Abstract
Stomata are microscopic valves on the plant epidermis that played a critical role in the evolution of land plants. Studies in the model dicot Arabidopsis thaliana have identified key transcription factors and signaling pathways controlling stomatal patterning and differentiation. Three paralogous Arabidopsis basic helix-loop-helix proteins, SPEECHLESS (SPCH), MUTE, and FAMA, mediate sequential steps of cell-state transitions together with their heterodimeric partners SCREAM (SCRM) and SCRM2. Cell–cell signaling components, including putative ligands, putative receptors, and mitogen-activated protein kinase cascades, orient asymmetric cell divisions and prevent overproduction and clustering of stomata. The recent availability of genome sequence and reverse genetics tools for model monocots and basal land plants allows for the examination of the conservation of genes important in stomatal patterning and differentiation. Studies in grasses have revealed that divergence of SPCH-MUTE-FAMA predates the evolutionary split of monocots and dicots and that these proteins show conserved and novel roles in stomatal differentiation. By contrast, specific asymmetric cell divisions in Arabidopsis and grasses require unique molecular components. Molecular phylogenetic analysis implies potential conservation of signaling pathways and prototypical functions of the transcription factors specifying stomatal differentiation.
INTRODUCTION
The evolution of stomata (Greek for mouths; singular, stoma) was a crucial adaptation occurring some 400 million years ago that made it possible for plants to thrive on land, which they began to colonize some 450 million years ago. To survive in the dry atmosphere, plants must protect against desiccation yet allow for the gas exchange necessary for photosynthesis and respiration. A watertight epidermis with closable valves, the stomata, was the solution. Stomata are composed of paired, symmetric guard cells that operate by changing turgor pressure, a system that forms an effective fail-safe against dehydration, since low turgor closes stomata. Transpiration, or water loss through stomata, promotes upward and outward water movement in plants and cools the plant surface via thermal dissipation. The distribution of stomata is far from random; they are evenly distributed, an adaptation allowing gases to reach interior tissues effectively, and they adhere closely to the one-cell spacing rule (Sachs, 1991), which means that stomata are always found with at least one nonstomatal epidermal cell separating them. This one-cell spacing rule is important for proper opening and closing of the stomatal aperture, which requires efficient exchange of water and ions with neighboring nonstomatal cells. Stomatal development must be tightly controlled to accomplish these goals.
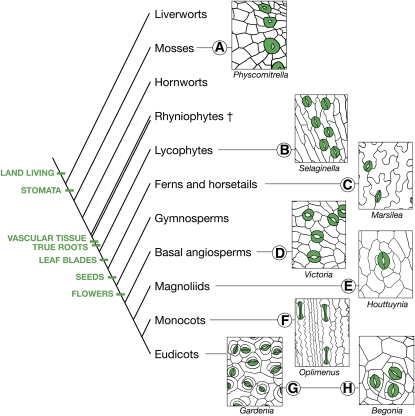
The fundamental physiological importance of stomata is underscored by the observation that their evolution predates that of flowers, leaves, roots, and even vascular systems (Freeman, 2008) (Figure 1). The fossil record indicates that the earliest nonvascular and vascular plants, such as Cooksonia, Rhynia, and Aglaophyton, possessed a simple architecture of bifurcated stems with apical spore capsules (Edwards et al., 1998), very different from modern land plants. Yet these ancient plants possessed stomata that were strikingly similar to those of extant land plants, such as Arabidopsis thaliana (Edwards et al., 1998; Taylor et al., 2005). In Figure 1, mature stomata and their neighboring epidermal cells over a large phylogenetic distance are presented to demonstrate the evolutionarily ancient derivation of the stoma.
Figure 1.
Diversity of Stomata across Land Plant Taxa.
A phylogenetic tree of extant and extinct (†) land plants includes evolutionary traits supporting success on land. The wide diversity of stomatal complexes among these groups is represented by epidermal tracings of P. patens ([A]; nonvascular), Selaginella kraussiana ([B]; lycophyte, vascular), Marsilea macropoda ([C]; fern, vascular), Victoria amazonica ([D]; Nymphaeaceae, basal angiosperm), Houttuynia cordata ([E]; Piperales, magnoliid), Oplimenus hirtellus ([F]; Poales, monocot grass), Gardenia taitensis ([G]; Gentianales, eudicot angiosperm), and Begonia rex-cultorum ‘Roberta’ ([H]; Cucurbitales, eudicot angiosperm). Stomata are colored green. Note that the stomata of Physcomitrella have a single GC, while the GCs of Oplimenus, a grass, have a dumbbell shape. Only Houttuynia, Gardenia, and Begonia show evidence of asymmetric amplifying divisions within the stomatal lineage. Physcomitrella is traced from a scanning electron microscopy image by L. Pillitteri.
If the evolution of stomata was indeed a single vital event in land plant evolution, what are the underlying genes and molecular mechanisms that gave rise to stomata? What are the molecular bases that explain the diversity of stomatal patterns seen today? Recent discoveries of key genes controlling stomatal development in the model plant Arabidopsis now provide molecular tools with which to tackle these questions. In this review, we first introduce Arabidopsis genes controlling stomatal development. We further describe the evolutionary conservation and uniqueness of stomatal development as well as the functions of orthologous genes in other plant species as an attempt to promote interest and discussion among the diverse disciplines of botany, paleobotany, and evolutionary biology.
STOMATAL DEVELOPMENT IN ARABIDOPSIS
Cell-State Transitions
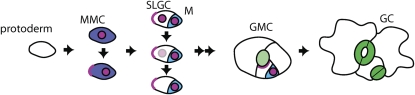
Stomata in Arabidopsis develop through a process of asymmetric cell division (Figure 2) that produces the anisocytic stomatal complexes commonly seen in Brassicaceae, in which each stoma has one smaller and two larger neighboring cells arising from asymmetric divisions within the stomatal lineage (Esau, 1977). First, in the protoderm, a population of cells initiates the stomatal lineage and divides asymmetrically to form a stomatal precursor cell called a meristemoid. The meristemoid reiterates asymmetric divisions several times, usually from one to three, creating sister cells called stomatal lineage ground cells (SLGCs) of decreasing size while renewing the meristemoid state (Figure 2). The meristemoid then differentiates into a guard mother cell (GMC). The GMC divides once symmetrically to form a pair of cells that develop the thickened walls and mature chloroplasts characteristic of stomatal guard cells (GCs). SLGCs eventually become pavement cells but may divide asymmetrically as well, producing a meristemoid away from the original stoma (Figure 2). The stomatal complex may be formed exclusively through multiple asymmetric divisions, in which case it is clonal, or if fewer asymmetric divisions occur the stoma may be adjacent to the lineage of another protodermal cell (Serna et al., 2002).
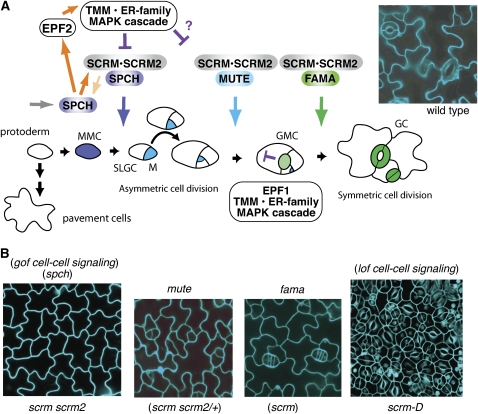
Figure 2.
Stomatal Development in Arabidopsis.
(A) Schematic diagram of stomatal development. The cell states of stomatal precursors are driven by three paralogous bHLH transcription factors, which likely dimerize with SCRM and SCRM2 as a mechanism for coordinated action. Initial specification of the stomatal cell lineage, in which a protodermal cell becomes a meristemoid mother cell (MMC), is controlled by SPCH. Protodermal cells not entering the stomatal lineage differentiate into pavement cells. The MMC divides asymmetrically to form a meristemoid (M) and SLGC and may reiterate similar divisions several times. MUTE controls the cell-state transition from M to GMC, and FAMA is required for correct division of the GMC into GCs forming a functional stoma. It is proposed that a MAP kinase signaling cascade following putative ligands EPF1 and EPF2 (EPF1 expressed in GMC, light green, and EPF2 expressed in MMC, blue, and M, cyan) perceived by TMM and the ER family of RLKs acts to suppress stomatal identity in cells adjacent to developing stomata; new meristemoids can differentiate at least one cell away, as shown near GMC. An image of wild-type epidermis is shown at the top right.
(B) Epidermal phenotypes of stomatal differentiation mutants. Shown are the rosette leaf epidermis of (from left) scrm scrm2, mute, fama, and scrm-D. scrm scrm2 produces epidermis solely composed of pavement cells, a phenotype identical to that of spch as well as gain-of-function mutants in stomatal cell–cell signaling genes. mute and fama produce epidermis with arrested stomatal precursor cells similar to scrm scrm2/+ and scrm, respectively. scrm-D produces epidermis solely composed of stomata, a phenotype similar to loss of function in stomatal signaling genes. Images are reproduced from Kanaoka et al. (2008).
Two groups of basic helix-loop-helix (bHLH) transcription factors regulate the major cell-state transitions through stomatal differentiation (Figure 2). The first group is encoded by the paralogs (closely related genes arising from duplication within a genome) SPEECHLESS (SPCH), MUTE, and FAMA, which operate sequentially to regulate the cellular identity of each step in the developmental process (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007b). SPCH controls the initial asymmetric division with which protodermal cells enter the stomatal lineage; spch mutants are entirely without stomatal lineage cells, having only puzzle piece–shaped pavement cells interlocking over the entire shoot surface (MacAlister et al., 2007; Pillitteri et al., 2007b). Next to act is MUTE, which governs the meristemoid's cessation of asymmetric divisions and its transition to GMC identity. Plants without functional MUTE also develop no stomata, but rather than lacking the stomatal lineage entirely they show rosette-like patterns of inwardly spiraling meristemoid cell divisions with aborted meristemoids in the center (MacAlister et al., 2007; Pillitteri et al., 2007b, 2008). Lastly, FAMA appears to control the switch from the GMC to GC identity; in fama mutants, the stomatal precursors halt at the GMC state, resulting in “caterpillars” of excessively symmetrically divided cells instead of mature GCs from a single symmetric division (the many “false mouths” of the Roman goddess of rumor, Fama) (Ohashi-Ito and Bergmann, 2006).
Regulating the sequential action of the master regulators are the second group of bHLH proteins, encoded by the broadly expressed paralogs SCRM and SCRM2, which have partially overlapping functions. The originally discovered gain-of-function mutation, scrm-D, causes all epidermal cells to take on stomatal fate, while successive loss of SCRM and SCRM2 creates phenotypes recapitulating spch, mute, and fama mutants (Kanaoka et al., 2008). The bHLH proteins function through dimerization (Massari and Murre, 2000), and SCRM and SCRM2 are capable of interacting with SPCH, MUTE, and FAMA in vivo (Figure 2) (Kanaoka et al., 2008). This suggests a possible mode of regulation in which transiently expressed stomatal lineage–specific bHLH proteins heterodimerize with widely expressed bHLH partner proteins to control a developmental progression (Pillitteri and Torii, 2007).
Other factors also act to promote stomatal development. Formation of satellite (secondary) meristemoids is controlled by microRNA miR824, which targets AGAMOUS-LIKE16 (AGL16) mRNA for degradation (Kutter et al., 2007). An AGL16 mRNA that no longer matches miR824 due to silent mutations causes additional reentry of SLGC into the stomatal lineage, resulting in more satellite meristemoids, a phenotype opposite to that of the agl16-1 dosage reduction mutant (Kutter et al., 2007).
Loss of function in a MYB transcription factor, FOUR LIPS (FLP), causes parallel pairs of stomata (Yang and Sack, 1995). Its paralog, At MYB88, is partially redundant with FLP: in flp myb88 double mutants, massive fama-like stacks of cells are frequently formed, although some are able to differentiate terminally into GCs (Lai et al., 2005). It is therefore likely that FLP and MYB88 together restrict the symmetric division of GMC, though unlike FAMA, they do not necessarily specify terminal differentiation into the GC state. Consistent with their redundant roles in GC differentiation, FLP and MYB88 share an amino acid substitution that is unique among known plant MYBs (Lai et al., 2005). The roles of FLP and MYB88 echo the ongoing pattern in which a gene and its paralog have redundant or related functions, which is consistent with evolution through gene duplication.
Patterning and Signaling
A repressive cell–cell signaling pathway in Arabidopsis maintains proper stomatal patterning as defined by the one-cell spacing rule (Figure 2A). Interactions among the partially redundant ERECTA (ER) family of leucine-rich repeat receptor-like kinases (LRR-RLKs) and the LRR receptor-like protein TOO MANY MOUTHS (TMM) affect stomatal spacing and density (Shpak et al., 2005). The function of TMM is organ dependent and complex: in tmm mutants, leaves show clusters of stomata, but stems develop no stomata at all despite the entry of some cells into the stomatal lineage (Bhave et al., 2009). Similarly, the three ER family RLKs exhibit combination-specific interactions with TMM in specific organ types, implying that different combinations of receptor complexes inhibit particular steps of stomatal differentiation. Consistent with the receptor model, two putative ligands of TMM and ER family RLKs, EPIDERMAL PATTERNING FACTOR1 (EPF1) (Hara et al., 2007) and EPF2 (Hara et al., 2009; Hunt and Gray, 2009) restrict the later and the earlier steps of stomatal cell fate specifications, respectively (Figure 2A). The cell–cell signals in turn appear to activate a mitogen-activated protein (MAP) kinase signaling cascade including the MAPKKK YODA (Bergmann et al., 2004), MPKK4/5, and MAPK MPK3/6 (Wang et al., 2007), to suppress stomatal development in neighboring cells. In general, MAP kinase cascades target transcription factors and regulate their activities via phosphorylation. Recently, SPCH was shown to be a MAP kinase target, thus providing a mechanism by which stomatal suppression signaling might affect stomatal cell state identity directly (Lampard et al., 2008). Recently, additional signaling ligands have been discovered: STOMAGEN/EPF-LIKE9 (EPFL9), which promotes stomatal differentiation through TMM, and CHALLAH/EPFL6, which plays a role in tissue-specific stomatal patterning (Abrash and Bergmann, 2010; Kondo et al., 2010; Sugano et al., 2010). These additional players highlight the roles of the EPF family of signaling ligands in various aspects of stomatal development and may clarify the complex actions of TMM/ER family RLKs.
STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) also acts as a negative regulator of stomatal formation (Berger and Altmann, 2000), but although it has been shown to require TMM for function (von Groll et al., 2002), it seems to be independent of EPF1 (Hara et al., 2007), EPF2 (Hara et al., 2009; Hunt and Gray, 2009), STOMAGEN/EPFL9 (Kondo et al., 2010; Sugano et al., 2010), and CHALLAH/EPFL6 (Abrash and Bergmann, 2010). Although SDD1 encodes a membrane-associated subtilisin-type protease (Berger and Altmann, 2000), it appears unlikely that it directly processes any known member of the EPF family of stomatal ligands.
Innate Polarity
Asymmetric cell division within the stomatal lineage is highly polar: The meristemoid divides at angles of exactly 60° from each previous division, forming an orderly inward spiral of SLGCs with a triangular meristemoid inside (Serna et al., 2002; Pillitteri et al., 2007b). When spacing divisions occur, the satellite meristemoids always divide away from any neighboring stoma to maintain the one-cell spacing rule. While cell–cell signaling and bHLH transcription factors determining the state of asymmetrically dividing stomatal precursors are well characterized, factors directing the innate polarity that allows spacing to emerge from the asymmetric cell division process have remained elusive, until the recent discovery of a unique protein, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) (Dong et al., 2009). The BASL protein fused with green fluorescent protein (GFP) is localized to the nuclei of dividing stomatal lineage cells and also to the cell cortex opposite the site of the most recent asymmetric division, which appears to be BASL's main site of activity (Dong et al., 2009) (Figure 3). The ectopic overexpression of BASL confers abnormal cell polarization. Interestingly, the loss of asymmetry in a basl mutant resulted in adoption of the stomatal fate, as depicted by the accumulation of a MUTE:nuclear GFP reporter fusion protein in both daughter cells (Dong et al., 2009). This observation implies that the asymmetric divisions in stomatal lineage cells are necessary for specifying the fate of the SLGC, but not the meristemoid, and a rapid cell polarization via a BASL-mediated mechanism is required to do so. This basl phenotype is the opposite of tmm er, where both daughter cells of a meristemoid adopt the SLGC fate (Shpak et al., 2005). While genetic studies suggest that BASL operates in a separate pathway from the TMM/ER family (Dong et al., 2009), the latter is required for proper positioning of GFP-BASL. Understanding the mechanism that achieves a peripheral localization of BASL may illuminate the intersection of signaling and innate asymmetry.
Figure 3.
Localization of BASL.
BASL acts as a molecular signal instructing stomatal lineage cells to divide away from it. BASL protein appears initially in the nuclei of meristemoid mother cells (MMC), which differentiate from protodermal cells. The protein then localizes in a second location at the cell periphery opposite the site of the future asymmetric division. Following that division, BASL remains at the cell periphery but fades away from the nucleus of the larger daughter cell (SLGC), which loses stomatal lineage identity; it remains in the nucleus of the meristemoid (M), which may further asymmetrically divide. BASL is not found in later stomatal lineage cells, such as (GMCs or GCs. However, when satellite meristemoids are formed by SLGCs that resume stomatal lineage fate, BASL appears at the SLGC periphery next to the stomatal lineage cell, providing a mechanism for maintenance of the one-cell spacing rule. (Based on data presented in Dong et al. [2009].)
Stomatal Patterning and Differentiation in Grasses
In grasses, stomatal complexes develop from nonclonal cells through a polarized division process (Figure 4). The cells are organized in cell files throughout the plant, which are visible on the macro scale as the typical parallel venation of the monocot (Figure 1F). Spacing is maintained by the specification of nonadjacent cell files to develop stomata, which begin with an asymmetric division toward the leaf apex for all cells in the file. One of the cells produced by the asymmetric division differentiates directly into a GMC, which before dividing again signals the neighboring cells in adjacent cell files to divide asymmetrically toward the GMC. These divisions produce subsidiary cells (SCs) important for stomatal action (Figure 4). Subsequently, the GMC makes a single symmetric division and differentiates two GCs in a distinctive dumbbell shape (Gallagher and Smith, 2000). GCs with this shape require less water to open than those with a kidney shape (Raschke, 1979), so they may be an adaptation to drought conditions.
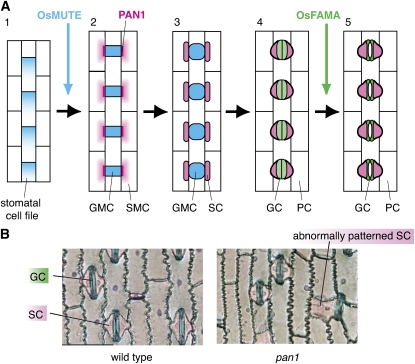
Figure 4.
Stomatal Development in Grasses.
(A) Schematic diagram of stomatal development in grasses. (1) During early epidermal development in grasses, stomatal and nonstomatal cell files are specified, and cell division polarity is established in the stomatal cell file. This polarity will ensure that the one-cell spacing rule is maintained. (2) Os MUTE controls a single asymmetric division toward the leaf apex in the stomatal cell file, which creates GMCs (blue). Neighboring cell files (SMC; pink gradient) receive a signal via putative receptor PAN1 (magenta), which localizes at the area of GMC contact and polarize in preparation for division. (3) SMCs divide asymmetrically toward PAN1 to form SCs (pink), which will act as ion reservoirs for the operation of mature stomata. (4) GMCs divide once symmetrically to form GCs (light green). (5) Finally, GC and SC terminally differentiate, forming mature dumbbell-shaped stomata (dark green). Os FAMA is required for the differentiation of GCs, though not their symmetric division.
(B) Leaf epidermal peel from the wild type (left) and pan1 mutant maize (right). GC and SC are stained in blue and pink, respectively. Unlike the wild type, the pan1 mutant occasionally fails to produce proper asymmetric divisions that give rise to SC, resulting in abnormal SC patterning. Images kindly provided by Laurie Smith (University of California, San Diego).
The recent availability of genome sequences and reverse-genetics tools for model monocot plants, rice (Oryza sativa) and maize (Zea mays), allows for the examination of the conservation of genes important in stomatal patterning and differentiation. Grasses contain orthologs (genes in different species that evolved from a common ancestor) of the master regulatory genes of Arabidopsis stomatal differentiation, SPCH, MUTE, and FAMA (Liu et al., 2009). Although the transcripts of the two Os SPCH genes were detected only in coleoptiles, one of them showed a decrease in entry into the stomatal lineage when mutated (Liu et al., 2009). This suggests that in rice, at least one of the SPCH genes controls initial cell lineage divisions; it may have additional functions that are redundant with the second Os SPCH, in which a mutation has not yet been found. Os MUTE and maize Zm MUTE were expressed earlier in development than their Arabidopsis ortholog, at the time of cell file specification. Ectopic overexpression of Os MUTE and Zm MUTE in Arabidopsis caused conversion of epidermal cells into stomata to varying degrees, a phenotype similar to that of At MUTE overexpression (Liu et al., 2009), and they were able to partially rescue the Arabidopsis mute mutant when expressed under the native Arabidopsis promoter (Liu et al., 2009). In mutants of Os FAMA, GCs fail to differentiate into the dumbbell shape of wild-type GCs, implying that Os FAMA controls GC fate, though there are none of the extra cell divisions seen in Arabidopsis fama plants (Liu et al., 2009).
This series of studies provides interesting implications. First, the SPCH-MUTE-FAMA bHLH genes diverged before the evolutionary split between monocots and dicots. Second, they have maintained their overall functions as regulators of stomatal differentiation across these widely separated taxa. Third, the orthologs have differences in their specific functions that reflect the differences in the specifics of stomatal ontogeny between dicots (Arabidopsis) and monocots (grasses). For instance, while both At fama and Os fama mutants fail to differentiate mature GCs, only the At fama mutant forms characteristic tumors of supernumerary GMC-like cells (Ohashi-Ito and Bergmann, 2006; Liu et al., 2009). These differences could be reconciled if we assume that, in Arabidopsis, a transition from a proliferative state to terminal differentiation of stomata requires a strong brake to halt cell division machinery. There is no actively dividing meristemoid state during stomatal differentiation in grasses; consequently, Os FAMA may not have acquired (or retained) the specific function to restrict cell division. The role of Os MUTE earlier in development, relative to At MUTE, may also suggest that these bHLH proteins acquired specific functions reflecting the uniqueness of stomatal development in grasses.
Formation of stomatal complexes in grasses (Figure 4) involves asymmetric cell divisions in subsidiary mother cells (SMCs), members of GMC-neighboring cell files that give rise to SCs, which have a highly specialized function as ion reserves for GC movement. Such an asymmetric cell division outside the stomatal lineage does not exist in Arabidopsis. Therefore, it is of great interest to unravel the molecular identities of the genes controlling subsidiary cell division and polarity in grasses. Loss-of-function mutations in PANGLOSS1 (PAN1) lead to aberrant asymmetric cell division and SC patterning (Gallagher and Smith, 2000). Recently, PAN1 was cloned and shown to encode an atypical group III LRR-RLK without kinase activity (Cartwright et al., 2009). It follows that PAN1 might associate with other RLKs with functional kinase domain to transmit signals. In Arabidopsis, an atypical LRR-RLK without kinase activity is known to regulate developmental processes (Chevalier et al., 2005). Interestingly, PAN1 protein is localized specifically at the plasma membrane of the SMC at the sites of contact with GMC. This is consistent with the hypothesis that PAN1 receives GMC signals to regulate the site of cytokinesis. Such polar localization has not been observed for the Arabidopsis stomatal LRR receptor TMM (Nadeau and Sack, 2002), and no member of LRR group III in Arabidopsis has been found to affect stomatal development. Thus, PAN1's function may highlight the unique regulation of stomatal patterning in grasses. Maize PAN2 is required for proper subcellular localization of PAN1 (Cartwright et al., 2009). The molecular identity of PAN2 is not known, so we cannot speculate yet on whether this protein and its function may be unique to grasses.
Diversity of Stomata and Stomatal Development
In extant land plant species, stomata appear on the sporophyte shoot epidermis of nearly every lineage. For most plants, stomata are evenly distributed in the shoot areas where they appear, and they adhere closely to the one-cell spacing rule (Sachs, 1991), allowing them to open and close effectively. Therefore, the diversity of stomata consists of variations on a theme: stomata are spaced at least one cell apart, but the specifics differ. Classic anatomical studies suggest that differences in stomatal patterning reflect the types, positions, and numbers of cell divisions that give rise to the stomatal complex (Esau, 1977; Payne, 1979). In this section, we describe stomatal development in other plant taxa and speculate regarding their possible alterations of regulatory mechanisms known in Arabidopsis.
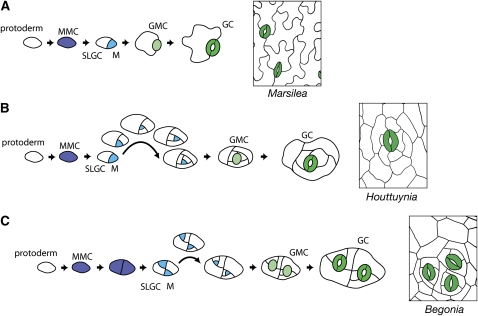
In mosses and lycophytes (Figures 1A and 1B), stomata develop through a simple process of a single asymmetric division followed by differentiation of a GMC, then GCs (Payne, 1979). We could speculate that the ancient function of SPCH-MUTE-FAMA family proteins may be to initiate the stomatal cell lineage and differentiate into GMC directly. In ferns, MMC in a cell file undergo one, or sometimes two, asymmetric division toward the leaf apex, then differentiate into GMC and GC (Apostolakos et al., 1997) (Figure 5A). This implies cell polarity mechanisms different from Arabidopsis.
Figure 5.
Developmental Hypotheses for Stomatal Complex Diversity.
(A) Stomata in the fern Marsilea appear to develop through a process lacking amplifying divisions of a meristemoid (cf. Arabidopsis in Figure 2A).
(B) Houttuynia (magnoliid) stomata are surrounded by a spiral arrangement of cells that suggests a large number of amplifying divisions.
(C) In Begonia, a eudicot, stomata arise in groups that can be explained by early division of a stomatal precursor, such as an MMC, and retention of MMC identity by the daughter cells.
In basal angiosperms, such as Victoria (Figure 1D), stomata appear to arise without amplifying divisions. (For terminology and evolution of stomatal complexes among basal angiosperms, see Carpenter [2005].) By contrast, the magnoliid Houttuynia shows a clear and extended inward spiral of cells around each stoma, which we hypothesize arise through an extended process of asymmetric division resulting from delayed MUTE activity (Figure 5B).
Gardenia and Begonia (Figures 1G and 1H) show some of the diversity found within eudicots, a category that also includes Arabidopsis (Figure 2). In Gardenia, it appears that most, if not all, protodermal cells enter the stomatal lineage, implying that the unknown mechanism by which MMCs arise operates at higher density than in Arabidopsis. In Begonia, small groups of stomata are separated by pavement cells. Figure 5C shows a hypothetical developmental process giving rise to such a stomatal complex: Instead of dividing asymmetrically, the MMC divides symmetrically one or more times and all daughter cells retain MMC identity. Asymmetric division then proceeds as in Arabidopsis, allowing the one-cell spacing rule to be maintained, and the mature Begonia epidermis has groups of up to four stomata (Figure 5C).
Phylogenetic Perspective on Stomata Development
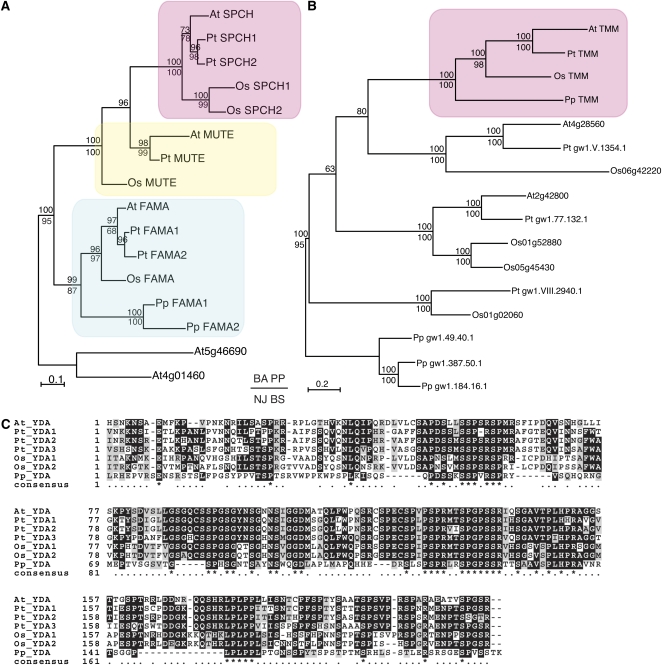
The recent availability of diverse land plant genomes, such as the moss Physcomitrella patens and model crop monocot plants rice and maize, allows for examination of the origin and conservation of genes important in stomatal patterning and spacing. Since stomata evolved only once in the ancestor of the mosses (Raven, 2002), it is expected that plants that arose from different basal land plant lineages, such as P. patens, would also possess some if not all of the genes found to be essential for stomatal patterning and differentiation in Arabidopsis. Stomata in moss form through a single asymmetric cell division to produce a GMC, which will divide partially or completely to produce a single GC or a pair of GCs surrounding a pore (Payne, 1979). Congruent with this relatively simplified developmental sequence to mature GC formation, the moss P. patens does not have genes encoding SPCH or MUTE. P. patens, however, has two FAMA-like genes (Figure 6A; see Supplemental Data Set 1 online), which likely function in moss as they do in Arabidopsis, rice, and maize: to control the final differentiation step to GCs. Based on our SPCH-MUTE-FAMA gene tree and taxonomic sample, it is not clear whether there was only a single stomatal master regulatory bHLH gene in the ancestor of land plants or if SPCH and MUTE have been subsequently lost in P. patens. It is fascinating, however, to speculate that a single stomatal bHLH protein in early land plants may have had a function to create stomatal-lineage cells and immediately trigger GC differentiation. Later, as this gene duplicated, its roles may have subfunctionalized and even acquired new functions to mediate amplifying and spacing asymmetric cell divisions. The evolution of the leaf blade (Figure 1) in vascular plants perhaps benefited from additional mechanisms to space stomata correctly during leaf laminar expansion, such as amplifying divisions of meristemoids and recruiting cell–cell signaling components to impinge directly on the stomatal differentiation process. In this regard, it is interesting that At SPCH possesses a specific MAP kinase target domain not found in At FAMA (Lampard et al., 2008).
Figure 6.
Stomatal Patterning and Spacing Genes Are Conserved among Embryophytes.
(A) A gene tree of master regulatory bHLH transcription factors (SPCH, MUTE, and FAMA) in stomatal development. Amino acid sequences from bHLH and ACT domains were aligned and the tree constructed using Bayesian and neighbor-joining methods. The Bayesian phylogram is shown. The pink, yellow, and cyan rectangles highlight the SPCH, MUTE, and FAMA clades, respectively. Posterior probability values are indicated above the nodes, and bootstrap values over 50 (100,000 replicates) are indicated below the nodes.
(B) A gene tree of TMM, which encodes an LRR receptor-like protein necessary for proper stomatal spacing in Arabidopsis. The entire amino acid sequence was aligned and the tree constructed as in (A). The Bayesian phylogram is shown. The pink rectangle highlights the single copy TMM clade. Posterior probability values are indicated above the nodes, and bootstrap values over 50 (100,000 replicates) are indicated below the nodes.
(C) Amino acid alignment of the N-terminal portion of YODA (YDA), a MAPKKK, which is essential to regulate the activity of the protein in Arabidopsis. YDA is also required for appropriate stomatal spacing. At, Arabidopsis thaliana; Pt, Populus trichocarpa; Os, Oryza sativa; Pp, Physcomitrella patens. See Supplemental Methods and Supplemental Table 1 online for detailed bioinformatic and phylogenetic methods and gene ID numbers.
Land plants also share common signaling components for the spacing of stomata. TMM, a member of the LRR-RLP family, functions as part of a signaling complex that regulates the spacing of stomata in Arabidopsis (Nadeau and Sack, 2002). TMM is present in a single copy in poplar, grass, and moss genomes, which may reflect its integral role in enforcing correct stomatal spacing (Figure 6B; see Supplemental Data Set 2 online). In Arabidopsis, TMM is proposed to associate with ER family receptors, which are Ser/Thr kinases (Shpak et al., 2005). TMM is specific to the stomatal lineage, whereas ER family members play additional roles in various developmental processes in Arabidopsis (Shpak et al., 2004; Pillitteri et al., 2007a; Hord et al., 2008; van Zanten et al., 2009, 2010). Thus, TMM may be necessary for recognition of ligands or receptor complex activation specific to the stomatal lineage in all land plants.
YODA is a MAPKKK thought to act downstream of TMM and is present in multiple copies in rice and poplar and in single copy in Arabidopsis and P. patens (Figure 6C). Although MAPKKKs comprise a large gene family, YODA can be identified by its unique N-terminal Ser-rich regulatory region (see Supplemental Data Set 3 online). Closely related MKKKs in Arabidopsis appear to be lacking this region (see Supplemental Data Set 3 online), and if it is deleted in YODA, the protein becomes constitutively active, resulting in a stomata-less phenotype (Lukowitz et al., 2004). Given the high level of sequence conservation in the region of this protein in P. patens relative to Arabidopsis, we predict that the N-terminal region regulates its YODA activity as well. Like ER family RLKs, YODA regulates multiple developmental processes in Arabidopsis in addition to stomatal patterning, such as embryo polarity/patterning, inflorescence elongation, and flower development (Lukowitz et al., 2004). Therefore, it would be interesting to see whether YODA might also be an essential component in the regulation of stomatal spacing in moss.
Although close anatomical descriptions of stomatal development exist for many taxa, little is known of the molecular and evolutionary basis of stomatal formation across land plants. Valuable insights into ancestral and conserved mechanisms of stomatal development will no doubt be gained from further studies on basal land plants. In the moss sporophyte, stomatal development is relatively more similar to that of grasses than that of eudicots, in that one asymmetric division gives rise to a GMC directly, which then divides symmetrically once to form paired GCs (Ziegler, 1987). However, unlike grasses, there are no subsidiary cells formed from outside the stomatal lineage. To reach a deeper understanding of novel mechanisms responsible for generating stomatal diversity (Figure 1), comparative studies of closely related, yet morphologically divergent, stomatal phenotypes are needed.
Future Perspectives
With recent advances in understanding the molecular processes controlling stomatal development in the model eudicot Arabidopsis, alongside the completion of several genome projects in monocot crop plants and in basal land plants, we are beginning to find that many of the molecular mechanisms that control stomatal development in Arabidopsis are part of an ancient machinery. As shown by PAN1 in maize and BASL in Arabidopsis, some of the unique aspects of stomatal development relate to the type and number of asymmetric cell divisions associated with the formation of the stomatal complex. Further identification of orthologs of stomatal regulator genes in phylogenetically and evolutionarily important species and analysis of their developmental functions may unravel the molecular basis of the evolution of stomatal development.
Several exciting questions remain: For instance, how is the stomatal lineage first established? What initiates SPCH expression, and what are the cellular dynamics that set up the entry into asymmetric cell divisions of the stomatal lineage? When did amplifying cell divisions come to play a key role in stomatal development, and is that innovation correlated with specific features of MUTE?
A second line of questions revolves around the intersection of environmental signaling and stomatal development. Some recent work is beginning to explain how light signals integrate with the stomatal development program (Boccalandro et al., 2009; Casson et al., 2009; Kang et al., 2009), and other work is examining how CO2 regulates stomatal density (Gray et al., 2000). In addition, while a direct role between temperature and stomatal patterning has not been determined, SCRM, which is also known as ICE1, is an important transcriptional activator of cold response genes (Chinnusamy et al., 2003). During the evolution of land plants, when and how did such network connections of environmental signaling pathways to stomatal development emerge? Clearly, we now have the molecular-genetic tools in hand to explore the evolution of this landmark developmental innovation.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Gene Names and IDs Used for the Phylogenetic Analysis.
Supplemental Data Set 1. Text File of Amino Acid Alignment of the bHLH Domain and the C-Terminal ACT-Like Domain of the SPCH, MUTE, and FAMA Orthologs Used for the Phylogenetic Analysis Presented in Figure 6A.
Supplemental Data Set 2. Text File of Amino Acid Alignment of the Full-Length At TMM and Its Orthologs Used for the Phylogenetic Analysis Presented in Figure 6B.
Supplemental Data Set 3. Text File of Amino Acid Alignment of the N-Terminal End of YODA and Related Genes Presented in Figure 6C.
Supplemental Methods.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Laurie Smith (University of California, San Diego) for the images of maize wild-type and pan1 epidermis (Figure 4B) and Lynn Pillitteri for the scanning electron microscopy image of P. patens stomata (Figure 1A). Our research is supported by grants from the National Science Foundation (IOB-0744892 and MCB-0855659), by the U.S. Department of Energy (DE-FG02-03ER15448), and by PREST, Japan Science and Technology Agency, to K.U.T. K.M.P. is a National Science Foundation Graduate Research Fellow.
References
- Abrash E.B., Bergmann D.C. (2010). Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 [DOI] [PubMed] [Google Scholar]
- Apostolakos P., Panteris E., Galatis B. (1997). Microtubule and actin filament organization during stomatal morphogenesis in the fern Asplenium nidus. 1. Guard cell mother cell. Protoplasma 198: 93–106 [DOI] [PubMed] [Google Scholar]
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bhave N.S., Veley K.M., Nadeau J.A., Lucas J.R., Bhave S.L., Sack F.D. (2009). TOO MANY MOUTHS promotes cell fate progression in stomatal development of Arabidopsis stems. Planta 229: 357–367 [DOI] [PubMed] [Google Scholar]
- Boccalandro H.E., Rugnone M.L., Moreno J.E., Ploschuk E.L., Serna L., Yanovsky M.J., Casal J.J. (2009). Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiol. 150: 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K.J. (2005). Stomatal architecture and evolution in basal angiosperms. Am. J. Bot. 92: 1595–1615 [DOI] [PubMed] [Google Scholar]
- Cartwright H.N., Humphries J.A., Smith L.G. (2009). PAN1: A receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323: 649–651 [DOI] [PubMed] [Google Scholar]
- Casson S.A., Franklin K.A., Gray J.E., Grierson C.S., Whitelam G.C., Hetherington A.M. (2009). Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol. 19: 229–234 [DOI] [PubMed] [Google Scholar]
- Chevalier D., Batoux M., Fulton L., Pfister K., Yadav R.K., Schellenberg M., Schneitz K. (2005). STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 9074–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D., Kerp H., Haas H. (1998). Stomata in early land plants: An anatomical and ecophysiological approach. J. Exp. Bot. 49: 255–278 [Google Scholar]
- Esau K. (1977). Stomata. Anatomy of Seed Plants (New York: John Wiley & Sons; ), pp. 88–99 [Google Scholar]
- Freeman S. (2008). Green plants. Biological Sciences, 3rd ed (San Francisco, CA: Pearson Education; ), pp. 626–663 [Google Scholar]
- Gallagher K., Smith L.G. (2000). Roles for polarity and nuclear determinants in specifying daughter cell fates after an asymmetric cell division in the maize leaf. Curr. Biol. 10: 1229–1232 [DOI] [PubMed] [Google Scholar]
- Gray J.E., Holroyd G.H., van der Lee F.M., Bahrami A.R., Sijmons P.C., Woodward F.I., Schuch W., Hetherington A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is auto-regulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hord C.L.H., Sun Y.J., Pillitteri L.J., Torii K.U., Wang H., Zhang S., Ma H. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.Y., Lian H.L., Wang F.F., Huang J.R., Yagn H.Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., Masuda Y., Irie K., Tanaka Y., Takada S., Kakimoto T., Sakagami Y. (2010). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51: 1–8 [DOI] [PubMed] [Google Scholar]
- Kutter C., Schob H., Stadler M., Meins F., Jr., Si-Ammour A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19: 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L.B., Nadeau J.A., Lucas J., Lee E.K., Nakagawa T., Zhao L., Geisler M., Sack F.D. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17: 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K., Bergmann D.C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Massari M.E., Murre C. (2000). Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne W.W. (1979). Stomatal patterns in embryophytes - Their evolution, ontogeny and interpretation. Taxon 28: 117–132 [Google Scholar]
- Pillitteri L.J., Bemis S.M., Shpak E.D., Torii K.U. (2007a). Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in ovule development. Development 134: 3099–3109 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Bogenschutz N.L., Torii K.U. (2008). The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 49: 934–943 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007b). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2007). Breaking the silence: Three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays 29: 861–870 [DOI] [PubMed] [Google Scholar]
- Raschke K. (1979). Movements of stomata. Physiology of Movements, Encyclopedia of Plant Physiology, Vol. 7, Haupt W., Feinleib M.E., (Berlin: Springer-Verlag; ), pp. 383–441 [Google Scholar]
- Raven J. (2002). Selection pressures on stomatal evolution. New Phytol. 153: 371–386 [DOI] [PubMed] [Google Scholar]
- Sachs T. (1991). Stomata as an example of meristmoid development. Pattern Formation in Plant Tissues (Cambridge, UK: Cambridge University Press; ), pp. 101–117 [Google Scholar]
- Serna L., Torres-Contreras J., Fenoll C. (2002). Clonal analysis of stomatal development and patterning in Arabidopsis leaves. Dev. Biol. 241: 24–33 [DOI] [PubMed] [Google Scholar]
- Shpak E.D., Berthiaume C.T., Hill E.J., Torii K.U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Sugano S.S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Taylor T.N., Kerp H., Hass H. (2005). Life history biology of early land plants: Deciphering the gametophyte phase. Proc. Natl. Acad. Sci. USA 102: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Basten Snoek L., van Eck-Stouten E., Proveniers M.C., Torii K.U., Voesenek L.A., Peeters A.J., Millenaar F.F. (2010). Ethylene-induced hyponastic growth in Arabidopsis thaliana is controlled by ERECTA. Plant J. 61: 83–95 [DOI] [PubMed] [Google Scholar]
- van Zanten M., Snoek L.B., Proveniers M.C., Peeters A.J. (2009). The many functions of ERECTA. Trends Plant Sci. 14: 214–218 [DOI] [PubMed] [Google Scholar]
- von Groll U., Berger D., Altmann T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Sack F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler H. (1987). The evolution of stomata. Stomatal Function, Zeiger E., Farquhar G.D., Cowan I.R., (Stanford, CA: Stanford University Press; ), pp. 29–57 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.