Abstract
DNA adenine methylase (Dam−) mutants of Salmonella enterica are attenuated in the mouse model and present multiple virulence-related defects. Impaired interaction of Salmonella Dam− mutants with the intestinal epithelium has been tentatively correlated with reduced secretion of pathogenicity island 1 (SPI-1) effectors. In this study, we show that S. enterica Dam− mutants contain lowered levels of the SPI-1 transcriptional regulators HilA, HilC, HilD, and InvF. Epistasis analysis indicates that Dam-dependent regulation of SPI-1 requires HilD, while HilA, HilC, and InvF are dispensable. A transcriptional hilD∷lac fusion is expressed at similar levels in Dam+ and Dam− hosts. However, lower levels of hilD mRNA are found in a Dam− background, thus providing unsuspected evidence that Dam methylation might exert post-transcriptional regulation of hilD expression. This hypothesis is supported by the following lines of evidence: (i) lowered levels of hilD mRNA are found in Salmonella Dam− mutants when hilD is transcribed from a heterologous promoter; (ii) increased hilD mRNA turnover is observed in Dam− mutants; (iii) lack of the Hfq RNA chaperone enhances hilD mRNA instability in Dam− mutants; and (iv) lack of the RNA degradosome components polynucleotide phosphorylase and ribonuclease E suppresses hilD mRNA instability in a Dam− background. Our report of Dam-dependent control of hilD mRNA stability suggests that DNA adenine methylation plays hitherto unknown roles in post-transcriptional control of gene expression.
DEOXYADENOSYL methyltransferases are common in bacteria, and most of them are part of restriction/modification systems (Marinus 1996; Wion and Casadesus 2006). In addition, many bacterial genomes contain solitary DNA adenine methylases, not involved in protecting DNA from a restriction enzyme companion. Two of these enzymes, the Dam methylase of gamma-proteobacteria and the CcrM methylase of alpha-proteobacteria, are paradigms of evolutionary processes that have turned DNA adenine methylation into an epigenetic signal for DNA–protein interactions (Reisenauer et al. 1999; Løbner-Olesen et al. 2005; Casadesus and Low 2006; Wion and Casadesus 2006).
In Escherichia coli and Salmonella enterica, Dam methylation controls chromosome replication, nucleoid organization, chromosome segregation, mismatch repair, and expression of certain genes (Marinus 1996; Løbner-Olesen et al. 2005; Wion and Casadesus 2006; Heusipp et al. 2007; Low and Casadesus 2008). Because of its multiple roles in bacterial physiology, loss of Dam methylation causes pleiotropic defects in certain species (e.g., E. coli and S. enterica) and inviability in others (e.g., Vibrio cholerae and certain strains of Yersinia enterocolitica) (Wion and Casadesus 2006).
DNA adenine methylase (Dam−) mutants of S. enterica are severely attenuated in the mouse model and present a plethora of virulence-related defects, both at the intestinal stage of infection and during systemic infection (Garcia-Del Portillo et al. 1999; Heithoff et al. 1999). Lack of Dam-dependent mismatch repair sensitizes Dam− mutants to the DNA-damaging action of bile salts (Prieto et al. 2004). Envelope instability may also contribute to bile sensitivity in Salmonella Dam− mutants (Pucciarelli et al. 2002). Lack of Dam methylation perturbs also the interaction of Salmonella with the intestinal epithelium. Impaired invasion of epithelial cells by Dam− mutants has been confirmed in tissue cultures and has been tentatively correlated with reduced secretion of invasion effectors encoded on Salmonella pathogenicity island 1 (SPI-1) (Garcia-Del Portillo et al. 1999). High-throughput analysis of gene expression has confirmed that SPI-1 is transcribed at lowered levels in Dam− mutants (Balbontin et al. 2006).
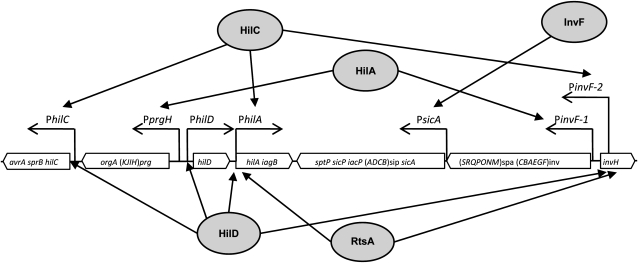
SPI-1 is an ∼40-kb gene cluster containing at least 37 genes (Lostroh and Lee 2001; Altier 2005; Jones 2005), located at centisome 63 on the S. enterica chromosome (McClelland et al. 2001). SPI-1 encodes a type 3 secretion system and secreted effectors that interact with proteins inside epithelial cells in the animal intestine (Galan and Curtiss 1989). SPI-1 genes are organized in seven or more transcriptional units, whose expression is under the control of four SPI-encoded transcription factors: HilA, HilC, HilD, and InvF (Lostroh and Lee 2001). HilA, a member of the OmpR/ToxR family (Lee et al. 1992; Bajaj et al. 1995), activates transcription of SPI genes that encode components of the secretion apparatus as well as the gene for the InvF transcriptional regulator (Bajaj et al. 1996). In association with SicA, InvF is necessary to boost transcription of the sicA and sipBCDA transcriptional units (Darwin and Miller 1999; Eichelberg and Galan 1999). HilC and HilD are members of the AraC/XylS family and activate transcription from the pinvF and psicA promoters in an apparently redundant manner (Akbar et al. 2003). Transcriptional activation by HilC and HilD relieves repression of the hilA promoter by the nucleoid proteins H-NS and Hha (Olekhnovich and Kadner 2006). Furthermore, HilC and HilD can activate inv/sicA transcription in the absence of HilA (Rakeman et al. 1999; Akbar et al. 2003). A transcription factor located outside SPI-1, RtsA, is also involved in transcriptional control of SPI-1 (Ellermeier and Slauch 2003). A diagram of SPI-1 transcriptional regulation is presented in Figure 1. Besides the regulatory actions described above, positive feedback loops are involved in the control of hilD, hilC, and rtsA transcription (Ellermeier et al. 2005).
Figure 1.—
Diagram showing the transcriptional units of Salmonella enterica SPI-1 and the regulatory circuits under the control of transcription factors HilA, HilD, HilC, RtsA, and InvF (adapted from Lostroh and Lee 2001; Ellermeier and Slauch 2003; Altier 2005; Jones 2005).
SPI-1 expression is under the control of additional regulators located outside the island. The ferric uptake regulatory protein, Fur, and the BarA/SirA two-component system are SPI-1 activators (Fortune et al. 2006; Ellermeier and Slauch 2008). In turn, HilE (Fahlen et al. 2000) and Lon (Takaya et al. 2003; Boddicker and Jones 2004) are negative regulators of SPI-1.
In this study, we show that Dam-dependent regulation of SPI-1 has a single target, the hilD gene. However, we present evidence that Dam methylation regulates hilD expression at the post-transcriptional level. Because Dam methylase is not known to have functions other than GATC methylation, a reasonable interpretation is that Dam methylation may control transcription of a post-transcriptional regulator of hilD expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and strain construction:
The S. enterica strains listed in Table 1 belong to serovar Typhimurium and derive from ATCC 14028. For simplicity, S. enterica serovar Typhimurium is often abbreviated as S. enterica. Luria–Bertani (LB) broth was used as liquid medium. Solid LB broth contained agar at 1.5% final concentration. Green plates (Chan et al. 1972) contained methyl blue (Sigma, St. Louis) instead of aniline blue. The indicator for monitoring β-galactosidase activity in plate tests was 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (“X-gal”; Sigma, 40 μg/ml). Antibiotics were used at the concentrations described previously (Torreblanca et al. 1999). Targeted gene disruption was achieved using plasmid pKD13 (Datsenko and Wanner 2000). Antibiotic resistance cassettes introduced during strain construction were excised by recombination with plasmid pCP20 (Datsenko and Wanner 2000). The oligonucleotides used for disruption (labeled “UP” and “DO”) are listed in supporting information, Table S1, together with the oligonucleotides (labeled “E”) used for allele verification by the polymerase chain reaction. Disruption of the rne gene, which encodes ribonuclease E, was performed with primers that eliminate the C-terminal region (Viegas et al. 2007). For the construction of transcriptional and translational lac fusions in the Salmonella chromosome, FRT sites generated by excision of Kmr cassettes (Datsenko and Wanner 2000) were used to integrate either plasmid pCE37 or pCE40 (Ellermeier et al. 2002). Unless specified otherwise, all lac fusions used in this study are translational. Addition of 3× FLAG and HA epitope tags to protein-coding DNA sequences was carried out using plasmids pSUB11 (Kmr, 3× FLAG) and pSU314 (Cmr, HA) as templates (Uzzau et al. 2001). Transductional crosses using phage P22 HT 105/1 int201 (Schmieger 1972; G. Roberts, unpublished data) were used for strain construction operations involving chromosomal markers. The transduction protocol was described elsewhere (Garzon et al. 1995). To obtain phage-free isolates, transductants were purified by streaking on green plates. Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5. Reconstruction of chromosomal duplications by P22 HT transduction was performed as previously described (Camacho and Casadesus 2001).
TABLE 1.
Strains of Salmonella enterica serovar Typhimurium
| Strain designation | Genotype | Reference or source |
|---|---|---|
| 14028 | Wild type | ATCC |
| SV5264 | Δdam-231 | This study |
| SV5278 | Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5279 | Δdam-231 Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5284 | Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5285 | Δdam-231 Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5286 | Φ(hilD-lacZ) | This study |
| SV5288 | Δdam-231 Φ(hilD-lacZ) | This study |
| SV5293 | Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5294 | Δdam-231 Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5297 | Φ(invF′-lacZ+)(Hyb) | This study |
| SV5298 | Δdam-231 Φ(invF′-lacZ+)(Hyb) | This study |
| SV5301 | Φ(invH′-lacZ+)(Hyb) | This study |
| SV5302 | Δdam-231 Φ(invH′-lacZ+)(Hyb) | This study |
| SV5308 | Δdam-231 ΔhilA Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5310 | Δdam-231 ΔhilC Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5312 | Δdam-231 ΔhilD Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5314 | ΔinvF Δdam-231 Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5316 | ΔhilA Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5318 | ΔhilC Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5320 | ΔhilD Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5322 | ΔinvF Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5335 | PtetA-hilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV5336 | Δdam-231 PtetA-hilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV5382 | Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5383 | Δdam-231 Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5384 | Φ(hilC′-lacZ+)(Hyb) | This study |
| SV5385 | Δdam-231 Φ(hilC′-lacZ+)(Hyb) | This study |
| SV5386 | ΔhilD Φ(hilC′-lacZ+)(Hyb) | This study |
| SV5387 | Δdam-231 ΔhilD Φ(hilC′-lacZ+)(Hyb) | This study |
| SV5399 | ΔhilD Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5400 | Δdam-231 ΔhilD Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5401 | ΔhilC Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5402 | Δdam-231 ΔhilC Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5403 | ΔhilA Φ(invF′-lacZ+)(Hyb) | This study |
| SV5404 | Δdam-231 ΔhilA Φ(invF′-lacZ+)(Hyb) | This study |
| SV5405 | ΔhilC Φ(invF′-lacZ+)(Hyb) | This study |
| SV5406 | Δdam-231 ΔhilC Φ(invF′-lacZ+)(Hyb) | This study |
| SV5407 | ΔhilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV5408 | Δdam-231 ΔhilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV5415 | ΔhilD Φ(invH′-lacZ+)(Hyb) | This study |
| SV5416 | Δdam-231 ΔhilD Φ(invH′-lacZ+)(Hyb) | This study |
| SV5417 | ΔhilC Φ(invH′-lacZ+)(Hyb) | This study |
| SV5418 | Δdam-231 ΔhilC Φ(invH′-lacZ+)(Hyb) | This study |
| SV5419 | ΔhilA Φ(invH′-lacZ+)(Hyb) | This study |
| SV5420 | Δdam-231 ΔhilA Φ(invH′-lacZ+)(Hyb) | This study |
| SV5455 | hilC∷3× FLAG | This study |
| SV5456 | hilA∷3× FLAG | This study |
| SV5457 | invF∷3× FLAG | This study |
| SV5540 | ΔrtsA Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5541 | Δdam-231 ΔrtsA Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5542 | ΔrtsA Φ(invF′-lacZ+)(Hyb) | This study |
| SV5543 | Δdam-231 ΔrtsA Φ(invF′-lacZ+)(Hyb) | This study |
| SV5592 | DUP[(purG)*MudP*(argG)] Φ(hilD-lacZ) | This study |
| SV5594 | DUP[(purG)*MudP*(argG)] ΔhilD Φ(hilD-lacZ) | This study |
| SV5596 | Δdam-231 DUP[(purG)*MudP*(argG)] Φ(hilD-lacZ) | This study |
| SV5598 | Δdam-231 DUP[(purG)*MudP*(argG)] ΔhilD Φ(hilD-lacZ) | This study |
| SV5624 | hilD∷HA | This study |
| SV5625 | Δdam-231 hilD∷HA | This study |
| SV5646 | Δhfq∷cat | M. Jakomin |
| SV5826 | PtetA-hilD Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5827 | Δdam-231 PtetA-hilD Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5828 | PtetA-hilD | This study |
| SV5829 | Δdam-231 PtetA-hilD | This study |
| SV5847 | Δdam-231 Δhfq∷cat | This study |
| SV5848 | Δhfq∷cat Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5849 | Δdam-231 Δhfq∷cat Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5850 | Δhfq∷cat Φ(invF′-lacZ+)(Hyb) | This study |
| SV5851 | Δdam-231 Δhfq∷cat Φ(invF′-lacZ+)(Hyb) | This study |
| SV5852 | Δhfq∷cat Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5853 | Δdam-231 Δhfq∷cat Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5854 | Δhfq∷cat Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5855 | Δdam-231 Δhfq∷cat Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5856 | Δhfq∷cat Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5857 | Δdam-231 Δhfq∷cat Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5873 | Δdam-231 hilC∷3× FLAG | This study |
| SV5874 | Δdam-231 hilA∷3× FLAG | This study |
| SV5875 | Δdam-231 invF∷3× FLAG | This study |
| SV5876 | Δhfq∷cat PtetA-hilD Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5877 | Δdam-231 Δhfq∷cat PtetA-hilD Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5878 | Δhfq∷cat PtetA-hilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV5879 | Δdam-231 Δhfq∷cat PtetA-hilD Φ(invF′-lacZ+)(Hyb) | This study |
| SV5961 | Δrne∷cat | This study |
| SV5962 | Δdam-231 Δrne∷cat | This study |
| SV5963 | Δpnp∷cat | This study |
| SV5964 | Δdam-231 Δpnp∷cat | This study |
| SV5965 | Δrne∷cat Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5966 | Δdam-231 Δrne∷cat Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5967 | Δrne∷cat Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5968 | Δdam-231 Δrne∷cat Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5969 | Δrne∷cat Φ(invF′-lacZ+)(Hyb) | This study |
| SV5970 | Δdam-231 Δrne∷cat Φ(invF′-lacZ+)(Hyb) | This study |
| SV5971 | Δrne∷cat Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5972 | Δdam-231 Δrne∷cat Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5973 | Δrne∷cat Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5974 | Δdam-231 Δrne∷cat Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5975 | Δpnp∷cat Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5976 | Δdam-231 Δpnp∷cat Φ(hilA′-lacZ+)(Hyb) | This study |
| SV5977 | Δpnp∷cat Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5978 | Δdam-231 Δpnp∷cat Φ(sicA′-lacZ+)(Hyb) | This study |
| SV5979 | Δpnp∷cat Φ(invF′-lacZ+)(Hyb) | This study |
| SV5980 | Δdam-231 Δpnp∷cat Φ(invF′-lacZ+)(Hyb) | This study |
| SV5981 | Δpnp∷cat Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5982 | Δdam-231 Δpnp∷cat Φ(sipB′-lacZ+)(Hyb) | This study |
| SV5983 | Δpnp∷cat Φ(sipC′-lacZ+)(Hyb) | This study |
| SV5984 | Δdam-231 Δpnp∷cat Φ(sipC′-lacZ+)(Hyb) | This study |
Construction of strain SV5828:
Strain SV5298 was transduced with a Tn10dTc pool prepared as previously described (Cano et al. 2002). Transductants were selected on LB plates supplemented with tetracycline and X-gal. Independent Lac+ transductants were sought and purified on green plates. Individual isolates were then patched on LB broth with X-gal and LB broth with X-gal and tetracycline. An isolate that was Lac+ in LB broth + X-gal + tetracycline and Lac− in LB broth + X-gal was used as donor in a P22 HT transductional cross to introduce the Tn10dTc insertion in a wild-type background. A transductant of this kind was propagated as SV5828. Two-strand DNA sequencing of the Tn10dTc element of SV5828 revealed that insertion had occurred in a GGG/GCT motif upstream of hilD, with the tetA promoter pointing out toward hilD. The insertion had thus generated a conditional, tetracycline-dependent hilD allele. Additional details about this allele are given in Figure S1 and Figure S2.
Protein extracts and Western blot analysis:
Total protein extracts were prepared from bacterial cultures grown at 37° in LB medium until stationary phase (final OD600 ∼ 2.5). Bacterial cells contained in 0.2 ml of culture were collected by centrifugation (16,000 × g, 2 min, 4°) and suspended in 50 μl of Laemmli sample buffer [1.3% SDS, 10% (v/v) glycerol, 50 mm Tris-HCl, 1.8% β-mercaptoethanol, 0.02% bromophenol blue, pH 6.8]. Proteins were resolved by Tris-Tricine-PAGE, using 12% gels. Conditions for protein transfer have been described elsewere (Jakomin et al. 2008). Primary antibodies were anti-Flag M2 monoclonal antibody (1:5000, Sigma), anti-HA HA.11 monoclonal antibody (1:1000; Covance, Princeton, NJ), and anti-GroEL polyclonal antibody (1:20,000, Sigma). Goat anti-mouse horseradish peroxidase-conjugated antibody (1:5000; Bio-Rad, Hercules, CA) or goat anti-rabbit horseradish peroxidase-conjugated antibody (1:20,000; Santa Cruz Biotechnology, Heildelberg, Germany) was used as secondary antibody. Proteins recognized by the antibodies were visualized by chemoluminescence using the luciferin–luminol reagents.
Quantitative reverse transcriptase PCR and calculation of relative expression levels:
RNA was extracted from S. enterica stationary phase cultures (OD600 ∼ 2.5), using the SV total RNA isolation system (Promega, Madison, WI) as described at http://www.ifr.ac.uk/safety/microarrays/protocols.html. The quantity and quality of the extracted RNA were determined using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). To diminish genomic DNA contamination, the preparation was treated with DNase I (Turbo DNA free; Applied Biosystems/Ambion, Austin, TX). An aliquot of 0.6 μg of DNase I-treated RNA was used for cDNA synthesis using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). Quantitative reverse transcriptase (RT)–PCR reactions were performed in an Applied Biosystems 7500 Fast Real-Time PCR System. Each reaction was carried out in a total volume of 25 μl on a 96-well optical reaction plate (Applied Biosystems) containing 12.5 μl Power SYBR Green PCR Master Mix (Applied Biosystems), 11.5 μl cDNA (1/10 dilution), and two gene-specific primers at a final concentration of 0.2 μm each. Real-time cycling conditions were as follows: (i) 95° for 10 min and (ii) 40 cycles at 95° for 15 sec and 60° for 1 min. No-template control was included for each primer set. Melting curve analysis verified that each reaction contained a single PCR product. Gene expression levels were normalized to transcripts of ompA or gmk, two housekeeping genes that served as internal controls. Gene-specific primers, designed with PRIMER3 software (http://primer3.sourceforge.net), are listed in Table S1.
Analysis of hilD mRNA decay:
Use of quantitative RT–PCR to monitor mRNA decay has been previously described (Baker et al. 2007). An overnight LB culture of the strain under study was diluted 50-fold and incubated at 37° with shaking until an OD600 ∼ 2.5. Transcription initiation was stopped by adding 500 μg/ml rifampicin and shaking vigorously during 10 sec. Cultures were kept at 37°. Aliquots were extracted at 1-min intervals and treated with a phenol (5%)–ethanol (95%) mixture. Each aliquot was immediately immersed in liquid N2 and kept frozen until RNA extraction. RNA was extracted using the standard protocol described above. Four independent quantitative (q)RT–PCR reactions, all using primers for the 5′ region of hilD mRNA, were used.
β-Galactosidase assays:
Levels of β-galactosidase activity were assayed using the CHCl3-sodium dodecyl sulfate permeabilization procedure (Miller 1972).
RESULTS
Levels of the SPI-1 transcription factors HilA, HilC, HilD, and InvF in Dam+ and Dam− hosts:
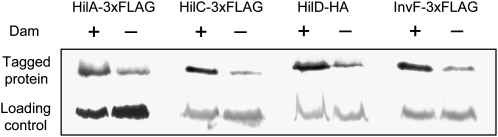
We examined the effect of Dam methylation on the levels of the main SPI-1 regulatory proteins: HilA, HilC, HilD, and InvF. For this purpose, we used HilA, HilC, and InvF protein variants tagged with the 3× FLAG epitope and a HilD variant tagged with the HA epitope. Western blot analysis in extracts from isogenic Dam+ and Dam− strains indicated that all four regulators were less abundant in Dam− hosts (Figure 2). This observation confirmed that SPI-1 expression is entirely under Dam methylation control as previously proposed (Balbontin et al. 2006), but did not provide any hint about the target(s) of Dam-dependent regulation. In silico examination of GATC site distribution in or near the hilA, hilC, hilD, and invF genes was likewise uninformative (data not shown).
Figure 2.—
Levels of HilA, HilC, HilD, and InvF in protein extracts from Dam+ and Dam− isogenic strains. Epitope-tagged proteins were detected by Western blotting with either anti-FLAG or anti-HA commercial antibodies, as appropriate. The charge control was GroEL in all cases. Strains were SV5456 (hilA∷3× FLAG), SV5874 (hilA∷3× FLAG Dam−), SV5455 (hilC∷3× FLAG), SV5873 (hilC∷3× FLAG Dam−), SV5624 (hilD∷HA), SV5625 (hilD∷HA Dam−), SV5457 (invF∷3× FLAG), and SV5875 (invF∷3× FLAG Dam−).
Dam-dependent regulation of SPI-1 is transmitted via HilD:
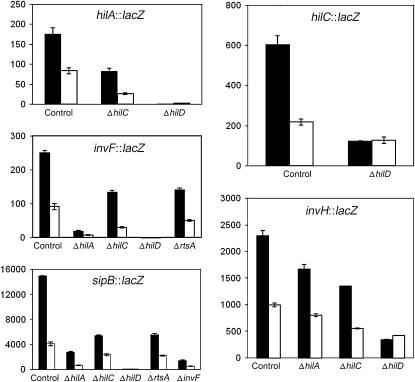
In an attempt to identify the SPI-1 regulator(s), if any, involved in transmission of Dam-dependent control to SPI-1, we examined the involvement of the SPI-1 “general” transcription factors HilA, HilC, and HilD and the sip-specific transcription factor InvF (Darwin and Miller 1999; Eichelberg and Galan 1999). RtsA, a general SPI-1 transcription factor encoded outside SPI-1 (Ellermeier and Slauch 2003), was also included in the survey. SPI-1 expression was monitored in a set of mutants, each lacking one SPI-1 transcription factor. Epistasis analysis took advantage of two well known traits of SPI-1 expression. One is regulatory redundancy by certain transcription factors (e.g., HilC and HilD) (Altier 2005; Jones 2005). The other is that lack of a single transcription factor does not completely abolish expression in certain transcriptional units (Ellermeier et al. 2005). Expression of SPI-1 transcriptional units was monitored by measuring β-galactosidase activities of lac fusions in representative genes. Only those regulators that are known to control a specific SPI-1 transcriptional unit were included in the analysis. For instance, expression of hilC in the absence of HilA was not tested because hilC is not regulated by hilA (Rakeman et al. 1999; Lostroh et al. 2000). In turn, expression of the hilA in the absence of InvF was omitted because InvF is downstream from HilA in the SPI-1 regulatory cascade (Eichelberg et al. 1999) (Figure 1). The results of these surveys are shown in Figure 3 and can be summarized as follows:
Dam-dependent regulation of hilA was not abolished in the absence of HilC. No information was obtained, however, on the potential involvement of HilD on Dam-dependent hilA regulation, since a hilD mutation completely abolished expression of the hilA∷lac fusion (Figure 3). In an analogous fashion, Dam-dependent regulation of invF was still observed in HilA−, HilC−, and RtsA− backgrounds, and no information was obtained in a HilD− background (Figure 3). Similar observations were made for sipB, which remained under Dam methylation control in HilA−, HilC−, RtsA−, and InvF− backgrounds. As above, absence of sipB expression in both HilD− Dam+ and HilD− Dam− hosts prevented any conclusion about Dam methylation dependence (Figure 3). However, these experiments provided evidence that none of the HilA, HilC, RtsA, and InvF transcription factors is involved in Dam-dependent control of SPI-1.
Expression of a hilC∷lac fusion was not completely abolished in a HilD− background (Figure 3), and similar levels of β-galactosidase activity were detected in cultures of HilD− Dam+ and HilD− Dam− hosts. Similar results were obtained for an invH∷lac fusion, which remained under Dam methylation control in HilA− and HilC− hosts, but not in a HilD− background (Figure 3). The epistatic effect of a hilD mutation over a dam mutation thus provided evidence that Dam-dependent regulation of SPI-1 requires a functional hilD gene.
Figure 3.—
β-Galactosidase activities of hilA∷lac, invF∷lac, sipB∷lac, hilC∷lac, and invH∷lac fusions in the presence and in the absence of individual transcription factors involved in SPI-1 control. Solid histograms represent β-galactosidase activities measured in a Dam+ background. Open histograms represent β-galactosidase activities measured in a Dam− background. Strains were SV5284 (hilA∷lac), SV5285 (hilA∷lac Dam−), SV5401 (hilA∷lac HilC−), SV5402 (hilA∷lac HilC− Dam−), SV5399 (hilA∷lac HilD−), SV5400 (hilA∷lac HilD− Dam−), SV5297 (invF∷lac), SV5298 (invF∷lac Dam−), SV5403 (invF∷lac HilA−), SV5404 (invF∷lac HilA− Dam−), SV5405 (invF∷lac HilC−), SV5406 (invF∷lac HilC− Dam−), SV5407 (invF∷lac HilD−), SV5408 (invF∷lac HilD− Dam−), SV5542 (invF∷lac RtsA−), SV5543 (invF∷lac RtsA− Dam−), SV5382 (sipB∷lac), SV5383 (sipB∷lac Dam−), SV5316 (sipB∷lac HilA−), SV5308 (sipB∷lac HilA− Dam−), SV5318 (sipB∷lac HilC−), SV5310 (sipB∷lac HilC− Dam−), SV5320 (sipB∷lac HilD−), SV5312 (sipB∷lac HilD− Dam−), SV5540 (sipB∷lac RtsA−), SV5541 (sipB∷lac RtsA− Dam−), SV5322 (sipB∷lac InvF−), SV5314 (sipB∷lac InvF− Dam−), SV5384 (hilC∷lac), SV5385 (hilC∷lac Dam−), SV5386 (hilC∷lac HilD−), SV5387 (hilC∷lac HilD− Dam−), SV5301 (invH∷lac), SV5302 (invH∷lac Dam−), SV5419 (invH∷lac HilA−), SV5420 (invH∷lac HilA− Dam−), SV5417 (invH∷lac HilC−), SV5418 (invH∷lac HilC− Dam−), SV5415 (invH∷lac HilD−), and SV5416 (invH∷lac HilD− Dam−). Data are averages and standard deviations from three experiments.
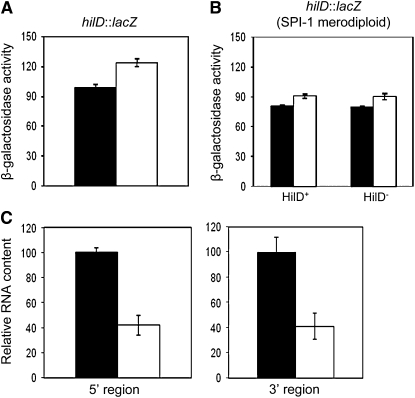
Dam methylation regulates the level of hilD mRNA:
In an attempt to confirm that Dam methylation regulates hilD expression, the activity of a hilD∷lac transcriptional fusion was monitored in Dam+ and Dam− hosts. To our surprise, no difference was found (Figure 4). However, these experiments left one possibility open. Transcription of hilD is under the control of an autogenous, positive feedback loop by the HilD product (Ellermeier et al. 2005; Ellermeier and Slauch 2008). Hence, use of a hilD∷lac fusion might prevent the observation of differences, if any, between Dam+ and Dam− hosts, simply because the hilD∷lac strain is HilD−. To circumvent this potential problem, the hilD∷lac fusion was transduced to isogenic Dam+ and Dam− strains carrying a chromosomal duplication that includes SPI-1 (Camacho and Casadesus 2001). β-Galactosidase activities were then monitored in Dam+ HilD+/hilD∷lac and Dam− HilD+/hilD∷lac merodiploids. No difference was found (Figure 4), thus ruling out the possibility that similar levels of hilD expression in Dam+ and Dam− hosts resulted from disruption of the HilD feedback loop. Evidence that transcription of the hilD gene is not under Dam methylation control (Figure 4) was in stark contrast with Western blot experiments showing different levels of HilD protein in Dam+ and Dam− hosts (Figure 2).
Figure 4.—
(A) β-Galactosidase activity of a hilD∷lac transcriptional fusion in Dam+ (SV5286) and Dam− (SV5288) isogenic hosts. Data are averages and standard deviations from three experiments. (B) β-Galactosidase activity of the same hilD∷lac transcriptional fusion in Dam+ HilD+ (SV5592), Dam+ HilD− (SV5594), Dam− HilD+ (SV5596), and Dam− HilD− (SV5598) isogenic merodiploids (averages of three experiments). (C) Relative amounts of hilD mRNA in Dam+ (ATCC 14028) and Dam− (SV5264) strains, normalized to ompA mRNA. Two primer pairs, complementary to 5′ and 3′ hilD regions, were used. Histograms represent the averages from three independent experiments.
Analysis of hilD mRNA content in Dam+ and Dam− hosts (ATCC 14028 and SV5264, respectively) was performed by quantitative reverse transcriptase PCR, using primer pairs complementary to both the 5′ and the 3′ regions of hilD. A lower level of hilD mRNA was found in the Dam− background (Figure 4). Hence, decreased levels of both hilD mRNA and HilD protein were found in Salmonella Dam− hosts (Figures 2 and 4), even though a hilD∷lac transcriptional fusion did not show Dam-dependent control (Figure 4).
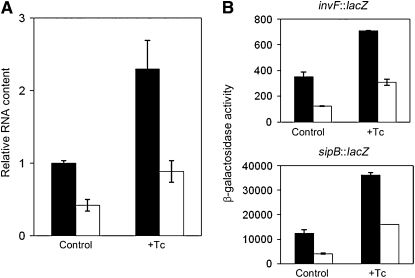
Expression of hilD from a heterologous promoter is Dam dependent:
The failure of a hilD∷lac transcriptional fusion to show Dam-dependent regulation admits a number of explanations, artifactual or not. Hence, we considered the possibility that hilD regulation by Dam methylation might be in fact transcriptional. If such was the case, we reasoned, Dam-dependent hilD regulation should be no longer observed when hilD expression was driven from a heterologous promoter. In contrast, Dam dependence in a hilD gene driven from a heterologous promoter would provide evidence for post-transcriptional control. On these grounds, we examined whether hilD expression remained Dam dependent in strain SV5828. This strain, whose construction is described in materials and methods, carries a conditional hilD mutation that renders the strain HilD− in the absence of tetracycline and HilD+ in the presence of either tetracycline or autoclaved chlortetracycline. Using this strain and its isogenic Dam− derivative SV5829, we compared hilD mRNA levels in Dam+ and Dam− hosts in the presence and in the absence of tetracycline. Expression of hilD was Dam dependent in the presence of tetracycline (Figure 5), thus indicating that a hilD transcript driven by the tetA promoter remained under Dam methylation control like wild-type hilD mRNA. As a validation for this conclusion, we observed that expression of invF∷lac and sipB∷lac fusions remained under Dam methylation control when hilD expression was tetracycline dependent (Figure 5). These results supported the view that Dam methylation might not regulate hilD transcription but might regulate hilD mRNA stability. This possibility was puzzling, because Dam methylation is a DNA modification function, not known to interact with nucleic acid molecules other than double-stranded DNA (Marinus 1996; Wion and Casadesus 2006).
Figure 5.—
(A) Relative amounts of hilD mRNA in Dam+ (solid histograms) and Dam− (open histograms) isogenic strains expressing hilD from a heterologous, tetracycline-dependent promoter. Levels of hilD mRNA were normalized to ompA mRNA, as above. Strains were SV5828 (PtetA-hilD), and SV5829 (dam PtetA-hilD). Data are averages and standard deviations from three independent experiments. (B) Transcription levels of two SPI-1 genes under HilD control (invF and sipB) in Dam+ (solid histograms) and Dam− (open histograms) strains that express hilD from a heterologous, tetracycline-dependent promoter. Strains were SV5297 (invF∷lac), SV5298 (invF∷lac Dam−), SV5335 (PtetA-hilD invF∷lac), SV5336 (PtetA-hilD invF∷lac Dam−), SV5382 (sipB∷lac), SV5383 (sipB∷lac Dam−), SV5826 (PtetA-hilD sipB∷lac), and SV5827 (PtetA-hilD sipB∷lac Dam−). Data are averages and standard deviations from three independent experiments.
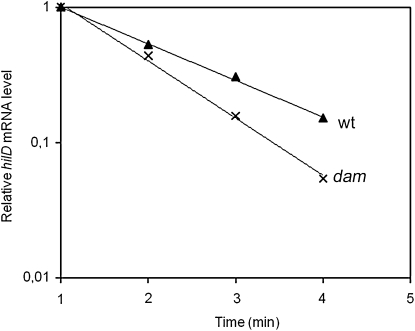
Dam methylation regulates hilD mRNA stability:
To compare hilD mRNA stability in Dam+ and Dam− hosts, stationary cultures (OD600 = 2.5) were treated with rifampicin to stop transcription. RNA samples were extracted at 1-min intervals and subjected to quantitative RT–PCR primed by two oligonucleotides of the 5′ region of hilD. In all RNA preparations, hilD mRNA was found to decay in a linear manner from 1 to 4 min after rifampicin addition, and a substantial difference in the decay rate was observed between the RNA preparations from a Dam+ strain and those from a Dam− mutant (Figure 6). The half lives of hilD mRNA were calculated as 67 sec in a Dam+ host and 47 sec in a Dam− host. These experiments provided direct evidence that hilD mRNA is less stable in the absence of Dam methylation. Because increased turnover of RNA is not a trait of Salmonella Dam− mutants (Balbontin et al. 2006), we interpret that hilD mRNA may undergo different post-transcriptional regulation in Dam+ and Dam− hosts.
Figure 6.—
Stability of hilD mRNA in Dam+ (ATCC 14028) and Dam− (SV5264) isogenic hosts. Values are averages from four independent qRT–PCR reactions. Error bars are not shown because the standard deviations were extremely small.
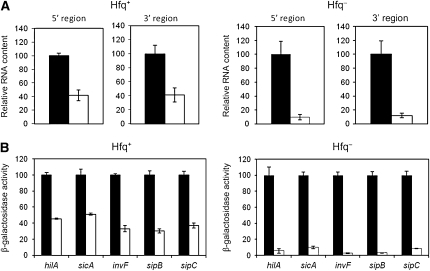
Lack of Hfq enhances hilD mRNA instability in Salmonella Dam− mutants:
The evidence that hilD mRNA undergoes post-transcriptional control led us to test the involvement of Hfq, an RNA chaperone that is known to interact with multiple RNA molecules including hilD mRNA (Sittka et al. 2008). To investigate whether lack of Hfq affected hilD mRNA stability, analysis of hilD mRNA content was performed in isogenic Dam+ Hfq+, Dam− Hfq+, Dam+ Hfq−, and Dam− Hfq− isogenic strains. Oligonucleotides complementary to both the 5′ and the 3′ regions of hilD were used to prime quantitative RT–PCR. In a Dam− background, the hilD mRNA level decreased 2.5-fold in the presence of Hfq and >10-fold in the absence of Hfq (Figure 7). Hence, lack of Hfq enhances the hilD mRNA instability caused by a dam mutation. A recent study has suggested that binding of Hfq to the AU-rich hilD mRNA might be peculiar, in the sense that Hfq might not bind one or more specific RNA regions but the entire mRNA molecule (Sittka et al. 2008). This binding pattern might contribute to the Hfq protective effect.
Figure 7.—
(A) Enhancement of hilD mRNA instability in the absence of Hfq. Solid histograms are for Dam+ strains, and open histograms are for their Dam− derivatives. RNA levels were normalized to either ompA mRNA or gmk mRNA. Strains were ATCC 14208 (wild type), SV5264 (Dam−), SV5646 (Hfq−), and SV5847 (Hfq− Dam−). Values are averages and standard deviations from three independent experiments. (B) Enhancement of the SPI-1 expression defect of S. enterica Dam− mutants by hfq null mutations. Solid histograms are for Dam+ strains, and open histograms are for their Dam− derivatives. To facilitate visual perception of differences, the β-galactosidase activities of individual lac fusions in Dam+ hosts have been normalized to 100. Strains were as follows: SV5284 (hilA∷lac), SV5285 (hilA∷lac Dam−), SV5278 (sicA∷lac), SV5279 (sicA∷lac Dam−), SV5297 (invF∷lac), SV5298 (invF∷lac Dam−), SV5382 (sipB∷lac), SV5383 (sipB∷lac Dam−), SV5293 (sipC∷lac), SV5294 (sipC∷lac Dam−), SV5848 (hilA∷lac Hfq−), SV5849 (hilA∷lac Hfq− Dam−), SV5856 (sicA∷lac Hfq−), SV5857 (sicA∷lac Hfq− Dam−), SV5850 (invF∷lac Hfq−), SV5851 (invF∷lac Hfq− Dam−), SV5852 (sipB∷lac Hfq−), SV5853 (sipB∷lac Hfq− Dam−), SV5854 (sipC∷lac Hfq−), and SV5855 (sipC∷lac Hfq− Dam−). Data are averages and standard deviations from three experiments.
Lack of Hfq enhances the SPI-1 expression defect of Salmonella Dam− mutants:
The effect of an hfq null mutation on Dam-dependent SPI-1 expression was examined in five SPI-1 genes, selected on the basis of their strong HilD dependence. β-Galactosidase activities were measured in Dam+ Hfq+, Dam− Hfq+, Dam+ Hfq−, and Dam− Hfq− isogenic strains carrying hilA∷lac, sicA∷lac, invF∷lac, sipB∷lac, and sipC∷lac fusions. Raw data are shown in Table S2. Figure 8 is an elaboration of Table S2 data that outlines the differences between Dam− Hfq+ and Dam− Hfq− mutants. Because lac fusions in individual SPI-1 genes have disparate β-galactosidase activities, the activity of each fusion has been normalized to 100 in the Dam+ background. Lack of Hfq caused a decrease in SPI-1 expression (Table S2), as previously described (Sittka et al. 2007). For the purpose of our study, however, the noteworthy result was that an hfq mutation enhanced the SPI-1 expression defect of Dam− mutants (Figure 7).
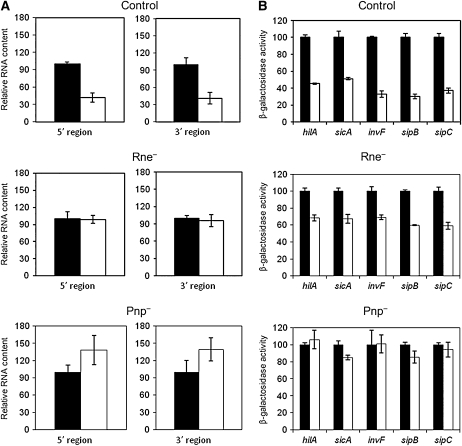
Figure 8.—
(A) Suppression of hilD mRNA instability in the absence of degradosome components ribonuclease E (Rne) and polynucleotide phosphorylase (Pnp). Solid histograms are for Dam+ strains, and open histograms are for their Dam− derivatives. RNA levels were normalized to either ompA mRNA or gmk mRNA. Strains were ATCC 14028 (wild type), SV5264 (Dam−), SV5961 (Rne−), SV5962 (Rne− Dam−), SV5963 (Pnp−), and SV5964 (Pnp− Dam−). Values are averages and standard deviations from three independent experiments. (B) Suppression of the SPI-1 expression defect of S. enterica Dam− mutants by rne and pnp mutations. Solid histograms are for Dam+ strains, and open histograms are for their Dam− derivatives. To facilitate visual perception of differences, the β-galactosidase activities of lac fusions in individual SPI-1 genes in Dam+ hosts have been normalized to 100. Strains were as follows: SV5284 (hilA∷lac), SV5285 (hilA∷lac Dam−), SV5278 (sicA∷lac), SV5279 (sicA∷lac Dam−), SV5297 (invF∷lac), SV5298 (invF∷lac Dam−), SV5382 (sipB∷lac), SV5383 (sipB∷lac Dam−), SV5293 (sipC∷lac), SV5294 (sipC∷lac Dam−), SV5965 (hilA∷lac Rne−), SV5966 (hilA∷lac Rne− Dam−), SV5967 (sicA∷lac Rne−), SV5968 (sicA∷lac Rne− Dam−), SV5969 (invF∷lac Rne−), SV5970 (invF∷lac Rne− Dam−), SV5971 (sipB∷lac Rne−), SV5972 (sipB∷lac Rne− Dam−), SV5973 (sipC∷lac Rne−), SV5974 (sipC∷lac Rne− Dam−), SV5975 (hilA∷lac Pnp−), SV5976 (hilA∷lac Pnp− Dam−), SV5977 (sicA∷lac Pnp−), SV5978 (sicA∷lac Pnp− Dam−), SV5979 (invF∷lac Pnp−), SV5980 (invF∷lac Pnp− Dam−), SV5981 (sipB∷lac Pnp−), SV5982 (sipB∷lac Pnp− Dam−), SV5983 (sipC∷lac Pnp−), and SV5984 (sipC∷lac Pnp− Dam−). Data are averages and standard deviations from three experiments.
Dam-dependent expression of SPI-1 was also affected by an hfq mutation when hilD was expressed from a heterologous promoter. In the experiments summarized in Figure S3, we compared the expression of lac fusions in two SPI-1 genes, invF and sipB, in isogenic Hfq+ Dam+, Hfq+ Dam−, Hfq− Dam+, and Hfq− Dam− hosts, all expressing hilD under the control of the tetA promoter. Lack of Hfq enhanced the SPI-1 expression defect of Salmonella Dam− mutants (Figure S3). Hence, an hfq mutation enhances the hilD mRNA instability associated to lack of Dam methylation, irrespective of the promoter that drives hilD expression.
Lack of degradosome components polyribonucleotide phosphorylase and ribonuclease E suppresses hilD mRNA instability in Salmonella Dam− mutants:
If lack of Dam methylation decreases hilD mRNA stability, we reasoned, mutations that reduce RNA turnover might suppress the SPI-1 expression defect of Dam− mutants. On these grounds, we constructed mutants lacking either ribonuclease E (Rne) or polynucleotide phosphorylase (Pnp), two components of the bacterial degradosome (Carpousis 2002). Ribonuclease E had been previously described as a SPI-1 regulator (Fahlen et al. 2000). For construction of an Rne− mutant, only a portion at the 3′ end of the rne coding sequence was eliminated (Viegas et al. 2007). Analysis of hilD mRNA content was performed in two sets of experiments. In the first set, Dam+ Rne+, Dam− Rne+, Dam+ Rne−, and Dam− Rne− isogenic strains were used. In the second set, we employed Dam+ Pnp+, Dam− Pnp+, Dam+ Pnp−, and Dam− Pnp− isogenic strains. Oligonucleotides complementary to both the 5′ and the 3′ regions of hilD (Table S1) were used to prime quantitative RT–PCR. Both rne and pnp mutations restored the hilD mRNA level of Salmonella Dam− mutants to levels similar to those found in a Dam+ strain (Figure 8A). Hence, lack of either Rne or Pnp suppresses the hilD mRNA instability caused by a dam mutation.
Lack of degradosome components Rne and Pnp suppresses the SPI-1 expression defect of Salmonella Dam− mutants:
The effect of rne and pnp mutations on Dam-dependent SPI-1 expression was examined in five SPI-1 genes strongly dependent on HilD (as above). β-Galactosidase activities were measured in two sets of isogenic strains. One set carried hilA∷lac, sicA∷lac, invF∷lac, sipB∷lac, and sipC∷lac fusions in Dam+/Dam− Rne+/Rne− backgrounds. The second set carried the same fusions in Dam+/Dam− Pnp+/Pnp− backgrounds. Raw data are shown in Table S2. Figure 8B is a normalized presentation of Table S2 data that outlines the differences between Dam− Rne+ and Dam− Rne− mutants, as well as those found between Dam− Pnp+ and Dam− Pnp− mutants. In the Dam− background, lack of ribonuclease E increased expression of all SPI lac fusions about twofold (Figure 8B). In turn, lack of polyribonucleotide phosphorylase completely restored the wild-type level of expression in the five lac fusions used to monitor SPI-1 expression (Figure 8B). Partial suppression by an rne mutation and complete supression by a pnp mutation further strengthen the evidence that the SPI-1 expression defect of Salmonella Dam− mutants is post-transcriptional.
DISCUSSION
Lowered levels of all SPI-1-encoded transcriptional regulators (HilA, HilC, HilD, and InvF) are found in Salmonella Dam− mutants (Figure 2), thereby confirming that the entire SPI-1 is under Dam-dependent control. Epistasis analysis indicates that SPI-1 activation by Dam methylation requires HilD, while the remaining SPI-1 transcriptional activators (HilA, HilC, RtsA, and InvF) are dispensable for Dam-dependent control (Figure 3). Hence, the first conclusion of this study is that Dam methylation activates SPI-1 expression by sustaining high levels of the HilD transcription factor. In the absence of Dam methylation, the HilD level is lower, and SPI expression decreases. This defect may contribute to the reduced capacity of Salmonella Dam− mutants to invade epithelial cells (Garcia-Del Portillo et al. 1999).
Because the methylation state of critical GATC sites can control binding of RNA polymerase and transcription factors, differences in gene expression between Dam+ and Dam− hosts usually provide evidence for transcriptional regulation (Roberts et al. 1985; Kücherer et al. 1986; Blyn et al. 1989; Torreblanca and Casadesus 1996; Haagmans and Van Der Woude 2000; Camacho and Casadesus 2002; Waldron et al. 2002; Balbontin et al. 2006; Jakomin et al. 2008). However, several lines of evidence suggest that Dam-dependent regulation of hilD expression is not transcriptional: (i) a transcriptional hilD∷lac fusion is expressed at similar levels in Dam+ and Dam− hosts (Figure 4); (ii) reduced levels of both hilD mRNA and HilD protein are, however, found in Dam− mutants (Figures 2 and 4); (iii) reduced amounts of hilD mRNA are found in a Dam− mutants when the hilD gene is expressed from a heterologous promoter (Figure 5); (iv) SPI-1 remains under Dam-dependent control when hilD transcription is activated by tetracycline (Figure 5); and (v) lack of DNA adenine methylation results in hilD mRNA instability (Figure 6). Therefore, the second, unsuspected conclusion from this study is that Dam methylation does not regulate hilD transcription but does regulate hilD mRNA turnover.
The hypothesis, at first sight odd, that Dam methylation is a post-transcriptional regulator of SPI-1, receives further support from the nature of mutations that act either as enhancers or as suppressors of hilD mRNA instability. Lack of the Hfq RNA chaperone enhances the SPI-1 expression defect of Salmonella Dam− mutants (Figure 7) and increases hilD mRNA instability (Figure 7). In turn, lack of degradosome components ribonuclease E or polynucleotide phosphorylase (Carpousis 2002) suppresses the SPI-1 expression defect of Salmonella Dam− mutants (Figure 8). Hfq has been previously shown to stabilize hilD mRNA (Sittka et al. 2008), and our observations indicate that absence of Hfq results in increased hilD mRNA degradation in a Dam− background (Figure 7). Binding of Hfq to hilD mRNA is unusual, and a tentative explanation is that Hfq may “coat” the entire hilD transcript (Sittka et al. 2008). Hence, Hfq binding might slow down hilD mRNA turnover. This possibility is supported by a previous study in E. coli, indicating that Hfq protects AU-rich RNA molecules from degradation by ribonuclease E and polynucleotide phosphorylase (Folichon et al. 2003).
The occurrence of Dam-dependent post-transcriptional control of hilD stability fits well in the current view that hilD mRNA may be the target for integration of multiple signals that regulate SPI-1 expression (Lucas and Lee 2001; Ellermeier and Slauch 2008; Kage et al. 2008). However, with the potential exception of FliZ (Kage et al. 2008) and CsrA (Altier et al. 2000; Ellermeier and Slauch 2007), post-transcriptional regulators of hilD seem to affect either the HilD protein level (Takaya et al. 2005; Matsui et al. 2008) or HilD protein activity (Baxter et al. 2003; Ellermeier and Slauch 2008). In contrast, Dam methylation regulates hilD mRNA turnover.
Because no evidence exists that Dam methylase can interact with RNA molecules, conceivable models to explain Dam-dependent control of hilD mRNA stability are either that Dam+ hosts produce a factor that stabilizes hilD mRNA or that Dam− mutants produce a hilD mRNA destabilizing factor. Such hypothetical factor(s) might be, for instance, an Hfq-independent sRNA or an RNA-binding protein. None of the RNA metabolism proteins investigated in this study (Hfq, ribonuclease E, and polynucleotide phosphorylase) is under transcriptional control by Dam methylation, as indicated by qRT–PCR experiments shown in Figure S4.
Additional cases in which Dam methylation appears to exert post-transcriptional control of gene expression are found in the literature. Dam− mutants of enterohemorrhagic E. coli (EHEC) synthesize elevated levels of three virulence proteins (intimin, Tir, and EspFU). However, the corresponding mRNA levels remain unaltered (Campellone et al. 2007), suggesting the possibility that Dam-dependent regulation is translational. In Y. enterocolitica, overproduction of Dam methylase alters the composition of the O antigen, increasing the amount of lipid A core. However, the transcript levels in the O antigen cluster remain unaltered in Dam-overproducing strains, thus raising the possibility that Dam-dependent regulation is post-transcriptional (Falker et al. 2007). Another intriguing case involves the E. coli DNA repair endonuclease Vsr. The vsr gene is cotranscribed with the DNA cytosine methylase gene, dcm (Bell and Cupples 2001). In stationary cultures of E. coli Dam− mutants, Vsr synthesis is reduced while Dcm synthesis is not (Bell and Cupples 2001). Hence, differential mRNA translation and/or differential degradation of the dcm-vsr transcript may occur in Dam− hosts. Like the hilD mRNA stability control presented in this study, those cases from the literature remain to be deciphered at the molecular level. However, their very existence is interesting since it indicates that Dam methylation has additional, hitherto unsuspected physiological functions. Their identification is therefore a challenge for future studies.
Acknowledgments
We are grateful to Dick D'Ari, Clara García-Calderón, Ignacio Cota, Ana Serna, and Roberto Balbontín for helpful discussions and to Modesto Carballo of the Servicio de Biología (Centro de Tecnología e Innovación de la Universidad de Sevilla, Universidad de Sevilla) for help in experiments performed at the facility. This study was supported by grants BIO2007-67457-CO2-02 and CSD2008-00013 from the Spanish Ministry of Science and Innovation (MCINN) and the European Regional Fund. J.L.G. holds a Formación del Profesorado Universitario fellowship from the MCINN.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.108985/DC1.
References
- Akbar, S., L. M. Schechter, C. P. Lostroh and C. A. Lee, 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47 715–728. [DOI] [PubMed] [Google Scholar]
- Altier, C., 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43 85–92. [PubMed] [Google Scholar]
- Altier, C., M. Suyemoto and S. D. Lawhon, 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68 6790–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj, V., C. Hwang and C. A. Lee, 1995. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18 715–727. [DOI] [PubMed] [Google Scholar]
- Bajaj, V., R. L. Lucas, C. Hwang and C. A. Lee, 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22 703–714. [DOI] [PubMed] [Google Scholar]
- Baker, C. S., L. A. Eory, H. Yakhnin, J. Mercante, T. Romeo et al., 2007. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 189 5472–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontin, R., G. Rowley, M. G. Pucciarelli, J. Lopez-Garrido, Y. Wormstone et al., 2006. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188 8160–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, M. A., T. F. Fahlen, R. L. Wilson and B. D. Jones, 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, D. C., and C. G. Cupples, 2001. Very-short-patch repair in Escherichia coli requires the dam adenine methylase. J. Bacteriol. 183 3631–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. H. Rolfson and D. A. Low, 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker, J. D., and B. D. Jones, 2004. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 72 2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, E. M., and J. Casadesus, 2001. Genetic mapping by duplication segregation in Salmonella enterica. Genetics 157 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, E. M., and J. Casadesus, 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44 1589–1598. [DOI] [PubMed] [Google Scholar]
- Campellone, K. G., A. J. Roe, A. Lobner-Olesen, K. C. Murphy, L. Magoun et al., 2007. Increased adherence and actin pedestal formation by dam-deficient enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 63 1468–1481. [DOI] [PubMed] [Google Scholar]
- Cano, D. A., G. Dominguez-Bernal, A. Tierrez, F. Garcia-Del Portillo and J. Casadesus, 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis, A. J., 2002. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30 150–155. [PubMed] [Google Scholar]
- Casadesus, J., and D. Low, 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70 830–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R. K., D. Botstein, T. Watanabe and Y. Ogata, 1972. Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology 50 883–898. [DOI] [PubMed] [Google Scholar]
- Darwin, K. H., and V. L. Miller, 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K. A., and B. L. Wanner, 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 90 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg, K., and J. E. Galan, 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect. Immun. 67 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg, K., W. D. Hardt and J. E. Galan, 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33 139–152. [DOI] [PubMed] [Google Scholar]
- Ellermeier, C. D., and J. M. Slauch, 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185 5096–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier, C. D., A. Janakiram and J. M. Slauch, 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290 153–161. [DOI] [PubMed] [Google Scholar]
- Ellermeier, C. D., J. R. Ellermeier and J. M. Slauch, 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57 691–705. [DOI] [PubMed] [Google Scholar]
- Ellermeier, J. R., and J. M. Slauch, 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10 24–29. [DOI] [PubMed] [Google Scholar]
- Ellermeier, J. R., and J. M. Slauch, 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlen, T. F., N. Mathur and B. D. Jones, 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28 25–35. [DOI] [PubMed] [Google Scholar]
- Falker, S., J. Schilling, M. A. Schmidt and G. Heusipp, 2007. Overproduction of DNA adenine methyltransferase alters motility, invasion, and the lipopolysaccharide O-antigen composition of Yersinia enterocolitica. Infect. Immun. 75 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon, M., V. Arluison, O. Pellegrini, E. Huntzinger, P. Regnier et al., 2003. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 31 7302–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune, D. R., M. Suyemoto and C. Altier, 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 74 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J. E., and R. Curtiss, III, 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86 6383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Del Portillo, F., M. G. Pucciarelli and J. Casadesus, 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 96 11578–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon, A., D. A. Cano and J. Casadesus, 1995. Role of Erf recombinase in P22-mediated plasmid transduction. Genetics 140 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans, W., and M. van der Woude, 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35 877–887. [DOI] [PubMed] [Google Scholar]
- Heithoff, D. M., R. L. Sinsheimer, D. A. Low and M. J. Mahan, 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284 967–970. [DOI] [PubMed] [Google Scholar]
- Heusipp, G., S. Fälker and M. A. Schmidt, 2007. DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297 1–7. [DOI] [PubMed] [Google Scholar]
- Jakomin, M., D. Chessa, A. J. Baumler and J. Casadesus, 2008. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA and HdfR. J. Bacteriol. 190 7406–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, B. D., 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43 110–117. [PubMed] [Google Scholar]
- Kage, H., A. Takaya, M. Ohya and T. Yamamoto, 2008. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J. Bacteriol. 190 2470–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kücherer, C., H. Lother, K. Kölling, M. A. Schauzu and W. Messer, 1986. Regulation of transcription of the chromosomal dnaA gene of Escherichia coli. Mol. Gen. Genet. 205 115–121. [DOI] [PubMed] [Google Scholar]
- Lee, C. A., B. D. Jones and S. Falkow, 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 89 1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen, A., O. Skovgaard and M. G. Marinus, 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8 154–160. [DOI] [PubMed] [Google Scholar]
- Lostroh, C. P., and C. A. Lee, 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3 1281–1291. [DOI] [PubMed] [Google Scholar]
- Lostroh, C. P., V. Bajaj and C. A. Lee, 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37 300–315. [DOI] [PubMed] [Google Scholar]
- Low, D. A., and J. Casadesus, 2008. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 11 106–112. [DOI] [PubMed] [Google Scholar]
- Lucas, R. L., and C. A. Lee, 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183 2733–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus, M. G., 1996. Methylation of DNA, pp. 782–791 in Escherichia coli and Salmonella: Cellular and Molecular Biology, edited by F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low et al. ASM Press, Washington, DC.
- Matsui, M., A. Takaya and T. Yamamoto, 2008. Sigma32-mediated negative regulation of Salmonella pathogenicity island 1 expression. J. Bacteriol. 190 6636–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille et al., 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413 852–856. [DOI] [PubMed] [Google Scholar]
- Miller, J. H., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Olekhnovich, I. N., and R. J. Kadner, 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357 373–386. [DOI] [PubMed] [Google Scholar]
- Prieto, A. I., F. Ramos-Morales and J. Casadesus, 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciarelli, M. G., A. I. Prieto, J. Casadesus and F. Garcia-del Portillo, 2002. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148 1171–1182. [DOI] [PubMed] [Google Scholar]
- Rakeman, J. L., H. R. Bonifield and S. I. Miller, 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181 3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer, A., L. S. Kahng, S. McCollum and L. Shapiro, 1999. Bacterial DNA methylation: A cell cycle regulator? J. Bacteriol. 181 5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, D., B. C. Hoopes, W. R. McClure and N. Kleckner, 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43 117–130. [DOI] [PubMed] [Google Scholar]
- Schmieger, H., 1972. Phage P22 mutants with increased or decreased transducing abilities. Mol. Gen. Genet. 119 75–88. [DOI] [PubMed] [Google Scholar]
- Sittka, A., V. Pfeiffer, K. Tedin and J. Vogel, 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63 193–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka, A., S. Lucchini, K. Papenfort, C. M. Sharma, K. Rolle et al., 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4 e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya, A., M. Suzuki, H. Matsui, T. Tomoyasu, H. Sashinami et al., 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect. Immun. 71 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya, A., Y. Kubota, E. Isogai and T. Yamamoto, 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55 839–852. [DOI] [PubMed] [Google Scholar]
- Torreblanca, J., and J. Casadesus, 1996. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics 144 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreblanca, J., S. Marques and J. Casadesus, 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau, S., N. Figueroa-Bossi, S. Rubino and L. Bossi, 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98 15264–15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas, S. C., V. Pfeiffer, A. Sittka, I. J. Silva, J. Vogel et al., 2007. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35 7651–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron, D. E., P. Owen and C. J. Dorman, 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44 509–520. [DOI] [PubMed] [Google Scholar]
- Wion, D., and J. Casadesus, 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]