Abstract
Motivation: Transcription factors (TFs) are crucial during the lifetime of the cell. Their functional roles are defined by the genes they regulate. Uncovering these roles not only sheds light on the TF at hand but puts it into the context of the complete regulatory network.
Results: Here, we present an alignment- and threshold-free comparative genomics approach for assigning functional roles to DNA regulatory motifs. We incorporate our approach into the Gomo algorithm, a computational tool for detecting associations between a user-specified DNA regulatory motif [expressed as a position weight matrix (PWM)] and Gene Ontology (GO) terms. Incorporating multiple species into the analysis significantly improves Gomo's ability to identify GO terms associated with the regulatory targets of TFs. Including three comparative species in the process of predicting TF roles in Saccharomyces cerevisiae and Homo sapiens increases the number of significant predictions by 75 and 200%, respectively. The predicted GO terms are also more specific, yielding deeper biological insight into the role of the TF. Adjusting motif (binding) affinity scores for individual sequence composition proves to be essential for avoiding false positive associations. We describe a novel DNA sequence-scoring algorithm that compensates a thermodynamic measure of DNA-binding affinity for individual sequence base composition. Gomo's prediction accuracy proves to be relatively insensitive to how promoters are defined. Because Gomo uses a threshold-free form of gene set analysis, there are no free parameters to tune. Biologists can investigate the potential roles of DNA regulatory motifs of interest using Gomo via the web (http://meme.nbcr.net).
Contact: t.bailey@uq.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The regulation of gene expression is crucial in the development and functioning of cells. DNA-binding proteins called transcription factors (TFs) are one cog in the regulatory machinery shared by all cellular and multi-cellular organisms. The human genome, for instance, is estimated to contain up to 3000 such TFs, of which only about 1000 are annotated as such, and only 62 have experimentally verified in vivo DNA-binding and regulatory activity (Vaquerizas et al., 2009). For the vast majority of TFs in higher organisms, the set of genes they regulate, as well as the biological functions they are involved in, is largely unknown.
However, for a growing number of TFs, models of their DNA-binding propensities are known. The advent of protein binding microarrays (PBMs) in particular is rapidly making DNA-binding affinity data available for large numbers of TFs in many species (Berger and Bulyk, 2009). Another source of DNA-binding affinity data for TFs is chromatin immunoprecipitation followed by deep sequencing (ChIP-seq; Barski and Zhao, 2009). Both of these types of data can be used to construct a ‘position weight matrix’ (PWM; Stormo, 2000) model of the DNA-binding affinity of a given TF. Such models are herein referred to as motifs, and can also model the DNA-binding affinity of other molecules, including microRNAs. Gene expression data can also be used to discover DNA-binding motifs utilizing ab initio motif discovery from the promoters of sets of co-expressed genes (Roven and Bussemaker, 2003). In contrast to the PBM and ChIP-seq approaches, motif discovery in sets of co-expressed genes usually results in DNA-binding motifs for which the binding molecule (e.g. TF or microRNA) is unknown.
One application of DNA-binding motifs is the in silico prediction of the regulatory targets of the TFs. To predict the targets of a TF, its binding motif is used to score promoter regions of genes for their potential to bind the TF protein. It is well-known that such predictions are not very specific, and many false positives must be tolerated if all regulatory targets of a TF are to be detected (Wasserman and Krivan, 2003). However, as we have previously shown, even such noisy TF target predictions contain sufficient information to allow us to make useful predictions of the biological roles of the TF (Bodén and Bailey, 2008). The focus of the current work is to improve the sensitivity of computational methods that make TF role predictions using DNA-binding motifs.
Our original method for predicting the roles of TFs starts with a PWM motif describing the DNA-binding affinity of the TF. We use the PWM to score the promoter region of each gene in the genome for its likelihood to be bound by the TF. We then use the resulting ‘affinity’ scores to test each term in the Gene Ontology (GO; Ashburner et al., 2000) for association with high-scoring genes. In contrast to other approaches (e.g. Sinha et al., 2008) that use an user-specified affinity score threshold to separate TF target genes from non-targets, we use the Mann–Whitney U-test (Mann and Whitney, 1947), also known as Wilcoxon rank sum test, to determine if the genes associated with a particular GO term have significantly high scores. This method was implemented in the original Gomo algorithm (Bodén and Bailey, 2008), which reports GO terms with significant rank sum P-values, after adjusting for multiple tests.
One obvious place to look for improvement in TF role prediction is in the quality of the PWM-based function used by Gomo to score promoter regions for their potential to be targets of the TF. In a similar application, other researchers (Sinha et al., 2008) used the likelihood function of a motif-based hidden Markov model (HMM) to score promoters. They then reran the HMM 100 times on shuffled versions of the promoter in order to convert the likelihood to a P-value, which they used as their final target function. This scoring function is computationally expensive compared with the ‘average motif affinity’ (Ama) function used by Gomo (Fig. 2 in Supplementary Material 1). It does, however, reduce the significance of binding affinity scores for promoters whose GC content is similar to that of the motif, which has been suggested to be important when using DNA-binding scores in gene set enrichment analyses (Sinha et al., 2008). In the current work, we examine the importance of GC content compensation for binding affinity scores, and develop a more computationally efficient scoring algorithm.
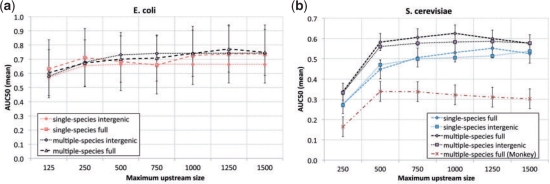
Fig. 2.
Multiple-species Gomo prediction accuracy. Each point shows the average AUC50 of TF–GO term association predictions made by Gomo in the key species E.coli (a) or S.cerevisiae ( b). Points labeled ‘multiple-species’ use promoter sequences from the key species and three related species; Monkey results use Monkey (Moses et al., 2004) minimum P-value scores instead of Ama scores (Supplementary Material 1). Points labeled ‘single-species’ use promoter sequences from the key species only, and are shown for comparison. The AUC50 is computed using a single TF, then averaged over TFs. The X-axis shows the upstream extent of promoter sequences (‘full’), or the maximum upstream extent when they are truncated at the first ORF (‘intergenic’). For clarity, standard error bars are shown for the ‘full’ promoter sequence set only; standard error bars for the ‘intergenic’ promoter set are similar.
A second obvious approach to consider for improving TF role prediction algorithms is to use comparative genomics. Such an approach assumes that functional TF binding sites are to some degree conserved in the promoters of orthologous genes from related species. Based on this assumption, numerous methods for motif-based TF binding site and TF target gene prediction have been developed that utilize sets of orthologous sequences from multiple species. The validity of the assumption is evident from the success of such methods, which includes phylogenetic footprinting and phylogenetic motif modeling (Hawkins and Bailey, 2008). We could chose to use one of these methods to score genes for their likelihood as targets of a given TF, rather than one of the single-species binding affinity-based motif scores used by Gomo or Sinha et al. (2008). However, the above phylogenetic scoring methods require multiple alignments of orthologous genes from species being compared, and they suffer when the alignments contain inaccuracies or when the location or orientation of the TF binding sites has not been conserved (Moses et al., 2006). Phylogenetic motif modeling has the additional problem of not scaling well to more than about five related species (Hawkins and Bailey, 2008).
In this work, we present a comparative genomics extension to Gomo (‘multiple-species Gomo’) that does not require multiple alignments. The approach requires sets of orthologous gene sequences, but does not require (nor use) alignments of the sequences. Instead, our method estimates the association between the query TF and a GO term independently for each species, and then combines these single-species association scores into a single score for the TF–GO term pair. As our method requires only a single GO map, only one of the species need functional annotation. We use multiple-species Gomo to assign functional roles to TFs in bacteria, fungi and mammals, and validate the accuracy of these predictions using known sets of regulatory targets for a number of TFs. We further validate the predictions made by multiple-species Gomo by conducting a false discovery rate (FDR) analysis of the predicted TF–GO term associations. The enhanced version of Gomo is available for download and Gomo 's functionality has been fully integrated with the Meme motif discovery tool (Bailey et al., 2009; http://meme.nbcr.net), so that motifs discovered by Meme can be sent with a single mouse click to Gomo for analysis.
2 METHODS
2.1 Incorporating comparative genomics into TF role prediction
Our alignment-free approach for improving the motif-based prediction of the roles of TFs is quite straightforward. In a nutshell, it works as follows. The input consists of a DNA-binding motif, a GO annotation map and the promoter sequences for n genomes. Using the method described in Bodén and Bailey (2008) (and see below), we compute an association score between the (putative) targets of the input TF motif and each GO term in the GO map. We do this separately n times, each time using the promoter sequences from a different genome. We then combine the n scores for each TF–GO term pair into a single score. The final output of the method is, for each TF–GO term pair, the q-value of the combined score. The details of the algorithm are given below, and illustrated in Figure 5 in Supplementary Material 1.
The final score of our method combines the evidence for a TF–GO term association from each of the species. This is done by combining the single-species scores (P-values) for a single TF–GO pair by taking their geometric mean, which is a simple way to combine evidence from multiple P-values. Thus, if St,i is the association score for GO term t computed using genome i∈[1,…, n], then the overall association score for term t is defined as
| (1) |
The distribution of this score is not easy to estimate analytically due to the non-independence of the P-values being combined. Therefore, we use a permutation test to assign statistical significance (q-values) to the multiple-species association scores, St, as we describe below.
The permutation test we use for computing the statistical significance of St, the association score for GO term t with respect to the current TF motif, is based on essentially the same null model as the rank sum test. The rank sum test null model assumes that the order of the gene names, when sorted by motif affinity score, is random. Therefore, our test permutes the assignment of gene names to scores. Because the binding affinity scores for orthologous genes are highly correlated, we permute the gene names for each species in exactly the same way. Failure to do this results in a null model that overestimates the significance of some TF–GO term associations (data not shown). As illustrated in Supplementary Material 1, Figure 4, after each permutation of the gene-name-to-score relationships, we compute null scores for all terms t. In this study, we repeat this process for 100 permutations, resulting in 100 null scores for every GO term, t. For a given TF, different GO terms have very similar null score distributions (data not shown). So, in order to increase the statistical power of the permutation test, we treat all sampled scores for a single TF motif as samples from a single null distribution. This gives us 100x null scores for estimating the significance of St, where x is the total number of GO terms. We compute empirical P-values for each real St by counting the number of null scores that are smaller than St and then dividing by the total number of null samples. These P-values are then adjusted for multiple tests by conversion to q-values using the method of Storey et al. (2004).
Our previous implementation of Gomo uses the ‘Ama’ score to rank promoters as (putative) targets of a TF. However, because the base composition of promoters in higher eukaryotes is highly variable (Sinha et al., 2008), binding affinity scoring methods that ignore this variability might predict TF–GO term associations that are not biologically meaningful. For this study, we therefore developed a new version of the Ama algorithm (part of the MEME Suite of tools; Bailey et al., 2009) that analytically estimates P-values for Ama scores based on a zero-order Markov model of the particular promoter sequence being scored (described in Fig. 1 in Supplementary Material 1). For reasons of computational efficiency, this is implemented assuming that the sequence has equal G and C content on a given strand, i.e. Pr(G) = Pr(C), and likewise for A and T. The method computes analytical Ama score distributions for a range of GC contents, and uses linear interpolation to estimate the P-value of the Ama score of a sequence, based on its actual GC content. Ama can also compute P-values that are not compensated for the GC content of individual sequences, but are based on a single, zero-order Markov model of all the promoters in a genome. Compared to the motif-based HMM (Hmm0) introduced by Sinha et al. (2008), which calculates empirical P-values for each sequence, our GC-compensated version of Ama, which calculates analytical P-values, is almost an order of magnitude faster. The P-values computed by the two methods have a median correlation coefficient of 0.92 for the yeast motifs (Fig. 2 in Supplementary Material 1). This speedup is important, because it makes a web-based version of Gomo feasible. Unless noted, in this study all results are based on GC-corrected Ama scores.
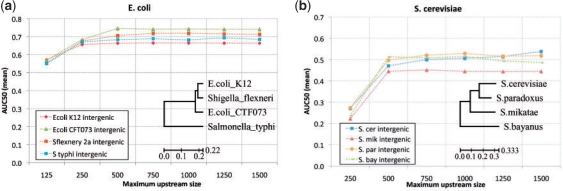
Fig. 1.
Single-species Gomo prediction accuracy using transferred GO maps. Each point shows the average AUC50 of TF–GO term associations predicted by Gomo using the E.coli (a) or S.cerevisiae (b) GO map and TFs, and promoter sequences from the single given species. The AUC50 is computed using a single TF, then averaged over TFs. The X-axis shows the maximum upstream extent of promoter sequences, which are truncated at the first ORF. The inset shows the phylogenetic tree of the corresponding species. Branch lengths denote average substitutions per site.
2.2 Evaluation methods and datasets
We study the ability of our prediction method (Gomo) to correctly identify associations between the target genes of a TF and GO functional categories, given only the DNA-binding motif of the TF. We focus on three relatively well-studied species: Escherichia coli, Saccharomyces cerevisiae, Homo sapiens. For each species, we utilize its GO annotation, the sequences of its promoters, the sequences of promoters of orthologous genes from three additional species and a species-specific set of TF binding motifs. To evaluate prediction accuracy, we create two sets of reference TF–GO associations based on the known targets of the TFs in E.coli and S.cerevisiae, respectively (Supplementary Material 2). (We do not create a reference set of associations for H.sapiens due to the relatively small number of known gene targets for human TFs.) We measure the accuracy of the associations predicted by Gomo with these reference associations in terms of the area under the ROC curve (AUC). As a second measure of prediction reliability, we use FDR analysis.
2.2.1 Evaluation using known TF–GO term associations
To create our reference sets of TF–GO associations for E.coli and S.cerevisiae, we apply the approach described in our previous study (Bodén and Bailey, 2008). For each organism, we first obtain a set of known gene targets for TFs. We obtain the known gene targets of TFs from RegulonDB v6.2 ((Gama-Castro et al., 2008; http://regulondb.ccg.unam.mx/) for E.coli and from MacIsaac et al. (2006) for S.cerevisiae. We then perform gene set enrichment analysis by applying the Fisher's exact test (Fisher, 1958) to the intersection of the set of known targets of a single TF and the set of genes annotated with a given GO term. We include a TF–GO term pair in our reference set for the given organism if, after adjusting for multiple tests, the enrichment is significant at 0.01 level. Consistent with previous researchers (e.g. Sinha et al., 2008), we do not include any TF–GO pairs containing non-specific GO terms—terms that are annotated to >20% of genes in the given genome. Our E.coli reference set of TF–GO associations contains 87 TF–GO pairs, and our S.cerevisiae set has 503 pairs.
We utilize our TF–GO association reference sets to measure the accuracy of predictions made by Gomo. We treat the TF–GO pairs in the reference set for a given species as ‘positives’, and all other TF–GO pairs as ‘negatives’. Our accuracy metric is AUC50, AUC up to the 50th false positive (Gribskov and Robinson, 1996), when all TF–GO terms are sorted by increasing score St. This metric is appropriate because it emphasizes differences in accuracy among prediction methods where it matters to biologists—in the short list of most confident predictions made by a prediction method. We compute the AUC50 for each TF represented by at least one TF–GO pair in the reference set for a given species. The AUC50 value will be 1 if Gomo assigns lower St scores to all the GO terms associated with the TF according to reference set (‘positive’ GO terms) than it does to other GO terms (‘negative’ GO terms). It will be zero if 50 (or more) ‘negative’ GO terms have lower St scores than the GO terms associated with the TF according to reference set. Our final accuracy measure for the species is the average of the AUC50 values of the TFs that have GO terms in the reference set.
2.2.2 Evaluation using FDR
To further evaluate the reliability of TF–GO term associations predicted made by Gomo, we also perform FDR analysis. FDR analysis allows us to estimate the fraction of predicted associations that are statistically significant, but cannot guarantee the biological significance of predictions. Nonetheless, FDR analysis has been widely used for estimating the accuracy of both TF role predictions (Sinha et al., 2008) and TF binding site predictions (Kheradpour et al., 2007) when only incomplete or noisy validation sets are available. As discussed in Section 2.1, Gomo computes the q-values of all TF–GO term associations from their empirical P-values. The q-value of a TF–GO pair represents the minimum FDR at which that association would be considered significant. Therefore, we report the number of associations detected at a q-value of 0.05. When computing q-values, we combine the P-values of all TF–GO pairs for a single organism across all TFs used as queries in order to adjust for all of the multiple tests conducted. As a further check on our FDR estimates, we verify that no significant predictions are reported when the input sequences are permuted.
2.2.3 Binding motifs
We perform our study using position-specific probability representations of TF binding motifs taken from the following sources. For E.coli, we use 85 of the 88 TF motifs from the Prodoric database release 8.9 (http://prodoric.tu-bs.de/; Münch et al., 2003). (We discard three TF motifs—MX000203, MX000181 and MX000160—because they are highly similar to other motifs.) For S.cerevisiae, we use the 124 yeast TF binding motifs from MacIsaac et al. (2006). For H.sapiens, we use the 56 H.sapiens TF motifs contained in the JASPAR CORE database release 2008 (Sandelin et al., 2004). We use all of the above motifs in the FDR analysis, and the subsets referenced in the TF–GO association reference sets for E.coli and S.cerevisiae in the AUC50 accuracy analysis. When the original source gives the motif in terms of observed ‘counts’, we convert them to position-specific probability PWMs by adding ‘pseudocounts’ of 0.01 times the average base frequencies in the organism's promoter sequences, B, before normalizing to probabilities.
2.2.4 Promoter sequences
We create sets of promoter sequences for each of our key species, E.coli, S.cerevisiae and H.sapiens. Then, for each key species, we identify the orthologous genes in each of three related species and construct three additional sets of promoter sequences. Critically, in the related-species promoter sets, we use the gene name from the orthologous gene in the key species as the gene name for a promoter. This allows us to use the GO map for the key species when we compute the association scores for the related species. Our related species for E.coli (K12) are E.coli (CTF073), Salmonella typhimurium and Shigella flexneri 2a. Our S.cerevisiae related species are S.paradoxus, S.mikatae and S.bayanus. For H.sapiens, our related species are Mus musculus, Canis familiaris and Equus caballus.
Our definition of what a promoter is depends on the key species. For S.cerevisiae and H.sapiens, we define the promoter to be the upstream region [relative to the transcription start site (TSS) of a gene]. Because prokaryotes organize their genes into transcriptional units and operons that are transcribed together, for E.coli we define promoters to be the sequence upstream of operons, rather than of genes. We take operon information for E.coli K12 from RegulonDB v6.2 (Gama-Castro et al., 2008).
To identify orthologous genes in species related to E.coli, we use the Enterobacter Genome Browser (http://engene.fli-leibniz.de/) to search for best pairwise Blast hits to E.coli K12 genes. For simplicity, we assume that the operons are not altered across the species, i.e. the genes and their order stay the same in an operon across closely related species. To identify orthologous genes in S.cerevisiae relatives, we use the mappings from Kellis et al. (2003). To identify genes orthologous to H.sapiens genes in related species, we use one-to-one ortholog gene maps obtained from Biomart (Smedley et al., 2009).
To create the promoter sequence sets for E.coli and S.cerevisiae and related species, we use the RSAT sequence extraction tool (Thomas-Chollier et al., 2008). We study varying the size of the upstream region, as well as allowing it to overlap upstream open reading frames (ORFs). We refer to the truncated promoters as the ‘intergenic’ set, and to the promoters that (may) overlap upstream ORFs as the ‘full’ set. For H.sapiens and related species, we define the promoter to be the 1000 bp upstream of the TSS, and extract them using Biomart (Smedley et al., 2009).
2.2.5 GO annotation
To create GO maps for the three key species, we use the E.coli GO annotation file v1.5, S.cerevisiae v1.1411 and H.sapiens v1.12, respectively. From each of these files, we create a GO map file that lists, for each GO term, the gene names annotated with it. Note that for E.coli, our promoters are upstream of operons, not genes, so our E.coli GO map maps GO terms to operons. To create this map, we first use the GO annotation file for E.coli to assign to each operon the union of all GO terms associated with any gene contained in the operon.
3 RESULTS
3.1 Successful transfer of GO annotation to related species
Our method for incorporating comparative genomics into TF role prediction depends on GO annotation being reliable when mapped from a gene in the key species to its ortholog in another species. The validity of this assumption is borne out for our choice of related species for E.coli and S.cerevisiae by the results shown in Figure 1, which is based on running Gomo with promoters from a single species. For a wide range of size definitions of the promoter regions, the accuracy (mean AUC50) of TF–GO term associations predicted by Gomo for the related species is similar to the accuracy using the key species. Indeed, for E.coli, the measured accuracy is slightly higher using the promoters from its related species (Fig. 1a). The E.coli reference set contains only 87 TF–GO term pairs, thus its accuracy measurements are based on a fairly small sample. The S.cerevisiae reference set is much larger (503 TF–GO term pairs), and the prediction accuracy using S.cerevisiae promoters is very similar to that using promoters from two of its related species (Fig. 1b). Somewhat surprisingly, using promoters from S.mikatae yields lower accuracy than using those from S.bayanus, even though S.mikatae appears evolutionarily closer to S.cerevisiae based on multiple alignments of all orthologous intergenic regions (Kellis et al., 2003). The phylogenetic trees shown in Figure 1 are for reference only—our method does not use them. We created the phylogenetic tree for enterobacter from our 1500 bp promoter sequences using the topology from Elena et al. (2005). The tree for yeast is from Kellis et al. (2003) and is based on intergenic sequences.
3.2 Appropriate size for promoters
The (maximum) size of the upstream region defined to be the promoter of a gene affects the accuracy of predictions made by Gomo, as seen in Figure 1. For both enterobacter and yeast, prediction accuracy drops sharply if promoters are limited to upstream regions <500 bp. Increasing the maximum promoter size seems to confer little or no increase in the accuracy of Gomo predictions for enterobacter species. However, for yeast species the optimal promoter size may be closer to 750–1000 bp, which is in agreement with the observation made by Thomas-Chollier et al. (2008) that 99% of known regulatory elements in promoters are found in regions within 800 bp upstream of the TSS.
3.3 Benefits of our comparative genomics approach
We now assess the benefit of using the proposed method of incorporating comparative genomics into TF role prediction. To do this, we assess the accuracy of predictions made by single- and multiple-species Gomo in two ways. First, for E.coli and S.cerevisiae we utilize ‘gold standard’ sets of TF–GO term relationships for each of these organisms. We realize that these reference TF–GO term sets are extremely incomplete due to the current lack of knowledge about the functions of many TFs. Consequently, although useful for comparing the accuracy of algorithms, these gold standards will label many true relationships as ‘false positives’. Therefore, we also perform FDR analyses of predictions on E.coli, S.cerevisiae and H.sapiens, and compare the number of statistically significant TF role predictions made by Gomo when using a single species or when using multiple species. As a further check, we also determine that Gomo makes no predictions judged by FDR to be statistically significant (q≤0.05) when given shuffled promoters as input.
Compared with using a single species, Gomo using multiple species gives a substantial increase in prediction accuracy (mean AUC50) for the yeast species, and a slight increase for the enterobacter species (Fig. 2; e.g. compare curves labeled ‘single-species intergenic’ and ‘multiple-species intergenic’). For yeast, the increase in accuracy is statistically significant using promoters defined as 500, 750 or 1000 bp upstream regions (P<0.05, two-tailed, paired t-test). This is true both for promoter defined as upstream regions of the given length (‘full’) and for regions truncated upon reaching an upstream ORF (‘intergenic’). The improvement in accuracy is due to the use of multiple genomes—the mean AUC50 is 0.63 using the four yeast species compared with only 0.53 using the single species (19% improvement, 1000 bp, ‘full’ promoters). Although using the ‘full’ yeast promoter regions yields slightly better accuracy than using the truncated regions, this difference is not statistically significant. With enterobacter, the increase in accuracy using multiple species is smaller than with yeast, and the relatively small size of the set of known enterobacter TF–GO term associations (only 87 compared with 503 pairs) causes the error bars to be large. However, in our FDR analysis below, only using multiple species allows Gomo to discover any statistically significant TF–GO term associations in E.coli at all.
The multiple-species prediction accuracy results shown in Figure 2a suggest that the optimal approach for identifying TF–GO term associations in E.coli is to define promoters as ‘full’ (non-truncated) upstream regions of length 1250 bp. However, the reference set for E.coli contains only 87 TF–GO pairs, so it is not possible to draw any strong conclusions about the optimal size for upstream regions to use as enterobacter promoters. Nonetheless, the results for S.cerevisiae (Fig. 2b), which are based on a much larger reference set, support a similar promoter definition. Multiple-species S.cerevisiae predictions are most accurate using ‘full’ promoters of length 1000 bp. Longer regions appear to decrease the accuracy of both single- and multiple-species Gomo, as would be expected if regions farther than 1000 bp from the TSS were less likely to contain TF binding sites. Since the Ama score averages the motif affinity along the entire defined promoter region, the signal-to-noise ratio decreases when the region is made too long.
As a further evaluation of the plausibility of TF–GO term predictions made by Gomo, we perform a FDR analysis. As the first step in our analysis, we follow Sinha et al. (2008) and perform a test where we scramble all of the promoter sequences and input them to Gomo. For all three key species, both single- and multiple-species versions of Gomo using GC-compensated Ama P-values report zero predictions with q≤0.05. This provides a negative control on the reliability of the q-values reported by Gomo. However, when we score the scrambled yeast species promoters using non-GC-compensated Ama P-values, we get 41 significant predictions using multiple-species Gomo. For mammalian species, the number of significant predictions using scrambled promoters is 935 (H.sapiens) and 1403 (multiple-species Gomo). This makes it clear that normalizing the TF binding affinity scores for the base content of the promoter sequences is important for accurate estimation of the FDR. In what follows, we use GC-compensated Ama P-values as input to Gomo.
Having established that Gomo reports no significant (q≤0.05) TF–GO term pairs when given scrambled promoters as input, we now count the number of significant pairs reported when using the real promoters. Rather than counting all significant TF–GO term pairs, we only count significant pairs for the most specific GO term. In other words, if for a given TF, the GO term ‘neuroblast division’ and its parent term ‘neurogenesis’ are both deemed significant by Gomo, we only include the former term in our count. Counting in this way is appropriate because GO is a hierarchy with the least specific terms at the base. If a TF is associated with a GO term, it is implicitly associated with all the parents of that term. Since a method that detects associations between a TF and highly specific GO terms is more useful than one that reports only general GO terms, we also measure the average depth of the most specific GO terms predicted by Gomo. These results are summarized for both single- and multiple-species Gomo in Table 1.
Table 1.
Improvement in TF role prediction using comparative genomics
|
Escherichia coli K12 |
Saccharomyces cerevisiae |
Homo sapiens |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Single | Multiple | Increase | Single | Multiple | Increase | Single | Multiple | Increase | |
| species | species | (%) | species | species | (%) | species | species | (%) | |
| Significant TF–GO term pairs | 0 | 14 | NA | 420 | 733 | 75 | 371 | 1112 | 200 |
| GO terms per TF tested | 0 | 0.16 | NA | 3.4 | 5.9 | 75 | 6.6 | 19.8 | 200 |
| Covered TFs | 0 | 9 | NA | 99 | 113 | 14 | 48 | 56 | 17 |
| Term specificity | 0 | 4.0 | NA | 4.5 | 4.6 | 2 | 3.8 | 4.2 | 11 |
| TFs tested | 85 | 124 | 56 | ||||||
The table shows FDR (q≤0.05) results for single- and multiple-species Gomo. The results shown are the total number of most-specific significant pairs (‘significant TF–GO term pairs’), the average number of most-specific GO terms per TF tested (‘GO terms per TF tested’), the number of TFs with at least one significant TF–GO term pair (‘covered TFs’), the average depth in the GO hierarchy of significant GO terms (‘term specificity’), and the total number of TFs in each experiment (‘TFs tested’). All results are for GC-compensated Ama scores and ‘full’ promoters of 500, 750 and 1000 bp for enterobacter, yeast and mammals, respectively. NA, not applicable.
Substantially, more significant TF–GO term pairs for E.coli, S.cerevisiae and H.sapiens are predicted by multiple-species Gomo compared with single-species Gomo (Table 1, ‘significant TF–GO term pairs’). We observe increases of 75 and 200% in the number of significant pairs for S.cerevisiae and H.sapiens, respectively. For E.coli, there are actually no significant pairs (q≤0.05) using the single-species approach, but 14 pairs are significant when we use all four enterobacter promoter sets as input.
The average number of significant GO terms predicted for TFs by Gomo increases accordingly (Table 1, ‘GO terms per TF tested’). Using four yeast species, multiple-species Gomo predicts about six most-specific GO terms per TF; using the four mammal species, multiple-species Gomo predicts 20 most-specific GO terms per TF. Very few predictions are made in E.coli when using the four enterobacter species (0.16 terms per TF tested), indicating that the sensitivity of multiple-species Gomo in enterobacter is very low.
Using multiple species also increases the chance that Gomo will predict that at least one GO term is significantly associated with a given TF. No TF–GO term associations are predicted by Gomo using E.coli promoters alone, whereas using the four enterobacter species, multiple-species Gomo predicts significant associations for nine (out of 85) TFs (Table 1, ‘covered TFs’). The number of TFs with at least one significant GO term increases by 14% for S.cerevisiae and 17% for H.sapiens when we apply our multiple-species approach. In S.cerevisiae and H.sapiens, single-species Gomo is more successful at finding at least one significant GO term for each TF than it is in E.coli, ‘covering’ 99 (out of 124) TFs in S.cerevisiae, and 48 (out of 56) in H.sapiens. However, using multiple species results in improvement in this regard as well for S.cerevisiae and H.sapiens. Multiple-species Gomo identifies at least one significant GO term for almost all yeast TFs (113 out of 124), and for all 56 H.sapiens TFs tested.
Importantly, the specificity of predicted GO terms for a given TF increases when using our multiple species with Gomo (Table 1, ‘term specificity’). The increase in specificity (as measured by the minimum distance from the predicted term to the root of the GO hierarchy) increases marginally using four species for S.cerevisiae (2%), and somewhat more (17%) when using four mammal species for H.sapiens, compared to using a single species. When we compare the sets of predictions made on H.sapiens using the single- and multiple-species approach, respectively, we find that only 72 significant predictions from the most specific set in the single species have no or only less specific counterparts in the multiple-species results. In contrast, 950 of the multiple-species results are more specific than corresponding single-species predictions, or are not captured by the single-species approach at all. Given that the total numbers of significant predictions are 371 for single-species and 1112 for multiple-species (Table 1), we observe a substantial increase in both the number of significant predictions and the specificity of GO terms when using multiple-species Gomo. Since more specific GO terms convey more detailed biological insight, the predictions made by multiple-species Gomo are more informative than those made using a single species.
Finally, Gomo can be used to create ‘role-centric’ regulatory maps, where TF motifs are connected via the predicted GO terms considered significant (Supplementary Material 1, 3 and 4). This representation of the data facilitates the identification of groups of TFs that are collectively involved in a particular biological process. Role-centric maps can also shed light on when secondary binding motifs (Berger and Bulyk, 2009) are functionally distinct from the primary binding motif.
4 DISCUSSION
We have presented a comparative genomics approach for assigning biological roles to sequence motifs using a form of gene set enrichment analysis. The approach does not require multiple alignments because role predictions are made independently for each comparative genome and then combined across genomes. This means that the method does not assume that the location and orientation of binding sites is conserved across species. The method does require that orthologous genes be identified in the species being utilized, as would also be the case for a method employing multiple alignments. The approach also assumes that the functions of orthologous genes, and the DNA-binding affinity of the regulatory molecule, have been conserved in the species being used. Although the above assumptions are no doubt sometimes violated, our comparative genomics approach nonetheless substantially improves the sensitivity of TF role predictions.
Our principal result is that it is possible to substantially improve the prediction of the association of a DNA-binding motif with an annotation term by mapping the annotation from a key species to related species, and combining the association scores for a single motif and term across species. The approach requires only that we are able to compute the P-value of the motif-term association in each species, and we combine across species by taking the geometric mean of the P-values. A simple permutation test then assigns significance values to the combined motif-term score. Since our method is alignment-free, it avoids problems that would be caused by imperfect alignments or ‘motif drift’ (Moses et al., 2006).
Our FDR analysis has shown the importance of compensating motif affinity scores for the base content of the sequences being scored. We have demonstrated that our GC-compensated Ama scores pass a ‘shuffled sequence test’, yielding no spurious significant predictions using such random data. We have also shown that our implementation can compute such GC-compensated scores fast enough to make a web-based service feasible. It should be noted that Gomo is not limited to Ama-derived gene scores but can work on any sequence scoring scheme. For example, Hmm0 (Sinha et al., 2008) can compute motif affinity scores using more than one TF motif at a time, which enables Gomo to investigate the role of synergistic TFs.
For the particular case of predicting TF–GO term associations, we have shown that our method is not particularly sensitive to the size of the upstream region designated as the promoter of a gene. For species as diverse as enterobacter and mammals, using regions of 1000 bp upstream of the TSS will probably work about as well as any other reasonable definition. Our results indicate that one should not truncate putative promoters at the nearest upstream ORF, but include overlapped ORF sequence up to the 1000 bp (or other) limit. This seems to suggest that closely spaced genes may reciprocally harbor regulatory sequence elements.
We have shown that using a H.sapiens TF motif with our multiple-species version of Gomo almost always results in at least one significant prediction of an associated GO term, and in 20 significant terms on average. Most yeast TF motifs also yield at least one significant prediction, but very few E.coli motifs do. The relative failure of even our multiple-species approach on enterobacter might be due to the way we aggregate gene annotations for all genes in an operon, or it might be due to the relative sparsity of GO annotation for bacteria due to their simple cellular structure. However, other factors than just the number of terms in the GO hierarchies for bacteria, fungi and mammals (1654, 4578 and 9409, respectively) may be at play here.
Although we have focused specifically on utilizing this approach with TF motifs and GO annotation via the Gomo software, the general method should be equally applicable to other types of sequence motifs (e.g. microRNA-binding motifs) and other types of annotation (e.g. metabolic pathways or gene sets from analyses of expression data). Gene set enrichment analysis approaches similar to single-species Gomo have previously been shown to be useful with these and other types of motifs and annotation sources (e.g. Sinha et al., 2008). Since nothing in the implementation of Gomo is specific to TFs and GO annotation, multiple-species Gomo can perform analyses using any type of DNA-binding motif expressed as a PWM and any mapping of gene names to functional terms (e.g. pathway names or tissue types). In the future, we will apply multiple-species Gomo to annotate more extensive collections of motifs with additional (non-GO) types of functional annotation.
ACKNOWLEDGEMENTS
The authors like to thank James Johnson for his skillful help during the implementation of multiple-species Gomo and for developing the web server under the supervision of T.L.B. and F.A.B.
Funding: T.L.B and M.B. are funded by NIH/NCRR award R01 RR021692 and by the ARC Centre of Excellence in Bioinformatics. F.A.B and D.C.B are funded by UQ International Research Tuition Award.
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Zhao K. Genomic location analysis by chip-seq. J. Cell Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodén M, Bailey TL. Associating transcription factor-binding site motifs with target GO terms and target genes. Nucleic Acids Res. 2008;36:4108–4117. doi: 10.1093/nar/gkn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, et al. Genomic divergence of Escherichia coli strains: evidence for horizontal transfer and variation in mutation rates. Int. Microbiol. 2005;8:271–278. [PubMed] [Google Scholar]
- Fisher RA. Statistical Methods for Research Workers. Oliver & Boyd; 1958. Chapter 4. [Google Scholar]
- Gama-Castro S, et al. RegulonDB (version 6.0): gene regulation model of E scherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M, Robinson NL. Use of receiver operating characteristic (ROC) analysis to evaluate sequence matching. Comput. Chem. 1996;20:25–33. doi: 10.1016/s0097-8485(96)80004-0. [DOI] [PubMed] [Google Scholar]
- Hawkins JC, Bailey TL. The statistical power of phylogenetic motif models. In: Vingron M, Wong L, editors. Research in Computational Molecular Biology, 12th Annual International Conference, RECOMB 2008. Vol. 4955. Heidelberg: Springer; 2008. pp. 112–126. [Google Scholar]
- Kellis M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, et al. Reliable prediction of regulator targets using 12 D rosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, et al. An improved map of conserved regulatory sites for S accharomyces cerevisiae. BMC Bioinform. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947;18:50–60. [Google Scholar]
- Moses AM, et al. MONKEY: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, et al. Large-scale turnover of functional transcription-factor binding sites in Drosophila. PLoS Comput. Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R, et al. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 2003;31:266–269. doi: 10.1093/nar/gkg037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roven C, Bussemaker HJ. REDUCE: an online tool for inferring cis-regulatory elements and transcriptional module activities from microarray data. Nucleic Acids Res. 2003;31:3487–3490. doi: 10.1093/nar/gkg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, et al. JASPAR: an open-access database for eukaryotic transcription-factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, et al. Systematic functional characterization of cis-regulatory motifs in human core promoters. Genome Res. 2008;18:477–488. doi: 10.1101/gr.6828808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, et al. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, et al. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. R. Stat. Soc. B. 2004;66:187–205. [Google Scholar]
- Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, et al. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Krivan W. In silico identification of metazoan transcriptional regulatory regions. Naturwissenschaften. 2003;90:156–166. doi: 10.1007/s00114-003-0409-4. [DOI] [PubMed] [Google Scholar]