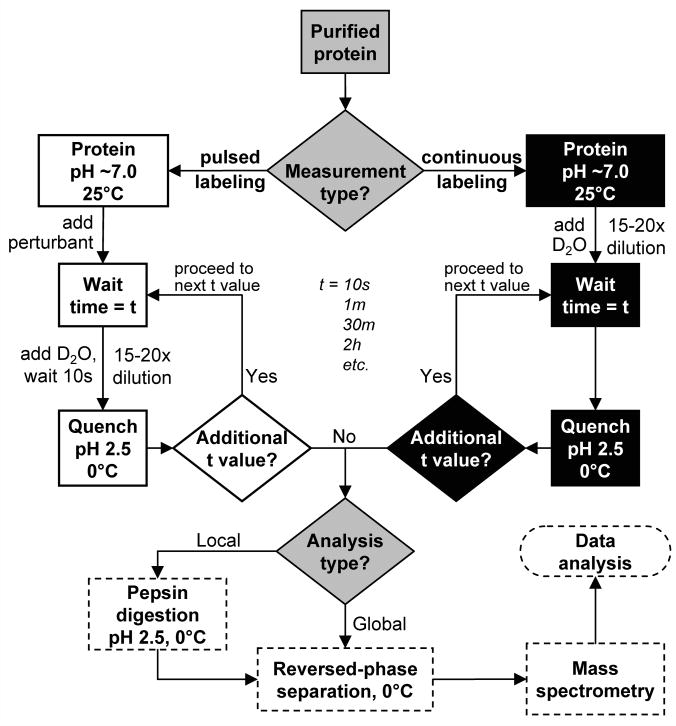
A flow-chart which highlights the steps from labeling to data analysis is shown in Figure 4. The details of each step throughout this flow chart are described in the following sections.
Figure 4.
Flow chart for hydrogen exchange mass spectrometry. Decision points are indicated with diamonds. The shading indicates which type of work is being performed: continuous labeling (black), pulsed labeling (white) and mass and data analysis (dashed border).
Protein preparation
Preparation of the protein for labeling is a step sometimes overlooked. The buffer system of the protein must be compatible with the mass spectrometer and the protein concentration should be known so that in the subsequent dilution steps, one is sure that there is still enough material to give a good signal in the mass spectrometer.
Deuterium addition
The first and foremost task in HX MS is to introduce the protein, normally in an all H2O environment, to D2O so labeling can begin. The primary method for introducing deuterium is by dilution. Typically, a solution of protein in a protiated buffer is diluted with a deuterated buffer that has a deuterium content of 99% or more. Dilutions of 15-fold or greater will produce final deuterium concentrations of >95% in the labeling solution. This serves to force the labeling reaction (k2) in one direction (see equation 3). However, with this labeling method, the original protein sample is diluted so care must be taken that such a dilution will be compatible with the sample quantity requirements of the mass spectrometer. An alternative to the dilution technique is to carry out a rapid buffer switch with small gel filtration spin columns [described in (Jeng et al., 1990; Zhang et al., 1996; Engen and Smith, 2000)]. Although the buffer switch technique introduces deuterium more slowly than the dilution method, the protein is not nearly as diluted.
Continuous labeling
Continuous labeling is the simplest labeling method. In this method, a fully protiated protein in a buffer containing 100% H2O is typically introduced to deuterium by dilution (15-20 fold is typical) with an identical buffer containing >99% D2O and the deuterium incorporation is then monitored as a function of time. Continuous labeling proceeds until the maximum amount of exchange time desired has passed, for example 24 hours. At various times along the way (see Figure 4), aliquots are removed from the labeling solution, the labeling is quenched and the resulting labeled protein is analyzed by mass spectrometry.
Continuous labeling experiments are usually performed under conditions (pH, ionic strength, etc.) where a protein is in its native state. Thus, the monitored exchange rate at various incubation times provides information on the conformational dynamics of a protein under equilibrium conditions. For any given protein, the natural fluctuations of the native state population could mean that, for example, 95% of the molecules exist in a given conformation at any one moment and the remaining 5% are in transition to another folded state. Once a protein has made a transition from a folded to an unfolded state, it becomes labeled with D2O and the mass increases. The deuterium level in the protein sample at any point in the course of the labeling experiment integrates the number of molecules in the sample that have become labeled up to that point (Miranker et al., 1993). Given enough time, all proteins should become totally deuterated during a continuous labeling experiment if protein motions and protein breathing expose all residues to exchange competent conditions. Transitioning between folded and unfolded species may be the result of natural motions of the protein but can also be induced in response of ligand binding, protein-protein complex formation, or by the addition of chaotropic agents (mild denaturing conditions). Continuous labeling is very useful for monitoring slow unfolding transitions [e.g., (Engen et al., 1997; Wales and Engen, 2006a)].
Pulsed labeling
Pulse labeling is not nearly as common as continuous labeling and for that reason will not be covered in as much detail. In pulsed labeling experiments, protein is exposed to deuterium for a very brief period of time, typically 10 seconds or less (depending on pH), after the protein has been forced to undergo structural changes via addition of a perturbant (Deng et al., 1999b; Konermann and Simmons, 2003), see Figure 4. Perturbants are usually chaotropic agents (urea, guanidine hydrochloride), but can also be binding partners, changes in pH or temperature. The goal of adding a perturbant is to establish a new, different equilibrium of populations, each presumably/possibly with a different conformation. Each of these populations may exhibit a different hydrogen exchange pattern in which case the resulting deuterium levels then indicate the instantaneous population of folded and unfolded molecules. Pulse labeling experiments have been used to study protein folding mechanisms by mass spectrometry as well as to identify transient intermediate states [for example (Yang and Smith, 1997; Deng and Smith, 1998; Deng and Smith, 1999; Deng et al., 1999b; Mazon et al., 2004; Pan et al., 2004)].
Quench
At the completion of the desired amount of labeling time, the pH is adjusted to “quench” the exchange reaction. As eluded to above, HX MS is made possible by the connection between pH and exchange rate. Protein labeling normally occurs at physiological pH (pH 7.0-8.0) where the majority of proteins are in their most biologically relevant conformation(s). To reduce the exchange rate so that the amount and location of deuterium can be determined, the pH of the labeling solution is reduced to the minimum value for exchange, pH 2.5 (as in Figure 2). By reducing the pH from 7.0 to 2.5, the exchange rate for the average backbone amide hydrogen is reduced about 10,000 fold [although this is not true for all sequences, see (Zhang and Smith, 1993; Smith et al., 1997)]. To make sure the analysis is reproducible, it is critical to always label and quench with the same pH values as alterations as small as 0.1 pH units can change the rate of exchange significantly. One good way to ensure a reproducible and reliable pH is to use a buffer with a pK value near the labeling or quenching pH (7.0 and 2.5, respectively). Phosphate buffers are particularly convenient in this regard because there is a pK1 at 2.15 for the quench and a pK2 at 6.82 for the label [see (Engen and Smith, 2000)]. Once the exchange reaction has been “quenched”, the samples must then either be analyzed immediately (“on-the-fly”) or quickly frozen by exposure to liquid nitrogen or dry ice. In many cases, the quenched sample can be stored at -80 °C for several days before analysis. However, this is also a function of each individual protein as some proteins are not able to withstand a single freeze-thaw cycle.
Analysis: General considerations
What follows next depends on what type of mass spectrometry will be employed. Generally, we have advised that LC-MS (using electrospray) be used as other types of mass spectrometry (such as MALDI) may appear easier at the outset, but in the end make things more complicated (see below). What we will describe in the following sections applies to electrospray MS.
A major concern is the temperature of all analysis steps post-quench. The reason this is an issue is that positions that have become deuterated during the labeling step can revert back to hydrogen before mass analysis can occur. Such “back-exchange” can be an issue if not properly controlled. However, when it is controlled and understood, it becomes more of an annoyance than a prohibitive factor.
Deuterium in a protein will revert to hydrogen if the concentration of deuterium in the surrounding environment becomes small. This reduction in deuterium concentration will occur in the 100% H2O environment of an HPLC solvent system, the H2O environment of a traditional MALDI matrix, or even the H2O-containing air surrounding a fast atom bombardment solids probe (David Smith, personal communication). At pH 2.5, if a deuterated protein were to suddenly find itself in a 100% H2O environment, the reversion of the deuterium label back to hydrogen would occur with a half-life of about 10-15 minutes. To reduce the rate of reversion by another order of magnitude, the temperature of the quenched sample is lowered to 0 °C. The half-life for back-exchange under quench conditions of 0 °C and pH 2.5 is between 30-120 min, depending on the sequence (Englander and Kallenbach, 1983; Bai et al., 1993; Smith et al., 1997). Consequently, the analysis steps must still be completed as quickly as possible to maximize the amount of deuterium that is recovered. The quench conditions of pH 2.5 and 0 °C must be maintained at all times once the quench has been initiated.
We advocate setting up experiments so that no back-exchange correction is needed. To do this, one must first understand the back-exchange characteristics of their particular experimental setup (i.e., how much deuterium is lost during each step of the process, values determined by running a series of totally deuterated control proteins). Once the back-exchange in the experimental system is understood, it becomes possible to make relative deuterium measurements, as described in detail in (Wales and Engen, 2006b). One compares two or more forms of a protein (e.g., wild-type vs. mutant, bound vs. free, active vs. inactive, etc.), determines the deuterium incorporation in both samples using identical experimental conditions, and compares where levels are similar or different. On the other hand, if the absolute deuterium level is of greater interest or if there are not two states that can be compared, back-exchange adjustment is required and several controls must be analyzed along with each sample set so an adjustment can be made for back-exchange. To arrive at an absolute number of deuteriums that exchanged at each time point, an adjustment equation is applied (Zhang and Smith, 1993). Although several variations to the original adjustment formula have been described (Hoofnagle et al., 2004), the original adjustment method has found the most use in HX MS experiments.
Global analysis
As shown in Figure 4, after labeling is finished and the quench completed, one must choose how to characterize the deuterium incorporation. The most basic way to do this is to measure the deuterium incorporated into a whole protein as a function of time, or what we term “global analysis”. Global analysis was the first type of analysis reported for HX MS (Chowdhury et al., 1990; Katta and Chait, 1991) and is the simplest to implement. Global analysis is also a good first experiment when working with a new protein, for several reasons. First, it is a good test to see if the protein of interest is stable through the HX MS procedure of labeling, quenching, LC and so on. Second, the results of global analysis can provide an indication of how structured (or unstructured) the protein is.
To determine the global incorporation of deuterium, the intact, labeled protein is analyzed by LC and electrospray MS. As there is really no separation required in global analysis of a single protein, there is often just a trap column used for the LC step. Several important things happen during this LC step and therefore it must be performed prior to MS analysis. First, sample concentration occurs which can be useful particularly if a dilute protein solution was used for labeling. Second, desalting removes buffer salts that will negatively impact the electrospray process and suppress the signal of the protein. Third, and most importantly, the LC step serves to wash away all deuterium that had exchanged into sidechain positions. Because the chromatography is performed with protiated solvent (all H2O), back exchange begins the moment the deuterated sample is injected. Any deuterium that had exchanged into side-chain positions, which have very fast exchange rates even under quench conditions (Figure 2) (Englander et al., 1985; Bai et al., 1993), will revert back to hydrogen during the chromatography step. Therefore, the deuterium incorporation that is measured is a result of deuteration at backbone amide positions only. This substantially simplifies the data interpretation step because the maximum number of deuterium atoms that can be found in each amino acid (except proline) will be one. Note that because arginine has a side chain amide hydrogen (ε-NH) with an exchange rate minimum near that of the backbone amide hydrogens (Figure 2), proteins rich in arginine can complicate the analysis because all side-chain deuterium may not be washed away during the analysis steps.
So while back-exchange of side chain positions is desirable, back-exchange of backbone amide hydrogens is not desirable. As described above, the injection valve, loop, trap column and all associated tubing must be maintained at 0 °C. By maintaining 0 °C and pH 2.5 throughout the chromatography step, as well as using a very short gradient (e.g., 5-40% organic in under 3 minutes), back-exchange from the backbone amide positions can be reduced to as little as 10-15% deuterium loss.
Mass measurement in global analysis only requires that the mass spectrometer be capable of accurately measuring the mass of whole proteins. We have found that time of flight analyzers work well for this purpose for the widest variety of proteins, although other mass analyzers can provide suitable results in many cases. The ionization parameters must be controlled so as not to elevate back-exchange losses during the electrospray process and subsequent desolvation. Control experiments with totally deuterated peptides and proteins can help determine the exact settings, but it is recommended that the source temperature be high enough to encourage good desolvation but not so high that back-exchange is elevated. In a Waters Z-Spray electrospray source, for example, an optimal source temperature is 80 °C with a desolvation gas temperature of 150-175 °C.
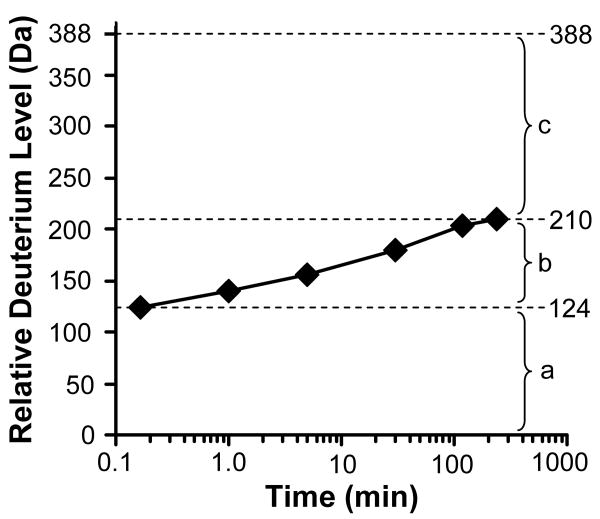
Once the spectra are obtained, the deuterium incorporation for each labelling time is determined. The increase in mass is determined by subtracting the mass measured for an undeuterated control sample from the mass at each deuterium exchange-in time point. For this calculation, the average mass is used, either from the deconvolved spectrum or from a particular charge state. The resulting data are plotted as the time in deuterium (on a log scale, x-axis) versus the increase in mass (Relative Deuterium Level, y-axis), see Figure 5. Several valuable pieces of information can be extracted from such a graph of whole protein HX. First, the number of rapidly exchanging amide hydrogens in the protein can be estimated if the first labeling time point is sufficiently short (such as 10 seconds or less). The amide hydrogens that exchange during such a short labeling time are those which must be highly exposed to solvent and probably not involved in hydrogen bonds within secondary structural elements (Dharmasiri and Smith, 1996). In the example shown in Figure 5, this number is 124. Second, the number of amide hydrogens that do not exchange at all during the course of the experiment can be determined. These hydrogens are those that are most protected and likely in areas inaccessible to solvent or strongly protected by hydrogen bonds. In Figure 5 this value is 178, or the total number of exchangeable backbone amide hydrogens in the protein minus the number of exchanged hydrogens at the longest timepoint. Third, the number of amide hydrogens that undergo some kind of localized unfolding or fluctuations during the labeling time course are those that contribute to the increase in mass between the shortest and longest timepoints (86 in the data shown in Figure 5). Although not discussed in this chapter, additional information about protein dynamics and kinetic mechanisms can be derived from the isotopic distribution profile of the mass spectrum [see (Wales and Engen, 2006b; Weis et al., 2006b; Brier and Engen, 2008) for further information].
Figure 5.
Example data for deuterium exchange into an intact protein. The maximum of the y-axis is equal to the maximum number of exchangeable backbone amide hydrogens. Three general regions can be defined in a graph of this type. Region (a) represents those amide hydrogens that exchanged before the first time point. These amide hydrogens are typically highly solvent exposed and not engaged in intraprotein hydrogen bonding. In region (b), the amide hydrogens that become exchange competent either through EX2 or EX1 kinetics (see Figure 3) become deuterated. These amide hydrogens are generally those that are in dynamic regions where local unfolding can occur during the deuteration time course. Region (c) contains those amide hydrogens that did not exchange even after the longest exchange timepoint in the experiment, here 4 hours. These amide hydrogens that are slow to exchange are often located within the core of the protein protected from solvent or are engaged in strong hydrogen bonds in structural elements.
Local analysis
Another option for measuring the incorporation of deuterium is local analysis (Figure 4). Its called local analysis because the outcome is a much more specific detailing of where the deuterium has been incorporated. While in global analysis the total amount of incorporation in the whole protein over time is measured, in local analysis the location and the amount of the deuterium incorporation can be refined to short stretches of amino acids. In order to accomplish local analysis, the protein is broken into pieces after the labeling is quenched.
The local analysis technique was first reported by Rosa and Richards (Rosa and Richards, 1979) who used T2O for labeling followed by enzymatic digestion. The Rosa and Richards method was later adopted, modified, and combined with mass spectrometry analysis by Smith and colleagues (Zhang and Smith, 1993). It is important to realize that fragmentation occurs after protein labeling is completed, when the deuteration reaction has already been quenched. Because the quench effectively “freezes” the deuteration of the native, pH 7 state of the protein, in later steps the deuterium levels of short pieces are directly related to the deuteration levels in the folded, intact protein. Because quench conditions must be maintained during the enzymatic digestion of the labelled, quenched proteins, acid proteases that are active at pH 2.5 must be employed. The resulting peptide fragments can then be separated by liquid chromatography (Figure 4, Figure 6).
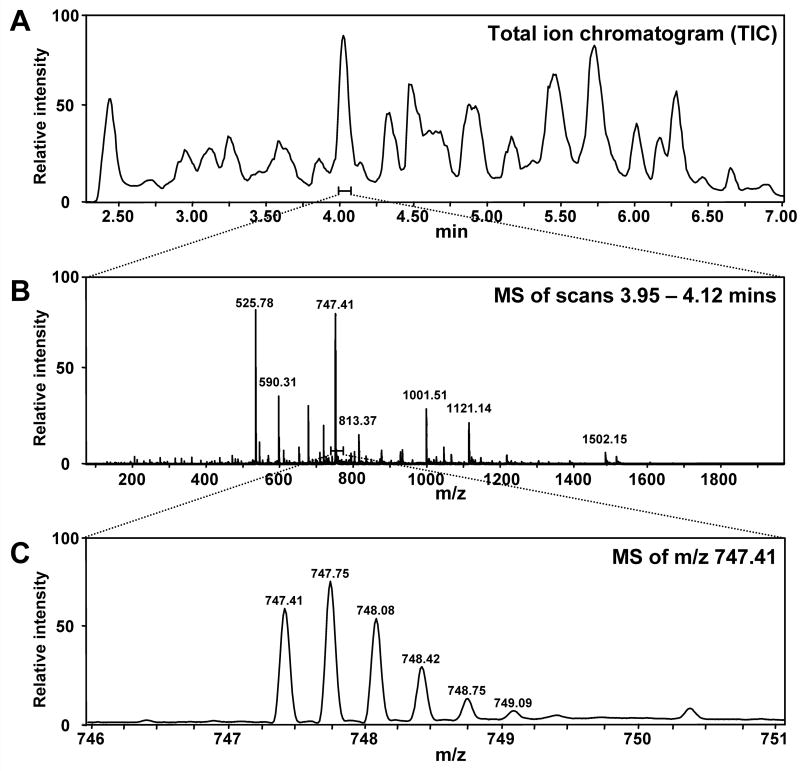
Figure 6.
Representative chromatograms and electrospray mass spectra to illustrate the LC separation and mass spectra for local analysis. A. Typical UPLC trace for separation of the peptic peptides from a 27 kDa protein [methods described in (Wales et al., 2008)]. The amount of time required for separation of the peptides is relatively short (~5 min). B. Full scan mass spectrum of the ions eluting between 3.95-4.12 minutes. While there are multiple ions, they are well resolved in the m/z scale. C. Zoom of the ion at m/z 747.41 illustrating the typical isotopic pattern and mass resolution observed. These results were obtained with a Waters Q-Tof Premier.
The most common protease used in HX MS is pepsin. As seen in Figure 2, pepsin is highly active at quench conditions (pH 2.5). Pepsin is a relatively “non-specific” protease in that it generally cleaves at hydrophobic residues but does not hold fast to this rule [see also (Hamuro et al., 2008)]. It does have some cleavage specificity but unfortunately cleavage products cannot be predicted from sequence alone as is the case for other enzymes such as trypsin. Although pepsin fragments are not predictable, cleavage is highly reproducible under identical conditions (pH, temp, time, and concentration). Each peptic peptide in the digestion must be identified, usually with a combination of MS/MS and exact mass analyses. Peptic digestion results in many, typically small (10-15 residues), and overlapping peptides which can serve to increase ones ability to localize deuterium to short stretches of amino acids or in the best case, to individual amino acids (Garcia et al., 2004). The addition of guanidine hydrochloride to the quench buffer has the effect of enhancing the digestion as pepsin can tolerate concentrations as high as 3 M (Wang and Smith, 2005). Other acid proteases such as aspergillopepsin I from Aspergillus saitoi (EC 3.4.23.18) have been shown to be an acceptable alternative to porcine pepsin (EC 3.4.23.1) as they are functional at quench pH (Figure 2). Localization of deuterium to shorter and shorter stretches of amino acids can be greatly enhanced when multiple proteases are used to digest the same protein (Cravello et al., 2003).
Digestion may be done in two ways. Offline digestion is carried out by preparation of a concentrated pepsin solution (typically 10 mg/mL in H2O, pH ~5.0) and addition of 1:1 pepsin:substate by weight to the quenched sample (Zhang et al., 1996; Engen and Smith, 2000). Digestion is typically carried out for 5 minutes or less at 0 °C. For offline digestion, the LC setup is similar to that of the intact mass analysis discussed above with the addition of a chromatography column and an increase in analysis time. Alternatively, digestion may be performed online by the use of a slightly more complex system involving an immobilized pepsin column (Wang et al., 2002). An injection valve is used to load quenched protein onto the pepsin column. Proteins enter one side of the column, peptides come out the other side and are trapped on a peptide trap inline with the pepsin column [see (Wang et al., 2002; Wu et al., 2006b)]. A switching valve then diverts the trapped peptides onto an analytical column for separation. As in intact analysis, the entire process of online digestion must be maintained at 0 °C and done in as short a time as possible.
As was the case for global analysis, an LC step is used for local analysis also. For local analysis, however, it serves a much more important role as there is actually something to be separated besides just whole proteins and buffer salts. Figure 6 shows an example of a separation of the peptides from pepsin digestion of a 27 kDa protein. Although analysis of deuteration with other mass spectral techniques such as MALDI has been reported (Mandell et al., 1998), such methods suffer from the lack of a separation step as is afforded with LC and electrospray MS. The separation step allows for detailed analysis of the location of deuteration even for very large proteins (upwards of 200 kDa), while simultaneously washing away deuterium from the sidechains. Unfortunately, the analysis requirements of 0 °C compromise the efficiency of conventional reversed-phase HPLC. For small proteins (and therefore not a lot of peptic peptides) peptides are resolved on the m/z scale even with poor chromatography. However, better chromatography (Figure 6, for example) significantly improves the entire MS analysis portion of local analysis. Switching from conventional HPLC to UPLC has the advantage of vastly improved separations, shorter separation times, and improved mass spectral response when compared with analysis of the identical sample with HPLC (Wu et al., 2006a; Wales et al., 2008).
The mass analysis portion of a local analysis HX MS experiment is similar to the mass analysis that would be done for any mixture of peptides, with a few exceptions. There must be special attention paid, as for global analysis, to the temperature of the source and desolvation regions. Similar to what was discussed above for intact proteins (see also Figure 5), the mass increase of each peptic peptide is determined by subtracting the centroid mass of the undeuterated peptides from the centroid mass of each deuterated peptide (Figure 7A,B) (Engen and Smith, 2000). Even in instruments with modest resolution (~2000), the isotopic peaks of peptides will be resolved (Figure 6C and 7A) and selecting which peaks to include in the centroid determination is fairly straightforward. These data processing steps are repeated for all the peptic peptides produced during the digestion. The analysis is then repeated for each time point in the time course and the results are plotted in the same fashion as stated above for intact analysis (Figure 7C). There are now a variety of software packages which offer varying degrees of automation for processing peptide data [e.g., (Weis et al., 2006a; Nikamanon et al., 2008; Pascal et al., 2009)].
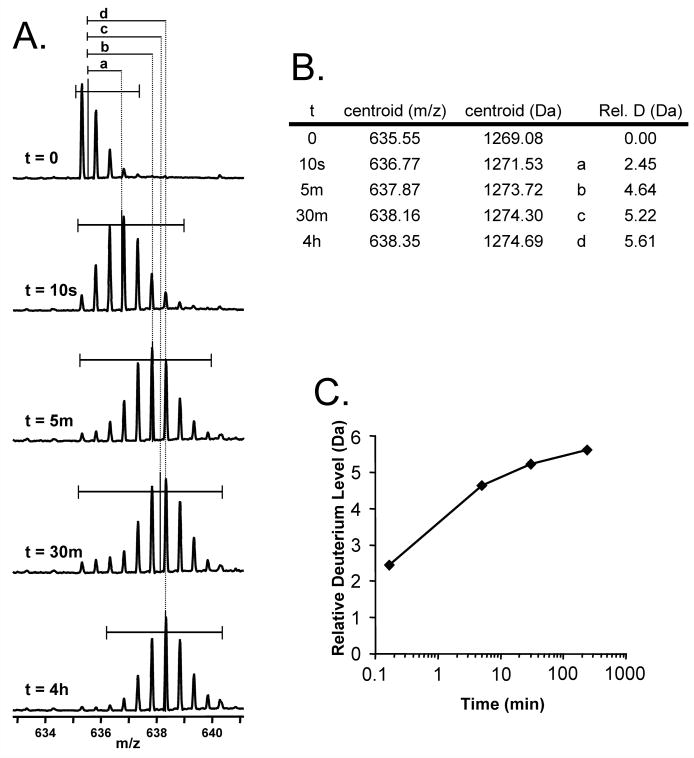
Figure 7.
An illustration of the steps required to convert the data in raw mass spectra from a time course of deuteration into a deuterium uptake graph. A. Raw mass spectra for an ion at m/z 635.32, produced by labeling a protein for a certain time, digesting the protein with pepsin and then carrying out UPLC/MS (as shown in Figure 6). The time spent in deuterium is indicated, with the undeuterated protein at the top. The horizontal bars indicate what part of each mass spectrum was used to determine the centroid mass of each isotopic distribution. The centroid values are named a,b,c, or d as shown. B. A table that tallies the time spent in deuterium, the centroid value and the mass increase of each time point relative to the undeuterated control. C. A graph of the data in panel B.
DATA INTERPRETATION
The final step in a hydrogen exchange experiment is to interpret the deuterium incorporation data. To visualize the data, the deuterium uptake is often converted into a graph, or a deuterium uptake curve (as in Figure 5, Figure 7C, Figure 8). If no adjustment has been made for back-exchange during analysis (discussed above), the y-axis will be labeled as relative deuterium level.
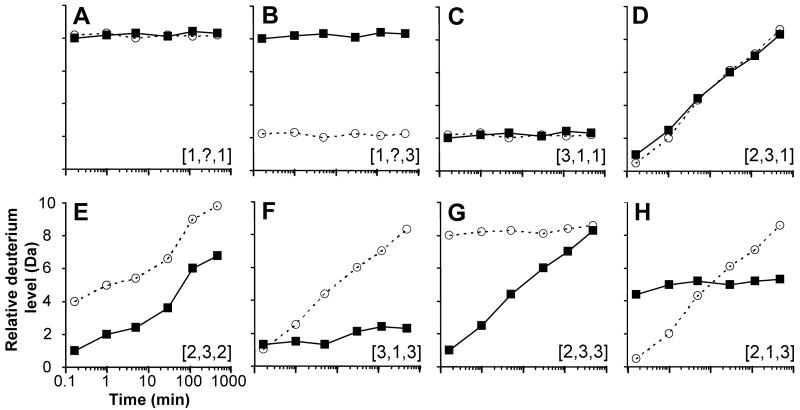
Figure 8.
Peptide hydrogen exchange data that may be encountered. Eight regions (A-H) from a hypothetical protein that has different physiological conformation and dynamics are described for wild-type protein by itself (solid symbols, solid lines) and the same protein when stimulated (open symbols, dotted lines). Stimulation means one of many things, including: ligand binding, mutation, activation, post-translational modification, protein-protein interactions, denaturant, etc. In these graphs, the maximum number of exchangeable hydrogens is plotted as the maximum of the y-axis. Each panel has been assigned a ranking with the following format: [x,y,z,] where x is the protection/structure, y is the dynamics, and z is the change upon stimulation. A rank of 1 is low, 2 is medium and 3 is high. The rank is based on the wild-type protein transitioning to the stimulated form. The details of each panel are discussed in the text.
To illustrate the different types of deuterium incorporation graphs that are possible, we have created a series of hypothetical curves for peptic peptides derived from a hypothetical protein with different exchange properties (Figure 8). The goal with this discussion is to understand what various shapes in deuterium uptake graphs mean at the protein biophysics and protein dynamics level. Each panel of Figure 8 (A-H) shows one potential result when a peptide from a wild-type protein under physiological conditions (black lines) is compared to the same peptide from the same protein after exposure to some external stimulus (dotted lines). By stimulus, we mean a generic term referring to any of a number of circumstances including mutation, ligand binding, denaturant, activation, protein-protein complex formation, etc. The data are plotted as Relative Deuterium Level (Da) as a function of time. In these graphs, we have adopted the common practice of making the maximum of the y-axis the maximum number of backbone amide hydrogens for the peptide in that graph. Further, we have ranked three parameters for each panel: [x,y,z] where x is protection/structure, y is dynamics, and z is change upon stimulation. A value of 1-3 is given for each parameter with 1 being low and 3 being high.
The protein region illustrated by the data in Figure 8A takes up nearly the maximum amount of deuterium possible by the earliest time point and remains at the same deuterium level for the duration of the experiment. Such results are an indication of a protein region with high solvent accessibility and/or a lack of hydrogen bonding. These data are consistent with a part of a protein that is unstructured and very easily deuterated. Upon stimulus (dotted line), there is no change to the deuteration, meaning nothing happened to the conformation of the protein in this region upon stimulation. The region in panel A receives a ranking of [1,?,1]; it has low protection, we are unsure about dynamics because its fully deuterated by the first labeling point in the experiment and it has low changes upon stimulation. In contrast, for the region shown in Figure 8B, although in the wild-type protein the data are the same as in panel A, upon stimulus, there is a drastic change in the deuteration in this region. This regions ranks [1,?,3] and the conclusions from results like panel B are that while the region was highly exposed and rapidly deuterated in the wild-type protein, when the protein stimulus occurred (say for example it bound a small molecule), the region became significantly protected from exchange. Such changes in deuteration are highly indicative of conformational rearrangements in response to some kind of stimulus.
In Figure 8C, the data indicate that this region of the protein was protected from exchange and the amount of deuterium that could exchange into the protein in this region is very low. There was no change to the deuteration upon stimulus. Deuteration curves like panel C indicate regions that are highly protected, generally not dynamic (no significant breathing motions or unfolding) and probably in very stable structural elements. The ranking is [3,1,1]. A few positions exchange immediately (in the case shown, 2 residues are deuterated at the first timepoint) and those might be backbone amide hydrogens at one end of the peptide that are not as structured. A different situation is shown in Figure 8D where there is significant protection at the beginning of the time course, but the region is rapidly deuterated during the time course. Data with this shape indicate regions of a protein that are dynamic, breathing in solution and visiting the exchange-competent state many times during the labeling experiment. By the end of the timecourse, there have been so many motions that the entire region has become deuterated. Again, in Figure 8D as in Figure 8A and C, there has been no change to the deuteration upon stimulus, indicating that these regions are not altered during the stimulation event. The rank for the data in panel D is [2,3,1]; that is, moderate protection/structure, high dynamics and low change on stimulation.
Other possible types of data follow in Figure 8 panels E-H. In panel E, the region is partially protected/structured, moderately dynamic and changes upon stimulation; the ranking is [2,3,2]. Since the change upon stimulation in panel E happens from the first time point and persists throughout the time course with almost the same difference between wild-type and stimulated, it is likely that the stimulus only perturbed rapidly exchanging amide hydrogens that become deuterated before the first time-point of the experiment. In panel F, the region is highly protected prior to stimulation but after stimulation it becomes highly dynamic and much less protected from exchange for a ranking of [3,1,3]. The protein region in panel G is one that is protected initially (similar to panel D), but upon stimulation becomes very unprotected and exchanges rapidly (like the region in panel A). The ranking of the region in panel G is [2,3,3] and such data are consistent with parts of a protein that become unstructured as a result of stimulation. Finally, in panel H we show an interesting case where a region is partially protected but not very dynamic initially; upon stimulation, some parts get more protected and other parts get deprotected. The rank for this region is [2,1,3] based on the unstimulated form.
The various types of deuteration curves that could be observed, as shown in Figure 8, provide clues about conformation and dynamics. If a crystal structure is available for the protein being investigated, the data interpretation can continue with the known structure as a guide. The deuterium incorporation information can be plotted onto the crystal structure rendering of the protein, considered in light of the hydrogen bonds found in the crystal structure and interpreted along with the structural elements found in each peptic peptide (recall that the deuterium exchange reaction captures the conformational information of the protein prior to digestion). We see crystal and NMR structures as complementary and a very good starting point as a model with which to interpret the HX MS results. However, it should be kept in mind that the solution form of a protein being investigated by HX MS might not be consistent with the structure revealed by crystallography.
Using the pepsin digestion methodology for local analysis, exchange cannot be localized in most cases to individual amide positions. Therefore, the interpretation of the results must consider that, for example, changes involving 2-3 deuteriums in a peptide of 20 amino acids do not occur throughout the entire peptide but are confined to only a few amino acids within that peptide. The identity of those few residues remains unknown without further experiments (see also next section).
PRACTICAL POINTERS
The following guidelines will work for most proteins and protein systems. As with all analyses of proteins, care should be taken with each protein as they all have their own unique characteristics and may not fall under these general guidelines in all cases. The limitations below are mainly due to the limitations of the LC-MS, and not due to the HX reaction itself.
Types of proteins
Any soluble protein can be used in routine HXMS experiments. Proteins that require detergents to exist in solution will not work as the detergents are detrimental to the mass spectrometry analyses in the later steps of the experiments. Up to 10% glycerol can be tolerated as well as certain non-ionic detergents like dodecyl maltoside. Membrane proteins/peptides are still very challenging for this method and successful analysis of these proteins requires removal of the lipid before MS analysis.
Size limitations
The typical protein being analyzed with HXMS at present is something smaller than ~70 kDa. Larger proteins and more complex systems can be analyzed as well but the amount of time required to process all the data that is generated can become prohibitive for extremely large systems. We have analyzed systems containing over 250 kDa of unique sequence in our laboratories. Proteins less than ~30 kDa are considered trivial to analyze.
Quantity requirements
The amount of material needed is highly dependent on the type and sensitivity of the mass spectrometer used. For the QTof-type instruments that we use in our laboratory, we recommend 20-30 pmoles of protein for experiments involving pepsin digestion and 100-150 pmoles of protein for intact protein experiments. These requirements will vary depending on the ionization efficiency of the peptides/proteins, but this is a good starting point. These values are for each injection. If a time course of 10 time points is needed, the total consumption will be 200-300 pmoles for peptide-level analyses and 1000-1500 pmoles for intact protein work.
Concentrations
Protein concentrations of 10-30 μM are where we typically operate. The concentration can be as low as 0.1 μM and the sample rapidly concentrated during the HPLC step prior to mass analysis. One must always inject the amount of material required by the MS system (see previous point on quantity). We have used up to 2.0 mL injections of dilute proteins with rapid loading of the solution onto the HPLC column for concentration. More typical conditions would be injections of 25-100 μL.
Ligand binding
Because non-covalent ligands are in equilibrium with their binding partners, a significant molar excess of ligand may be required to force binding of the partner of interest to a sufficient level (>60%). The greater the molar excess of ligand the greater percent bound but that in turn makes the analysis more difficult as the signal from the ligand can easily swamp out the signal from the protein. Care must be taken to calculate the effect such concentrations of ligand may have on the dynamic range of the analysis.
System complexity
The limitations of system complexity are related to the amount of data processing and the peak capacity of the HPLC and MS systems. Current systems of 2-3 proteins not totalling more than ~250 kDa are feasible but challenging. Complicated systems involving whole protein machines with multiple subunits cannot be analyzed with this technique as it stands now. Future improvements should allow for more and more complex systems.
Useful websites: HX Express software: http://www.hxms.com/HXExpress/; HD Desktop software: http://hdx.florida.scripps.edu/; general tutorial on HXMS: http://www.hxms.com.
FUTURE PERSPECTIVES
It would be most desirable to be able to monitor deuterium exchange at individual amino acids with mass spectrometry. Attempts have been made to use CID to fragment deuterium labeled peptic peptides into shorter pieces (b/y ions) and thereby find each deuterated residue (Deng et al., 1999a; Demmers et al., 2000; Kim et al., 2001; Demmers et al., 2002; Hoerner et al., 2004). It was originally observed that b ions from high-energy CID with argon as the collision gas yielded deuterium levels that were consistent with NMR measured deuterium levels (Deng et al., 1999a; Kim et al., 2001). However, later work showed that scrambling appears to be 100% in both b and y ions when CID is used for fragmentation (Jorgensen et al., 2005). Other methods of fragmentation offer a solution to the scrambling issue. It was recently shown that electron transfer dissociation (ETD) can be used to fragment solution deuterated peptides and that gas-phase scrambling can be minimized with specific ionization conditions (Zehl et al., 2008). In the coming years, it seems likely that ETD fragmentation will routinely provide deuteration information for individual amino acids.
Another area that will see growth in the future is the automation of data processing. It is surprisingly difficult to automate the processing of deuterium exchange data. Progress in this area has been slow, but major strides have been made in the last few years [e.g., (Pascal et al., 2009)]. With the combination of good chromatography, such as that from UPLC, ETD fragmentation for improved localization and rapid data processing, the rate at which proteins can be interrogated by this method will rapidly increase, as will the amount of useful information about proteins.
SUMMARY
As was illustrated in this chapter, mass spectrometry can be used for much more than just measuring the molecular weight of proteins. With the labeling method of hydrogen-deuterium exchange, protein conformation and changes in conformation can be investigated with mass spectrometry. Because so many proteins remain uncharacterized and are not amenable to other types of structural analyses, it is almost certain that HX MS will make substantial contributions towards understanding proteins and their functions.
Acknowledgments
This work was supported in part by NIH grant R01-GM070590 and is contribution number 938 from the Barnett Institute.
LITERATURE CITED
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Protein stability parameters measured by hydrogen exchange. Proteins. 1994;20(1):4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- Bohak Z. Purification and characterization of chicken pepsinogen and chicken pepsin. J Biol Chem. 1969;244(17):4638–4648. [PubMed] [Google Scholar]
- Brier S, Maria G, Carginale V, Capasso A, Wu Y, Taylor RM, Borotto NB, Capasso C, Engen JR. Purification and characterization of pepsins A1 and A2 from the Antarctic rock cod Trematomus bernacchii. Febs J. 2007;274(23):6152–6166. doi: 10.1111/j.1742-4658.2007.06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier S, Engen JR. Hydrogen exchange mass spectrometry: Principles and capabilities. In: Chance M, editor. Mass spectrometry analysis for protein-protein interactions and dynamics. New York: Wiley-Blackwell; 2008. pp. 11–43. [Google Scholar]
- Chowdhury SK, Katta V, Chait BT. Probing conformational changes in proteins by mass spectrometry. J Am Chem Soc. 1990;112(24):9012–9013. [Google Scholar]
- Cravello L, Lascoux D, Forest E. Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins. Rapid Commun Mass Spectrom. 2003;17(21):2387–2393. doi: 10.1002/rcm.1207. [DOI] [PubMed] [Google Scholar]
- Demmers JA, Haverkamp J, Heck AJ, Koeppe RE, 2nd, Killian JA. Electrospray ionization mass spectrometry as a tool to analyze hydrogen/deuterium exchange kinetics of transmembrane peptides in lipid bilayers. Proc Natl Acad Sci U S A. 2000;97(7):3189–3194. doi: 10.1073/pnas.050444797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmers JA, Rijkers DT, Haverkamp J, Killian JA, Heck AJ. Factors affecting gas-phase deuterium scrambling in peptide ions and their implications for protein structure determination. J Am Chem Soc. 2002;124(37):11191–11198. doi: 10.1021/ja0125927. [DOI] [PubMed] [Google Scholar]
- Deng Y, Smith DL. Identification of unfolding domains in large proteins by their unfolding rates. Biochemistry. 1998;37:6256–6262. doi: 10.1021/bi972711o. [DOI] [PubMed] [Google Scholar]
- Deng Y, Pan H, Smith DL. Selective isotope labeling demonstrates that hydrogen exchange at individual peptide amide linkages can be determined by collision-induced dissocation mass spectrometry. J Am Chem Soc. 1999a;121:1966–1967. [Google Scholar]
- Deng Y, Smith DL. Rate and equilibrium constants for protein unfolding and refolding determined by hydrogen exchange-mass spectrometry. Anal Biochem. 1999;276(2):150–160. doi: 10.1006/abio.1999.4347. [DOI] [PubMed] [Google Scholar]
- Deng Y, Zhang Z, Smith DL. Comparison of continuous and pulsed labeling amide hydrogen exchange/mass spectrometry for studies of protein dynamics. J Am Soc Mass Spectrom. 1999b;10(8):675–684. doi: 10.1016/S1044-0305(99)00038-0. [DOI] [PubMed] [Google Scholar]
- Dharmasiri K, Smith DL. Mass spectrometric determination of isotopic exchange rates of amide hydrogens located on the surfaces of proteins. Anal Chem. 1996;68(14):2340–2344. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Identification and localization of slow, natural, cooperative unfolding in the hematopoietic cell kinase SH3 domain by amide hydrogen exchange and mass spectrometry. Biochemistry. 1997;36(47):14384–14391. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- Engen JR, Smith DL. Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol Biol. 2000;146:95–112. doi: 10.1385/1-59259-045-4:95. [DOI] [PubMed] [Google Scholar]
- Englander JJ, Rogero JR, Englander SW. Protein hydrogen exchange studied by the fragment separation method. Anal Biochem. 1985;147:234–244. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW, Poulsen A. Hydrogen-tritium exchange of the random chain polypeptide. Biopolymers. 1969;7:379–393. [Google Scholar]
- Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Garcia RA, Pantazatos D, Villarreal FJ. Hydrogen/deuterium exchange mass spectrometry for investigating protein-ligand interactions. Assay Drug Dev Technol. 2004;2(1):81–91. doi: 10.1089/154065804322966342. [DOI] [PubMed] [Google Scholar]
- Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun Mass Spectrom. 2008;22(7):1041–1046. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
- Hoerner JK, Xiao H, Dobo A, Kaltashov IA. Is there hydrogen scrambling in the gas phase? Energetic and structural determinants of proton mobility within protein ions. J Am Chem Soc. 2004;126(24):7709–7717. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- Hoofnagle AN, Resing KA, Ahn NG. Practical methods for deuterium exchange/mass spectrometry. Meth Mol Biol. 2004;250:283–298. doi: 10.1385/1-59259-671-1:283. [DOI] [PubMed] [Google Scholar]
- Hvidt A, Linderstrom-Lang K. Exchange of hydrogen atoms in insulin with deuterium atoms in aqueous solutions. Biochim Biophys Acta. 1954;14(4):574–575. doi: 10.1016/0006-3002(54)90241-3. [DOI] [PubMed] [Google Scholar]
- Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- Jeng M-F, Englander SW, Elove GA, Wand AJ, Roder H. Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR. Biochemistry. 1990;29(46):10433–10437. doi: 10.1021/bi00498a001. [DOI] [PubMed] [Google Scholar]
- Jorgensen TJ, Gardsvoll H, Ploug M, Roepstorff P. Intramolecular migration of amide hydrogens in protonated peptides upon collisional activation. J Am Chem Soc. 2005;127(8):2785–2793. doi: 10.1021/ja043789c. [DOI] [PubMed] [Google Scholar]
- Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom. 1991;5(4):214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- Kim K-S, Woodward C. Protein internal flexibility and global stability: Effect of urea on hydrogen exchange rates of bovine pancreatic trypsin inhibitor. Biochemistry. 1993;32:9609–9613. doi: 10.1021/bi00088a013. [DOI] [PubMed] [Google Scholar]
- Kim MY, Maier CS, Reed DJ, Deinzer ML. Site-specific amide hydrogen/deuterium exchange in E. coli thioredoxins measured by electrospray ionization mass spectrometry. J Am Chem Soc. 2001;123(40):9860–9866. doi: 10.1021/ja010901n. [DOI] [PubMed] [Google Scholar]
- Konermann L, Simmons DA. Protein-folding kinetics and mechanisms studied by pulsed-labeling and mass spectrometry. Mass Spectrom Rev. 2003;22:1–26. doi: 10.1002/mas.10044. [DOI] [PubMed] [Google Scholar]
- Li R, Woodward C. The hydrogen exchange core and protein folding. Protein Sci. 1999;8(8):1571–1590. doi: 10.1110/ps.8.8.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Falick AM, Komives EA. Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal Chem. 1998;70(19):3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- Mazon H, Marcillat O, Forest E, Smith DL, Vial C. Conformational dynamics of the GdmHCl-induced molten globule state of creatine kinase monitored by hydrogen exchange and mass spectrometry. Biochemistry. 2004;43(17):5045–5054. doi: 10.1021/bi049965b. [DOI] [PubMed] [Google Scholar]
- Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262(5135):896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11(2):150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- Nikamanon P, Pun E, Chou W, Koter MD, Gershon PD. “TOF2H”: a precision toolbox for rapid, high density/high coverage hydrogen-deuterium exchange mass spectrometry via an LC-MALDI approach, covering the data pipeline from spectral acquisition to HDX rate analysis. BMC Bioinformatics. 2008;9:387. doi: 10.1186/1471-2105-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Raza AS, Smith DL. Equilibrium and kinetic folding of rabbit muscle triosephosphate isomerase by hydrogen exchange mass spectrometry. J Mol Biol. 2004;336(5):1251–1263. doi: 10.1016/j.jmb.2003.12.076. [DOI] [PubMed] [Google Scholar]
- Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20(4):601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa JJ, Richards FM. An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- Smith DL, Deng Y, Zhang Z. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J Mass Spectrom. 1997;32(2):135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Tello-Solis SR, Hernandez-Arana A. Effect of irreversibility on the thermodynamic characterization of the thermal denaturation of Aspergillus saitoi acid proteinase. Biochem J. 1995;311(Pt 3):969–974. doi: 10.1042/bj3110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui Y, Wintrode PL. Hydrogen/deuterium exchange-mass spectrometry: a powerful tool for probing protein structure, dynamics and interactions. Curr Med Chem. 2007;14(22):2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006a;357(5):1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006b;25(1):158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees celsius. Anal Chem. 2008;80(17):6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1(2):132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- Wang L, Smith DL. Capsid structure and dynamics of a human rhinovirus probed by hydrogen exchange mass spectrometry. Protein Sci. 2005;14(6):1661–1672. doi: 10.1110/ps.051390405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis DD, Engen JR, Kass IJ. Semi-Automated Data Processing of Hydrogen Exchange Mass Spectra Using HX-Express. J Am Soc Mass Spectrom. 2006a;17(12):1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006b;17(11):1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Woodard SL, Mayor JM, Bailey MR, Barker DK, Love RT, Lane JR, Delaney DE, McComas-Wagner JM, Mallubhotla HD, Hood EE, Dangott LJ, Tichy SE, Howard JA. Maize (Zea mays)-derived bovine trypsin: characterization of the first large-scale, commercial protein product from transgenic plants. Biotechnol Appl Biochem. 2003;38(Pt 2):123–130. doi: 10.1042/BA20030026. [DOI] [PubMed] [Google Scholar]
- Woodward C, Simon I, Tuchsen E. Hydrogen exchange and the dynamic structure of proteins. Mol Cell Biochem. 1982;48(3):135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]
- Wu Y, Engen JR, Hobbins WB. Ultra Performance Liquid Chromatography (UPLC) Further Improves Hydrogen/Deuterium Exchange Mass Spectrometry. J Am Soc Mass Spectrom. 2006a;17(2):163–167. doi: 10.1016/j.jasms.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kaveti S, Engen JR. Extensive deuterium back-exchange in certain immobilized pepsin columns used for H/D exchange mass spectrometry. Anal Chem. 2006b;78(5):1719–1723. doi: 10.1021/ac0518497. [DOI] [PubMed] [Google Scholar]
- Yang H, Smith DL. Kinetics of cytochrome c folding examined by hydrogen exchange and mass spectrometry. Biochemistry. 1997;36(48):14992–14999. doi: 10.1021/bi9717183. [DOI] [PubMed] [Google Scholar]
- Zehl M, Rand KD, Jensen ON, Jorgensen TJ. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J Am Chem Soc. 2008;130(51):17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2(4):522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Post CB, Smith DL. Amide hydrogen exchange determined by mass spectrometry: application to rabbit muscle aldolase. Biochemistry. 1996;35(3):779–791. doi: 10.1021/bi952227q. [DOI] [PubMed] [Google Scholar]