Abstract
Talin is a 270-kDa protein that activates integrins and couples them to cytoskeletal actin. Talin contains an N-terminal FERM domain comprised of F1, F2 and F3 domains, but it is atypical in that F1 contains a large insert and is preceded by an extra domain F0. Although F3 contains the binding site for β-integrin tails, F0 and F1 are also required for activation of β1-integrins. Here, we report the solution structures of F0, F1 and of the F0F1 double domain. Both F0 and F1 have ubiquitin-like folds joined in a novel fixed orientation by an extensive charged interface. The F1 insert forms a loop with helical propensity, and basic residues predicted to reside on one surface of the helix are required for binding to acidic phospholipids and for talin-mediated activation of β1-integrins. This and the fact that basic residues on F2 and F3 are also essential for integrin activation suggest that extensive interactions between the talin FERM domain and acidic membrane phospholipids are required to orientate the FERM domain such that it can activate integrins.
Keywords: focal adhesions, integrins, NMR, phospholipids, talin
Introduction
Cell adhesion to the extracellular matrix is of fundamental importance to the development of multicellular organisms (Aszodi et al, 2006) and requires the coordinated assembly of the integrin family of adhesion receptors into complexes containing cytoplasmic proteins with scaffolding, adaptor, regulatory and mechanotransduction functions (Hynes, 2002; Legate et al, 2006). Complex formation can be initiated either by ligand binding to the extracellular domain of integrins or by activation of intracellular signalling pathways that regulate the affinity and avidity of integrins for ligands (Luo et al, 2007). The cytoskeletal protein talin has both a regulatory and scaffolding function in these events by switching integrins from a low to high-affinity state (Calderwood, 2004b), by promoting integrin clustering (Saltel et al, 2009) and by coupling integrins directly to the actin cytoskeleton (Critchley and Gingras, 2008).
Talin is a 270-kDa protein (Figure 1A) consisting of a 47-kDa N-terminal head and an elongated 220 kDa C-terminal rod composed of amphipathic α-helical bundles (Critchley, 2009; Roberts and Critchley, 2009), the most C-terminal of which contains a conserved actin-binding site (Gingras et al, 2008). The talin head incorporates a FERM (Band 4.1, ezrin, radixin, moesin) domain made up of F1, F2 and F3 domains, and a binding site for the cytoplasmic tails of β-integrin subunits has been localized to F3 (Calderwood et al, 2002; Garcia-Alvarez et al, 2003), although a second integrin-binding site (IBS2) has been identified in the talin rod (Xing et al, 2001; Moes et al, 2007; Gingras et al, 2009). However, only F3 (Calderwood et al, 2002) and not IBS2 (Tremuth et al, 2004) was able to activate αIIbβ3-integrin expressed in CHO cells. Both crystallographic and NMR studies reveal an extensive-binding interface between F3 and β3-integrin tails that includes both the membrane-proximal NPxY motif and the membrane-proximal helical region (Garcia-Alvarez et al, 2003; Wegener et al, 2007; Anthis et al, 2009). The results indicate that F3 activates β3-integrins by disrupting the salt bridge between the α and β tails that normally keeps the integrin locked in a low-affinity state. Moreover, the association of basic patches on both F2 and F3 that bind acidic phospholipids are also required for β3-integrin activation (Anthis et al, 2009) and clustering (Saltel et al, 2009).
Figure 1.
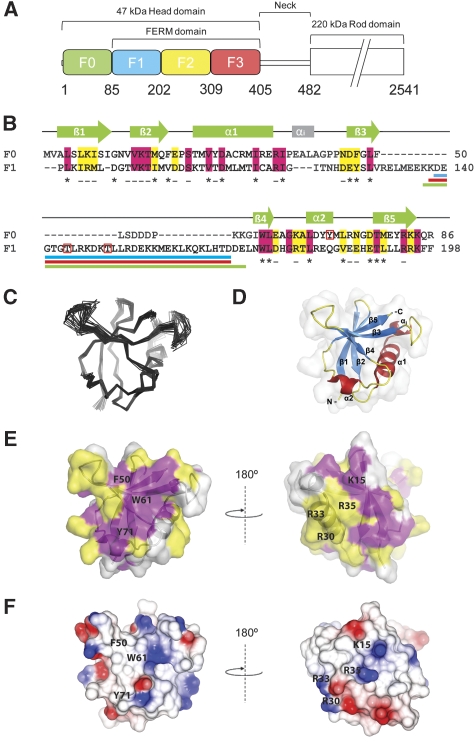
Solution structure of the N-terminal F0 domain of talin1. (A) Domain structure of the talin1 head; residue numbers indicate domain boundaries. (B) Amino-acid sequence alignment and secondary structure of the F0 and F1 domains. Invariant residues are in magenta and conserved residues in yellow. Buried residues are marked ‘–' (<80% exposed) and ‘*' (<20% exposed). Phosphorylation sites are indicated by red boxes. Loop deletion regions are shown by coloured lines under the sequence; Δ30 (blue), Δ31 (red) and Δ34 (green). (C, D) Solution structure of the F0 domain; (C) superposition of the 20 lowest energy structures. (D) Ribbon diagram of the F0 structure. (E, F) Surface representations of the talin1 F0 domain. (E) Surface representation showing the position of residues conserved among talins. The sequences used in the conservation analysis are shown in Supplementary Figure 2A, colour scheme as in (B). A conserved surface comprising helix 1 and the loop between helix 1 and strand 3 (shown in the right panel) is present in all talin isoforms. (F) Electrostatic surface of talin1 F0. Residues discussed in the text are marked.
Interestingly, cell biological studies suggest that other regions of the talin head are also required for efficient integrin activation. Thus, talin F2F3 is not sufficient to activate β1A-integrins, although it binds to β1A and β3 tails with similar affinity (Bouaouina et al, 2008). However, expression of the whole talin head (residues 1–405) was able to support β1A-integrin activation, and this required residues 1–85, which precede the FERM domain (referred to as the F0 domain), plus the F1 FERM domain and the integrin-binding F3 domain (Bouaouina et al, 2008). Moreover, the talin head was found to activate β3-integrins more efficiently than F2F3 alone. These results implicate F0 and F1 in β1- and β3-integrin activation, although F0 or F0F1 alone are unable to support integrin activation (Bouaouina et al, 2008).
Here, we report the solution structure of the talin F0, F1 and F0F1 domains. Both F0 and F1 have similar ubiquitin-like folds, and the two domains have a fixed orientation stabilized by a well-defined set of hydrophobic and charge–charge interactions. We identify a large loop within the F1 domain and show (i) that it has a propensity to adopt a helical structure in which basic residues are clustered on one surface and (ii) that it interacts with vesicles containing acidic phospholipids. Furthermore, the loop and basic residues within the loop are required for talin-mediated β1-integrin activation.
Results
Talin FERM domain is atypical
The N-terminal talin head contains a FERM domain (residues 86–400), but it is atypical in that the F1 domain includes a large ∼30-residue insert and is preceded by an earlier uncharacterized region referred here as F0 (Figure 1A). Sequence alignments show that F0 is homologous (22% identity, 40% similarity) to the talin FERM F1 domain (Figure 1B) that is predicted to have a ubiquitin-like fold, although the structure of the F1 domain has not earlier been determined. Interestingly, the F0 domain is not identified by database servers such as SMART (Schultz et al, 1998), which may explain the lack of attention to this region. The F0 domain is highly conserved in all talin isoforms including talin-1 and 2 and the two Dictyostelium discoideum talins, indicating that it has an important function. Indeed, we have recently shown that F0 is essential for the activation of β1-integrins and enhances the activation of β3-integrin (Bouaouina et al, 2008).
Structure of the talin F0 domain
The NMR spectra (Supplementary Figure 1A) of the recombinant F0 domain (residues 1–86) show that it forms a stable globular structure, as the resonances are highly dispersed and have uniform line width. We have determined the solution structure of the F0 domain from 2349 distance and 108 dihedral angle restraints identified using [13C,15N]-labelled protein. The statistics of the structure determination are presented in Supplementary Table 1, and the structures are shown in Figure 1C and D. As predicted, F0 has a ubiquitin-like β-grasp fold (topology β1, β2, α1, β3, β4, α2, β5). The secondary structure consists of a five-stranded twisted β-sheet: β1 8–10, β2 13–19, β3 47–52, β4 60–63 and β5 78–83, an α-helix α1 24–35 and a short 310 helical turn 68–71 (Figure 1B and D). The loop between the helix α1 and strand β3, as well as the N-terminal part of the loop between strands β3 and β4 are unstructured, whereas the rest of the β3–β4 loop and all other loops are well ordered. There is an extra helical turn present in the α1–β3 loop, designated αi in Figure 1B. This turn is not a common feature of ubiquitin-like folds, but the sequence is conserved in all talins.
The F0 domain of talin is required for maximal integrin activation (Bouaouina et al, 2008), suggesting that it must interact with other FA components, although no binding partner for F0 has yet been identified. Sequence alignment across species (Supplementary Figure 2A) shows that a large proportion of the conserved residues are buried and stabilize the protein fold. The conserved surface exposed residues from two distinct clusters on opposite faces of F0 (Figure 1E). One of these coincides with the area of contact between F0 and F1 and is discussed later. The second conserved cluster involves strand β2, the β1–β2 loop and the N-terminal part of the α1–β3 loop in the region of the αi helix. Intriguingly, this area is similar to the Ras-binding site in RalGDS (Huang et al, 1998), a structural homologue of F0. Similar to the Ras-binding surface of RalGDS and other Ras/Rap1-binding domains, the conserved surface of F0 has a high concentration of positive charges (Figure 1F). The charged residues K15, R30 and R35 of F0 are located in similar positions to the important positively charged residues of Ras/Rap1-binding domains that form salt bridges with the negatively charged groups on Ras/Rap1. In F0, K7 and R33 introduce additional positive charges that are also observed in some Ras/Rap1-binding domains (Kiel et al, 2005) (Supplementary Figure 2). Ha-Ras has been shown to bind to the talin head (Goldfinger et al, 2007) and Rap1 is implicated in talin activation (Han et al, 2006; Lee et al, 2009). However, Ha-Ras does not bind to F0 and GMP.PNP-Rap1 binds only weakly (Kd=140 μM), although the interaction is GTP dependent (Supplementary Figure 2E and F).
Talin F0 and F1 structures are similar
The solution structure of F1 was calculated from 2306 distance and 56 dihedral angle restraints measured using [13C,15N]-labelled protein. The statistics of the structure determination are presented in Supplementary Table 2 and the structures are shown in Figure 2A and B. Similar to F0, the F1 domain adopts a ubiquitin-like β-grasp fold composed of a twisted five-stranded β-sheet with boundaries β1 86–92, β2 95–102, β3 128–132, β4 171–174 and β5 190–194, an α-helix1 109–119 and a short helical turn 180–185. One interesting feature of F1 was the presence of four well-defined signals at 15N chemical shifts of ∼84 p.p.m. in the [1H,15N]-HSQC spectrum assigned to the NɛH signals of Arg86, Arg91, Arg131 and Arg178 (Supplementary Figure 3A). Each of these resonances gave extensive NOEs in the 15N-edited NOESY-HSQC indicating that the side chains of these residues were in well-defined positions, forming electrostatic contacts with other residues. The insert in F1 is located between strands β3 and β4 as part of a large 35-residue loop (residues 133–170) that is unstructured and highly dynamic, as shown by intense sharp signals in the [1H,15N]-HSQC spectra, and the presence of only weak intra-residue and sequential NOE interactions within the loop. Using the assignments of F1, it was possible to identify the resonances of the F1 loop in the [1H,15N]-HSQC spectrum of the whole talin head (F0F1F2F3, residues 1–400) (Supplementary Figure 4). The sharp signals of the loop region in the isolated F1 domain remain sharp and intense in the spectrum of the whole head, confirming our earlier conclusion that the loop remains unstructured even in the full-talin head and does not interact with any of the other domains (Barsukov et al, 2003).
Figure 2.
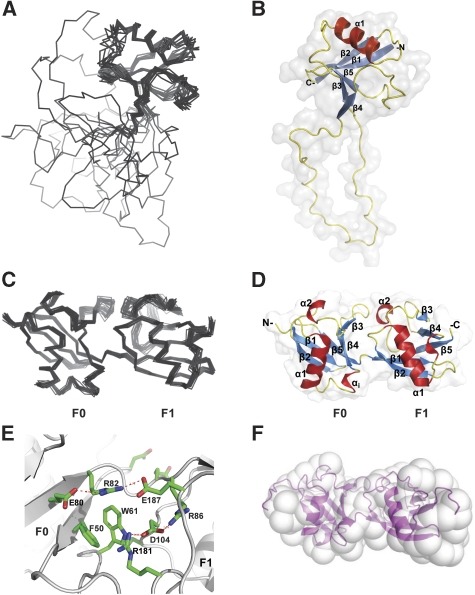
Solution structure of the talin F1 domain and the F0F1 double domain. (A) Superposition of the 20 lowest energy structures of F1. (B) Ribbon diagram of the F1 structure. (C, D) Solution structure of F0F1(Δ30) minus the F1 loop. (C) Superposition of the 20 lowest energy structures. (D) Ribbon diagram of the F0F1(Δ30) structure. (E) The interface between F0 (left) and F1 (right). (F) Small angle X-ray scattering envelope reconstruction with GASBOR, showing the best fit with the F0F1(Δ30) solution structure.
The structures of the F0 and F1 domains are remarkably similar, as can be seen from comparison of Figures 1D and 2B (r.m.s.d.=1.7 Å for the secondary structure regions). The loop inserted in the F1 domain forms a natural extension of the smaller partially disordered β3–β4 loop in F0. The helical turn αi of F0 is absent in F1 because of the three-residue deletion in the sequence (Figure 1B). The majority of residues conserved between F0 and F1 (22 out of 34) (Figure 1B) are buried as determined by NACCESS (Hubbard and Thornton, 1993) and are involved in stabilization of the ubiquitin fold. To identify members of the ubiquitin family with the closest structural similarity to F0 and F1, we used the F0 structure, as it lacks the large insertion that could potentially distort the comparison. The closest structural homologues of F0 based on a DALI database search (Holm and Sander, 1995) are interferon-induced ubiquitin cross-reactive protein ISG15 (1z2m; Z-score=9.5; r.m.s.d.=5.9 Å) (Narasimhan et al, 2005), ubiquitin (1ubi; Z-score=8.5; r.m.s.d.=2.8 Å) (Ramage et al, 1994) and the Ras-binding domain of RalGDS (1lfd; Z-score=8.1; r.m.s.d.=2.6 Å) (Huang et al, 1998) (Supplementary Figure 5).
To facilitate further structural studies of the talin head, we designed a construct in which the F1 loop was removed with minimal disturbance to the F1 structure. The pattern of NOEs across the β3–β4 hairpin clearly shows the absence of inter-strand interactions in the region L133–E170. To define the minimal link required between the β-strands, we tested three different F1 deletion constructs: Δ30 (residues D139-D168 removed), Δ31 (residues K138-D168 removed) and Δ34 (residues K137-E170 removed) indicated in Figure 1B. Comparison of the [1H,15N]-HSQC spectra of the deletion mutants (Supplementary Figure 6) shows large spectral changes when the number of residues connecting the β3 and β4 strands is less than eight, but a construct (Δ30) with eight connecting residues showed minimal spectral differences from the wild-type F1, showing the absence of structure distortion. Interestingly, the corresponding β3–β4 loop in the F0 domain is also eight residues in length. The Δ30 deletion was, therefore, introduced into other talin head fragments.
F0 and F1 form a pair of closely packed ubiquitin-like domains
Comparison of the [1H,15N]-HSQC spectra of the F0, F1 and F0F1 constructs showed localized chemical shift differences that map on continuous surface regions of the isolated domains adjacent to the inter-domain connection (Supplementary Figure S1A and B), suggesting close contact between the domains. Mixing the isolated F0 and F1 domains in solution led to similar changes in the individual spectra showing a strong interaction between the two domains (data not shown). Such a close interaction is unusual, as most of the other known structures of tandem ubiquitin-like domains show no contacts between the two domains (Narasimhan et al, 2005). The solution structure of the double domain with the F1 loop removed (F0F1(Δ30)) was determined from 6606 distance, 124 dihedral angle restraints and 66 backbone 1H,15N-RDCs measured using [13C,15N]-labelled protein. The statistics of the structure determination are presented in Supplementary Table 3 and the structures are shown in Figure 2C and D. The conformations of the individual domains within the double domain are essentially the same as those of the isolated domains (r.m.s.d.=1.05 and 1.86 Å for the F0 and F1 domains, respectively). A short three-residue linker joins the C-terminus of strand β5 in F0 and the N-terminus of strand β1 in F1. The relative orientation of the domains is well defined, and is supported by the large number of inter-domain NOEs and by RDC values that are consistent with both domains fitting to a single alignment tensor (Supplementary Figure 7A).
The interface between the domains shows a striking match between positively charged side chains of arginines and negatively charged side chains of aspartates and glutamates, creating a network of salt bridges across and around the interface (Figure 2E). All the interface arginine side chains show intense cross-peaks in the [1H,15N]-HSQC corresponding to NɛH-groups (see Supplementary Figure 3A), confirming the formation of the salt bridges. Large numbers of intra- and inter-domain NOEs were detected from the NɛH-groups (Supplementary Figure 3B), determining the positions of the charged groups. Notably, the NɛH-group of R181 is not observed in the spectrum of F1 alone. However, in the double domain, this signal becomes clearly visible, and shows a large number of NOEs (Supplementary Figure 3B) showing its involvement in the interface. In addition, the interface incorporates the aromatic ring of W61, which is wedged flat between the F0 and F1 domains, providing a close fit (Figure 2E). The involvement of W61 in the inter-domain interaction is supported by a network of inter- and intra-domain NOEs (Supplementary Figure 3B). The orientation of the ring is maintained through a stacking interaction with F50 in F0 (Figure 2E). The extensive hydrophobic surface of the W61 ring, in combination with the electrostatic charge match, provides an effective locking mechanism fixing the orientation of the F0 and F1 domains. An additional inter-domain contact is formed between K57 at the tip of the β3–β4 loop of F0 and E182 from the helical turn in the β3–β4 loop of F1. As the β3–β4 loop is largely unstructured in the isolated F0 domain, this contact is likely to have only a minor effect on the inter-domain orientation. Cross-species comparisons (Supplementary Figure 2A) show that the area of contact between F0 and F1 is highly conserved including W61 and F50. Interestingly, Y71, which is close to the interface, is completely conserved in all talins and has been shown to be phosphorylated in platelet talin (Ratnikov et al, 2005).
Independent evidence for a fixed mutual orientation of the domains was derived from small angle X-ray scattering. The experimental scattering profile (Supplementary Figure 7B) corresponds to a maximum linear dimension (Dmax) of 62 (±3) Å for the F0F1(Δ30) double domain with a radius of gyration, Rg of 20.1 (±0.1) Å. Both values are consistent with the conclusion that the F0F1 double domain has a compact elongated structure. The ab initio shape reconstruction with GASBOR (Svergun et al, 2001) agrees well with the NMR structure (Figure 2F).
F1 loop is important for talin-mediated integrin activation
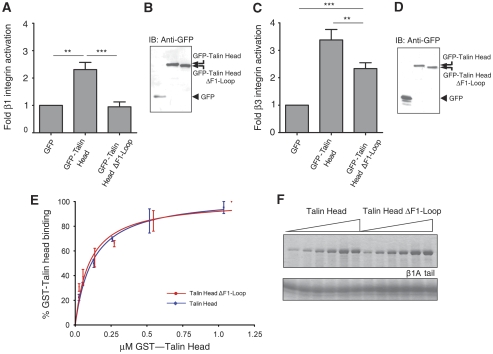
An unusual feature of the talin FERM domain is the large insert in F1, which has a high content of charged residues (∼60%) evenly distributed along the sequence. The majority of these residues are conserved across the species (Supplementary Figure 8A). It also contains two threonine residues (T144 and T150) that have been shown to be highly phosphorylated in platelet talin (Ratnikov et al, 2005). We have earlier shown that the F1 domain of talin is required for detectable talin-mediated activation of α5β1-integrins, and that the F1 domain also contributes to αIIbβ3 activation (Bouaouina et al, 2008). We, therefore, assessed the ability of GFP-talin head and GFP-talin head minus the F1 loop (residues 133–165; ΔF1 loop) to activate endogenous α5β1 in CHO cells. Integrin activation in GFP-positive cells was assessed in flow cytometric assays by measuring the binding of a recombinant soluble integrin-binding fragment of fibronectin, composed of fibronectin type III repeats 9 to 11 (FN9-11), (Hughes et al, 2002; Tadokoro et al, 2003; Calderwood et al, 2004). FN9-11 binding is specific as it can be inhibited by function blocking anti-α5β1 antibodies, RGD peptides, small molecule α5β1 inhibitors or EDTA and stimulated by the β1-activating antibody 9EG7, or Mn2+ (Hughes et al, 2002; Tadokoro et al, 2003; Calderwood et al, 2004; Bouaouina et al, 2008). Integrin expression in an equivalent population of transfected cells was measured in parallel using antibodies that bind in an activation-independent manner, allowing correction for any changes in integrin expression induced after transient expression of the talin head constructs. As shown in Figure 3A, and consistent with earlier reports (Bouaouina et al, 2008), the talin head significantly increased α5β1 activation, although talin head ΔF1 loop was unable to do so. This was not due to differences in protein expression levels as both populations were gated to have equivalent GFP signals, and western blotting showed that both talin head constructs were expressed at the expected molecular weights (Figure 3B).
Figure 3.
The F1 loop is required for integrin activation by the talin head. (A) CHO cells were transfected with empty pEGFP vector or cDNAs encoding GFP-tagged talin1 head, and α5β1-integrin activation was assessed by FN9-11 binding. Activation indices of transfected cells were calculated (see Supplementary data). The results represent mean±s.e. (n⩾4). (B) Expression of recombinant GFP-tagged talin1 head constructs was checked by immunoblotting cell lysates. IB, immunoblot. (C) CHO cells stably expressing αIIbβ3-integrin were transfected with empty pEGFP vector or cDNAs encoding GFP-tagged talin1 head. Cells were harvested and αIIbβ3-integrin activation in transfected cells assessed using PAC1 antibody (see Supplementary data). The results represent mean±s.e. (n⩾4). (D) Expression of recombinant GFP-tagged talin1 head constructs was checked by immunoblotting cell lysates. Statistical significance was defined as **P<0.01 and ***P<0.001. (E, F) The F1 loop is not required for talin head binding to β1A-integrin tail. (E) Binding of increasing amounts of purified GST-talin head or GST-talin head ΔF1loop to β1A tail proteins was assessed in pull-down assays followed by protein staining. Binding was quantified by scanning densitometry and the amount bound (expressed as per cent maximal binding for each protein) was plotted against the input concentration. Non-linear curve fitting was performed with a one-site-binding model. The results represent mean±s.e. GST-talin head (n⩾3; R2=0.95), GST-talin head ΔF1loop (n⩾3; R2=0.91). (F) Representative experiment of GST-talin head and GST-talin head ΔF1loop binding to β1A tail. We did not detect either GST-talin head or GST-talin head ΔF1loop binding to αIIb tails (data not shown).
Our earlier data indicated that, unlike α5β1, αIIbβ3 can be activated by the F3 or F2F3 region of talin, but that inclusion of the F1 or F0F1 region enhances talin-mediated αIIbβ3 activation (Calderwood et al, 2002; Bouaouina et al, 2008). We, therefore, assessed the ability of the talin head minus the loop to activate αIIbβ3-integrins stably expressed in CHO cells (O'Toole et al, 1994). The activation state of αIIbβ3-integrins was assessed by measuring the binding of the ligand-mimetic anti-αIIbβ3 mAb PAC1 in three-colour flow cytometric assays as described earlier (Calderwood et al, 1999, 2004; Tadokoro et al, 2003). As expected, expression of GFP-talin head induced robust αIIbβ3 activation, whereas deletion of the F1 loop significantly reduced talin head-mediated αIIbβ3 activation (Figure 3C). Again the difference between GFP-talin head and GFP-talin head ΔF1 loop could not be explained by different expression levels, as gating ensured that the mean GFP fluorescence was the same in each measured population, and western blotting indicated expression of comparable levels of intact fusion proteins (Figure 3D). In the case of αIIbβ3-integrins, GFP-talin ΔF1 loop did induce some activation consistent with our earlier results showing that smaller fragments of the talin head lacking the entire F1 domain are capable of activating αIIbβ3, albeit to a lesser extent than the intact talin head (Bouaouina et al, 2008).
Direct talin head/β-integrin tail interactions are required for talin-mediated integrin activation (Calderwood, 2004a). To test whether the inability of GFP-talin head ΔF1 loop to activate α5β1-integrins could be explained by a defect in integrin binding, we compared the binding of purified recombinant GST-talin head and GST-talin head ΔF1 loop to recombinant models of β1A-integrin cytoplasmic tails (Pfaff et al, 1998; Calderwood et al, 1999; Lad et al, 2007; Bouaouina et al, 2008). As shown in Figure 3E and F, both GST-talin head and GST-talin head ΔF1 loop produced very similar dose-dependent saturable-binding curves. Curve fitting using a one-site-binding model yielded apparent Kd-values of 124 (±19) nM for the β1 tail-talin head and 99 (±20) nM for the β1 tail-talin head ΔF1 loop interactions, respectively. These data indicate that loss of the F1 loop does not significantly alter the affinity of the talin head for β1 tails.
F1 loop has a propensity to form an α-helix
The F1 loop has an important function in talin head-mediated integrin activation, but it does not bind integrin directly. This suggests that the loop interacts with another component of the system, a conclusion supported by the conservation of the loop size and composition across species, and between mammalian talin 1 and 2 (Supplementary Figure 8A). Although the F1 loop is highly dynamic and unstructured even in the full-talin head, the NOESY spectra of the corresponding peptide exhibit a characteristic pattern of sequential HN-HN NOEs that suggests transient helix formation predominantly in the C-terminal part of the loop (residues 155–161) (Supplementary Figure 8C and D). This is also supported by CD measurements (see later).
Although the loop region has a clear propensity to form a helix, the low abundance of the helical state in aqueous buffer prevents structural characterization. To stabilize the intrinsic structure of the loop peptide, we used trifluoroethanol (TFE), a structure-stabilizing solvent widely used in peptide characterization. The addition of TFE stabilizes secondary structure favoured by the primary sequence, and the formation of both α-helices and β-strands have been observed (Zhong and Johnson, 1992). The addition of 35% TFE led to changes in the NMR spectra of the loop peptide characteristic of the formation of a stable helical structure. These include increased resonance line width, chemical shift dispersion and NOE cross-peak intensity accompanied by detection of medium-ranged NOEs characteristic for an α-helix. Two continuous stretches of medium-range NOE contacts between residues separated by helical turns are identified in the loop region (Supplementary Figure 8C and D). The C-terminal region 154–167 exhibits a high density of NOE contacts, and the Hα proton chemical shifts have values characteristic of a helical structure, whereas for the N-terminal region (residues 144–152), the NOE density is lower and the Hα proton chemical shifts are closer to random-coil values. These NMR data indicate formation of a stable α-helix in the C-terminal part of the F1 loop and a more transient helical structure in the N-terminal region, correlating well with the higher helical propensity in the C-terminal region in the absence of TFE. The middle region 152–154 has a smaller number and lower intensity of NOE contacts, indicating lower structure stability in that region.
The structure of the loop peptide (Figure 4A and B) has been calculated from 266 NOE distance restraints identified in 2D NOESY spectra in the presence of TFE, and the superposition of the structures is shown in Figure 4A. As expected from the direct analysis of the NMR data above, two helical regions are formed within the loop, separated by a partially structured region. The C-terminal part of the helix (residues 154–167) is better defined and longer than the N-terminal one (residues 144–152), and the preceding region 139–144 is unstructured. The connecting region displays significant structure variation, although it is predominantly helical, resulting in a nearly linear arrangement of the two stable helices. Thus, the F1-loop region has a propensity to form helical structures in the regions 144–152 and 154–167, separated by a less stable region near the centre. The structure is transient, but can be stabilized under appropriate solvent conditions.
Figure 4.
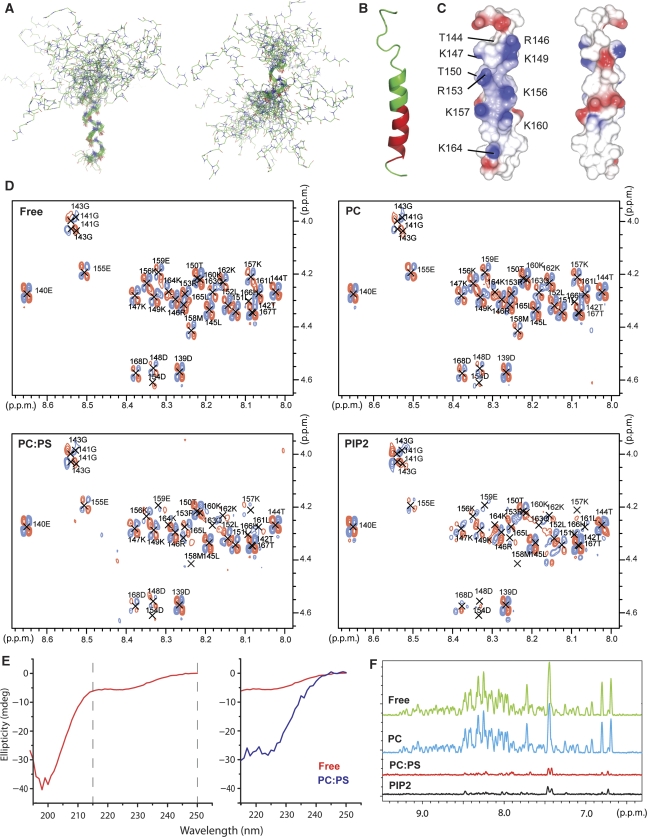
Structure of the F1 loop and its function in membrane binding. (A) Superposition of the 20 lowest energy structures calculated for the F1-loop peptide in 35% TFE. Superposition was performed separately on the C-terminal helix (residues 154–167, left) and the N-terminal helix (residues 144–152, right). The middle part of the helix is not well defined in the ensemble of the calculated structure because of its lower stability, introducing a variable kink in the peptide structure. (B) Ribbon diagram of a low energy structure of the F1-loop peptide in 35% TFE corresponding to a continuous helix. Residues experiencing resonance broadening on the addition of 1:4 POPS:POPC SUVs are highlighted in red. (C) Surface charge distribution for the structure in (B). The views differ by a 180° rotation and are optimized to show the positively charged face. (D) HN/Hα region of the 2D DQF-COSY spectra of the loop peptide alone (Free) and in the presence of 2 mM SUVs composed of POPC (PC), 1:4 POPS:POPC (PC:PS) and 1:19 PIP2:POPC (PIP2) SUVs illustrating resonance broadening on addition of negatively charged lipids. (E) Secondary structure analysis of the F1-loop (145–168) peptide by circular dichroism (CD). Left—free peptide; right—superposition of the CD spectra of the free peptide (red) and the peptide in the presence of 1:4 POPS:POPC SUVs (blue) in the 215–250 nm region. Strong contribution from the SUVs to the spectrum below 215 nm prevents the detection of the peptide signal. (F) Superposition of 1H projections of F1 HSQC spectra illustrating the reduction in resonance intensities in the presence of POPS (red) and PIP2 (black), but not pure POPC (blue).
F1 loop interacts with negatively charged lipid bilayers
The talin F1 loop contains a high proportion of positively charged residues, most of which are conserved (Supplementary Figure 8A). These residues are mainly localized in the regions of high helical propensity, and their distribution in the sequence makes one face of the helix highly positively charged, whereas the opposite face contains only negative charges (Figure 4C; Supplementary Figure 8B). This distribution is clearest when an uninterrupted helix is formed from residues 144 to 167, as seen in some of the calculated lowest energy structures (Figure 4A–C). As the high concentration of positive charges suggests a possible interaction with a negatively charged membrane surface, we tested binding of the loop peptide to lipid bilayers. Addition of small unilamellar vesicles (SUV) containing negatively charged 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine (POPS) or L-α-phosphatidylinositol-4,5-bisphosphate (PIP2) led to selective broadening of NMR resonances in the region of residues 155–166, which has the highest helical propensity, as can be clearly seen in 2D COSY spectra (Figure 4D). The broadening is accompanied by small chemical shift changes, suggesting that it is caused by an exchange between the peptide in solution and a complex with the vesicles. The residues affected by the lipids form a continuous stretch that overlaps with the region of helical propensity (Figure 4B), consistent with an interaction involving formation of an α-helix on the membrane surface. No effect was observed with neutral 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) vesicles (Figure 4D), highlighting the critical function of ion-pairs in the membrane interaction.
The stabilization of a helix by interaction with negatively charged membranes was directly detected by CD spectroscopy. To reduce the contribution from the unstructured N-terminal part of the loop, we used a peptide corresponding to amino-acids 145–168 of talin that contains all the residues affected by membrane association. The CD spectrum of the free peptide is dominated by a strong negative band near 200 nm, characteristic of a disordered polypeptide (Figure 4E). In addition, the spectrum has a small contribution from a negative band in the 205–230 nm region, characteristic of helix formation, and consistent with the transient helix identified by NMR. The helical contribution increases significantly in the presence of the negatively charged SUVs, showing that the interaction is mediated by formation of a helical structure in the loop peptide.
In agreement with the experiments on the isolated F1-loop peptide, we observed strong interaction between the F1 domain and negatively charged membranes. Incubation of F1 with vesicles containing either POPS or PIP2 resulted in co-sedimentation of the majority of the protein with SUVs, whereas pure POPC vesicles had no effect (Figure 5A). In addition, we observed severe broadening of all NMR signals in [1H,15N]-HSQC spectra of F1 in the presence of negatively charged SUVs containing either POPS or PIP2, and no effect from pure POPC vesicles (Figure 4F; Supplementary Figure 9). The residual signals in the spectra in the presence of POPS or PIP2 correspond to the most intense resonances from a low concentration of the unbound protein. The fraction of unbound protein depends on the incubation time as can be seen from the comparison of the two co-sedimentation experiments in Figure 5A, showing a relatively slow conversion of F1 into the fully lipid-associated state. Similarity in the interaction with POPS or PIP2 shows that the membrane association of F1 is controlled through a general interaction with negatively charged lipid head groups.
Figure 5.
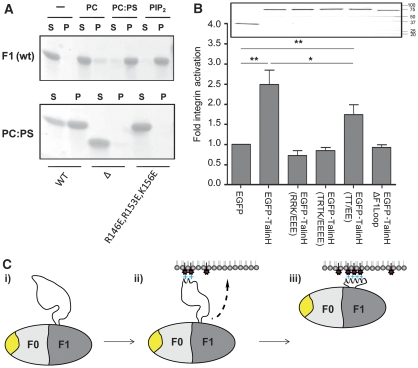
Function of the F1 loop. (A) Association of F1 and its mutants with SUVs as measured by a co-sedimentation assay. Top—effect of negatively charged phospholipids on binding. Lanes 1, 2—no lipid; 3, 4—POPC; 5, 6—1:4 POPS:POPC; 7, 8—1:19 PIP2:POPC. Incubation time was 1.5 h. Supernatant (S) and (P) pellet. Bottom—effect of F1 mutations on association with 1:4 POPS:POPC SUVs. Lanes 1, 2—wild type; 3, 4—F1(Δ30); 5, 6—F1(R146E,R153E,K156E). Incubation time 1 h. (B) Charge substitution within the F1 loop impairs talin head-mediated β1A-integrin activation. CHO cells were transfected with empty pEGFP vector or cDNAs encoding WT or mutant GFP-tagged talin1 head domains, and α5β1-integrin activation was assessed by assaying FN9-11 binding. The results represent mean±s.e. (n⩾4). The constructs used were wild-type talin1 head—TalinH 1–433; TalinH(RRK/EEE)—(R146E,R153E,K156E); TalinH(TRTK/EEEE)—(T144E,R146E,T150E,K156E); TalinH(TT/EE)—(T144E,T150E) and TalinHΔF1loop (residues 139–168 deleted). (inset) Expression of recombinant talin1 head constructs was checked by immunoblotting SDS–PAGE-fractionated cell lysates. Statistical significance was defined as *P<0.05 and **P<0.01. (C) Suggested function of the F1 loop—the fly-casting model: (i) on its own, the F1 loop is predominantly unstructured with only a small population in a helical state; (ii) on contact with negatively charged phospholipids, the helical state is favoured and the loop begins to fold into a helix, resulting in cluster of positive charges on one side of the helix and (iii) shortening of the loop draws the talin F0F1 domain towards the membrane. The conserved surface on F0 (shown in yellow) is exposed and accessible for an interaction with a membrane-associated protein.
To identify the residues critical for membrane binding, we conducted co-sedimentation and NMR experiments on a series of F1 mutants. Deletion of the F1 loop completely abolished binding (Figure 5A), showing the dominant function of the loop in the interaction of F1 with SUVs. The highest density of positive charges is located at the N-terminal, less stable, part of the helix, and involves the near neighbours R146, K149, R153 and K156 (Figure 4C). A triple mutant R146E/R153E/K156E effectively reverses the surface charge of a large part of the helix, making it unfavourable for interaction with negatively charged membranes. In agreement with this, we did not detect any co-sedimentation of the F1 mutant in the presence of POPS (Figure 5A), and saw no effect of negatively charged SUVs on the [1H,15N]-HSQC spectra of the F1 mutant (data not shown). Similarly, an F1(T144E/R146E/T150E/K156E) mutant in which we introduced two additional negative charges to reflect phosphorylation of T144 and T150 also showed no interaction with negatively charged SUVs.
The charge reversal mutations show the crucial function of positive charges in the F1 loop for membrane binding. We next tested the effects of mutations that disrupt helix formation. To minimize any direct effect on binding, we replaced residues on the helix surface opposite that containing positively charged residues. Introduction of Gly or Ser residues with low helical propensity had negligible effect on the co-sedimentation behaviour F1(M158G/E159G and M158S/E159S/L161S) mutants (Supplementary Figure 10E). At the same time, a significantly larger fraction of non-bound F1(M158G/E159G/L161G) was detected by NMR compared with wild type (Supplementary Figure 10A, C), showing a reduction in affinity. Introduction of a destabilizing Pro residue into the more stable C-terminal part of the helix (M158P and L161P mutants) led to a significant reduction of bound protein in the co-sedimentation assays (Supplementary Figure 10E). Moreover, the NMR spectra of the L161P mutant on addition of SUVs displayed large broadening and chemical shift changes for only a small number of signals, with the majority of the resonances remaining unaffected (Supplementary Figure 10B, D and F). Such an effect corresponds to an exchange of the protein between the free and membrane bound state, with the residues associated with the affected signals involved in a direct interaction with the membrane. Among them are the positively charged residues R146, K156, K157 and K164 identified by structural analysis and mutagenesis as crucial for the interaction. The helix destabilizing mutants show that helix stability is important for membrane binding, further supporting a model in which F1 interacts with negatively charged membranes through formation of a transient helix in the loop region.
We also observed a major contribution from the loop in the interaction of the full-talin head with different negatively charged phospholipids spotted on nitrocellulose membranes (Supplementary Figure 11). The intact talin head bound strongly to a range of negatively charged phospholipids including PIP2 and phosphatidic acid, whereas deletion of the loop resulted in a substantial reduction in binding. The residual interaction with the lipids correlates with the reported binding of talin F2 and F3 to acidic phospholipids (Anthis et al, 2009; Saltel et al, 2009).
To determine whether the basic residues in the F1 loop and/or the two potential phosphorylation sites have a function in integrin activation, we assessed the effects of mutations of these residues on the ability of GFP-talin head to activate α5β1-integrins in CHO cells. As shown in Figure 5B, charge reversal mutations at R146, R153 and K156 completely inhibited the ability of talin head to trigger α5β1 activation. Substituting T144 and T150 by two glutamates also partially impaired the ability of talin head to activate α5β1. These data reveal that specific sites within the F1 loop are important for integrin activation, and raise the possibility that lipid binding through positively charged residues within the F1 loop contributes to the ability of the talin head to activate integrins. Moreover, phosphorylation of T144/T150 in the talin F1 loop may modulate integrin activation.
Discussion
The structure of the F2F3 region of the talin FERM domain has been determined and the mechanism by which the F3 domain binds β-integrin tails is well understood (Garcia-Alvarez et al, 2003; Wegener et al, 2007; Anthis et al, 2009). However, it has become increasingly apparent that the ability of the talin head to activate integrins requires additional interactions mediated by the talin F0 and F1 domains (Bouaouina et al, 2008), although the molecular basis for this effect was unknown. The structural, biochemical and cell biological studies reported here provide new insights into the mechanisms involved. The F0 and F1 domains both have ubiquitin-like folds that stack against each other in a fixed orientation through an extensive charged interface. We show that the large loop in the F1 domain is essential for β1-integrin activation and for maximal activation of β3-integrins. The loop has a tendency to form a helical structure that positions basic residues on one side of the helix, and we show that these basic residues are essential both to the ability of the F1 domain to bind acidic phospholipids and to β1-integrin activation by the talin head.
A function for the talin F3 domain in integrin activation is well established and structural, biochemical and genetic analyses show that a direct integrin β tail-talin F3 domain interaction is central to this function (Calderwood et al, 2002; Garcia-Alvarez et al, 2003; Tadokoro et al, 2003; Petrich et al, 2007; Wegener et al, 2007). However, it is now clear that additional talin domains have important functions, and recent structural studies of a talin2 F2F3 domain-β1D-integrin tail complex show that F2F3 contributes to integrin activation in at least two ways (Anthis et al, 2009). First, an ionic interaction between K327 in the F3 domain of talin2 and D759 in the β1D-integrin tail has the potential to disrupt the salt bridge between the α- and β-integrin tails that normally maintains the integrin in an inactive state. Second, clusters of basic residues on the membrane-facing surface of both F2 and F3 are also essential to integrin activation. The authors present a model in which binding of these basic residues to acidic membrane phospholipids re-orientates the talin head and, therefore, the integrin β-subunit, leading to separation of the transmembrane helices of the two integrin (Lau et al, 2009) and subsequent integrin activation.
We suggest that the charged F1 loop, which is also essential to integrin activation, is similarly orientated towards the membrane and that its large size and dynamic properties allow it to sample the environment by a so-called ‘fly-casting mechanism' (Shoemaker et al, 2000). An encounter between the loop and a cluster of negatively charged phospholipids on the cytoplasmic face of the membrane would induce the loop to adopt a folded helical structure in which basic residues on one side of the helix are bound to the membrane. This folding would decrease the length of the loop and draw the F1 domain closer to the membrane, as illustrated schematically in Figure 5C. The analysis of mutants that destabilize the transient helix in the F1 loop support the two-step-binding model presented. The L161P mutation prevents helix formation and blocks the transition to the fully membrane-associated state. In the absence of helix assembly, F1 is connected to the membrane through a small subset of positively charged residues at the end of the highly dynamic F1 loop, corresponding to the intermediate-binding stage of the model. In agreement with this, there was no effect on the NMR resonances of the folded region in the L161P mutant, whereas the resonances of the positively charged residues associated with the membrane experienced dramatic broadening. The F1(M158G/E159G/L161G) mutant has a reduced helical propensity, although there are no steric restrictions on helix formation. The lower affinity of this mutant for membranes is likely associated with the reduced efficiency of the transition to the fully bound state (Figure 5C).
The F1 association would bring the whole talin head closer to the membrane, facilitating interactions between the basic patch on F2 and F3 and acidic membrane phospholipids. Phosphorylation at T144/T150 in the F1 loop could disrupt membrane association either by reducing the overall positive charge in the loop, or by decreasing the propensity of the loop to form a helix. It is notable that these phosphorylation sites are located in the region of lower helix stability. The interaction of the F1 loop with the membrane is compatible with the conserved surface on F0 remaining exposed. The fact that this surface overlaps the site in F0 that interacts with GTP-bound Rap1, albeit with low affinity, raises the possibility that F0 binds to another small GTP-binding protein, providing an additional membrane anchor, although no candidates have yet been identified.
All earlier characterized FERM domains contain three lobes (Hamada et al, 2000; Bretscher et al, 2002; Ceccarelli et al, 2006), and initially, the presence of an additional F0 domain, along with the insertion of a relatively large F1 loop, seemed unique to the talin FERM domain. However, we have recently found that the kindlin family of FERM domain proteins, which synergize with talin to activate integrins (Larjava et al, 2008; Moser et al, 2009; Plow et al, 2009), also contains an F0 domain and a large loop (86–120 residues) in the F1 domain (Goult et al, 2009). Moreover, the kindlin1 F1 loop also contains two phosphorylation sites (http://www.cellmigration.org). It will be important to establish whether FERM domains in other proteins contain similar features or whether this architecture is restricted to talin and kindlins, proteins important for regulating integrin activation.
In summary, we have described the structure of the earlier uncharacterized N-terminal portion of the talin head, revealing the domain boundaries and their architecture, as well as identifying a large loop in F1 that binds acidic phospholipids and is essential for integrin activation. Overall, the results suggest that integrin activation by the talin head involves a combination of relatively weak protein–protein and protein–lipid interactions, each of which alone is insufficient to trigger the full response. Although the talin F3 domain is solely responsible for binding to β-integrin tails, optimum activation of integrins by the talin head requires additional interactions between basic residues spread along the whole of the talin head and acidic phospholipids on the cytoplasmic face of the plasma membrane. The PIP-kinase type1γ isoform has been implicated in the translocation of talin to the plasma membrane and in the assembly of focal adhesions (Di Paolo et al, 2002; Ling et al, 2002), and may generate PIP2-rich microdomains that contribute to the talin-mediated activation of integrins.
Materials and methods
Expression of recombinant talin polypeptides
Proteins corresponding to murine talin residues 1–85 (F0), 86–202 (F1) and 1–202 (F0F1) were expressed in Escherichia coli and purified using standard affinity chromatography. Details are presented in Supplementary data.
Peptide synthesis and preparation
A peptide corresponding to residues 139–168 and 145–168 were synthesized and purified by high performance liquid chromatography by GL Biochem (Shanghai) Ltd. A 10-mg/ml stock solution was made by dissolving the peptide in H2O.
Lipid vesicle preparation
SUVs for the NMR titrations were prepared by sonication as detailed in Supplementary data. POPC, POPS and PIP2 used in the titrations were obtained from Avanti Polar Lipids (Alabaster, AL). Pure POPC, 1:4 POPS:POPC and 1:19 PIP2:POPC (molar ratio) vesicles were used for binding studies.
NMR spectroscopy
Protein structures were determined using 1 mM protein solution in phosphate buffer comprising 20 mM sodium phosphate pH 6.5, 50 mM NaCl, 2 mM DTT with 10% (v/v) of 2H2O were used for structure determination. Peptide structures were determined using 1 mM samples in 20 mM MES buffer, pH 6.1 containing either 5% (v/v) of 2H2O or 35% (v/v) TFE-d3 (Sigma-Aldrich). Peptide interactions with lipids were analysed using 0.1 mM solutions in 10 mM MES buffer, pH 6.1 containing 5% (v/v) of 2H2O in the presence of 2 mM lipid added as SUVs. The F1 interactions with lipids were analysed using a 0.01 mM solution of uniformly 15N-labelled F1 fragment or mutants thereof in 20 mM MES buffer, pH 6.1 containing 20 mM NaCl and 5% (v/v) of 2H2O. 2D [1H,15N]-HSQC spectra were recorded at 298 K for the free protein and the protein in the presence of 6 mM lipid added as SUVs. All experiments were conducted as 298 K. Additional details are presented in Supplementary data.
CD spectroscopy
CD spectroscopy is described in Supplementary data.
NMR structure calculations
Protein structures were calculated using a combination of CYANA (Herrmann et al, 2002) and Aria (Linge et al, 2001) software on the basis of NOE distance restraints, dihedral restraints (Φ/Ψ) were obtained with TALOS software (Cornilescu et al, 1999), hydrogen-bond restraints were incorporated based on the temperature dependence of NMR chemical shifts (Baxter et al, 1998) and residual 1H,15N dipolar couplings measured in the presence of 8 mg/ml Pf1 phage (Asla Ltd., Latvia). The structural statistics for each domain are presented in Supplementary Tables 1–3. Peptide structure was calculated using Aria software. Additional details are presented in Supplementary data.
Small angle X-ray scattering
All scattering data was collected at the Daresbury Synchrotron Radiation Source (Station 2.1) using a multiwire gas detector covering a momentum transfer range of 0.02 Å−1<q<0.70 Å−1 with q=4πsinθ/λ (where 2θ is the scattering angle and λ, the X-ray wavelength at 1.54 Å). Data collection was performed at 4°C, at protein concentrations of 2 and 10 mg/ml in the NMR buffer. Data were accumulated in 60-s frames. Before averaging, frames were inspected for X-ray-induced damage or aggregation. No protein aggregation was detected and the linearity of the Guinier plot indicated that the protein solutions were homogeneous. The background was subtracted using the scattering from the buffer solution alone. Data reduction was carried out with software provided at SRS Daresbury and subsequent analysis was carried out with programs from the ATSAS package (Konarev et al, 2006). Particle shapes at low resolution were reconstructed ab initio with the bead modelling program, GASBOR (Svergun et al, 2001), which represents the protein as a chain of dummy residues centred at the Cα positions.
Phospholipid binding
For co-sedimentation assays, 20 μM protein solution was incubated in the presence of SUVs at the total lipid concentration of 6 mM for 1–1.5 h at 30°C. The solution was centrifuged at 13 000 r.p.m. for 5 min, the pellet separated from the supernatant and re-suspended in the equal volume of the buffer. The protein distribution was analysed on a 10–20% gradient gel (Expedeon). Standard conditions were used for the PIP strip assay as detailed in Supplementary data.
Antibodies and DNAs
Ligand-mimetic anti-αIIbβ3 PAC1 (BD Biosciences), anti-hamster α5β1 PB1 (Developmental Studies Hybridoma Bank), Goat anti-GFP (Rockland) and anti-αIIbβ3 monoclonal antibody D57 (Diaz-Gonzalez et al, 1996) were used. GST-fibronectin type III repeats 9–11 (FN9–11) (Hughes et al, 2002) and GFP-mouse talin-1 (1–433) (Bouaouina et al, 2008) have been described earlier. Constructs encoding GFP- and GST-tagged mouse talin-1 (1–433)ΔF1 loop (lacking residues 133–165) were generated by PCR from a talin-1 cDNA and confirmed by DNA sequencing.
Pull-down assays with recombinant integrin tails
Pull-down assays with GST-talin fragments were performed using recombinant integrin tails bound to His-bind resin (Novagen) as described earlier (Calderwood et al, 2002; Lad et al, 2007). To estimate the apparent affinity constants, the binding of increasing amounts of purified GST-talin proteins to β1A tails was quantified by densitometry. Data were plotted as per cent maximal binding versus input concentration and fitted to a one-site-binding model (Y=Bmax·X/(Kd+X)) using GraphPad Prism version 4 for Windows (GraphPad Software).
Analysis of integrin activation
The activation state of endogenous CHO cell α5β1 was assessed by measuring the binding of a recombinant soluble integrin-binding fragment of fibronectin (FN9–11) in three-colour flow cytometric assays as described earlier (Tadokoro et al, 2003; Calderwood et al, 2004). α5β1-integrin expression was assessed in parallel by staining with PB1 (Brown and Juliano, 1985) and activation was normalized to α5β1 expression levels as described earlier—see Supplementary data for details. The activation state of αIIbβ3-integrins, stably expressed in CHO cells (O'Toole et al, 1994; Hughes et al, 1997), was assessed by measuring the binding of the ligand-mimetic anti-αIIbβ3 monoclonal antibody PAC1 in three-colour flow cytometric assays as described earlier (O'Toole et al, 1995; Calderwood et al, 1999, 2004; Tadokoro et al, 2003). For additional details see Supplementary data.
Accession numbers
Resonance assignments have been submitted to the BioMagResBank (http://www.bmrb.wisc.edu/), accession numbers 15458 (F0), 15616 (F1) and 15615 (F0F1(Δ30)). Twenty lowest energy structures have been deposited in the Protein Data Bank (http://www.rcsb.org) with the accession numbers 2kc1 (F0), 2kc2 (F1) and 2k34 (F0F1(Δ30)).
Supplementary Material
Acknowledgments
The work was supported by grants from the NIH Cell Migration Consortium (Grant U54 GM64346 from the National Institute of General Medical Sciences), NIH RO1 GM068600 and R21 HL089433, the Wellcome Trust, Cancer Research UK and BBSRC. We thank Professor A Wittinghofer (Department of Structural Biology, Max-Planck-Institut for Molecular Physiology, Dortmund, Germany) for the Rap1 construct.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID (2009) The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J 28: 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszodi A, Legate KR, Nakchbandi I, Fassler R (2006) What mouse mutants teach us about extracellular matrix function. Annu Rev Cell Dev Biol 22: 591–621 [DOI] [PubMed] [Google Scholar]
- Barsukov IL, Prescot A, Bate N, Patel B, Floyd DN, Bhanji N, Bagshaw CR, Letinic K, Di Paolo G, De Camilli P, Roberts GC, Critchley DR (2003) Phosphatidylinositol phosphate kinase type 1gamma and beta1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J Biol Chem 278: 31202–31209 [DOI] [PubMed] [Google Scholar]
- Baxter NJ, Hosszu LL, Waltho JP, Williamson MP (1998) Characterisation of low free-energy excited states of folded proteins. J Mol Biol 284: 1625–1639 [DOI] [PubMed] [Google Scholar]
- Bouaouina M, Lad Y, Calderwood DA (2008) The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate beta1 and beta3 integrins. J Biol Chem 283: 6118–6125 [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG (2002) ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol 3: 586–599 [DOI] [PubMed] [Google Scholar]
- Brown PJ, Juliano RL (1985) Selective inhibition of fibronectin-mediated cell adhesion by monoclonal antibodies to a cell-surface glycoprotein. Science 228: 1448–1451 [DOI] [PubMed] [Google Scholar]
- Calderwood DA (2004a) Integrin activation. J Cell Sci 117: 657–666 [DOI] [PubMed] [Google Scholar]
- Calderwood DA (2004b) Talin controls integrin activation. Biochem Soc Trans 32: 434–437 [DOI] [PubMed] [Google Scholar]
- Calderwood DA, Tai V, Di Paolo G, De Camilli P, Ginsberg MH (2004) Competition for talin results in trans-dominant inhibition of integrin activation. J Biol Chem 279: 28889–28895 [DOI] [PubMed] [Google Scholar]
- Calderwood DA, Yan B, de Pereda JM, Alvarez BG, Fujioka Y, Liddington RC, Ginsberg MH (2002) The phosphotyrosine binding-like domain of talin activates integrins. J Biol Chem 277: 21749–21758 [DOI] [PubMed] [Google Scholar]
- Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH (1999) The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem 274: 28071–28074 [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ (2006) Crystal structure of the FERM domain of focal adhesion kinase. J Biol Chem 281: 252–259 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Critchley DR (2009) Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys 38: 235–254 [DOI] [PubMed] [Google Scholar]
- Critchley DR, Gingras AR (2008) Talin at a glance. J Cell Sci 121: 1345–1347 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De Camilli P (2002) Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420: 85–89 [DOI] [PubMed] [Google Scholar]
- Diaz-Gonzalez F, Forsyth J, Steiner B, Ginsberg MH (1996) Trans-dominant inhibition of integrin function. Mol Biol Cell 7: 1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC (2003) Structural determinants of integrin recognition by talin. Mol Cell 11: 49–58 [DOI] [PubMed] [Google Scholar]
- Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, Grossmann JG, Liu H, Putz NS, Roberts GC, Volkmann N, Hanein D, Barsukov IL, Critchley DR (2008) The structure of the C-terminal actin-binding domain of talin. EMBO J 27: 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Bobkov AA, Joyce MG, Fasci D, Himmel M, Rothemund S, Ritter A, Grossmann JG, Patel B, Bate N, Goult BT, Emsley J, Barsukov IL, Roberts GC, Liddington RC, Ginsberg MH, Critchley DR (2009) Structural determinants of integrin binding to the talin rod. J Biol Chem 284: 8866–8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Han J, Haling JR, Sherman NE, Fox JW, Hunt DF, Ginsberg MH (2007) An experimentally derived database of candidate Ras-interacting proteins. J Proteome Res 6: 1806–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bouaouina M, Harburger DS, Bate N, Patel B, Anthis NJ, Campbell ID, Calderwood DA, Barsukov IL, Roberts GC, Critchley DR (2009) The structure of the N-terminus of kindlin-1: a domain important for alphaIIbbeta3 integrin activation. J Mol Biol 394: 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J 19: 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH (2006) Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol 16: 1796–1806 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24: 171–189 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1995) Dali: a network tool for protein structure comparison. Trends Biochem Sci 20: 478–480 [DOI] [PubMed] [Google Scholar]
- Huang L, Hofer F, Martin GS, Kim SH (1998) Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol 5: 422–426 [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM (1993) ‘NACCESS', Computer Program, Department of Biochemistry and Molecular Biology, University College London
- Hughes PE, Oertli B, Hansen M, Chou FL, Willumsen BM, Ginsberg MH (2002) Suppression of integrin activation by activated Ras or Raf does not correlate with bulk activation of ERK MAP kinase. Mol Biol Cell 13: 2256–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PE, Renshaw MW, Pfaff M, Forsyth J, Keivens VM, Schwartz MA, Ginsberg MH (1997) Suppression of integrin activation: a novel function of a Ras/Raf-initiated MAP kinase pathway. Cell 88: 521–530 [DOI] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L (2005) Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J Mol Biol 348: 759–775 [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Cryst 39: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad Y, Harburger DS, Calderwood DA (2007) Integrin cytoskeletal interactions. Methods Enzymol 426: 69–84 [DOI] [PubMed] [Google Scholar]
- Larjava H, Plow EF, Wu C (2008) Kindlins: essential regulators of integrin signalling and cell-matrix adhesion. EMBO Rep 9: 1203–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Kim C, Ginsberg MH, Ulmer TS (2009) The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J 28: 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH (2009) RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem 284: 5119–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate KR, Montanez E, Kudlacek O, Fassler R (2006) ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol 7: 20–31 [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420: 89–93 [DOI] [PubMed] [Google Scholar]
- Linge JP, O'Donoghue SI, Nilges M (2001) Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol 339: 71–90 [DOI] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA (2007) Structural basis of integrin regulation and signaling. Annu Rev Immunol 25: 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes M, Rodius S, Coleman SJ, Monkley SJ, Goormaghtigh E, Tremuth L, Kox C, van der Holst PP, Critchley DR, Kieffer N (2007) The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin beta subunits to the cytoskeleton. J Biol Chem 282: 17280–17288 [DOI] [PubMed] [Google Scholar]
- Moser M, Legate KR, Zent R, Fassler R (2009) The tail of integrins, talin, and kindlins. Science 324: 895–899 [DOI] [PubMed] [Google Scholar]
- Narasimhan J, Wang M, Fu Z, Klein JM, Haas AL, Kim JJ (2005) Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem 280: 27356–27365 [DOI] [PubMed] [Google Scholar]
- O'Toole TE, Katagiri Y, Faull RJ, Peter K, Tamura R, Quaranta V, Loftus JC, Shattil SJ, Ginsberg MH (1994) Integrin cytoplasmic domains mediate inside-out signal transduction. J Cell Biol 124: 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole TE, Ylanne J, Culley BM (1995) Regulation of integrin affinity states through an NPXY motif in the beta subunit cytoplasmic domain. J Biol Chem 270: 8553–8558 [DOI] [PubMed] [Google Scholar]
- Petrich BG, Marchese P, Ruggeri ZM, Spiess S, Weichert RA, Ye F, Tiedt R, Skoda RC, Monkley SJ, Critchley DR, Ginsberg MH (2007) Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J Exp Med 204: 3103–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff M, Liu S, Erle DJ, Ginsberg MH (1998) Integrin beta cytoplasmic domains differentially bind to cytoskeletal proteins. J Biol Chem 273: 6104–6109 [DOI] [PubMed] [Google Scholar]
- Plow EF, Qin J, Byzova T (2009) Kindling the flame of integrin activation and function with kindlins. Curr Opin Hematol 16: 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage R, Green J, Muir TW, Ogunjobi OM, Love S, Shaw K (1994) Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem J 299 (Pt 1): 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnikov B, Ptak C, Han J, Shabanowitz J, Hunt DF, Ginsberg MH (2005) Talin phosphorylation sites mapped by mass spectrometry. J Cell Sci 118: 4921–4923 [DOI] [PubMed] [Google Scholar]
- Roberts GC, Critchley DR (2009) Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys Rev 1: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltel F, Mortier E, Hytonen VP, Jacquier MC, Zimmermann P, Vogel V, Liu W, Wehrle-Haller B (2009) New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J Cell Biol 187: 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA 97: 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA (2003) Talin binding to integrin beta tails: a final common step in integrin activation. Science 302: 103–106 [DOI] [PubMed] [Google Scholar]
- Tremuth L, Kreis S, Melchior C, Hoebeke J, Ronde P, Plancon S, Takeda K, Kieffer N (2004) A fluorescence cell biology approach to map the second integrin-binding site of talin to a 130-amino acid sequence within the rod domain. J Biol Chem 279: 22258–22266 [DOI] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID (2007) Structural basis of integrin activation by talin. Cell 128: 171–182 [DOI] [PubMed] [Google Scholar]
- Xing B, Jedsadayanmata A, Lam SC (2001) Localization of an integrin binding site to the C terminus of talin. J Biol Chem 276: 44373–44378 [DOI] [PubMed] [Google Scholar]
- Zhong L, Johnson WC Jr (1992) Environment affects amino acid preference for secondary structure. Proc Natl Acad Sci USA 89: 4462–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.