Abstract
The smut genus Thecaphora contains plant parasitic microfungi that typically infect very specific plant organs. In this study, we describe a new species of Thecaphora from Oxalis lanata var. rosea (Oxalidaceae) in the Cape Floristic Region of South Africa. Molecular phylogenetic reconstructions based on large subunit ribosomal DNA sequence data confirmed the generic placement of the fungus and confirmed that it represents an undescribed species for which the name T. capensis sp. nov. is provided. The closest known sister species of the new taxon is T. oxalidis that infects the fruits of Oxalis spp. in Europe, Asia and the Americas. In contrast, T. capensis produces teliospores within the anthers of its host. This is the first documented case of an anther-smut from an African species of Oxalis and the first Thecaphora species described from Africa.
Keywords: anther-smut, Cape Floristic Region, Oxalis, phylogeny, Thecaphora
INTRODUCTION
The smut genus Thecaphora resides in the Glomosporiaceae (Bauer et al. 2001, Vánky et al. 2008) and includes c. 60 described species. Species of Thecaphora produce sori within various plant organs including seeds, flowers, leaves, stems and roots. The host range of these fungi is broad and includes various monocotyledonous and dicotyledonous families (Vánky et al. 2008). Only a single species, T. oxalidis is known on members of the Oxalidaceae. Hosts of T. oxalidis include the species Oxalis corniculata, O. dillenii, O. fontana and O. stricta (all in sect. Corniculatae) and O. laxa (sect. Alpinae) (Lourteig 1994, 1995, 2000) from Europe, Asia and the Americas.
Oxalis is the largest genus within the Oxalidaceae, and the c. 500 included species are concentrated mainly in South and Central America and southern Africa (Salter 1944, Lourteig 1994, 1995, 2000). The New World represents the larger centre of species diversity (c. 250 species) for Oxalis (Lourteig 1994, 1995, 2000), where the plants display diverse growth forms including geophytes (underground storage organ), annuals, stem succulent perennials and small trees. The majority of southern African species are confined to the winter rainfall region of the Western Cape Province (Oberlander et al. 2002). This area, known as the Cape Floristic Region (CFR, Goldblatt & Manning 2000), displays an exceptionally rich floristic diversity, and is considered as one of six global Floral Kingdoms (Good 1947, Takhtajan 1986). Oxalis is the seventh largest genus in the CFR (Goldblatt & Manning 2000) and the largest bulbous genus in the region. The CFR Oxalis spp. flower during the wet winter months (April to August), and escape the drier summer months below ground, only to emerge again at the onset of the next rainy season (Dreyer et al. 2006).
Thecaphora oxalidis is known only from Europe, Asia and the Americas. Although this species is well defined, its generic placement has been problematic. When it was first described, the fungus was placed in Ustilago based on morphology (Ellis & Tracy 1890). More recent advances in fungal taxonomy have shown that Ustilago includes only species associated with members of the plant family Poaceae (Bauer et al. 1997, 2001, Vánky 1999). Members of Ustilago associated with other host families were thus transferred to new genera (e.g. Bauerago for species associated with Cyperaceae; Vankya for species associated with the Liliaceae; and Microbotryum for species with violet spores commonly associated with the Caryophyllaceae) (Ershad 2000, Vánky 1998, 1999). Ustilago oxalidis was transferred to the Glomosporiaceae (Bauer et al. 2001) in the monotypic genus Kochmania (Piątek 2005). More recently, ultrastructural and DNA sequence data showed that Kochmania resides in the genus Thecaphora (Vánky et al. 2008). This taxonomic placement is followed in the present study.
The reddish brown teliospore masses of T. oxalidis are formed within the seeds of its hosts, while the anamorph stage resides within host anthers (Ellis & Tracy 1890). We are unaware of any described Thecaphora sp. that forms teliospores in the anthers of its hosts. During recent surveys, a smut fungus infecting the anthers of Oxalis lanata var. rosea in the CFR of South Africa was discovered (Fig. 2). The aim of this study was to identify the fungus and to consider its taxonomic placement based on morphology and phylogenetic reconstructions obtained from large subunit ribosomal DNA gene sequence data (LSU).
Fig. 2.
Light and electron micrographs of Thecaphora capensis and T. oxalidis on Oxalis sp. a. Healthy O. lanata var. rosea flower; b. Oxalis lanata flower showing mass of T. capensis teliospores replacing pollen in anthers; c. close-up of healthy anthers (petals removed); d. close-up of infected anthers (petals removed); e. light micrograph of T. capensis teliospores mounted in lactophenol; f, g. scanning electron micrographs of T. capensis teliospores; h. light micrograph of T. oxalidis teliospores mounted in lactophenol; i, j. scanning electron micrographs of T. oxalidis teliospores. — Scale bars = 10 μm.
MATERIALS AND METHODS
Specimens
Individuals of O. lanata var. rosea infected with an unidentified smut fungus were collected from the Jonkershoek Forestry Reserve (Assegaaibos area), Stellenbosch, South Africa during the course of botanical surveys in July and August 2007. Infections of anther smut were found on four specimens of O. lanata var. rosea. To obtain fresh material for analysis, whole plants were collected, potted and maintained under nursery conditions (reference number MO211) in the Stellenbosch Botanical Garden, University of Stellenbosch, Stellenbosch, South Africa. Herbarium specimens of the teliospores of the unknown fungus were deposited in the herbarium of the National Collection of Fungi, Pretoria, South Africa (PREM) and Herbarium Ustilaginales Vánky (HUV), Tübingen, Germany (Table 1).
Table 1.
List of isolates and LSU sequences used in this phylogenetic study with GenBank accession numbers. Isolates obtained in the present study are indicated in bold.
| Species | Vouchera | GenBank acc. no. | Reference |
|---|---|---|---|
| Doassansiopsis deformans | MP 2066 | AF009849 | Begerow et al. 1997 |
| Sporisorium sorghi | MP 2036a | AF009872 | Begerow et al. 1997 |
| Thecaphora affinis | TUB 015855 | EF647747 | Vánky et al. 2008 |
| T. alsinearum | HUV 10535 | EF200057 | Vánky & Lutz 2007 |
| HUV 11533 | EF200058 | Vánky & Lutz 2007 | |
| T. amaranthi | HUV 15882 | AF009873 | Begerow et al. 1997 |
| HUV 20727 | EF200038 | Vánky & Lutz 2007 | |
| T. capensis | HUV 21531 | ||
| PREM 60075 | EU660478 | This study | |
| PREM 60076 | EU660479 | ||
| PREM 60077 | EU660480 | ||
| PREM 60078 | EU660481 | ||
| T. haumanii | HUV 19965 | EF647749 | Vánky et al. 2008 |
| T. hedysari | HUV 13620 | EF647750 | Vánky et al. 2008 |
| T. hennenea | HUV 14434 | EF200039 | Vánky & Lutz 2007 |
| T. italica | HUV 20344 | EF200050 | Vánky & Lutz 2007 |
| HUV 20345 | EF200051 | Vánky & Lutz 2007 | |
| T. lathyri | HUV 11020 | EF647748 | Vánky et al. 2008 |
| T. leptidium | HUV 5916 | EF647745 | Vánky et al. 2008 |
| T. melandrii | HUV 13273 | EF200048 | Vánky & Lutz 2007 |
| HUV 12677 | EF200049 | Vánky & Lutz 2007 | |
| T. oxalidis | TUB 015854 | EF647746 | Vánky et al. 2008 |
| T. polymniae | HUV 17240 | EF647751 | Vánky et al. 2008 |
| T. saponariae | HUV 15015 | EF200042 | Vánky & Lutz 2007 |
| TUB 012794 | EF200041 | Vánky & Lutz 2007 | |
| T. schwartzmaniana | HUV 21117 | EF647752 | Vánky et al. 2008 |
| T. seminis-convolvuli | GD 1391 | AF009874 | Begerow et al. 1997 |
| T. solani | TS 5 | AY344049 | Andrade et al. unpubl. |
| TS 22 | AY344054 | Andrade et al. unpubl. | |
| T. spilanthis | HUV 21043 | EF647753 | Vánky et al. 2008 |
| AFTOL 1913 | DQ832241 | Matheny et al. 2006 | |
| T. thlaspeos | TUB 015857 | EF647754 | Vánky et al. 2008 |
| Urocystis colchici | AFTOL 1647 | DQ838576 | Matheny et al. 2006 |
a Acronyms: AFTOL = Assembling the Fungal Tree Of Life, http://aftol.org; GD = G. Dem; HUV = Herbarium Ustilaginales Vánky, Tübingen, Germany; MP = M. Piepenbring; TS = Instituto de Investigaciones Agropecuarias, Centro Regional de Investigación Carillanca, Chile; TUB = Herbarium of the Spezielle Botanik/Mykologie, Eberhard-Karls-Universität Tübingen, Germany; PREM = National Collection of Fungi, Pretoria, South Africa.
DNA phylogeny
Teliospores of the unknown fungus did not germinate on artificial media and DNA isolations were made directly from naturally infected tissue. Genomic DNA was extracted from fungal teliospores using a Sigma GenElute™ plant genomic DNA miniprep kit (Sigma-Aldrich Chemie CMBH, Steinheim, Germany) according to the manufacturer’s protocol. The primers LROR and LR5 (www.biology.duke.edu/fungi/mycolab/primers.htm) were used to amplify the nuclear LSU rDNA gene region. PCR reaction volumes (50 μL) consisted of: 32.5 μL ddH2O, 1 μL DNA, 5 μL (10×) reaction buffer (Super-Therm, JMR Holdings, USA), 5 μL MgCl2, 5 μL dNTP (10 mM of each nucleotide), 0.5 μL (10 mM) of each primer and 0.5 μL Super-Therm Taq polymerase (JMR Holdings, USA). DNA fragments were amplified using a Gene Amp®, PCR System 2700 thermal cycler (Applied Biosystems, Foster City, USA). PCR reaction conditions were: an initial denaturation step of 2 min at 95 °C followed by 35 cycles of 30 s denaturation at 95 °C, 30 s annealing at 55 °C and 1 min elongation at 72 °C. The PCR process terminated with a final elongation step of 8 min at 72 °C. Amplified PCR products were cleaned using the Wizard® SV gel and PCR clean-up system (Promega, Madison, Wisconsin, USA) following the manufacturer’s protocols. Purified fragments were sequenced using a Big Dye™ Terminator v. 3.0 cycle sequencing premix kit (Applied Biosystems). The fragments were analysed on an ABI PRISIM™ 3100 Genetic Analyser (Applied Biosystems).
The sequence data obtained were compared with accessions acquired from the NCBI’s GenBank nucleotide database (www.ncbi.nlm.nih.gov) using a parsimony, likelihood and Bayesian approach (Table 1). The species Doassansiopsis deformans, Sporisorium sorghi and Urocystis colchici were chosen as outgroup based on results of previous analyses (Vánky et al. 2008). Sequences were automatically aligned using the Clustal X (1.81) software package. For parsimony, a heuristic search (5 000 random addition sequence replicates) using the Phylogenetic Analysis Using Parsimony (PAUP), v. 4.0 beta 10 software package (Swofford 2000) was performed with tree-bisection-reconnection (TBR) branch swapping and characters treated as unordered and equally weighted. Starting trees were obtained through step-wise addition. All most parsimonious trees were combined into a consensus tree. One tree was saved per replicate to facilitate an optimal search of tree space. A total of 5 000 bootstrap replicates (Felsenstein 1985) were performed with the simple-stepwise addition option in order to estimate confidence levels.
For maximum likelihood analysis, likelihood settings were set to the GTR+I+G model as determined by Akaike Information Criteria (AIC) in Modeltest 3.06 (Posada & Crandall 1998). The data were analysed using a genetic algorithm to find the trees with the highest likelihood in the software program GARLI v. 0.951 (Zwickl 2006) using default values. Confidence values were estimated using bootstrap analysis (100 replicates), which were summarized as a 50 % majority rule consensus tree in PAUP.
Bayesian phylogenetic inference was implemented using the GTR+I+G (shape parameter with 4 rate categories) model and the Markov Chain Monte Carlo technique in the software package MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003). Two independent Markov chains of 5 000 000 generations each (sample frequency of 500) were initiated from a random starting tree. The first 500 000 generations were discarded as burnin and the remaining trees were pooled into a 50 % majority rule consensus tree. Bayesian analyses were repeated five times for improved sampling of tree space and to guard against local optima in searches.
Morphology
Teliospores of the unknown fungus and T. oxalidis (Herbarium of Dominik Begerow, reference number 684) were collected from infected plant organs and mounted in lactophenol on microscope slides and studied using a Nikon Eclipse E600 light microscope (Nikon Corporation, Tokyo, Japan) with differential interference contrast. Photographic images were captured using a Nikon DXM1200 digital camera (Nikon Corporation, Tokyo, Japan). In addition, spores were studied with a Leo 1430 VP7 scanning electron microscope (SEM, Leo Electronic Systems, Cambridge, UK). For SEM, spores were mounted on brass stubs using double-sided carbon tape, sputter coated with gold-palladium and viewed using standard methods. Measurements (n = 50) of all taxonomically informative characters were made.
RESULTS
DNA phylogeny
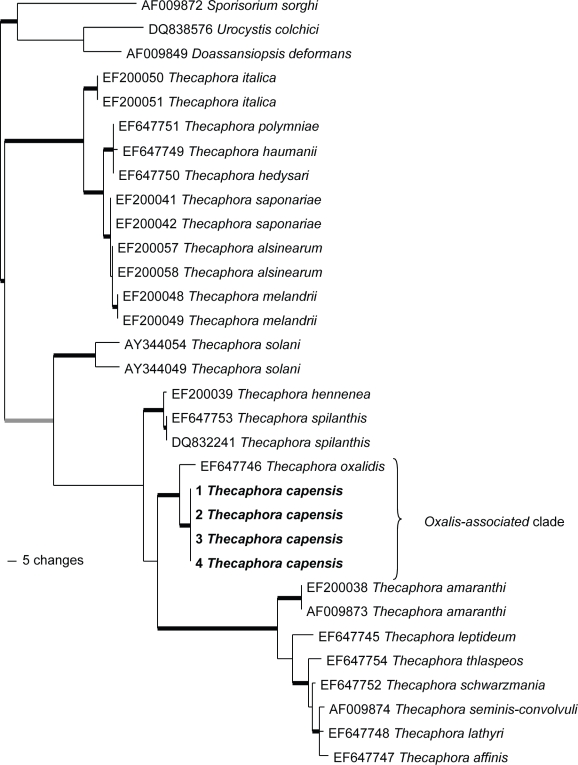
The aligned LSU rDNA sequence data matrix contained 32 taxa (including outgroups) and 1034 characters. Of these, 144 characters were parsimony informative and 137 were parsimony uninformative while the rest were constant. Parsimony analysis of the dataset resulted in four trees of 466 steps long, one of these was chosen for presentation (Fig. 1). The consistency- and retention indices were 0.7446 and 0.851, respectively, indicating low homoplasy. The trees resulting from the different analyses were very similar and did not differ markedly from the tree presented by Vánky et al. (2008). Aligned sequences have been deposited in TreeBase (accession number S2201).
Fig. 1.
One of four trees resulting from parsimony analysis of Thecaphora and closely related species, based on sequence data from the large subunit rDNA region. Thickened black lines indicate groups with strong support in all analysis (parsimony bootstrap > 80; Bayesian posterior probability > 0.95; Maximum likelihood bootstrap > 80). Thickened grey lines indicate groups with strong support using model based methods and moderate support for parsimony analysis (parsimony bootstrap between 70 and 80).
All included South African specimens had identical LSU rDNA sequences. In analysis the samples from the South African Oxalis specimens clustered together with strong support (Fig. 1). These samples grouped as sister to T. oxalidis within a strongly supported clade. This Oxalis-associated clade is strongly supported as a derived group within Thecaphora in all analyses (Fig. 1).
Taxonomy
The external morphology of Oxalis lanata specimens infected with this anther-smut did not differ significantly from apparently healthy specimens. Rather than presenting anthers at two different levels as in healthy plants (Salter 1944, Fig. 2c), all anthers of infected plants were carried at approximately the same height (Fig. 2d). The most conspicuous external symptom of infection was the reddish brown teliospore masses (Fig. 2b, d) which were distinct from the normally yellow, pollen-filled anthers. Flowers of infected plants appeared to live longer than those that were healthy.
Light- and scanning-electron microscope studies showed that sori lacked a peridium, columella and sterile cells. Spores were light yellowish brown in colour (Fig. 2e) and were produced singly rather than in spore-balls. The spore surfaces were finely verruculose (Fig. 2f, g). These morphological characteristics are typical of species of Thecaphora and they were similar to those of T. oxalidis (Fig. 2h–j).
Morphological comparisons and analyses of phylogenetic data provided strong support for the view that the specimens from Oxalis anthers in South Africa represent an undescribed species of Thecaphora. The fungus is, therefore, described as follows:
Thecaphora capensis Roets & L.L. Dreyer, sp. nov. — MycoBank MB508255; Fig. 2
Sori in antheris, vice pollinis massa sporarum pulveracea porphyrea. Sporae unicae globosae 14–17 × 16–18 μm, crocueae nec violaceae, dense irregulatimque subtiliterque microreticulatae, verruculosae, verrucis ad 0.75 μm altis, saepe ad basibus anastomosis. Anamorpha non visa.
Etymology. Name refers to the Cape region of South Africa.
Sori in anthers, replacing pollen with reddish brown powdery mass of spores. Spores single, globose, 14–17 × 16–18 μm, pale yellowish brown (Fig. 2e), lacking violet tints, surface densely, irregularly and finely micro-reticulate, verruculose, warts up to 0.75 μm high, often anastomosing at the bases (Fig. 2f, g). Anamorph not seen.
Specimens examined. South Africa, Western Cape Province, Jonkershoek forestry station (Assegaaibos), on flowers of Oxalis lanata var. rosea, July 2007, F. Roets & L.L. Dreyer, PREM 60075 holotype; HUV 21532 isotype; PREM 60076 paratype; PREM 60077 paratype; PREM 60078 paratype.
DISCUSSION
This study records the first Thecaphora species to have been discovered in Africa. The smut was shown to represent an undescribed species for which the name T. capensis has been provided. Thecaphora capensis is closely related to T. oxalidis which is also found on Oxalis species but is known only from Asia, Europe and the Americas. The two fungi are similar but they are morphologically distinct and are unlikely to be confused.
Although teliospore size-ranges of T. capensis and T. oxalidis overlap, these species can be readily be distinguished by their teliospore surface ornamentation. The teliospore surface-warts in T. oxalidis are much larger than those in T. capensis, resulting in teliospores of T. oxalidis having a rougher surface sculpture. The most distinct difference between these two species, however, is that the teliospores of T. oxalidis are formed within the fruits of its hosts (anamorph in anthers), while those of T. capensis are formed in the anthers. This study thus introduces the first known Thecaphora species to produce teliospores within the anthers of its host.
Interestingly, there are some groups of smut fungi, where a switch in organ specificity can be observed. Microbotryum is the most prominent example with multiple origins of sporulation in anthers (Kemler et al. 2006), but Antherospora was recently described with the same evolutionary trend and is closely related to Urocystis (Bauer et al. 2008). Thecaphora capensis represents only the second species in the genus associated with Oxalis and only the third species that produce solitary spores rather than spore balls. While T. capensis appears to be confined to O. lanata var. rosea in South Africa, T. oxalidis has been found on various Oxalis spp. and is distributed globally. This global distribution can be ascribed to the wide distribution (e.g. O. stricta) and/or the weedy nature (e.g. O. corniculata) of some of its host plants. It is also very likely that this species is overlooked in other countries in which these hosts occur.
The phylogenetic relationship between the hosts of T. oxalidis and T. capensis is interesting. Thecaphora oxalidis has been reported from various hosts in section Corniculatae (Oberlander pers. comm.), as well as from O. laxa in sect. Alpinae, which is not a close relative of species in sect. Corniculatae. In molecular phylogenetic reconstructions, the southern African species of Oxalis resolve together in a clade with strong support, and the hosts of T. oxalidis and T. capensis are thus phylogenetically distantly related (Oberlander pers. comm.). The O. lanata host of T. capensis resolves within a well-supported subclade of the southern African clade, characterised by the presence of well-developed above-ground stems. The remaining species in this clade have not been carefully inspected for infections by Thecaphora spp., but it seems probable that they would include additional hosts of the smut.
An interesting observation in this study was that all flowers of infected plants had anthers where the pollen was completely replaced by teliospores. This suggests that the fungus grows endophytically in infected plants after infection. All native South African Oxalis spp. are bulbous (Salter 1944) and they only produce stems and leaves during the rainy season (winter in winter rainfall species; summer in summer rainfall species). It would therefore be interesting to know whether this fungus also survives within these bulbs during the hot and dry summer months. If this is true, pollen and consequently seed production in infected populations of O. lanata would be compromised, as infected plants are rendered permanently sterile.
Propagules of T. capensis are most likely vectored between hosts by Oxalis pollinating-insects, as is true for various anther-infecting smut fungi such as Microbotryum violaceum (Roy 1994). Although the pollination biology of native South African Oxalis species is poorly documented, they are mostly believed to follow generalist pollination strategies (Dreyer, pers. obs.). The possible permanent infection of the host, coupled with the apparent ease of spore dispersal in T. capensis could severely limit the fitness and survival of O. lanata plants and will have a large role to play in the ecology of infected populations.
More than 200 species of Oxalis are endemic to South Africa (Salter 1944). Most of these are confined to the CFR of the Western Cape Province (Oberlander et al. 2002), a region that has been largely under-collected for fungi. It is thus possible that many more Thecaphora species and/or hosts await discovery in this region. Future studies should focus on elucidating these associations and consider the effect that these fungi have on host plant population dynamics.
Acknowledgments
This work was carried out with financial support from the National Research Foundation (GUN nr. 2053585) and the NRF/DST Centre of Excellence in Tree Health Biotechnology (CTHB). We thank the Western Cape Nature Conservation Board for issuing the necessary collecting permits. We further acknowledge Kenneth Oberlander for assistance with analyses and useful suggestions to improve the manuscript, Dr E.G.H. Oliver who translated the Latin description, Angela Schäfer for the SEM micrographs of T. oxalidis, and we appreciate the helpful discussions and the sequence data for T. oxalidis provided by Drs Kálmán Vánky and Matthias Lutz.
REFERENCES
- Bauer R, Begerow D, Oberwinkler F, Piepenbring M, Berbee ML. 2001. Ustilaginomycetes. In: Mclaughlin DJ, Mclaughlin EG, Lemke PA. (eds), The Mycota. 7. Systematics and Evolution Part B: 57–83 Springer Verlag, Berlin-Heidelberg, Germany: [Google Scholar]
- Bauer R, Lutz M, Begerow D, Piątek M, Vánky K, Bacigálová K, Oberwinkler F. 2008. Anther smut fungi on monocots. Mycological Research doi: 10.1016/j.mycres.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Bauer R, Oberwinkler F, Vánky K. 1997. Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany 75: 1237 – 1314 [Google Scholar]
- Begerow D, Bauer R, Oberwinkler F. 1997. Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Canadian Journal of Botany 75: 2045 – 2056 [Google Scholar]
- Dreyer LL, Esler KJ, Zietsman J. 2006. Climate influences flowering phenology of Oxalis – implications under climate change. South African Journal of Botany 72: 150 – 156 [Google Scholar]
- Ellis JB, Tracy SM. 1890. A few new fungi. Journal of Mycology 6: 76 – 77 [Google Scholar]
- Ershad D. 2000. Vankya, a new genus of smut fungi. Rostaniha 1: 65 – 72 [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenetics: an approach using the bootstrap. Evolution 39: 783 – 791 [DOI] [PubMed] [Google Scholar]
- Goldblatt P, Manning J. 2000. Cape plants. A conspectus of the Cape Flora of South Africa, Strelitzia 9 National Botanical Institute of South Africa, South Africa: [Google Scholar]
- Good R. 1947. The geography of flowering plants 4th edn Longman, UK: [Google Scholar]
- Kemler M, Göker M, Oberwinkler F, Begerow D. 2006. Implications of molecular characters for the phylogeny of the Microbotryaceae (Basidiomycota: Urediniomycetes). BMC Evolutionary Biology 6: 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourteig A. 1994. Oxalis L. subgénero Thamnoxys (Endl.) Reiche emend. Lourteig. Bradea. Boletim do Herbarium Bradeanum 7: 1 – 199 [Google Scholar]
- Lourteig A. 1995. Oxalis L. subgenus Trifidus Lourt. subgen. nov. Bradea 6: 389 – 392 [Google Scholar]
- Lourteig A. 2000. Oxalis L. subgénero Monoxalis (Small) Lourteig, Oxalis y Trifidus Lourteig. Bradea 7: 201 – 629 [Google Scholar]
- Matheny PB, Gossmann JA, Zalar P, Arun Kumar TK, Hibbett DS. 2006. Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Canadian Journal of Botany 84: 1794 – 1805 [Google Scholar]
- Oberlander KC, Dreyer LL, Esler KJ. 2002. Biogeography of Oxalis (Oxalidaceae) in South Africa: a preliminary study. Bothalia 32: 97 – 100 [Google Scholar]
- Piątek M. 2005. Kochmania, a new genus of smut fungi, and new records of cypericolous species from Poland and Ukraine. Mycotaxon 92: 33 – 42 [Google Scholar]
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817 – 818 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572 – 1574 [DOI] [PubMed] [Google Scholar]
- Roy BA. 1994. The use and abuse of pollinators by fungi. Trends in Ecology and Evolution 9: 335 – 339 [DOI] [PubMed] [Google Scholar]
- Salter TM. 1944. The genus Oxalis in South Africa. The Journal of South African Botany Suppl Vol 1 [Google Scholar]
- Swofford DL. 2000. PAUP* (Phylogenetic Analysis Using Parsimony), Version 4.0b10 Sinauer Associates, Sunderland, Massachusetts, USA: [Google Scholar]
- Takhtajan A. 1986. Floristic regions of the world University of California Press, USA: [Google Scholar]
- Vánky K. 1998. The genus Microbotryum (smut fungi). Mycotaxon 67: 33 – 118 [Google Scholar]
- Vánky K. 1999. The new classificatory system for smut fungi, and two new genera. Mycotaxon 70: 35 – 49 [Google Scholar]
- Vánky K, Lutz M. 2007. Revision of some Thecaphora species (Ustilaginomycotina) on Caryophyllaceae. Mycological Research 111: 1207 – 1219 [DOI] [PubMed] [Google Scholar]
- Vánky K, Lutz M, Bauer R. 2008. About the genus Thecaphora (Glomosporiaceae) and its new synonyms. Mycological Progress 7: 31 – 35 [Google Scholar]
- Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, The University of Texas, Austin, USA: [Google Scholar]