Abstract
Purpose
Accumulating evidence suggests that cancer associated stromal fibroblasts contribute to tumor growth by actively communicating with cancer cells. Our aim is to identify signaling pathways involved in tumor-stromal cell interactions in human pancreatic cancer.
Experimental Design
We established primary fibroblast cultures from human pancreatic adenocarcinomas and non-neoplastic pancreas tissues. To identify differentially expressed genes in CAFs, we performed gene expression profiling of human pancreatic CAFs and non-neoplastic pancreatic fibroblasts.
Results
The Hedgehog receptor Smoothened (SMO) was upregulated in cancer associated fibroblasts relative to control fibroblasts. CAFs expressing SMO could transduce the Shh signal to activate Gli1 expression, and siRNA knockdown of SMO blocked the induction of Gli1 in these cells. Stromal fibroblasts of human primary pancreatic adenocarcinomas overexpressed Smo compared to normal pancreatic fibroblasts.
Conclusions
These findings implicate overexpression of Smo as a mechanism for the activation of Hedgehog signaling in human pancreatic CAFs and suggest that stromal cells may be a therapeutic target for Smo antagonists in pancreatic cancer.
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the United States (1). It is one of the most highly invasive of the solid cancers and is characterized by an extensive desmoplastic stromal response (2). Mounting evidence suggests that cancer associated fibroblasts (CAFs), the predominant stromal cell type, actively communicate with and stimulate tumor cells, thereby contributing to tumor development and progression. Recent studies in multiple pancreatic cancer model systems have implicated the Hedgehog (Hh) signaling pathway in these tumor-stromal interactions (3, 4).
The Hedgehog signaling pathway, a crucial regulator of proliferation and differentiation during embryonic development, has been reported to be aberrantly activated in many solid tumors, including basal cell carcinoma (5-7), medulloblastoma (8), and, more recently, in several gastrointestinal cancers, including pancreatic cancer (9-12). Hedgehog proteins are secreted signaling molecules that can signal responsive cells at a significant distance from the producing cells. Three mammalian Hedgehog ligands have been described: Sonic hedgehog (Shh), Indian hedgehog (Ihh) and Desert hedgehog (Dhh). These ligands initiate Hedgehog signaling by binding to the Patched (Ptch) 12-transmembrane domain receptor. Ptch then activates Smoothened (Smo), a 7-transmembrane spanning protein and the central transducer of the Hedgehog signal. Activated Smo induces nuclear localization of the Gli family of transcription factors, resulting in transcription of hedgehog specific target genes, including Gli1 and Ptch.
Constitutive activation of the pathway results in cell proliferation and tumor formation and commonly occurs as a result of activating mutations in Smo (13, 14) or inactivating mutations in the tumor suppressor gene Ptch (5, 15). Mutations of Ptch or Smo have not been described in pancreatic cancer (16), but overexpression of the Shh ligand has been reported to occur in 70% of primary pancreatic adenocarcinomas (12) and has been implicated in the development and progression of pancreatic tumors. Forced overexpression of Shh during mouse development results in formation of lesions resembling pancreatic cancer precursor pancreatic intraepithelial neoplastic (PanIN) lesions (12, 17). Cell lines established from primary and metastatic pancreatic cancers retain the expression of several components of the Hedgehog signaling pathway (3, 12). The plant-derived teratogen cyclopamine, which inhibits Smo activity, suppresses growth of these cell lines both in vitro and in vivo (12). Furthermore, cyclopamine therapy inhibits development of tumor metastases in xenografted mice (10, 18) and prolongs survival in a mouse model of pancreatic cancer (19). These data support a functionally important role for Hedgehog signaling in pancreatic ductal tumorigenesis.
Previously, a cell-autonomous role for Hedgehog signaling has been described in tumor types driven by mutations in Hedgehog pathway components, such as medulloblastoma and basal cell carcinoma (20). However, an alternative mechanism, in which tumor cell-derived Hedgehog ligands stimulate neighboring stromal cells, has recently been described in mouse models of pancreatic cancer. To identify signaling pathways involved in tumor-stromal cell interactions in human pancreatic cancer, we have now established primary cancer associated fibroblast cultures from human pancreatic adenocarcinomas and non-neoplastic pancreas tissues. By performing global gene expression analysis of pancreatic CAFs vs. fibroblasts from non-neoplastic pancreas using Affymetrix Exon microarrays we identified the Hedgehog receptor SMO as overexpressed in human pancreatic CAFs. Overexpression of Smo protein was confirmed by immunohistochemical staining in stromal fibroblasts of primary human pancreatic adenocarcinomas. We also present evidence of Hedgehog pathway activity in stromal cells derived from primary pancreatic adenocarcinomas. Our results implicate overexpression of SMO as a mechanism for Hedgehog signaling in the stromal cells of pancreatic ductal adenocarcinomas.
MATERIALS AND METHODS
Culture of cell lines and establishment of fibroblast cultures
Primary cultures of stromal fibroblasts, designated cancer associated fibroblasts (CAFs) CAF11, CAF12, CAF13, CAF15, CAF16, CAF18, CAF19, CAF20, CAF21, CAF22, CAF25, CAF26, CAF27, CAF37, CAF38, CAF39, and CAF40, were established as previously described (21) from surgically resected pancreatic cancer tissue from 17 patients (8 males and 9 females with a mean ± standard deviation age of 64±12 years) with clinically sporadic pancreatic ductal adenocarcinoma. The cancers were all moderate to poorly differentiated with a mean tumor size of 3.4 cm. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. All CAFs were used at early passage numbers (passages 3-6). Nine CAFs were used for microarray analysis. Because these CAFs senesced after several passages additional primary CAFs were generated for use in subsequent experiments. The packaging cell line Phoenix A (kindly provided by Dr. C. Dang and Dr. T.C. Wu, Johns Hopkins University, Baltimore, MD) was maintained in DMEM containing 10% fetal bovine serum, 2% penicillin and streptomycin (Invitrogen). Human pancreatic ductal epithelial (HPDE) cells, generously provided by Dr. Ming-Sound Tsao (University of Toronto, Ontario Canada), were also maintained in DMEM containing 10% fetal bovine serum, 2% penicillin and streptomycin (Invitrogen) and were used as a control line for immunohistochemical staining (22). All samples were collected with approval from the Johns Hopkins Committee for Clinical Investigation.
Immortalization of pancreatic fibroblasts
The human pancreatic Nestin-expressing cells (HPNE) generously provided by Dr. Michel M. Ouellette (University of Nebraska Medical Center, Omaha, NE) were used as a control cell line. These cells were derived from normal pancreatic tissue and immortalized using hTERT (23).
Two immortalized control fibroblast cultures (SC2 and SC3) were also established from non-malignant pancreatic tissues. SC2 was derived from an intraductal papillary mucinous neoplasm (IPMN) of the pancreas, and SC3 was derived from resected chronic pancreatitis tissue. Immortalized fibroblasts (SC2 and SC3) were generated by transduction of primary normal fibroblast cultures with an hTERT-expressing retrovirus. The amphotropic packaging cell line Phoenix A was transiently transfected with 5μg of the pBABE-puro-hTERT (Addgene plasmid 1771) retroviral vector generated according to Counter et al. (24). After 24-hour transfection in Lipofectamine 2000 and OptiMEM (Invitrogen), the cells were incubated for 48 hours at 37°C in complete medium (DMEM containing 10% FBS). The viral supernatants were then filtered through a 0.45-μm filter and used to infect target cells in the presence of 8μg/mL polybrene (Sigma). A second round of viral infection was carried out 72 hours after the first infection. The infected cells were subsequently selected by growing them in the presence of 1μg/mL puromycin (Sigma).
Sample preparation and Affymetrix Exon Array analysis
Total RNA was isolated from CAF and control fibroblast cultures using a Qiagen kit (Qiagen,) or an Ambion mirVana miRNA kit (Ambion), according to the manufacturer’s instructions. The Affymetrix GeneChip® Human Exon 1.0 ST Array (Santa Clara, CA) platform was used to analyze gene expression patterns in CAFs and control fibroblasts on a whole-genome scale. Using the GeneChip Whole Transcript Sense Target Labeling Assay (Affymetrix), 1 μg of total RNA from each CAF culture or control fibroblast culture was processed, labeled, and hybridized to Affymetrix Human Exon ST 1.0 arrays according to the manufacturer’s recommendation (Affymetrix, Santa Clara, CA). In brief, 1 μg of total RNA was processed using a RiboMinus Human/Mouse Transcriptome Isolation Kit (Invitrogen, Carlsbad, CA) to remove ribosomal RNA. Following ribosomal reduction, double-stranded cDNA was synthesized from the RNA using a random hexamer incorporating a T7 promoter. The double-stranded cDNA was used as a template to generate antisense cRNA through an in vitro transcription reaction. Random primers were then used to reverse-transcribe the cRNA to single-stranded cDNA. The cDNA was then fragmented by restriction digestion and end-labeled with a biotinylated dideoxynucleotide. Biotinylated cDNA was hybridized to a Human Exon 1.0 ST GeneChip array and scanned by the Microarray Core Facility at Johns Hopkins using an Affymetrix GeneChip Scanner 3000. We are in compliance with the Minimum Information about a Microarray Experiment (MIAME) guidelines and have submitted our microarray data set to the Gene Expression Omnibus (GEO) repository.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA (1 μg) was reverse-transcribed using AMV Reverse Transcriptase and random primers (Promega, Madison, WI) according to the manufacturer’s instructions. The resulting cDNA was amplified on the ABI 7300 Real Time PCR thermocycler (Applied Biosystems) using Taqman Universal PCR Master Mix and recommended PCR conditions (Applied Biosystems) to quantitatively assess gene transcript levels in normal fibroblast and cancer associated fibroblast samples. The expression level of the housekeeping gene 18s rRNA (Applied Biosystems) was used for normalization. Results are expressed as normalized values relative to the indicated cell line (=2-ΔΔCt). All PCR reactions were performed in triplicate. The following primer sets were used for Taqman assays (Applied Biosystems): Taqman Gene Expression Assays Hs00170665_m1 (hSmo), Hs00179843_m1 (hShh), Hs00171790_m1 (hGli1), Hs00257977_m1 (hGli2), 18S rRNA VIC-MGB probe dye.
siRNA knockdown of Smoothened in CAF cultures
CAFs were plated in DMEM containing 10% FBS and 2% penicillin-streptomycin at a density of 5 × 104 cells/well in a 24-well plate and transfected with 2μM Smoothened or non-targeting control pool siRNA (Dharmacon) using the DharmaFECT4 transfection reagent (Dharmacon) in OptiMEM (Invitrogen). RNA was extracted 72 hours after transfection using Trizol (Invitrogen). RNA was reverse transcribed to cDNA using random primers and AMV Reverse Transcriptase (Promega).
ShhN ligand stimulation of CAF cultures
CAF or control fibroblast cells were plated in complete medium (DMEM + 10% FBS) at a density of 1 × 104 in a 24-well plate and grown to confluence. The cells were incubated for 24 hours in low-serum medium (DMEM + 0.5% FBS) containing 0, 0.5, 1.0, or 2.5 μg/mL recombinant human sonic hedgehog (ShhN) ligand (1845-SH, R&D Systems, Inc.). Recombinant ShhN ligand was added to the siRNA-transfected cells at 48 hours after transfection, and the cells were incubated for 24 hours in low-serum medium. RNA was extracted using Trizol (Invitrogen) and assayed for the expression of Shh target genes.
Pancreatic Adenocarcinoma Tissue Microarrays and SMO Immunohistochemistry
The expression of the SMO protein was examined by immunohistochemical labeling of formalin-fixed, paraffin-embedded whole tissue sections or tissue microarrays (TMAs) using a DAKO Autostainer (DAKO, Carpinteria, CA). Eight tissue microarrays containing a total of 156 different surgically resected pancreatic ductal adenocarcinomas, IPMNs, and chronic pancreatitis tissues were constructed as previously described (25). All specimens were collected and analyzed with the approval of the Johns Hopkins Committee for Clinical Investigation. Tissue sections were deparaffinized in xylene, hydrated in graded ethanol concentrations, and boiled for 20 minutes in epitope retrieval buffer. Immunostaining was then performed on the DAKO Autostainer using a rabbit polyclonal antihuman Smoothened antibody (ab72130, Abcam, Inc., Cambridge, MA) at a 1:800 dilution, a mouse monoclonal anti-vimentin antibody (clone V9, DAKO, Carpinteria, CA), at a 1:100 dilution, a mouse monoclonal anti-cytokeratin 19 antibody (Santa Cruz Biotechnology) at a 1:100 dilution, or a mouse monoclonal anti- α-smooth muscle actin antibody (clone IA4, DAKO, Carpinteria CA) at a 1:100 dilution for an incubation time of 60 minutes. Labeling was performed according to the manufacturer’s protocol using the Envision Plus Detection Kit (DAKO, Carpinteria, CA). Nuclei were counterstained with hematoxylin. For immunocytochemistry experiments, cells were cultured on chamber slides (BD Falcon) and grown until sub-confluent. Cells were then fixed in either 10% formalin (for Oil Red O staining) or in 70% methanol (for immunostaining) and subjected to Oil Red O staining for 15 minutes or to immunostaining on the DAKO Autostainer. To quantify Smo expression in stromal fibroblasts, three consecutive sections from one TMA were stained with antibodies against vimentin and alpha-smooth muscle actin to identify fibroblasts and activated fibroblasts, respectively, and against Smo. 61 cores from pancreatic cancer tissue and 24 cores from normal pancreatic tissue were scored by two independent researchers (SH and KW) for expression of Smo in stromal fibroblasts. An average of 40 fibroblasts from each individual core was scored, and the percentage of Smo positive fibroblasts was calculated. Cells with a weak positive signal were considered positive for Smo expression, and cores containing any areas of focal expression were considered positive for Smo expression.
Statistical Analysis
Descriptive statistical values and plots were generated using the Microsoft Excel software package, the SPSS Mann-Whitney (non-parametric test) and Pearson Chi-Square tests were performed using SPSS 17.0 software statistical program, or the Partek® Genomics Suite™ version 6.3 beta. For the qRT-PCR experiments, statistical analysis was performed with Microsoft Excel software using a paired Student’s t test (two-tailed). Differences were considered significant at P < 0.05, and values reported are means ± SD. For microarray experiments, gene expression analysis was performed using Partek Genomics Suite version 6.3 beta (Partek, Inc., St. Louis, MO). The Robust Multichip Average (RMA) method was used to normalize the raw intensity measurements of all probe sets. Gene expression values were then obtained using the one-step Tukey’s biweight method. Two-way ANOVA was performed to identify significant expression changes between CAFs and control fibroblasts, using a fold-change criterion of 2.8 -fold and a P-value of <0.05. Gene ontology analysis was performed using the Biological Interpretation feature of the Partek software (v6.5).
RESULTS
Characterization of pancreatic fibroblasts
The pancreatic fibroblasts isolated from benign and malignant primary pancreatic resection specimens expressed the mesenchymal marker vimentin by immunocytochemistry (supplemental Figure 1). Both the pancreatic CAFs and the fibroblast lines, SC2 and SC3, from non-neoplastic pancreas appear as myofibroblasts with both a spindle-shaped and stellate morphology (supplemental figure 2). Pancreatic CAFs contain lipid droplets and stain positive for vimentin and alpha-SMA, a marker for activated fibroblasts (supplemental figure 2).
SMO is upregulated in pancreatic CAFs
To identify genes that are differentially expressed in pancreatic CAFs in pancreatic CAFs relative to control fibroblasts, we compared the global gene expression profiles of nine CAFs (CAFs 11, 12, 13, 15, 16, 18, 19, 21, and 22) to control fibroblast lines derived from non-neoplastic pancreas (HPNE, SC3) and from an IPMN (SC2). The non-neoplastic pancreatic cell line, HPNE was used as a control fibroblast line as it has fibroblast morphology, expresses vimentin and does not express the ductal epithelial marker cytokeratin 19 (Supplemental figure 2). In addition, global gene expression analysis of HPNE reveals that it resembles fibroblasts and not pancreatic ductal cells by principal component analysis (data not shown).
Gene expression profiles were obtained using the Affymetrix Exon Array ST 1.0. We first examined the top 200 candidate genes that were expressed at significantly higher levels (~2.8-fold or greater and P < 0.05 by ANOVA) in CAFs compared to control fibroblasts. This criterion identified several genes that have previously been described in tumor-stromal interactions, such as POSTN (26), CXCL12 (27), ADAM12 (28), IGFBP3 (29), TIMP3 (30), and cytokines IL1 and IL6 (Supplemental Table 1). A gene ontology analysis revealed this list of genes upregulated in pancreatic CAFs to be enriched for genes involved in biological adhesion and development process functions, as indicated by enrichment scores of 30 and 36, respectively (Supplemental Figure 3).
Of particular interest among the genes upregulated in pancreatic CAFs was SMO, the receptor for Shh ligand. The hedgehog pathway has been recently implicated in tumor-stromal interactions in pancreatic cancer (3, 4), and overexpression of SMO could explain the mechanism of activation of this pathway in stromal cells (Fig. 1a).
Figure 1.
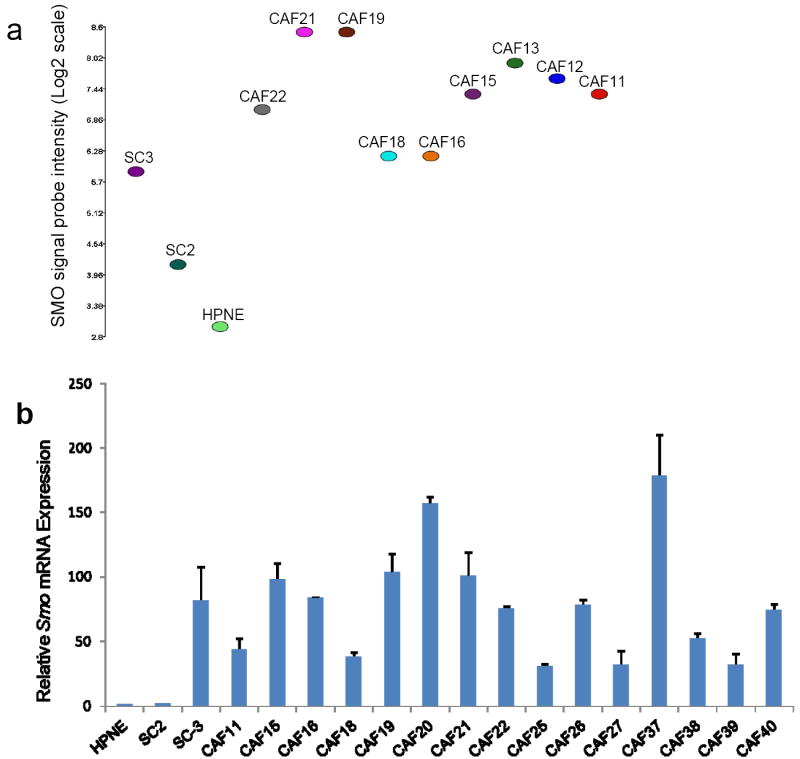
Analysis of Smo mRNA expression in fibroblast cultures. (a) Affymetrix exon array analysis of Smo mRNA expression in fibroblast cultures established from chronic pancreatitis tissue (SC3), IPMN tissue (SC2), normal pancreas (HPNE), and primary pancreatic adenocarcinomas (CAFs). Relative Smo expression values were obtained after using the Robust Multichip Average (RMA) method to normalize the raw intensity measurements of all probe sets. (b) Quantitative RT-PCR analysis of Smo mRNA expression in fibroblast cultures established from normal pancreas (HPNE), IPMN tissue (SC2), chronic pancreatitis tissue (SC3), and primary pancreatic adenocarcinomas (CAFs). Relative Smo mRNA levels after normalization to the corresponding 18S rRNA levels are expressed. Each assay was performed in triplicate. Data are means of three independent experiments; bars are SD values.
We confirmed the overexpression of SMO mRNA in pancreatic CAFs by using quantitative RT-PCR (qRT-PCR). SMO mRNA levels were low or undetectable in the control fibroblasts HPNE and SC2, and modest expression was detected in the control fibroblast line SC3. In contrast, higher levels of SMO mRNA were detected in all 15 CAFs tested (Fig.1b). Relative levels of SMO mRNA correlated closely with the results obtained by exon array analysis (Fig. 1a). Pancreatic CAFs also expressed the Hedgehog pathway components Ptch1, Gli1, and Gli2 mRNA (data not shown), but did not express Shh mRNA, consistent with previous studies of human (3, 4) and mouse fibroblasts (3, 4, 31).
Shh ligand induces expression of Gli1 mRNA in pancreatic CAFs
To determine if pancreatic CAFs are responsive to Shh signaling, we treated pancreatic CAFs with recombinant Shh ligand (ShhN, #1845-SH, R&D Systems, Inc. Because expression of the transcription factor GLI1 is a reliable marker of Hedgehog pathway activity, we measured Gli1 mRNA expression by qRT-PCR in response to ShhN treatment (Figure 1b). Since all the CAFs tested had overexpression of SMO, we selected 3 CAFs growing in culture (CAF25, CAF27, and CAF38) for ShhN ligand treatment. Exogenous ShhN ligand treatment for 24 hours resulted in a ~2-fold induction of Gli1 mRNA (Fig. 2).
Figure 2.
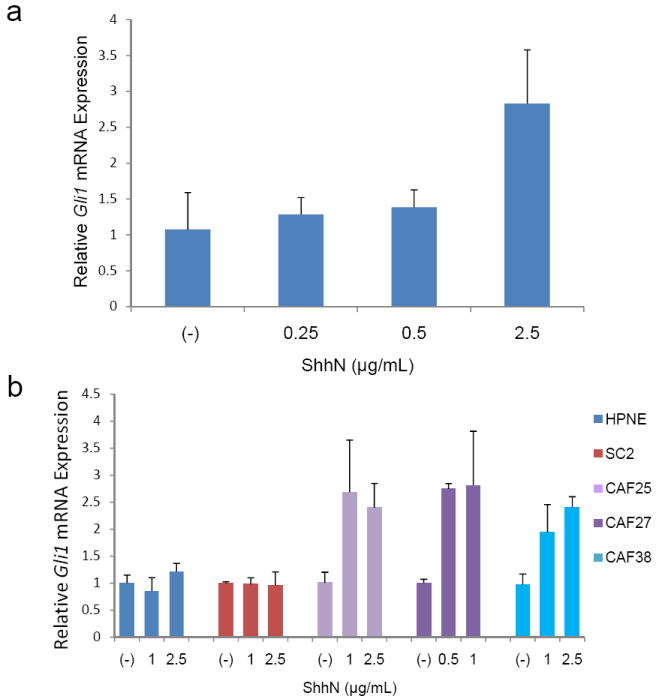
Effect of recombinant ShhN treatment on Gli1 mRNA expression in pancreatic CAFs. (a) Gli1 mRNA levels were assessed 24 hours after treatment of CAF27 cells with indicated concentrations of recombinant ShhN. (b) Comparison of ShhN treatment in control pancreatic fibroblasts and pancreatic CAFs. Gli1 mRNA levels were assessed 24 hours after treatment of HPNE, SC2, or CAF cells with indicated concentrations of recombinant ShhN. Relative Gli1 mRNA levels after normalization to the corresponding 18S rRNA levels are shown. Data are means of four independent experiments; bars are SD values.
In contrast, ShhN was unable to induce Gli1 expression in HPNE or SC2 cells which express low or undetectable levels of SMO.
siRNA knockdown of SMO expression blocks GLI1 induction in pancreatic CAFs
To further investigate the role of SMO expression in transducing the Shh signal, we transiently knocked down SMO expression in another overexpressing CAF, CAF26, and then stimulated them with ShhN ligand. Using siRNA we were able to knock down SMO with 90% efficiency (Fig.3a) which resulted in a 70% (P=0.007) reduction in Gli1 mRNA expression in CAF26 (Fig.3b). We also found that reducing SMO expression blocked the induction of Gli1 mRNA expression in response to ShhN treatment. ShhN treatment induced Gli1 mRNA expression in a dose-dependent manner in CAF27 cells that were either mock-transfected or transfected with non-targeting siRNA, but could not induce Gli1 mRNA expression in cells with siRNA-mediated Smo knockdown (Fig. 3c).
Figure 3.
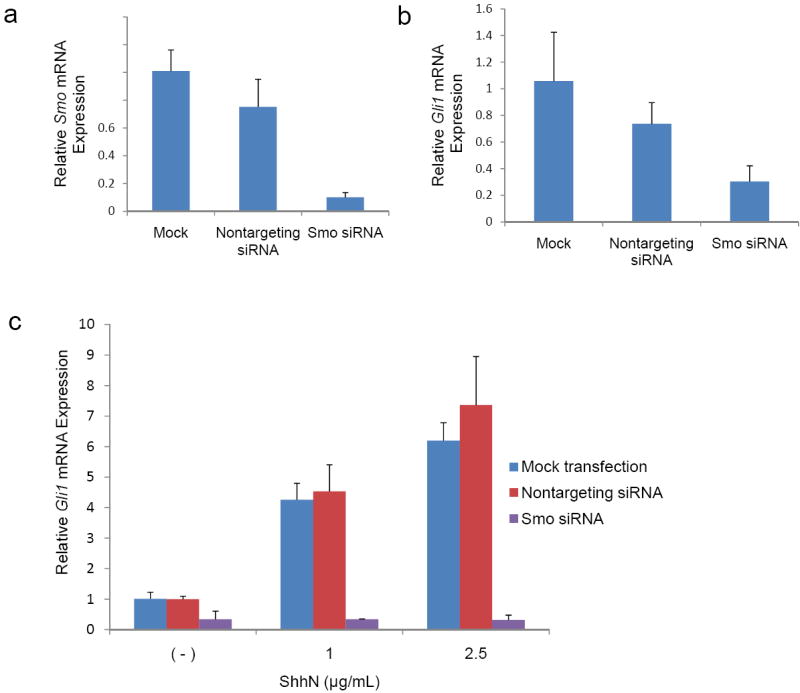
Effects of Smo siRNA and recombinant ShhN treatment on Gli1 mRNA expression in pancreatic CAFs. (a) Smo and (b) Gli1 levels were assessed by qRT-PCR in CAF26 cells transfected with 2μM non-targeting or Smo siRNA for 72 hours. (c) CAF27 cells were transfected with 2μM non-targeting or Smo siRNA and treated with the indicated concentrations of recombinant ShhN 48 hours later. Gli1 mRNA levels were assessed at 72 hours post-transfection. Relative Smo or Gli1 mRNA levels after normalization to the corresponding 18S rRNA levels are shown. Data represent the mean of three independent experiments; error bars are SD values.
Smo protein is expressed in stromal fibroblasts in primary human pancreatic tissues
Having verified that cultured pancreatic CAFs overexpress Smo mRNA, we performed immunohistochemical labeling of human primary pancreatic ductal adenocarcinomas to determine whether overexpression of SMO protein occurs in stromal fibroblasts in vivo (Fig. 4). We used Purkinje neurons in cerebellar sections of brain tissue as a positive control for Smo staining (Fig. 4a). Renal epithelial cells of the distal convoluted tubule also stained Smo (data not shown). No significant staining was observed with the secondary antibody alone (Fig. 4c). We analyzed Smo expression in normal and cancer-associated stromal fibroblasts from whole tissue sections of the pancreas. Immunohistochemical staining of consecutive tissue sections confirmed that the vimentin-positive fibroblasts surrounding the normal pancreatic duct lacked expression of Smo (Supplemental Figure 4), while the activated cancer associated fibroblasts expressing alpha-SMA stained positive for Smo (Supplemental Figure 5). We then analyzed 53 primary pancreatic cancers, 35 IPMN tumors, and 52 chronic pancreatitis tissue samples on tissue microarrays. Stromal fibroblasts surrounding the normal pancreatic duct were negative for SMO expression (Fig. 4a), and stromal fibroblasts in chronic pancreatitis tissue were weakly positive for SMO expression in 38 of 52 (73.0%) of cases (data not shown). This finding is consistent with our observation that SC3, a fibroblast cell line established from chronic pancreatitis tissue, weakly expresses Smo mRNA. It is possible that similar mechanisms are responsible for the fibroblast overexpression of Smo in chronic pancreatitis and in pancreatic cancer. We also frequently detected Smo expression in fibroblasts associated with IPMN tumors (28 of 35 or 80.0% of cases), although their expression levels were generally weaker than those in cancer associated fibroblasts (data not shown).
Figure 4.
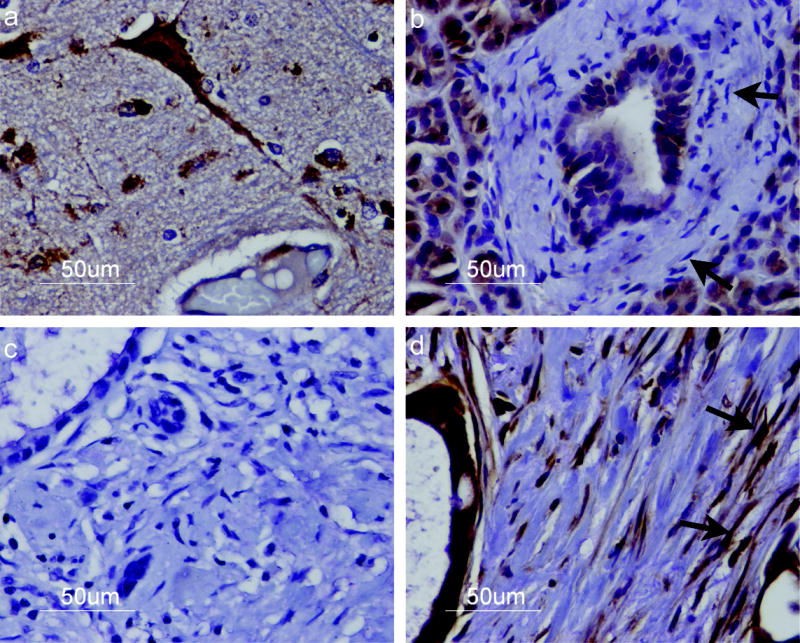
Immunohistochemical staining of Smo protein. Cells were stained with an anti-SMO antibody and an HRP-labeled secondary antibody (brown) then counterstained with hematoxylin and eosin. Staining revealed (a) strong Smo expression in Purkinje neurons of cerebellar tissue; (b) lack of Smo expression in normal pancreatic stromal fibroblasts (arrows) (c) no Smo staining in a primary pancreatic cancer section incubated with secondary antibody alone; and (d) strong SMO expression in pancreatic cancer associated stromal fibroblasts (arrows). Smo expression is also detected in pancreatic cancer cells. Representative samples from each type of tissue are shown. Magnification x40.
We observed a heterogeneous pattern of Smo expression in the stromal fibroblasts of pancreatic cancers. While fibroblasts surrounding the normal pancreatic duct lacked Smo expression (Fib. 4b), cancer associated fibroblasts stained positive for Smo, as indicated in Fig. 4d, a representative pancreatic adenocarcinoma strongly expressing Smo in the stromal fibroblasts. In the majority of pancreatic cancer cases we observed a variable pattern of staining throughout the tumor, with areas of minimal staining interspersed with focal areas of intense staining of Smo in the fibroblasts. 51 of 53 (96.2%) of pancreatic cancers evaluated contained areas with stromal expression of Smo. We observed Smo expression in pancreatic cancer cells (Figure 4d, supplemental figure 5b,d) and weak expression in normal ductal cells, consistent with previous reports that Smo is only occasionally detected in normal duct epithelium but is frequently detected in neoplastic epithelium (12, 32). We also observed weak staining of Smo in acinar cells, consistent with previous reports (12). We further quantified Smo expression in stromal fibroblasts by counting a percentage of Smo positive fibroblasts in cores from both normal pancreas and pancreatic cancer tissues. An average of 75.5% ±16.6% of cancer associated stromal fibroblasts and 10.5% ±13.9% of fibroblasts from normal pancreatic tissue were positive for Smo protein expression (P=9.9 × 10-19).
DISCUSSION
Using Affymetrix Exon arrays, we find that human pancreatic CAFs overexpress the Shh receptor Smoothened. This overexpression was confirmed in vivo since stromal fibroblasts in human primary pancreatic adenocarcinomas overexpress Smo protein relative to fibroblasts in normal pancreas. The hedgehog pathway has been identified as activated in cancer associated stromal fibroblasts in mouse models of pancreatic cancer. Our results implicate overexpression of Smo as a mechanism responsible for the activation of the hedgehog pathway in human pancreatic CAFs. Although it remains unclear how paracrine Hedgehog signaling of fibroblasts contributes to tumor growth, recent work suggests that targeting Smo in the tumor stroma may be an effective strategy in treating pancreatic cancer. Previous work identified Shh as a mediator of the desmoplastic response in pancreatic cancer and suggested that the stroma may serve as a barrier to delivery of therapeutic compounds (33). Indeed, Olive et al. recently reported that mice treated with the cyclopamine derivative and Smoothened inhibitor IPI-926 exhibited depletion of desmoplastic stroma and improved perfusion and delivery of chemotherapeutic drugs to pancreatic tumor cells, thereby increasing the survival time in these mice (34). Our immunohistochemical data indicate that these results are likely to be relevant to the treatment of human pancreatic cancer. We frequently observed Smo overexpression in the tumor stroma, suggesting that the stromal cells in human pancreatic cancers may be sensitive to Smoothened inhibition.
We demonstrated that pancreatic CAFs can actively transduce the Hedgehog signal to induce GLI expression. CAFs expressing SMO respond to exogenous Hedgehog ligand, whereas control fibroblasts lacking SMO expression are unresponsive to Hedgehog ligand, and downregulation of SMO in CAFs inhibits transduction of the Hedgehog signal. Our work is consistent with recent studies in mouse models of pancreatic cancer demonstrating a paracrine mechanism of Hedgehog signaling in cancer associated stromal cells. First, expression of an oncogenic allele of Smoothened (SmoM2) in the mouse pancreas was unable to activate the Hedgehog pathway in ductal epithelial cells, but resulted in Hedgehog signaling in adjacent stromal cells in several mouse models of pancreatic cancer (4). Second, co-culture of Hedgehog-producing pancreatic cancer cell lines with 10T1/2 fibroblasts resulted in GLI reporter activity in the fibroblasts, demonstrating the capacity of tumor cells to induce paracrine signaling (3). This mechanism was also observed in vivo in a xenograft model established from Hedgehog-expressing pancreatic cancer cell lines, in which Hedgehog pathway activation was detected in mouse stromal cells immediately adjacent to the xenografted tumors (3). Our findings that Gli1 expression can be induced in human pancreatic CAFs support these data and further demonstrate an intact canonical Hedgehog signaling pathway in human CAFs. Third, genetic deletion of Smo in mouse embryonic fibroblasts inhibited Gli1 induction in response to Hedgehog stimulation and resulted in decreased tumor growth of xenografts co-injected with these mouse embryonic fibroblasts (3). Similarly, we find that siRNA knockdown of SMO expression in CAFs results in decreased GLI1 expression, supporting a role for SMO overexpression in Hedgehog pathway activation. Taken together with these data, our work demonstrates ligand-dependent Hedgehog pathway activation in the stromal microenvironment and supports a paracrine mechanism of Hedgehog signaling in human pancreatic cancer.
Not surprisingly, we observed increased GLI activity in response to ShhN stimulation in cells with higher levels of SMO expression. Our finding that ShhN is unable to induce GLI1 expression in normal control fibroblasts which do not express SMO is consistent with previous reports that cells lacking SMO expression lack the ability to receive the Hedgehog signal (35). Furthermore, siRNA knockdown of SMO blocked the ability of CAFs to induce GLI1 expression, indicating that Smo directly transduces the Hedgehog signal in these cells. Interestingly, we observed GLI1 expression in SC2 (IPMN-derived) fibroblasts lacking SMO expression (data not shown), consistent with recent reports of SMO-independent GLI1 transcription through non-canonical activation of the Hedgehog pathway (31, 36). It is also interesting that although GLI1 transcription increases upon ShhN stimulation, CAFs overexpressing SMO also expressed detectable Gli1 levels in the absence of Hedgehog ligand (data not shown). This finding agrees with the observation that NIH-3T3 cells in which Smo is transiently overexpressed (37) as well as mouse pancreatic fibroblasts overexpressing SmoM2(4) have increased Gli expression in the absence of Shh.
In summary, we find that human pancreatic CAFs overexpress the Hedgehog receptor SMO. Increased SMO expression denotes increased Hedgehog pathway activity in these cells, and we provide evidence of Hedgehog pathway activity in pancreatic cancer associated stromal cells in vivo. These data contribute to a growing body of evidence that the Hedgehog pathway acts through a paracrine mechanism in human pancreatic cancer. SMO overexpression in the stromal compartment of human primary pancreatic adenocarcinomas suggests a tumor-stromal mechanism of Hedgehog pathway activation in vivo and may represent a therapeutic target in human pancreatic cancer.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute grant (CA62924, CA120432), and the Michael Rolfe Foundation.
Footnotes
Statement of Translational Relevance This paper describes the identification of differentially expressed genes in human pancreatic cancer associated fibroblasts compared to pancreatic fibroblasts from controls and identifies overexpression of the hedgehog pathway member, Smoothened. We further describe evidence of activation of the hedgehog pathway in pancreatic cancer associated fibroblasts that is dependent on smoothened expression. These findings suggest that smoothened overexpression is the mechanism of activation of the hedgehog pathway in human pancreatic cancer fibroblasts and support the rationale for using smoothened antagonists in the treatment of patients with pancreatic cancer.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Hartel M, Di Mola FF, Gardini A, et al. Desmoplastic reaction influences pancreatic cancer growth behavior. World J Surg. 2004;28:818–25. doi: 10.1007/s00268-004-7147-4. [DOI] [PubMed] [Google Scholar]
- 3.Yauch RL, Gould SE, Scales SJ, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–10. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 4.Tian H, Callahan CA, DuPree KJ, et al. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci U S A. 2009;106:4254–9. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–51. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 7.Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–81. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 8.Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–10. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 9.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–12. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 12.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–6. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reifenberger J, Wolter M, Weber RG, et al. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–803. [PubMed] [Google Scholar]
- 14.Xie J, Murone M, Luoh SM, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–2. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 15.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton JP, Mongeau ME, Klimstra DS, et al. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci U S A. 2007;104:5103–8. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldmann G, Fendrich V, McGovern K, et al. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther. 2008;7:2725–35. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldmann G, Habbe N, Dhara S, et al. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57:1420–30. doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–33. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 21.Walter K, Omura N, Hong SM, Griffith M, Goggins M. Pancreatic cancer associated fibroblasts display normal allelotypes. Cancer Biol Ther. 2008;7:882–8. doi: 10.4161/cbt.7.6.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N, Furukawa T, Kobari M, Tsao MS. Comparative phenotypic studies of duct epithelial cell lines derived from normal human pancreas and pancreatic carcinoma. Am J Pathol. 1998;153:263–9. doi: 10.1016/S0002-9440(10)65567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun. 2003;301:1038–44. doi: 10.1016/s0006-291x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 24.Counter CM, Hahn WC, Wei W, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–8. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Infante JR, Matsubayashi H, Sato N, et al. Peritumoral fibroblast SPARC expression and patient outcome with resectable pancreatic adenocarcinoma. J Clin Oncol. 2007;25:319–25. doi: 10.1200/JCO.2006.07.8824. [DOI] [PubMed] [Google Scholar]
- 26.Fukushima N, Kikuchi Y, Nishiyama T, Kudo A, Fukayama M. Periostin deposition in the stroma of invasive and intraductal neoplasms of the pancreas. Mod Pathol. 2008;21:1044–53. doi: 10.1038/modpathol.2008.77. [DOI] [PubMed] [Google Scholar]
- 27.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 28.Peduto L, Reuter VE, Sehara-Fujisawa A, Shaffer DR, Scher HI, Blobel CP. ADAM12 is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression. Oncogene. 2006;25:5462–6. doi: 10.1038/sj.onc.1209536. [DOI] [PubMed] [Google Scholar]
- 29.Massoner P, Haag P, Seifarth C, et al. Insulin-like growth factor binding protein-3 (IGFBP-3) in the prostate and in prostate cancer: local production, distribution and secretion pattern indicate a role in stromal-epithelial interaction. Prostate. 2008;68:1165–78. doi: 10.1002/pros.20785. [DOI] [PubMed] [Google Scholar]
- 30.Byrne JA, Tomasetto C, Rouyer N, Bellocq JP, Rio MC, Basset P. The tissue inhibitor of metalloproteinases-3 gene in breast carcinoma: identification of multiple polyadenylation sites and a stromal pattern of expression. Mol Med. 1995;1:418–27. [PMC free article] [PubMed] [Google Scholar]
- 31.Nolan-Stevaux O, Lau J, Truitt ML, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Tian X, Xie X, Zhuang Y, Wu W, Wang W. Expression and regulation of hedgehog signaling pathway in pancreatic cancer. Langenbecks Arch Surg Epub. 2009 doi: 10.1007/s00423-009-0493-9. [DOI] [PubMed] [Google Scholar]
- 33.Bailey JM, Swanson BJ, Hamada T, et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14:5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blair SS, Ralston A. Smoothened-mediated Hedgehog signalling is required for the maintenance of the anterior-posterior lineage restriction in the developing wing of Drosophila. Development. 1997;124:4053–63. doi: 10.1242/dev.124.20.4053. [DOI] [PubMed] [Google Scholar]
- 36.Arimura S, Matsunaga A, Kitamura T, Aoki K, Aoki M, Taketo MM. Reduced level of Smoothened suppresses intestinal tumorigenesis by down-regulation of Wnt signaling. Gastroenterology. 2009;137:629–38. doi: 10.1053/j.gastro.2009.04.059. [DOI] [PubMed] [Google Scholar]
- 37.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–7. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


