Abstract
Studies in mice have yielded invaluable insight into our understanding of the p53 pathway. Mouse models with activated p53, no p53, and mutant p53 have queried the role of p53 in development and tumorigenesis. In these models, p53 is activated and stabilized via redundant posttranslational modifications. On activation, p53 initiates two major responses: inhibition of proliferation (via cell-cycle arrest, quiescence, senescence, and differentiation) and induction of apoptosis. Importantly, these responses are cell-type and tumor-type-specific. The analysis of mutant p53 alleles has established a gain-of-function role for p53 mutants in metastasis. The development of additional models that can precisely time the oncogenic events in single cells will provide further insight into the evolution of tumors, the importance of the stroma, and the cooperating events that lead to disruption of the p53 pathway. Ultimately, these models should serve to study the effects of novel drugs on tumor response as well as normal homeostasis.
Studies of mice expressing mutant forms of the p53 tumor suppressor, or too much wild-type p53, reveal it has cell- and tumor-type-specific effects on cell proliferation, differentiation, senescence, and apoptosis in vivo.
The p53 tumor suppressor is a master regulator of transcription, positively and negatively regulating a plethora of genes with roles in cell-cycle arrest, quiescence, senescence, differentiation, and apoptosis. Because these activities are so potent at preventing cell proliferation or causing cell death, p53 is normally kept at very low levels. p53 levels and activity increase dramatically in response to DNA damage, oncogenic stress, and other stimuli that are viewed by a cell as anomalous. Animal models have allowed us to examine the in vivo effects of p53 activation, p53 deletion, and the role of p53 missense mutations in tumorigenesis. This article focuses on the important role of animal models in deciphering p53 activities and their implications for survival, development, and tumorigenesis.
IN VIVO REDUNDANCY IN UPSTREAM SIGNALS THAT STABILIZE AND ACTIVATE p53
The activity of p53 is carefully restrained in normal cells. In response to numerous signals, p53 becomes stabilized and activated (Horn and Vousden 2007) (Fig. 1). In response to these various signals, phosphorylation of p53 occurs at both amino and carboxyl termini of the protein (Appella and Anderson 2001). The phosphorylation events at the amino terminus occur in the p53 transactivation domain, some of which disrupt interactions with the p53 negative regulators Mdm2 and Mdm4. Phosphorylation also allows p53 to recruit additional transcriptional cofactors to coordinate transcription of its target genes. Molecularly, the ATM and Chk2 kinases phosphorylate murine p53 at serines 18 and 23, respectively (Lozano and Zambetti 2005). Mice lacking Atm or Chk2 still stabilize and activate p53 in response to γ-irradiation, although the response is delayed. These in vivo data suggest redundancy in the signals that activate p53.
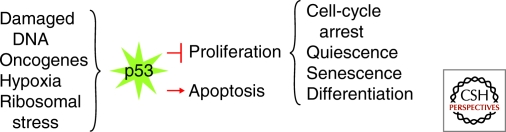
Figure 1.
The cell views DNA damage, inappropriate oncogene activation, hypoxia, and ribosomal stress as anomalous signals, which activate p53 to inhibit proliferation or induce apoptosis.
To directly test the hypothesis that phosphorylation of p53 is required to stabilize and activate its tumor-suppressive function, substitutions of serines 18 and 23 to alanines were made in the endogenous p53 locus (i.e., knockin alleles) (Sluss et al. 2004; Armata et al. 2007; MacPherson et al. 2004; Chao et al. 2006). Knockin alleles create alterations within the locus and thus preserve the normal regulatory components of the gene. Thymocytes from mice expressing p53S18A stabilize and activate p53 strongly upon γ-irradiation, similar to wild-type thymocytes, but show a partial defect in apoptosis (Sluss et al. 2004). B cells also have a partial defect in p53-dependent apoptosis (Armata et al. 2007). Consistent with these data, homozygous mutant mice for the p53S18A mutation develop late-onset tumors, the majority of which are B-cell lymphomas (Armata et al. 2007). In contrast, mutation of p53 serine 23 to alanine impairs both stabilization and activation of p53 following whole-body γ-irradiation, with a diminished apoptotic response in both B cells and thymocytes (MacPherson et al. 2004). p53S23A mice have a more pronounced tumor phenotype and develop tumors earlier than p53S18A mice, with a median survival similar to p53+/− mice. Again, the tumor spectrum of these mice was very different than that of p53+/− mice. p53S23A homozygous mutant mice do not develop T-cell lymphomas typical of p53+/− and p53−/− mice, but instead develop B-cell lymphomas that are positive for a B-cell-specific marker (MacPherson et al. 2004). In a third study, homozygous double-mutant mice with alanine substitutions at both serines 18 and 23 completely lack p53-dependent apoptosis in γ-irradiated thymocytes like those of p53-null mice, but are tumor prone with a similar survival curve to that of p53+/− mice (Chao et al. 2006). Thus, despite impaired apoptosis induction in thymocytes, the in vivo phenotype of mice with two alanine mutations was B-cell lymphoma. These data indicate that apoptosis assays in thymocytes do not predict tumor suppression in the whole mouse. Moreover, phosphorylation may be important in tumor suppression in only certain tissues (B cells, for example) and not others (T cells).
Other phosphorylation events occur at the carboxyl end of p53 in response to some DNA damage signals (Appella and Anderson 2001; Toledo and Wahl 2006). The carboxyl end of p53 has a dual function in tetramerization and in regulation of DNA binding. Acetylation of this domain of p53 also occurs in response to DNA damage. Mice with a serine-to-alanine mutation at p53 amino acid 389 have been made (Bruins et al. 2004). This serine is phosphorylated in response to UV but not γ-irradiation, suggesting a potential role in regulating downstream events (Kapoor and Lozano 1998; Lu et al. 1998). Disappointingly, these mice do not show a tumor phenotype. The lack of a tumor defect again supports the concept that redundancies in p53 activation exist such that posttranslational modifications at individual amino acids do not strongly affect p53 tumor suppressor activity. However, with exposure to ultraviolet light, skin tumors are more readily observed in p53S389A homozygous mutant mice than in wild-type mice (Bruins et al. 2004). These data suggest that when the mouse is challenged by exposure to a DNA damaging agent, lack of phosphorylation at amino acid 389 contributes to a tumor phenotype in mice.
The carboxyl domain of p53 is also modified via acetylation. Wahl and colleagues generated knockin mice with the seven acetylated lysines mutated to arginines to test the importance of acetylation on p53 tumor suppressor function (Krummel et al. 2005). Surprisingly, these mice lacked a tumor phenotype as well, suggesting that redundancies, perhaps at both ends of the p53 protein, and multiple backup signals stimulate p53 activity sufficiently to inhibit tumor development.
Together, these in vivo data suggest that multiple kinases phosphorylate p53, and that loss of phosphorylation at individual amino acids per se only modestly affects p53 activities and tumor suppression. Importantly, at least seven serines in the transactivation domain and two in the carboxyl end of p53 are phosphorylated and may serve to ensure some level of p53 phosphorylation and activation. The modest effects of alanine substitutions on p53 tumor suppressor function in vivo are also supported by the lack of specific mutations of these amino acids in human tumors (http://www-p53.iarc.fr). Another important concept derived from these studies is the tissue-specific nature of phosphorylation as mice that lack phosphorylation of p53 serines 18 and/or 23 develop a tumor spectrum different from that of other phosphorylation mutants. Additionally, though, the modest effects of these alterations on p53 activity may serve to modify a tumor phenotype in combination with other more potent tumor alterations.
THE IN VIVO EVIDENCE FOR THE ROLE OF p53 IN APOPTOSIS
The generation of p53-null mice led to a myriad of experiments to assess p53 functions in vivo by comparison to wild-type mice (Lozano and Liu 1998). That p53-null mice succumb to a tumor phenotype established the mouse as a viable model to study tumor suppressors (Donehower et al. 1992; Jacks et al. 1994; Clarke et al. 1993). As a possible mechanism of tumor suppression, the ability of p53 to initiate apoptosis was examined. In response to γ-irradiation, or drugs such as etoposide, normal thymocytes show a p53-dependent apoptosis response (Clarke et al. 1993; Lowe et al. 1993). The central nervous system of the developing embryo at E12.5 also shows a radiation- and p53-dependent cell-death phenotype (Lee et al. 2001), as does the intestine (Merritt et al. 1994; Clarke et al. 1994). These experiments established the role of p53 in initiating apoptosis in response to DNA damage, and predicted that loss of apoptosis would contribute to tumor development.
Further studies using the p53-null mouse highlight the significance of p53-dependent apoptosis in tumor suppression in vivo. Choroid plexus papillomas, initiated by inhibiting the activity of the retinoblastoma protein, develop rather slowly because of a high rate of apoptosis (Symonds et al. 1994). p53 loss eliminates apoptosis in these tumors and dramatically accelerates tumorigenesis. In another cell type, inactivation of the retinoblastoma protein in photoreceptors leads to massive cell death and degeneration of the retina (Howes et al. 1994). Again, deletion of p53 rescues the apoptotic defect and allows development of a poorly differentiated tumor of the photoreceptor cell. Thus, the p53-null mouse was instrumental in showing the importance of p53-dependent apoptosis in tumor suppression.
The data discussed above indicate that p53 induces apoptosis in response to DNA damage, and that p53-dependent apoptosis inhibits tumor growth. However, these data do not address whether the p53-dependent apoptosis that occurs after DNA damage contributes to tumor suppression. Although intuitively, the ability of p53 to initiate apoptosis after DNA damage would seem to be a clear tumor-suppressive mechanism, recent experiments in the mouse suggest differently (Christophorou et al. 2006). A p53-ER fusion was generated at the endogenous p53 locus so that p53 activity could be turned on and off at will using tamoxifen (Christophorou et al. 2005). Mice were irradiated and p53 activity transiently restored 1 week post-radiation. Mice lacking p53 activity develop lymphomas. Importantly, restoring p53 long after recovery from radiation delays the development of lymphomas and suggests that induction of apoptosis by radiation per se is not tumor suppressive. In these experiments, the p53-dependent apoptosis initiated by γ-irradiation is not necessary for tumor suppression.
p53 CELL-CYCLE ARREST/SENESCENCE FUNCTIONS CONTRIBUTE TO TUMOR SUPPRESSION
The first p53 target gene identified was p21, which encodes a cell-cycle inhibitor (el-Deiry et al. 1993; Harper et al. 1993; Noda et al. 1994). p21 overexpression in cells inhibits proliferation (Harper et al. 1993; el-Deiry et al. 1993) and induces senescence (Noda et al. 1994). These data suggest that the ability of p53 to induce cell-cycle arrest and/or senescence might also contribute to tumor suppression. A human tumor-specific mutation in the p53 gene that alters arginine 175 to proline produces a mutant protein that is unable to induce apoptosis but retains the ability to activate p21 and initiate a partial cell-cycle arrest (Rowan et al. 1996; Ludwig et al. 1996). p53 mutant knockin mice with the equivalent mutation at amino acid 172 clearly show an important role of p53 cell-cycle arrest function in tumor suppression (Liu et al. 2004). Thymocytes and cells of the developing central nervous system of homozygous mutant mice lack an apoptotic response after γ-irradiation and are thus indistinguishable from p53−/− mice, indicating that the p53R172P is defective in apoptosis. In contrast, mouse embryo fibroblasts from p53R172P mice retain a partial cell-cycle arrest phenotype. p53R172P-homozygous mice developed tumors with a median of 11.5 months as compared with p53-null mice that die with a median of 5.5 months, clearly indicating an important role of cell-cycle arrest in tumor suppression. The generation of p53S18/23A mice also support the importance of cell-cycle arrest in tumor suppression (Chao et al. 2006). Thymocytes from p53S18/23A mice lack apoptosis in response to γ-irradiation, but mouse embryo fibroblasts retain a partial cell-cycle arrest and senescent phenotype. These mice also show a delayed tumor phenotype as compared with p53-null mice, clearly indicating that p53 activities other than apoptosis also contribute to tumor suppression. An analysis of the lymphomas and sarcomas that eventually developed in p53R172P-homozygous mice showed that they remained diploid, suggesting that the ability of p53 to initiate cell-cycle arrest contributed to the maintenance of genomic stability in vivo (Liu et al. 2004).
The importance of p21 in p53-mediated tumor suppression and genomic stability was further explored in compound mutant mice homozygous for the p53R172P mutation that lacked p21 (Barboza et al. 2006). These mice develop tumors with almost the same survival curve as p53-null mice. Additionally, the lymphomas and sarcomas in these mice are aneuploid in this context. These data suggest an important role of p21 in p53-mediated tumor suppression and maintenance of chromosome stability. Thus, the ability of p53 to initiate both cell-cycle arrest and apoptotic programs contributes to tumor suppression.
The importance of p53-mediated senescence in tumor suppression also became clear from studies in other mouse models. Senescence contributes to treatment outcome as Eμ-Myc induced lymphomas that cannot undergo apoptosis because of expression of bcl2 initiate a senescence program on treatment with cyclophosphamide (Schmitt et al. 2002). Cyclophosphamide treatment produces a senescent phenotype that is tumor suppressive and p53- and p16-dependent. The importance of senescence as a tumor suppressive mechanism also became clear from the development of mouse models to restore p53 activity in various tumors. Restoration of p53 functions in sarcomas from p53-null mice and in hepatocellular carcinomas induced by HrasV12 results in tumor regression via a senescence phenotype (Ventura et al. 2007; Xue et al. 2007). On the other hand, restoration of p53 function in T-cell lymphomas lacking p53 and in B-cell lymphomas induced by the Myc oncogene results in apoptosis and tumor regression (Ventura et al. 2007; Martins et al. 2006). These data indicate that p53 loss is required for maintenance of the tumor phenotype, and that the myriad of changes that accumulate in tumors does not alter the vulnerability to p53 activation. Just as significant is the finding that specific tumors respond differently to reactivation of p53. These mouse models have thus identified several p53 activities that contribute to tumor suppression.
DIFFERENTIATION/DEVELOPMENT
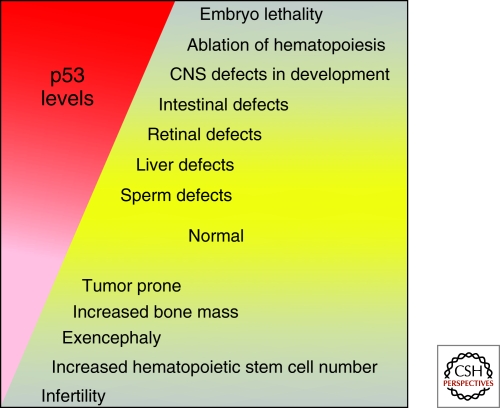
Analyses of p53-null mice have also led to an understanding of the importance of p53 in embryo development, and in differentiation and quiescence of particular cell types (Fig. 2). Although most p53-null mice are viable during development, p53-null females are highly susceptible to exencephaly, an outgrowth of the brain that results from failure of the neural tube to close (Armstrong et al. 1995; Sah et al. 1995). The process of neural tube closure is very sensitive to changes in cell proliferation and migration and this may account for the defect. Other cranial facial abnormalities that include fusion of upper incisors and ocular abnormalities of the retina and lens are also apparent in p53-null embryos (Armstrong et al. 1995).
Figure 2.
Too much or too little p53 in the mouse results in lethal phenotypes.
The first mouse generated that indicated a role of p53 in differentiation overexpressed wild-type p53 in mesenchymal cells of the developing kidney. Kidneys of transgenic mice are smaller and express markers of differentiation throughout the ureteric bud, suggesting increased differentiation of these cells (Godley et al. 1996). These mice eventually succumb to renal failure. In many other mouse models with elevated p53 activity (described later), the effects of p53 on induction of apoptosis and inhibition of cell proliferation are so profound that they preclude the possibility of examining effects on differentiation. Thus, targeted activation of p53 shows increased differentiation of mesenchymal cells in vivo.
Analysis of p53-null mice has also shown the importance of p53 in differentiation. p53-null mice have a slight increase in bone mineral density (Wang et al. 2006). Absence of p53 accelerates the differentiation of osteoblasts, the cells that make bone, leading to increased mineral deposition. Tissue culture data support these findings as treatment of mouse embryo fibroblasts with BMP2, an inducer of bone differentiation, leads to increased mineral deposition in p53-null cells as compared with wild-type cells (Lengner et al. 2006; Molchadsky et al. 2008). Similar in vitro experiments show the importance of p53 in inhibition of adipogenesis, but this role has not yet been analyzed in vivo (Molchadsky et al. 2008). These data may appear conflicting at first because increased p53 in the kidney allows cells to differentiate earlier than normal, whereas loss of p53 also increased differentiation of osteoblasts. The effect of p53 on differentiation is real and probably depends on the relationship of perturbed cells to surrounding normal cells and the timing of environmental cues that signal differentiation. Thus, differentiation of a cell with high p53 levels in a normal environment (kidney model) may occur before that cell reaches its proper location or before sufficient cell numbers have been generated. On the other hand, as in bone morphogenesis, p53 loss may increase proliferation of specific cell types that reach their destination more quickly and differentiate sooner than normal. Timing of p53 activation and tissue/cell fate specificity may thus regulate differentiation (Molchadsky et al. 2008). These ideas need to be addressed in vivo by precisely deleting p53 at different stages of differentiation at various times using conditional loss-of-function alleles.
Lastly, p53-null female mice also have infertility problems, but males are normal (Hu et al. 2007). The infertility is caused by the failure of blastocysts, regardless of their genotype, to implant into the p53−/− uterus. The clever decision to look for p53 binding sites in genes involved in reproduction led to the identification of a p53-responsive sequence in the promoter of the Lif gene. Lif, leukemia inhibitory factor, is an essential cytokine for implantation. Mechanistically then, p53 normally activates Lif in females, which then promotes embryo implantation. The infertility defect in p53-null females is rescued by the injection of Lif during pregnancy. The activation of Lif by p53 highlights the tissue-specific nature of the p53 response. The transcriptional activation of p53 target genes is likely a combination of p53 binding sites, the availability of other cooperating transcription factors, and the state of chromatin structure, the latter two of which may be tissue specific. These data highlight some of the subtle defects observed in p53-null mice (Fig. 2). Despite these minor abnormalities, the p53-null mouse is amazingly normal.
A ROLE FOR p53 IN STEM CELLS
A role for p53 in hematopoietic stem cells (HSCs) has also recently been identified by analysis of p53-null mice. The HSC consists of two stem-cell types called LT-HSC (long term-hematopoietic stem cell) and ST-HSC (short term-hematopoietic stem cell). The ST-HSC quickly matures into the common progenitor cells for myeloid (CMP) and lymphoid (CLP) pathways (Orkin and Zon 2008). HSCs are identified using numerous markers, and one population known as LSK cells are Lin−, Sca-1+, and c-Kit+. p53-null mice have a two- to threefold increase in LSK cells (Chen et al. 2008; Liu et al. 2009). An analysis of the proliferation status of LSK cells from bone marrow shows that 60% of p53-null cells incorporate bromodeoxyuridine in contrast to 30% for wild-type bone marrow cells (Liu et al. 2009). Thus, hematopoietic stem cells from p53-null mice appear to enter the cell cycle more readily, suggesting that p53 functions in stem cells as a block to cell-cycle entry. It would be interesting to determine if this difference in HSC composition affects life span of the animal. Unfortunately, the p53−/−mice die very young as they are predisposed to tumor development.
p53, in cooperation with Pten, also regulates neural stem cell (NSC) differentiation as mice lacking both genes have NSCs with increased proliferation and self-renewal abilities (Zheng et al. 2008). In culture, NSCs lacking p53 and Pten do not respond to differentiation signals and retained stem-cell properties consistent with a role of p53 in differentiation. It is interesting to note that two cell types that are very sensitive to increased p53 levels via general Mdm2 loss are hematopoietic cells and neural cells of the developing cerebellum (Terzian et al. 2007; Liu et al. 2007).
MOUSE MODELS WITH CONSTITUTIVELY ACTIVE p53
Most of the mouse models described previously are ones in which the p53 gene is deleted. Other models have been developed in which p53 levels are elevated. Given the role of p53 in inhibition of proliferation and induction of apoptosis, it is not surprising that mouse models that constitutively activate p53 are, in general, not viable. The best examples of the detrimental effects of high p53 activity on development are the Mdm2-null and Mdm4-null mice (Jones et al. 1995; Montes de Oca Luna et al. 1995; Parant et al. 2001; Migliorini et al. 2002). Both Mdm2 and Mdm4 are critical negative regulators of p53 (see Perry 2010). Loss of either of these genes leads to embryo lethal phenotypes that are caused by constitutive activation of p53 as concomitant deletion of p53 rescues these phenotypes. Conditional alleles of Mdm2 and Mdm4 have been used to delete these genes in numerous cell types (Marine et al. 2006). Deletion of Mdm2 in all cell types examined results in cell-lethal phenotypes. Loss of Mdm2 in the central nervous system causes hydraencephaly and embryo death (Francoz et al. 2006; Xiong et al. 2006). In cardiomyocytes, it leads to the absence of a heart (Grier et al. 2006). In smooth muscle cells of the mouse, Mdm2 loss leads to abnormalities of the stomach and intestine that result from irregularities in architecture of smooth muscle cells (Boesten et al. 2006). Mdm2 is also required in primitive erythropoiesis to inhibit p53-dependent apoptosis (Maetens et al. 2007). In the intestine, as well, Mdm2 loss results in cell death, but the animal survives because the organ has the capacity to increase proliferation (Valentin-Vega et al. 2008). The organ recovers as it selects against cells that lose Mdm2. In a model that restores p53 activity in the absence of Mdm2 throughout all cells of the adult mouse, Evan and colleagues show an almost complete ablation of radio-resistant tissues that include the bone marrow, thymus, spleen, small intestine, and colon via apoptosis that results in lethality (Ringshausen et al. 2006). In the testis and liver, defects in proliferation are observed without pathological changes probably because of short survival time of the animal.
The effects of Mdm4 loss are less severe in general. In the central nervous system, Mdm4 loss results in cell death and a proenchephalic phenotype (Francoz et al. 2006; Xiong et al. 2006). In erythropoiesis, Mdm4 is required during the rapidly growing phase of definitive erythropoiesis (Maetens et al. 2007). Mice lacking Mdm4 in the heart are born but die between 3 and 8 months of age because of a decrease in the number of cardiomyocytes, and a compensatory increase in muscle mass resulting in cardiomyopathy (Grier et al. 2006; Xiong et al. 2007). Minor effects of Mdm4 loss are seen in smooth muscle cells and in the intestinal epithelium (Boesten et al. 2006; Valentin-Vega et al. 2009). All phenotypes because of Mdm2 or Mdm4 loss are rescued by loss of p53. These data underscore the important physiological role of regulating p53 via Mdm2 and Mdm4 in numerous cell types and shows that even in the absence of DNA damage, p53 can be activated through loss of either of these two inhibitors. The data suggest that Mdm2 is the major inhibitor of p53 activities, but in some situations may need to partner with Mdm4 to maintain appropriate p53 levels. These observations have important implications for the use of drugs that have been designed to disrupt the p53-Mdm interactions in tumors with high levels of Mdm2 and/or Mdm4. Proliferating normal cells may be extremely sensitive to increased wild-type p53 activities via Mdm2 and Mdm4 inhibition.
Another knockin mouse generated that also conditionally expresses high levels of p53 is one that contains two amino acid changes in the p53 transactivation domain. This mutant has little transcriptional activity, and cannot bind Mdm2 (Johnson et al. 2005). In vivo, the p53L25Q W26S protein is selectively impaired for p53 transcriptional activation and shows a differential apoptotic response to different stimuli. Thus, although doxorubicin treatment or UV-C exposure did not induce apoptosis in p53L25Q W26S expressing cells as compared with wild-type cells, serum deprivation and hypoxia induce substantial levels of apoptosis. As a heterozygote, the p53L25Q W26S mutant is dead very early in embryogenesis. Either select p53 transcriptional activities coupled with stabilization is sufficient to induce embryonic lethality, or functions other than transcriptional activation are important for cell viability.
Although complete loss of Mdm2 and Mdm4 have dramatic effects on cell survival, heterozygous mice are normal. However, in combination with other tumor-specific events, these subtle modifications in the levels of Mdm2 and Mdm4 proteins affect tumor outcome. For example, Mdm2 haploinsufficiency significantly delays the development of Eμ-Myc driven B-cell lymphomas (Alt et al. 2003). The same is true for Mdm4 heterozygous mice (Terzian et al. 2007). In another tumor type, decreased Mdm2 levels lead to a delay of intestinal tumors in APC mice (Mendrysa et al. 2006). Thus, although the loss of one allele of either Mdm2 or Mdm4 does not affect normal development and life span, it does delay tumor development most likely through elevated p53 activities in response to abnormal cell proliferation. Mdm2/Mdm4 double heterozygous mice cannot be analyzed for effects on a tumor phenotype because they die by 15 days after birth due to ablation of hematopoiesis. That this is a p53-dependent phenotype was shown by survival of the Mdm2+/− Mdm4+/− mice by deletion of a single p53 allele (Terzian et al. 2007). These experiments highlight the exquisite balance between p53 and Mdm proteins in maintaining homeostasis. Too much p53 kills the cell and in some cases the mouse, whereas not enough p53 enhances tumorigenesis. In some cases, increased p53 activity causes an aging phenotype and these data are reviewed by Larry Donehower (Donehower 2009).
The p53R172P mutant described previously that has partial p53 tumor suppressor activity also has detrimental defects on cell survival in the absence of Mdm2 (Liu et al. 2007). p53R172P-homozygosity rescues the Mdm2-null embryo lethality but postnatal development is perturbed. p53R172P-homozygous mice null for Mdm2 are born at the expected Mendelian ratio and appeared normal at birth but became severely growth retarded and died by 2 weeks of age. These mice develop several abnormalities, which include ablation of hematopoiesis, cerebellar atrophy, severe atrophy of the retina, kidney defects, and vacuolation of the liver. These data thus identify additional tissues and cell types that are sensitive to increased p53 activity. In summary, it appears that actively proliferating cells are the most sensitive to activation of p53 via loss of Mdm2 or Mdm4.
MUTANT p53 FUNCTIONS
Because human cancers in general have p53 missense mutations and not deletions of p53, many investigators have postulated gain-of-function and dominant–negative activities for mutant p53 (see Oren and Rotter 2010). Numerous mouse models have been generated to study the consequences of missense mutations on p53 activities in vivo (Iwakuma and Lozano 2007; Donehower and Lozano 2009). Specifically, some of the missense mutations generated as knockin alleles mimic the hot spot mutations found in human p53 (Lang et al. 2004; Olive et al. 2004; Song et al. 2007). Two of these mutations, p53R172H and p53R270H, were generated in the mouse gene that correspond to amino acids 175 and 273 in human p53, respectively (Lang et al. 2004; Olive et al. 2004). The third mutation, p53R245W, was generated in the hupki mouse, which carries a mouse/human hybrid p53 gene (Song et al. 2007). These studies have led to major findings regarding mutant p53 activities.
First, all three mutants show a gain-of-function phenotype in vivo manifested as increased metastasis and genetic instability as compared with mice lacking p53. Secondly, the analyses of these mice indicates that mutant p53 is unstable in normal tissues, but is stabilized in tumor cells, and in cells under the stress of culture. These data connote that tumor-specific alterations stabilize mutant p53. This hypothesis was directly tested by deletion of Mdm2. Mdm2 deletion leads to stabilization of mutant p53R172H and enhancement of the metastatic gain-of-function phenotype (Terzian et al. 2008). This experiment is a proof of principle that stabilization of mutant p53 leads to gain-of-function phenotypes in vivo. Because Mdm2 loss does not occur in human cancers (quite the opposite), this experiment did not address what tumor-specific changes might regulate mutant p53 stability. The cell-cycle inhibitor, p16, dampens p53 activity via the Rb/E2F pathway (Sherr and McCormick 2002). Thus, loss of p16 results in stabilization of wild-type p53. In combination with p53R172H, p16 loss also stabilizes mutant p53R172H in some cells and enhances the metastatic phenotype (Terzian et al. 2008). Because Mdm2 and p16 loss stabilize and activate wild-type p53, these observations also indicate that mutant p53 is posttranslationally regulated like wild-type p53, such that the same signals that stabilize wild-type p53 may stabilize mutant p53. Indeed, γ-irradiation also stabilized mutant p53 (Terzian et al. 2008).
Thirdly, the analysis of p53 mutant mice also suggests that the dominant–negative phenotype associated with mutant p53 requires stabilization of mutant p53. For example, the dominant–negative phenotype is manifested upon γ-irradiation of cells of the central nervous system, but not by Mdm2 loss (Lang et al. 2004). However, once stabilized, mutant p53 has a longer half life than wild-type p53 and thus basically overwhelms wild-type p53 function (Terzian et al. 2008). Lastly, these p53 mutant alleles are true loss-of-function alleles as they have no wild-type p53 activity in numerous assays.
DISRUPTING THE p53 PATHWAY BY MULTIPLE MECHANISMS
The initial observation that p53+/− mice develop a wide range of tumors similar, although not identical, to Li-Fraumeni syndrome patients with p53 mutations contributed to our understanding of the importance of p53 in tumor suppression. It is now clear that multiple mechanisms cooperate to disrupt the p53 pathway in tumor development. The mouse has served as a model to directly examine some of those mechanisms.
A tumor suppressor gene is classically defined as the inheritance of one mutant allele and loss of the remaining wild-type allele (loss of heterozygosity [LOH]) during the process of tumor evolution (Knudson 2001). A detailed study of LOH in tumors from p53+/− mice indicates that 50% of the mice showed LOH when the mouse developed a tumor before 18 months of age, but only 15% in mice older than 18 months of age (Venkatachalam et al. 1998). These data indicate that although loss of both p53 alleles occurs in many tumors, loss of the wild-type p53 allele is not a requirement of tumorigenesis. More importantly, the data imply that older mice have more time to acquire additional alterations that undermine the p53 pathway.
What other changes contribute to inactivation of the p53 pathway? Many human tumors have high levels of p53 and Mdm2 (Valentin-Vega et al. 2007). Clearly, increased Mdm2 and Mdm4 levels should inactivate the p53 pathway in tumorigenesis given what we know about the regulation of p53 by Mdm2 and Mdm4 in mice. Data from mouse tumor models supports this hypothesis. Lymphomas from Mdm2/Eμ-myc double transgenic mice show a dramatic reduction in p53 alterations as compared with lymphomas in Eμ-myc single transgenic mice (Wang et al. 2008). An Mdm4 transgenic mouse also has a tumor phenotype that is exacerbated by p53 heterozygosity, emphasizing the cooperative nature of different molecular changes to inactivate the p53 pathway (Xiong and Lozano, in prep).
LOH in mice with a mutant p53 knockin allele ranges from 23% to 67% in the few animals that were studied (Lang et al. 2004; Olive et al. 2004). A more careful analysis should be performed to determine if those tumors with stable mutant p53 are those that do not need to lose the wild-type p53 allele because of p53 inactivation via dominant–negative interactions with mutant p53. The timing of the second event that stabilizes mutant p53 may be the determining factor of whether tumor cells lose the wild-type p53 allele or not.
CONCLUSION
p53 is a potent regulator of cell proliferation and death. Altering p53 levels in vivo has consequences on organismal survival. Increased p53 activity results in cell-cycle arrest, senescence, early differentiation, or apoptotic cell death. Survival of the organism depends on the extent of p53 activation, the cell type affected, and the timing of p53 activation. The ability of p53 to enact these functions is important for tumor suppression. On the other hand, cells tolerate p53 loss extremely well. Cells lacking p53 are for the most part normal and do not necessarily become tumorigenic. Given the total number of cells in a mouse, it is most surprising that p53-null mice do not develop many more tumors more rapidly. The DNA damage that accumulates in a cell in the absence of p53 is likely insufficient for tumor formation. Cooperating changes in cell behavior seem to be required for tumor formation. For example, low dose ionizing radiation and carcinogen exposure contribute to an enhanced tumor phenotype in p53-null mice (Kemp et al. 1994; Harvey et al. 1993), but the molecular events that cooperate and contribute to inactivation of the p53 pathway remain largely unknown. Additionally, tissue and tumor specificity of cooperating events are different. Not all p53 targets are activated in one specific tissue or in response to the same signal in different tissues. The reactivation of p53 in tumors induces senescence in some and apoptosis in others. The cell and tissue-specific nature of p53 function needs to be addressed in more detail. Determination of the events required for tumor maintenance must also be more fully explored. Lastly, the signals responsible for p53 stabilization in tumors must be understood more fully as it is stabilization that contributes to the gain-of-function and dominant–negative activities that cooperate in tumorigenesis. The mouse has only begun to shed light on the tissue specificity, timing, and order of events that contribute to tumorigenesis in general, and to inactivation of p53 in particular.
ACKNOWLEDGMENTS
I thank Drs. M.E. Perry and J.G. Jackson for helpful discussions and criticism of this work.
Footnotes
Editors: Arnold J. Levine and David P. Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Alt JR, Greiner TC, Cleveland JL, Eischen CM 2003. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. Embo J 22:1442–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E, Anderson CW 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268:2764–2772 [DOI] [PubMed] [Google Scholar]
- Armata HL, Garlick DS, Sluss HK 2007. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res 67:11696–11703 [DOI] [PubMed] [Google Scholar]
- Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR 1995. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol 5:931–936 [DOI] [PubMed] [Google Scholar]
- Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G 2006. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci 103:19842–19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten LS, Zadelaar SM, De Clercq S, Francoz S, van Nieuwkoop A, Biessen EA, Hofmann F, Feil S, Feil R, Jochemsen AG, et al. 2006. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ 13:2089–2098 [DOI] [PubMed] [Google Scholar]
- Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB, Miranda B, van Oostrom CT, van den Berg J, van den Aardweg GJ, et al. 2004. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 24:8884–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C, Herr D, Chun J, Xu Y 2006. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. Embo J 25:2615–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ellison FM, Keyvanfar K, Omokaro SO, Desierto MJ, Eckhaus MA, Young NS 2008. Enrichment of hematopoietic stem cells with SLAM and LSK markers for the detection of hematopoietic stem cell function in normal and Trp53 null mice. Exp Hematol 36:1236–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI 2005. Temporal dissection of p53 function in vitro and in vivo. Nat Genet 37:718–726 [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI 2006. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 443:214–217 [DOI] [PubMed] [Google Scholar]
- Clarke AR, Gledhill S, Hooper ML, Bird CC, Wyllie AH 1994. p53 dependence of early apoptotic and proliferative responses within the mouse intestinal epithelium following γ-irradiation. Oncogene 9:1767–1773 [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849–852 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215–221 [DOI] [PubMed] [Google Scholar]
- Donehower LA 2009. Using mice to examine p53 functions in cancer, aging, and longevity. Cold Spring Harb Perspect Biol 1:a001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LD, Lozano G 2009. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer 9:831–841 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825 [DOI] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC 2006. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad SciM 103:3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godley LA, Kopp JB, Eckhaus M, Paglino JJ, Owens J, Varmus HE 1996. Wild-type p53 transgenic mice exhibit altered differentiation of the ureteric bud and possess small kidneys. Genes Dev 10:836–850 [DOI] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G 2006. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 26:192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816 [DOI] [PubMed] [Google Scholar]
- Harvey M, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A, Donehower LA 1993. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet 5:225–229 [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH 2007. Coping with stress: Multiple ways to activate p53. Oncogene 26:1306–1316 [DOI] [PubMed] [Google Scholar]
- Howes KA, Ransom N, Papermaster DS, Lasudry JG, Albert DM, Windle JJ 1994. Apoptosis or retinoblastoma: Alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev 8:1300–1310 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ 2007. p53 regulates maternal reproduction through LIF. Nature 450:721–724 [DOI] [PubMed] [Google Scholar]
- Iwakuma T, Lozano G 2007. Crippling p53 activities via knock-in mutations in mouse models. Oncogene 26:2177–2184 [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4:1–7 [DOI] [PubMed] [Google Scholar]
- Johnson TM, Hammond EM, Giaccia A, Attardi LD 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet 37:145–152 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206–208 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Lozano G 1998. Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc Natl Acad Sci 95:2834–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Wheldon T, Balmain A 1994. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet 8:66–69 [DOI] [PubMed] [Google Scholar]
- Knudson AG 2001. Two genetic hits (more or less) to cancer. Nat Rev Cancer 1:157–162 [DOI] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM 2005. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci 102:10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119:861–872 [DOI] [PubMed] [Google Scholar]
- Lee Y, Chong MJ, McKinnon PJ 2001. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J Neurosci 21:6687–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN 2006. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol 172:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. 2009. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4:37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G 2004. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet 36:63–68 [DOI] [PubMed] [Google Scholar]
- Liu G, Terzian T, Xiong S, Van Pelt CS, Audiffred A, Box NF, Lozano G 2007. The p53-Mdm2 network in progenitor cell expansion during mouse postnatal development. J Pathol 213:360–368 [DOI] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847–849 [DOI] [PubMed] [Google Scholar]
- Lozano G, Liu G 1998. Mouse models dissect the role of p53 in cancer and development. Semin Cancer Biol 8:337–344 [DOI] [PubMed] [Google Scholar]
- Lozano G, Zambetti GP 2005. What have animal models taught us about the p53 pathway? J Pathol 205:206–220 [DOI] [PubMed] [Google Scholar]
- Lu H, Taya Y, Ikeda M, Levine AJ 1998. Ultraviolet radiation, but not γ radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc Natl Acad Sci 95:6399–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig RL, Bates S, Vousden KH 1996. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol 16:4952–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA, Venere M, Halazonetis TD, Bronson R, De Vries A, et al. 2004. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. Embo J 23:3689–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, Klingmuller U, Lozano G, Marine JC 2007. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood 109:2630–2633 [DOI] [PubMed] [Google Scholar]
- Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G 2006. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ 13:927–934 [DOI] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI 2006. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127:1323–1334 [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, O'Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME 2006. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev 20:16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt AJ, Potten CS, Kemp CJ, Hickman JA, Balmain A, Lane DP, Hall PA 1994. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res 54:614–617 [PubMed] [Google Scholar]
- Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 22:5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchadsky A, Shats I, Goldfinger N, Pevsner-Fischer M, Olson M, Rinon A, Tzahor E, Lozano G, Zipori D, Sarig R, et al. 2008. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One 3:e3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206 [DOI] [PubMed] [Google Scholar]
- Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR 1994. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res 211:90–98 [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119:847–860 [DOI] [PubMed] [Google Scholar]
- Oren M, Rotter V 2010. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2:a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI 2008. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29:92–95 [DOI] [PubMed] [Google Scholar]
- Perry ME 2010. The regulation of the p53-mediated stress response by MDM2 and MDM4. Cold Spring Harb Perspect Biol 2:a000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI 2006. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10:501–514 [DOI] [PubMed] [Google Scholar]
- Rowan S, Ludwig RL, Haupt Y, Bates S, Lu X, Oren M, Vousden KH 1996. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. Embo J 15:827–838 [PMC free article] [PubMed] [Google Scholar]
- Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T 1995. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet 10:175–180 [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109:335–346 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103–112 [DOI] [PubMed] [Google Scholar]
- Sluss HK, Armata H, Gallant J, Jones SN 2004. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol 24:976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y 2007. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 9:573–580 [DOI] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78:703–711 [DOI] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G 2008. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 22:1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G 2007. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol 27:5479–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6:909–923 [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Barboza JA, Chau GP, El-Naggar AK, Lozano G 2007. High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Hum Pathol 38:1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Box N, Terzian T, Lozano G 2009. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 77:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Okano H, Lozano G 2008. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ 15:1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, Donehower LA 1998. Retention of wild-type p53 in tumors from p53 heterozygous mice: Reduction of p53 dosage can promote cancer formation. Embo J 17:4657–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445:661–665 [DOI] [PubMed] [Google Scholar]
- Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, et al. 2006. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol 172:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM 2008. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 27:1590–1598 [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Fernandez-Garcia B, Lozano G 2007. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. Circulation 115:2925–2930 [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G 2006. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci 103:3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. 2008. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455:1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]