Abstract
Like their eukaryotic counterparts, bacterial cells have a highly organized internal architecture. Here, we address the question of how proteins localize to particular sites in the cell and how they do so in a dynamic manner. We consider the underlying mechanisms that govern the positioning of proteins and protein complexes in the examples of the divisome, polar assemblies, cytoplasmic clusters, cytoskeletal elements, and organelles. We argue that geometric cues, self-assembly, and restricted sites of assembly are all exploited by the cell to specifically localize particular proteins that we refer to as anchor proteins. These anchor proteins in turn govern the localization of a whole host of additional proteins. Looking ahead, we speculate on the existence of additional mechanisms that contribute to the organization of bacterial cells, such as the nucleoid, membrane microdomains enriched in specific lipids, and RNAs with positional information.
Geometric cues and restricted sites for self-assembly localize “anchor” proteins to specific regions of bacteria. These anchors in turn govern localization of many other proteins.
Our view of the organization of the bacterial cell has changed radically over the past two decades. Once seen as an amorphous vessel harboring a homogeneous solution of proteins, these primitive organisms are now known to have an intricate subcellular architecture in which individual proteins localize to particular sites in the cell, often in a dynamic manner. Of course, bacteria frequently show conspicuous morphological features, such as division septa, flagella, pili, and stalks, which implied a nonuniform, underlying distribution of proteins. But it was not until the early 1990s that it became clear that proteins can, and often do, have distinctive subcellular addresses. Among the earliest discoveries were: (1) the formation of a ringlike structure at the mid cell position by the cytokinetic protein FtsZ (Bi and Lutkenhaus 1991), (2) the clustering of chemotaxis proteins at the poles of cells (Alley et al. 1992), (3) the compartment-specific production of sporulation proteins and their assembly into shell-like structures (Driks and Losick 1991), and (4) the asymmetric distribution of proteins involved in actin polymerization along the cell surface (Goldberg et al. 1993; Kocks et al. 1993). These discoveries were initially made by immunoelectron and immunofluorescence microscopy with fixed cells, but the discovery of green fluorescent protein (GFP) and the demonstration that proteins could retain their proper subcellular localization as GFP fusions opened the way to visualizing proteins and their dynamic behavior in living cells, including, importantly, in bacteria (Arigoni et al. 1995).
Knowing where proteins are in the cell is often critical to understanding their function. Thus, the position of the aforementioned FtsZ ring (the Z-ring) dictates where cytokinesis will take place (Margolin 2005). The clustering of chemotaxis proteins plays an important role in the extraordinary gain in the responsiveness of chemotatic behavior to small changes in attractants (Ames and Parkinson 2006). Where sporulation proteins are produced and the way in which they assemble governs spore morphogenesis (Stragier and Losick 1996; Errington 2003). The asymmetric distribution of actin-polymerization proteins on the cell surface explains how certain pathogens harness host cytoskeletal proteins for their own motility (Smith et al. 1995). From these and other examples emerge a view of the bacterial cell as a dynamic, three-dimensional system in which protein localization and changes in protein localization over time orchestrate growth, the cell cycle, behavior, and differentiation.
Here, after an initial discussion of general principles governing the positioning of proteins within the cell, we consider five broad categories of subcellular localization: the divisome, polar assemblies, cytoplasmic clusters, cytoskeletal elements, and organelles. We end by looking ahead to exciting new aspects of bacterial cytology just emerging from current research. Our goal is not to be comprehensive but rather to focus on examples that are illustrative of general principles of protein localization. Comprehensive treatment of individual topics can be found in other articles on this topic.
DIFFUSION-AND-CAPTURE AND SELF-ASSEMBLY
What principles govern subcellular protein localization? We distinguish between two mechanisms. Perhaps the most pervasive and best established mechanism is diffusion-and-capture, whereby a protein diffuses in one dimension along the DNA until it encounters its DNA binding site, in two dimensions in the membrane or in three-dimensions through the cytoplasm until it encounters, and is captured by, a target protein to which it adheres. A clear example in which diffusion-and-capture mediates the localization of integral membrane proteins is the three-protein complex that governs the activation of the sporulation transcription factor σK (Rudner and Losick 2002). The complex localizes to a particular membrane that will become the outer membrane around the spore. One member of the complex serves as an anchor that is responsible for capturing the other two proteins, which reach their target by diffusion in the cytoplasmic membrane. A key observation is that under circumstances in which the target membrane has become topologically isolated from the cytoplasmic membrane, proteins in the cytoplasmic membrane cannot reach the target protein (Rudner et al. 2002).
Complementary evidence in support of the diffusion-and-capture mechanism comes from studies on the localization of the histidine kinase PleC to the cell pole in Caulobacter (Deich et al. 2004). Individual fluorescently labeled molecules of PleC were visualized in living cells, revealing a focus of stationary molecules at the cell pole and other molecules that were diffusing without a directional bias in the membrane. Thus, PleC molecules appear to diffuse in a random manner until they are captured at the pole.
Another example that is best understood in the context of diffusion-and-capture is the localization of proteins to the divisome, which is composed of components that bind to the scaffold of the Z-ring in an ordered sequence (Margolin 2005). Thus, each division protein (or protein subcomplex) diffuses until it encounters, and then adheres to, the Z-ring or to another protein that has already assembled into the divisome. As we will argue, it seems likely that diffusion-and-capture is the principal mechanism by which protein localization takes place in bacteria in that most proteins find their position in the cell by interacting with another protein (or protein complex). Of course, and as we shall see, diffusion-and-capture begs the question of the ultimate cue(s) by which the location of the target protein itself is determined.
A related mechanism that does not depend on a positional cue is self-assembly. A striking example is the self-assembly of the MinD and MinE proteins of Escherichia coli, which show a dynamic pattern of pole-to-pole oscillation in the cell (Raskin and de Boer 1999; Fu et al. 2001; Hale et al. 2001). Recent work has shown that MinD and MinE alone are sufficient to form dynamic waves on a planar surface (Loose et al. 2008). The frequencies of these waves are consistent with those observed in vivo when models for the dynamic behavior of the proteins are adjusted to account for the expected lower diffusion constants of the cytoplasm. Thus, key features of the dynamic behavior of MinD and MinE can be explained without invoking cues from the cell other than the ends of the cell, which restrict how far the waves can travel. In a sense, self-assembly is a variation on diffusion-and-capture in that the proteins reach their targets by diffusion and are then captured by the self-assembled complexes. Self-assembly differs from other examples of diffusion-and-capture to the extent that it does not rely on a separate anchor protein or other cellular cue.
ARE PROTEINS TARGETED IN BACTERIA?
The word “targeted” is often used to describe protein localization in bacteria. That is, proteins are said to be targeted to one or another location. But the word targeted denotes an active process in which the protein in question is somehow aimed at its destination. For example, the secretion of some proteins are said to be “targeted” to a particular location either because the secretory apparatus is located there or because the signal sequence directs secretion to a particular location. It is likely, however, in the first scenario, that the secretory apparatus localizes to a particular location either by diffusion to, and capture at, some cellular cue or by a process of self-assembly, as described previously. Likewise, in the second scenario, the signal sequence undoubtedly finds its proper location by a passive process in which by diffusion it eventually encounters a landmark that restricts its secretion to a particular location. Perhaps the use of the word targeted is best justified for plasmid segregation in which an active mechanism driven by protein polymerization (and depolymerization) pushes (or in some cases pulls) newly duplicated plasmid molecules apart toward opposite poles of the cell (Ebersbach and Gerdes 2005). Likewise, in yeast, the mRNA for a protein involved in mating type switching (Ash1) is directed to the daughter cell by an energy-driven process involving motor proteins (Takizawa et al. 1997). Here, too, it seems justified to refer to ASH1 mRNA as being targeted to the bud. It is our contention, however, that in general, protein localization in bacteria cannot properly be said to be targeted to a particular location.
ASSEMBLY AND PLACEMENT OF THE DIVISOME
The best studied and most conspicuous example of subcellular localization is the divisome, the cytokinetic structure that mediates, and dictates the site of, cell division (see deBoer 2010). The divisome is generally located at the mid cell position but in some circumstances it is near a pole (see later). The divisome poses two issues in protein subcellular localization: its assembly and its position. The divisome is composed of at least 10 proteins that show an elaborate web of interactions (Goehring and Beckwith 2005; Margolin 2005). The function of these proteins is to anchor the complex in the membrane and to remodel the cell envelope biosynthetic machinery so as to mediate constriction and cytokinesis. At the heart of the divisome is a ring-like structure composed of the tubulin-like protein FtsZ. This Z-ring is a scaffold on which the remaining divisome proteins assemble, although only some of them directly interact with the ring. This is therefore a simple diffusion-and-capture system (as far as we know) in which some proteins (e.g., FtsA, ZipA, ZapA) are captured by the Z-ring and they, in turn, capture additional members (e.g., FtsK) of the structure and so forth.
How does the Z-ring ring assemble? This is largely through self-assembly as was most vividly shown with a modified FtsZ that had been fused to a membrane anchor (Osawa et al. 2008). The membrane-anchored FtsZ was capable of assembling into ring-like structures in liposomes in the absence of any other proteins. However, in the cell, the dynamics of the FtsZ polymers are influenced by a variety of proteins that either stabilize or destabilize the ring, much as is in the case of microtubules in eukaryotes. How the division machinery once assembled mediates cytokinesis remains an outstanding and unsolved problem in bacterial cell biology.
Finally, what are the factors that dictate the site in the cell at which the Z-ring, and hence the entire divisome, assembles? Two principal processes normally limit Z-ring assembly to the mid cell position in E. coli and Bacillus subtilis. One is nucleoid occlusion, which restricts Z-ring formation to nucleoid-free regions of the cell, that is, space between newly duplicated and segregated chromosomes (Yu and Margolin 1999). Nucleoid occlusion in E. coli is mediated in part by SlmA, a protein that both binds to the nucleoid and interacts with FtsZ (Bernhardt and de Boer 2005). An unrelated but analogous DNA binding protein, Noc, functions in nucleoid occlusion in B. subtilis (Wu and Errington 2004). Interestingly, it was recently shown that Noc binds to sequence elements that are largely absent from the terminus region, suggesting that the organization of the chromosome plays an important role in nucleoid occlusion (Wu et al. 2009). The second process is destabilization of Z-rings near the poles, which in E. coli is mediated by MinC, MinD, and MinE (Margolin 2005). MinC, which is anchored to the membrane by MinD (Hu et al. 2003), destabilizes FtsZ polymers (Hu et al. 1999). MinC and MinD rapidly oscillate from pole to pole in a process that is mediated by MinE (Raskin and de Boer 1999; Fu et al. 2001; Hale et al. 2001). As a consequence of this oscillation, the time-averaged concentration of the Z-ring destabilizing protein MinC is highest near the poles and lowest at the mid cell position. As discussed in the introduction, the Min system is itself a remarkable example of dynamic self-assembly, which can be largely reconstructed in vitro in the absence of any other protein. Thus, at least in E. coli, positioning of the divisome is ultimately dictated by chromosome segregation and a self-assembly process.
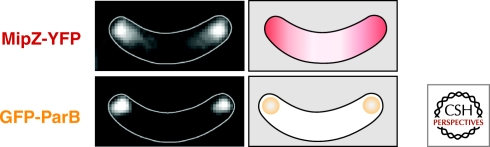
Interestingly, B. subtilis lacks MinE, and MinC and MinD do not oscillate. Instead, they are found statically at the poles and at mature divisomes (Marston et al. 1998; Gregory et al. 2008). As a consequence, Z-ring formation is normally excluded from the poles and at zones near existing divisomes. Yet another mechanism restricts Z-ring formation to the mid cell in Caulobacter, a bacterium in which the replication origin regions of the chromosome segregate to the extreme opposite poles of the cell (Jensen and Shapiro 1999). Caulobacter does not have the cell division inhibitor MinC. Instead, it has a protein MipZ, which shares similiarity to MinD and the partitioning protein ParA (Thanbichler and Shapiro 2006). MipZ localizes to the replication origin region (by binding to the chromosome segregation protein ParB) and directly inhibits Z-ring formation (Fig. 1). Thus, and in analogy to MinC, MipZ is thought to form a gradient with its highest concentration at the cell pole. This gradient is likely to involve nucleoprotein filaments of MipZ molecules that are ultimately anchored to the origin region (and hence the pole) via ParB.
Figure 1.
MipZ localization in C. crescentus. MipZ is anchored adjacent to the cell pole through its interaction with ParB (bound to the origin of replication) and appears to form a nucleoprotein gradient. The site of lowest concentration of the FtsZ inhibitor dictates future division site. Images show MipZ-YFP in a predivisional cell and GFP-ParB in a cell at a similar stage. Images kindly provided by M. Thanbichler.
In all of the examples considered earlier, the cell is, in effect, using inhibition (at the poles and atop the nucleoid in E. coli and B. subtilis) to dictate where the Z-ring can assemble. These negatively acting cues contrast with other cues we will consider that act positively to recruit proteins and promote localization.
Finally, we come to the unusual case of the asymmetrically positioned divisome of sporulating B. subtilis. On entry into sporulation, Z-ring formation switches from the midcell to sites near the poles, one of which mediates the formation of a division septum (Levin and Losick 1996). Polar Z-rings arise from a medial Z-ring in which FtsZ molecules appear to redistribute to the poles via a striking spiral intermediate (Ben-Yehuda and Losick 2002). Two factors govern this redistribution. One is an increase in FtsZ protein levels, which occurs as cells enter sporulation. The other is a sporulation protein called SpoIIE that interacts both with the membrane and with FtsZ (Lucet et al. 2000). That these two factors are sufficient to account for asymmetric placement of Z-rings is shown by experiments in which vegetative (nonsporulating) cells can be made to undergo asymmetric division by engineering the expression and overexpression of spoIIE and ftsZ, respectively, during growth (Ben-Yehuda and Losick 2002). How high levels of FtsZ decorated with SpoIIE overcomes nucleoid occlusion and the effects of MinC to favor polar assembly remains an outstanding question for the future.
POLAR ASSEMBLIES
Next, we turn our attention to the poles of the cell. What feature or features of the pole allow certain proteins to coalesce at the cell extremities? Three broad categories of models can be envisaged for how proteins localize to the extreme ends of cells, which are referred to as the geometric cue model, the division remnant model, and the self-assembly model. In what follows, we present examples of polar localization that are illustrative of these models.
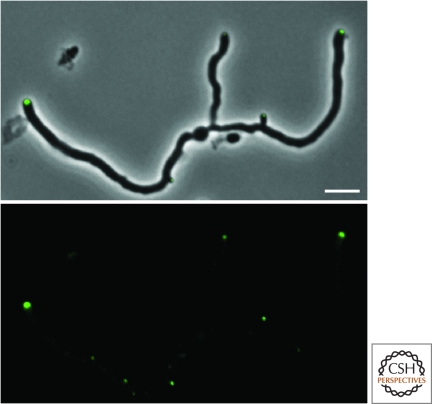
Perhaps the simplest is the geometric cue model, which we illustrate with the example of DivIVA of B. subtilis, which localizes both to the poles and even more strongly to mature division septa (Edwards and Errington 1997). Multiple functions have been ascribed to this highly conserved protein (Fig. 2) (Flardh 2003), among which is the recruitment of the replication origin region to the poles during sporulation (Thomaides et al. 2001). In B. subtilis, entry into sporulation is associated with the movement of the replication origin regions of the two chromosomes of the early-stage sporangium to the extreme opposite poles of the cell (Webb et al. 1997). Polar localization is mediated by RacA, a sporulation protein that binds to multiple sites in the origin-proximal region of the chromosome and directly or indirectly adheres to DivIVA, which is located at the poles (Ben-Yehuda et al. 2003). Interestingly, something analogous takes place in Caulobacter in which origin regions are propelled to extreme opposite poles during the cell cycle. In this case, the chromosome segregation protein ParB (which as we saw also interacts with the Z-ring inhibiting protein MipZ) directly binds to a DivIVA-like protein, PopZ, which is localized to the poles (Bowman et al. 2008; Ebersbach et al. 2008). Both DivIVA and PopZ, which are not homologous to each other, are thought to coat the inside surface of the cell pole to create surfaces that capture origin-region-bound RacA (Lenarcic et al. 2009) or ParB.
Figure 2.
Polarly localized DivIVA in Streptomyces coelicolor A3(2). S. coelicolor grows as a branching hyphae similar to filamentous fungi. Growth occurs at the hyphal tips in a manner dependent on DivIVA (Flardh 2003). Images show a DivIVA-GFP fusion. Lower panel: DivIVA-GFP foci. Upper panel: a merged image of DivIVA-GFP and phase contrast. Scale bar indicates 4 µm. Image kindly provided by K. Flardh.
What causes DivIVA (and PopZ) to localize to the cell poles and mature septa? Recent findings suggest that DivIVA recognizes the negative curvature of the membrane at the end of the cell as well as at the newly completed septum (Howard 2004; Lenarcic et al. 2009; Ramamurthi and Losick 2009). It is further suggested that DivIVA prefers the greater negative curvature of mature septa over the gentler curvature of the hemispherical poles. The chief evidence for these conclusions is: (1) when cytokinesis is blocked, DivIVA redistributes itself to the poles; (2) in aberrantly shaped cells created by use of a mutation in the gene for a cytoskeleton protein, DivIVA localizes preferentially to regions of negative curvature; and (3) in spherical cells (protoplasts) with uniform negative curvature, the protein shows a more or less uniform distribution (Lenarcic et al. 2009; Ramamurthi and Losick 2009). This compelling model awaits biochemical confirmation using purified DivIVA and lipid tubules. Conceivably, negative curvature is also a cue for the polar localization of PopZ. Of course, for both DivIVA and PopZ, other as yet unidentified proteins could assist in proper localization.
Next, we turn to the division remnant model, an example of which is TipN in Caulobacter crescentus (see Viollier 2010). TipN localizes to the divisome after constriction has initiated and is retained at this newly created pole after division is complete (Huitema et al. 2006; Lam et al. 2006). TipN then remains at the pole to create a mark or “birth scar,” which in turn recruits other proteins to the pole. TipN is required for the assembly of polar flagellum at the newly formed pole and is thought to serve as an organizational center for polar structures. In its absence, flagella are often assembled at the old cell pole instead of the new one. Once cytokinesis begins anew, TipN relocalizes from the soon to be old pole to the incipient new one.
It is not known what recruits TipN to the divisome or how it is retained at the new pole once division is completed. Conceivably, the geometry of the inside surface of the pole contributes both to the recruitment of TipN to the divisome and its retention at the pole. Alternatively, or additionally, TipN binds to the newly synthesized septal peptidoglycan. Once cytokinesis is complete and the divisome complex disassembles, it remains (a division remnant) anchored at the newly formed poles through interactions with the cell wall.
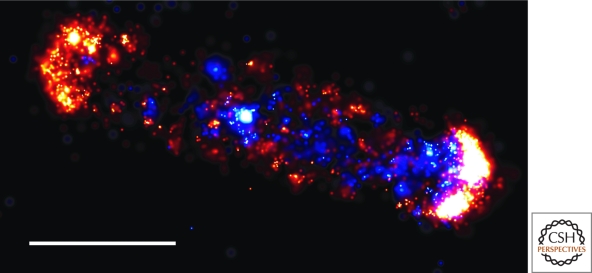
Finally, we illustrate polar localization by self-assembly with the case of the polar clustering of chemoreceptors at the cell pole of E. coli. When visualized by super-high resolution fluorescence microscopy (photoactivated localization microscopy or PALM), chemoreceptors fused to a photoactivatable fluorescent reporter protein are found to show an exponential distribution of cluster sizes, which are spread along the length of the cell with the largest clusters at the poles (Fig. 3) (Greenfield et al. 2009). The arrangement of these membrane clusters is consistent with a model in which a receptor is most likely to be captured by an existing cluster if one is nearby and to nucleate the formation of a new cluster when existing clusters are far (Wang et al. 2008). As a consequence, large clusters preferentially assemble as far as possible from each other, i.e., the two poles of the cell. If this model is correct, then self-assembly into clusters would be sufficient to explain the pattern of large polar clusters of chemoreceptors, without having to invoke the existence of an anchor or other cue at the pole. Of course, it is possible that a polar cue, such as negative curvature or a remnant protein from cell division, also contributes to the polar localization of chemoreceptors. Conversely, it is possible that self-assembly contributes to the localization of all of the proteins considered previously in this section.
Figure 3.
Chemoreceptors in E. coli show an exponential distribution of cluster sizes with the largest clusters at the poles. The image shows the Tar receptor fused to mEos visualized by super-high resolution fluorescence microscopy (photoactivated localization microscopy or PALM). Blue foci are from a PALM image in TIR-illumination. Red foci are from a PALM image in epi-fluourescence illumination (taken after Tar-mEos protein in the TIR region were bleached). Scale bar indicates 1 µm. Reprinted, with permission, from Greenfield et al. 2009.
One other case of polar localization merits comment because it does not seem to conform to any of the polar localization mechanisms we have considered so far. The virulence protein IcsA of Shigella flexneri localizes to the cell pole where it is incorporated into the outer membrane and mediates the assembly of an actin tail (Goldberg et al. 1993). The actin tail drives the movement of the bacterium through the cytoplasm of its mammalian host cell. Polar localization can occur in the absence of translocation across the inner membrane, a finding that reveals that IcsA localizes to the cytoplasmic face of the inner membrane at the pole before being secreted (Charles et al. 2001). Interestingly, IcsA localizes to the pole in E. coli as well as in Shigella, which suggests that the protein recognizes conserved features in both species. Using E. coli as a host, Goldberg and coworkers have made the discovery that under conditions in which cell division is blocked, IcsA localizes to future division sites (which will become cell poles after cytokinesis) in the resulting filaments. Remarkably, this localization does not appear to depend on FtsZ or the factors (nucleoid occlusion and MinCDE) that are known to govern the position of the divisome in E. coli (Janakiraman and Goldberg 2004). Evidently, IcsA is able to recognize some other as yet unknown feature of the future division site. Just what this feature is and how it is recognized by IcsA remain exciting and as yet unanswered questions.
PROTEIN CLUSTERS AND MICROCOMPARTMENTS
So far, we have considered examples of proteins or protein assemblies that are localized to the membrane. But some bacteria also produce clusters of proteins within the cytoplasm that lack any obvious membrane attachment. A striking example of such a cytoplasmic cluster is that formed by certain chemotaxis proteins in Rhodobacter sphaeroides (Wadhams et al. 2002). (R. sphaeroides also produces a separate cluster of chemotaxis proteins at the cell pole similar to that of many other bacteria.) The cytoplasmic chemotaxis proteins are seen as one large cluster, which gives rise to two clusters early in the cell cycle. The daughter clusters are located at the one-fourth and three-fourths positions of the cell, a pattern reminiscent of that seen for certain low-copy number plasmids. Remarkably, separation of a parental cluster into two daughter clusters requires an ortholog of the plasmid partitioning protein ParA (Thompson et al. 2006) (see Mullins 2010). Evidently, the same kinds of mechanisms that mediate plasmid separation in bacteria have also been harnessed to drive the separation and positioning of protein clusters. It will be interesting to see if these unusual ParA proteins take advantage of the nucleoid to segregate these cytoplasmic clusters as is hypothesized for plasmid segregation (Ebersbach and Gerdes 2005). In a sense, the positioning of the cytoplasmic chemotaxis cluster of R. sphaeroides is an example of protein targeting in that a ParA-like protein drives the daughter clusters apart to a characteristic position in the cell. It will be interesting to see whether other proteins or protein complexes take advantage of partitioning proteins for proper localization.
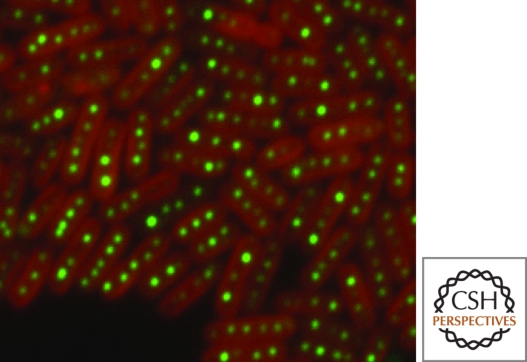
If so, an attractive candidate would be the carboxysome, a protein microcompartment in cyanobacteria that contains the carboxylase RuBisCO (Yeates et al. 2008). The carboxysome, which mediates the fixation of carbon dioxide, is believed to isolate RuBisCO from the cytoplasmic environment and thereby facilitate the incorporation of carbon dioxide into 3-phosphoglycerate. Recent studies in which carboxysome proteins in Synechococcus elongatus were fused to the green fluorescent protein revealed fluorescent foci in a strikingly even spacing along the length of the cell (Fig. 4) (D. Savage and P. Silver, pers. comm.). In light of the discovery that the spacing of cytoplasmic chemotaxis protein clusters in R. sphaeroides is governed by a ParA homolog, it is appealing to imagine that plasmid partitioning proteins might also contribute to the stereotyped distribution of carboxysomes in cyanobacteria.
Figure 4.
Carboxysomes in S. elongatus are found evenly spaced along the length of the cell. The image shows the carboxysome-localized enzyme RuBisCO fused to the green fluorescent protein. The native chlorophyll fluorescence (red) labels the peripheral membranes. Image kindly provided by D. Savage and P. Silver.
CYTOSKELETAL ELEMENTS
As alluded to earlier, proteins can also localize as polymeric assemblies inside the cell. These assemblies are called cytoskeletal elements, and the proteins that form them include the tubulin-like protein FtsZ, the actin-like protein MreB (Jones et al. 2001), and the Caulobacter protein Crescentin (Ausmees et al. 2003), which resembles an intermediate filament protein. As discussed earlier, FtsZ filaments assemble into a ring-like structure at mid-cell and the localization of this cytoskeletal element is dictated by the inhibitory activities of MinC/D and nucleoid. MreB forms helical filaments that extend the entire length of the cell and localizes adjacent to the cytoplasmic membrane. A linear polymer forming along the periphery of a cylinder will naturally trace out a spiral unless otherwise constrained. Accordingly, it is possible that MreB adopts this helical localization pattern without cellular cues. If this is the case, we can consider the localization of MreB as an example of self-assembly. Interestingly and importantly, in some bacteria, the MreB helical filament appears to coalesce into a ring-like structure at mid-cell during cytokinesis (in an FtsZ-dependent manner), suggesting that other proteins can influence its organization (Figge et al. 2004).
Many functions have been attributed to MreB filaments, the principal of which is cell shape determination. In its absence (or on depletion or inhibition), cells lose their rod-shaped appearance and become spherical before lysing (Jones et al. 2001; Gitai et al. 2005). Despite the word “skeleton” embedded within cytoskeleton and the helical localization pattern, MreB does not appear to serve as a bacterial skeleton. This job is the purview of the cell wall, the peptidoglycan, which acts as an exoskeleton, maintaining the cell’s shape. The current thinking is that MreB and its homologs participate in cell shape by organizing the peptidoglycan biosynthetic enzymes into a helical arrangement that is oriented along the long axis of the cell. In support of this idea, fluorescent probes that label sites of active cell wall synthesis have a spiral-like pattern (Daniel and Errington 2003; Tiyanont et al. 2006). Whether the reorganization of MreB at the mid-cell is involved in redirecting these enzymes toward peptidoglycan synthesis at or adjacent to the septum remains unclear. The localization of these biosynthetic enzymes to the MreB filament is presumably achieved by diffusion and capture, much like the localization of the divisome components to the cytoskeletal FtsZ ring. The emerging view is that MreB and other cytoskeletal components serve as major organizing centers inside the bacterial cell. One clear example is the arrangement of magnetosomes inside Magnetospirillum (to follow).
Yet a third variety of cytoskeletal elements exists in Caulobacter crescentus. These polymers are composed of a protein called Crescentin that resembles intermediate filaments found in eukaryotes (Ausmees et al. 2003). Crescentin is responsible for the curved crescent, comma-shape of Caulobacter. Cells lacking Crescentin are rod-shaped and expression of Crescentin in E. coli results in a transition from a rod to a crescent (Cabeen et al. 2009). Crescentin forms an extended filament that localizes adjacent to the cell envelope along the concave side of Caulobacter cells. This filament appears to govern cell shape by a different mechanism than the one proposed for MreB. Current data suggest that physical strain imparted by the Crescentin structure can alter the kinetics of cell wall insertion, such that less growth occurs along the Crescentin-containing side the of cell, resulting in curvature (Cabeen et al. 2009). Interestingly, the MreB cystoskeleton appears to help anchor the Crescentin filament to the cell envelope (Charbon et al. 2009).
ORGANELLES
We do not usually think of bacteria as having organelles, but some bacteria do have organelle-like structures that pose fascinating issues of protein localization and assembly. Indeed, the cell envelope of Gram-negative bacteria can be thought of as an organelle that is bound on the inside by the cytoplasmic membrane and on the outside by the outer membrane. Because an entire article is devoted to the envelope (see Silhavy et al. 2010), we do not consider it further here except to point out that proteins in every layer of the envelope have been found to localize to specific subcellular sites. For example, some periplasmic proteins specifically localize to the cytokinetic ring during cell division (Uehara et al. 2009), whereas others involved in cell wall remodeling localize in spiral-like structures reminiscent of cytoplasmic MreB spirals (Divakaruni et al. 2007).
A paradigm for a bacterial organelle is the magnetosome of Magnetospirillum (see Fig. 1 in Murat et al. 2010). The magnetosome is a chain of magnetite crystals that acts as a compass to passively orient the bacterium in a geomagnetic field (Komeili 2007; Schuler 2008). Each crystal is surrounded by a membrane and long chains of membrane-encased crystals are contained within a tubular membrane compartment that grows out by invagination of the cytoplasmic membrane. The crystals are organized in a linear array within the bacterium by a dedicated cytosekeleton composed of an MreB homolog called MamK (Komeili et al. 2006). The magnetosome is specified by a genomic island that consists of about 100 genes that are responsible for biogenesis of the magnetosome membrane and cytoskeleton and the formation of the crystals. Importantly, the genomic island encodes membrane and cytoplasmic proteins that localize uniquely to the magnetosome (Schubbe et al. 2003; Fukuda et al. 2006). Thus, the magnetosome raises fascinating and as yet unanswered questions about its biogenesis and the mechanisms that direct proteins to a membrane compartment.
A second example of an organelle-like body in a bacterium is the developing spore or, more properly, the endospore, of B. subtilis. The endospore is a special kind of dormant cell that is generated internally to its host cell (Stragier and Losick 1996; Errington 2003). Endospore formation takes place in two principal stages. First, a cell that has entered the pathway to sporulate undergoes asymmetric division to create a small compartment known as the forespore and an adjacent, large compartment, the mother cell. Next, in a process that resembles phagocytosis, the mother-cell membrane migrates around and engulfs the forespore, eventually pinching it off as a free protoplast in the mother-cell cytoplasm. In subsequent development, a shell composed of more than 70 proteins is produced in the mother cell and deposited around the forespore to create the spore coat (Kim et al. 2006).
Interestingly, engulfment resembles magnetosome formation in the sense that the engulfing membrane is initially continguous with the cytoplasmic membrane of the mother cell. (The magentosome membrane, as we saw, arises as an invagination of the cytoplasmic membrane.) Unlike the magnetosome, however, the forespore becomes topologically isolated from the cytoplasmic membrane when engulfment is complete (Sharp and Pogliano 1999).
How is this protein shell around the forespore assembled? As in the case of other multiprotein complexes we have considered, the spore coat poses two questions: What is the primary cue that dictates the location of coat protein deposition and how are the myriad proteins properly assembled? The first step in coat assembly is the localization of a small amphipathetic helix, SpoVM, to the outer membrane that surrounds the forespore (van Ooij and Losick 2003). How does this small protein discriminate between the outer forespore membrane and the cytoplasmic membrane of the mother cell? Evidence indicates that SpoVM preferentially binds to positively curved surfaces, thereby discriminating between the concave shape of the cytoplamic membrane and the convex shape of the outer forespore membrane (Ramamurthi et al. 2009). SpoVM, in turn, recruits a cytosolic protein called SpoIVA, which polymerizes into long filaments in a process that is driven by ATP hydrolysis (Ramamurthi et al. 2006; Ramamurthi and Losick 2008). These long filaments wrap around the forespore to create the basement layer of the coat. A complex web of still-poorly understood interactions involving scores of additional proteins drive the remaining steps in coat assembly. Thus, in the case of the protein coat, localization to the prospective spore appears to be achieved by geometric cues followed by the recruitment of additional proteins by diffusion-and-capture.
A second case of protein localization to the membranes surrounding the forespore merits comment because it does not conform to any of the mechanisms we have considered previously. In this case, protein localization is governed by the interaction of membrane proteins in adjacent daughter cells. A membrane protein (called SpoIIIAH) produced in the mother cell becomes anchored in the membranes surrounding the spore through its interaction with a membrane protein (SpoIIQ) synthesized in the forespore (Blaylock et al. 2004; Doan et al. 2005). The extracellular domains of these two proteins interact in the space between the double-membrane that separates them. Recent evidence suggests that these proteins are, in fact, part of a larger complex that serves as a channel or secretion apparatus that links the mother cell and forespore (Camp and Losick 2008; Meisner et al. 2008; Camp and Losick 2009; Doan et al. 2009). The SpoIIIAH-SpoIIQ complex also plays a key role in anchoring several other proteins in the forespore membranes (Doan et al. 2005; Jiang et al. 2005; Campo et al. 2008). In this unusual case of protein localization, the ultimate anchor is the interaction of two proteins across adjacent membranes that separate daughter cells. This type of localization mechanism is reminiscent of the interactions between cadherins or claudins in neighboring cells at tight junctions in eukaryotes (Tsukita et al. 2001) and of the interaction between pathogens and host cells at sites of adherence (Kenny et al. 1997).
CONCLUSIONS AND FUTURE PROSPECTS
As stated from the outset, we have only touched on a few representative examples of protein localization to highlight general principles. Many, many proteins (and clearly more to come) have been found to show specific patterns of subcellular localization. We suspect that most of these proteins attain their proper localization by diffusion-and-capture but anticipate more examples like the clusters of cytoplasmic chemotaxis proteins in R. sphaeroides, where proteins or protein complexes are actively “targeted” to specific sites within the cell.
One of the principal challenges for the future is defining the mechanisms by which proteins are anchored at specific subcellular positions. We have argued that geometric cues, self-assembly, and restricted sites of assembly are all exploited by the cell to specifically localize a subset of proteins. These “founder” proteins, in turn, govern (directly or indirectly) the localization of a whole host of others. We anticipate that many other anchor proteins will be identified that either recognize geometric cues or self-assemble. Nonetheless, we suspect that other mechanisms exist that anchor proteins at specific sites. If, for example, we look at the rich literature of cell biology in eukaryotes, it is not hard to imagine that the phospholipid bilayer of bacteria contains rafts enriched in specific lipids and/or cholesterol-like molecules that recruit and anchor a subset of integral membrane proteins (Brown and London 1998). In fact, there is growing evidence in support of lipid microdomains in bacteria (D. López and R. Kolter, Membrane rafts in bacteria, submitted; reviewed in Matsumoto et al. 2006). Of course, lipid rafts are yet another example of self-assembly, although where these membrane patches localize is thought, at least in some cases, to be dictated by geometric cues. It is also not hard to imagine, by analogy to eukaryotic cell biology, that specific mRNAs in bacteria localize to the sites where the proteins they encode are destined (Shav-Tal and Singer 2005). If such messages exist, protein localization in these cases could be governed by mRNA localization. Understanding how these mRNAs are themselves anchored at specific sites will likely reveal new and interesting biology.
We only briefly touched on the role of the largest macromolecule in the cell, the chromosome, in orchestrating protein localization. The example we highlighted was division site selection and the role of the nucleoid in preventing FtsZ assembly. We hypothesize that this highly organized macromolecule (bound by numerous proteins) exerts its influence on the subcellular organization of many proteins and protein complexes, the extent of which will be revealed in the coming years. Along these lines, we eagerly await definitive proof that membrane protein insertion is indeed coupled to transcription and translation. Once established, a future challenge will be to assess the role of the “transertion” process in localizing membrane proteins at specific sites and conversely in organizing and sculpting the nucleoid itself.
Finally, we anticipate that the ability to more precisely define the localization of proteins in the cell using super-high resolution microscopy will shed light on the organization of the nucleoid and other macromolecules and macromolecular complexes that have remained refractory to diffraction-limited microscopy (Bates et al. 2008). Super-resolution single-molecule tracking holds similar promise for characterizing the dynamic behaviors of proteins and protein complexes (Xie et al. 2008). It is our expectation that these new cytological approaches will reveal ultrastructure and protein dynamics in bacteria that parallel eukaryotic cells.
Footnotes
Editors: Lucy Shapiro and Richard Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Alley MR, Maddock JR, Shapiro L 1992. Polar localization of a bacterial chemoreceptor. Genes Dev 6:825–836 [DOI] [PubMed] [Google Scholar]
- Ames P, Parkinson JS 2006. Conformational suppression of inter-receptor signaling defects. Proc Natl Acad Sci 103:9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270:637–640 [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C 2003. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- Bates M, Huang B, Zhuang X 2008. Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr Opin Chem Biol 12:505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257–266 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell 18:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164 [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP Jr, Pogliano K 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev 18:2916–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, London E 1998. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol 14:111–136 [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C 2009. Bacterial cell curvature through mechanical control of cell growth. Embo J 28:1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69:402–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Marquis KA, Rudner DZ 2008. SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis. J Biol Chem 283:4975–4982 [DOI] [PubMed] [Google Scholar]
- Charbon G, Cabeen MT, Jacobs-Wagner C 2009. Bacterial intermediate filaments: In vivo assembly, organization, and dynamics of crescentin. Genes Dev 23:1131–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles M, Perez M, Kobil JH, Goldberg MB 2001. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc Natl Acad Sci 98:9871–9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J 2003. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- deBoer PA 2010. Cytokinesis and the min system. Cold Spring Harb Perspect Biol 2:a000323 [Google Scholar]
- Deich J, Judd EM, McAdams HH, Moerner WE 2004. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Natl Acad Sci 101:15921–15926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol 66:174–188 [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol 55:1767–1781 [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP Jr, Rudner DZ 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5:e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A, Losick R 1991. Compartmentalized expression of a gene under the control of sporulation transcription factor σ E in Bacillus subtilis. Proc Natl Acad Sci 88:9934–9938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K 2005. Plasmid segregation mechanisms. Annu Rev Genet 39:453–479 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134:956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol 24:905–915 [DOI] [PubMed] [Google Scholar]
- Errington J 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126 [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- Flardh K 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol 49:1523–1536 [DOI] [PubMed] [Google Scholar]
- Fu X, Shih YL, Zhang Y, Rothfield LI 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci 98:980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Okamura Y, Takeyama H, Matsunaga T 2006. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett 580:801–812 [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341 [DOI] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J 2005. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr Biol 15:R514–R526 [DOI] [PubMed] [Google Scholar]
- Goldberg MB, Barzu O, Parsot C, Sansonetti PJ 1993. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. Infect Agents Dis 2:210–211 [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J 2009. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol 7:e1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Pogliano K 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev 22:3475–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Meinhardt H, de Boer PA 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. Embo J 20:1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M 2004. A mechanism for polar protein localization in bacteria. J Mol Biol 335:655–663 [DOI] [PubMed] [Google Scholar]
- Hu Z, Saez C, Lutkenhaus J 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: Role of MinD and MinE. J Bacteriol 185:196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci 96:14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037 [DOI] [PubMed] [Google Scholar]
- Janakiraman A, Goldberg MB 2004. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc Natl Acad Sci 101:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci 96:10661–10666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rubio A, Chiba S, Pogliano K 2005. Engulfment-regulated proteolysis of SpoIIQ: Evidence that dual checkpoints control σ activity. Mol Microbiol 58:102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J 2001. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, Ferguson CC, Eichenberger P, Driks A 2006. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol 59:487–502 [DOI] [PubMed] [Google Scholar]
- Kocks C, Hellio R, Gounon P, Ohayon H, Cossart P 1993. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J Cell Sci 105:699–710 [DOI] [PubMed] [Google Scholar]
- Komeili A 2007. Molecular mechanisms of magnetosome formation. Annu Rev Biochem 76:351–366 [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245 [DOI] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124:1011–1023 [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW 2009. Localisation of DivIVA by targeting to negatively curved membranes. Embo J 28:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev 10:478–488 [DOI] [PubMed] [Google Scholar]
- Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P 2008. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320:789–792 [DOI] [PubMed] [Google Scholar]
- Lucet I, Feucht A, Yudkin MD, Errington J 2000. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. Embo J 19:1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W 2005. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6:862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12:3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Kusaka J, Nishibori A, Hara H 2006. Lipid domains in bacterial membranes. Mol Microbiol 61:1110–1117 [DOI] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP Jr 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci 105:15100–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins D 2010. Bacterial cytoskeleton: the function of FtsZ and ParA polymers. Cold Spring Harb Perspect Biol 2:a002550 [Google Scholar]
- Murat D, Byrne M, Komeili A 2010. Cell biology of prokaryote organelles. Cold Spring Harb Perspect Biol 2:a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R 2008. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell 31:406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci 106:13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62:1547–1557 [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R 2009. Geometric cue for protein localization in a bacterium. Science 323:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci 96:4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev 16:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci 99:8701–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbe S, Kube M, Scheffel A, Wawer C, Heyen U, Meyerdierks A, Madkour MH, Mayer F, Reinhardt R, Schuler D 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J Bacteriol 185:5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler D 2008. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol Rev 32:654–672 [DOI] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci 96:14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Singer RH 2005. RNA localization. J Cell Sci 118:4077–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Portnoy DA, Theriot JA 1995. Asymmetric distribution of the Listeria monocytogenes ActA protein is required and sufficient to direct actin-based motility. Mol Microbiol 17:945–951 [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30:297–341 [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389:90–93 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162 [DOI] [PubMed] [Google Scholar]
- Thomaides HB, Freeman M, El Karoui M, Errington J 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev 15:1662–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Wadhams GH, Armitage JP 2006. The positioning of cytoplasmic protein clusters in bacteria. Proc Natl Acad Sci 103:8209–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci 103:11033–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M 2001. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2:285–293 [DOI] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG 2009. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191:5094–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, Losick R 2003. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol 185:1391–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier P 2010. Polar structures and the proteins that regulate their biogenesis. Cold Spring Harb Perspect Biol 2:a000372 [Google Scholar]
- Wadhams GH, Martin AC, Porter SL, Maddock JR, Mantotta JC, King HM, Armitage JP 2002. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol Microbiol 46:1211–1221 [DOI] [PubMed] [Google Scholar]
- Wang H, Wingreen NS, Mukhopadhyay R 2008. Self-organized periodicity of protein clusters in growing bacteria. Phys Rev Lett 101:218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667–674 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J 2009. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. Embo J 28:1940–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XS, Choi PJ, Li GW, Lee NK, Lia G 2008. Single-molecule approach to molecular biology in living bacterial cells. Annu Rev Biophys 37:417–444 [DOI] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM 2008. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Microbiol 6:681–691 [DOI] [PubMed] [Google Scholar]
- Yu XC, Margolin W 1999. FtsZ ring clusters in min and partition mutants: Role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32:315–326 [DOI] [PubMed] [Google Scholar]