Abstract
PURPOSE
As an E2 conjugating enzyme for sumoylation, Ubc9 plays a critical role in sumoylation-mediated cellular pathways, ultimately impacting cell growth and cancer development. The aim of this study is to investigate the regulation of Ubc9 in cancer cells.
EXPERIMENTAL DESIGN
Immunohistochemistry (IHC) and Western blot were used to determine Ubc9 expression in paraffin-embedded tumor tissue and frozen specimens of the matched tumors from the same patient, respectively. To establish the causal relationship between miR-30e and Ubc9 expression, we overexpressed miR-30e and then determined the resultant effects on Ubc9 expression. To determine whether miR-30e directly targets Ubc9, we performed luciferase assays using luciferase reporters carrying the 3’-untranslated region (3’-UTR) of the Ubc9 gene.
RESULTS
We find that Ubc9 is upregulated in breast, head and neck, and lung cancer specimens. In addition, examination of 8 pairs of matched breast tumor specimens by Western blot analysis reveals that on average, the level of Ubc9 is a 5.7-fold higher in tumor than the matched normal breast tissue. Of interest, we present evidence that Ubc9 is subjected to the post-transcriptional regulation by microRNAs and the miR-30 family, such as miR-30e, negatively regulate Ubc9 expression. In contrast to Ubc9, miR-30e is underexpressed in tumors. Moreover, ectopic expression of miR-30e suppresses cell growth which can be partially reversed by Ubc9. Finally, using luciferase-Ubc9-3’-UTR reporters, we show that Ubc9 is a direct target for miR-30e by interactions with the putative miR-30e binding sites.
CONCLUSION
These results provide new insight into regulation of Ubc9 in cancer cells.
Keywords: Breast cancer, microRNA, miR-30, post-transcriptional regulation, tumorigenesis, Ubc9
Introduction
Post-translational modifications play an important role in protein function through the regulation of their activity, turnover and localization and/or interactions. One such modification involves the covalent attachment of the small ubiquitin-related polypeptide SUMO (small ubiquitin-like modifier) to different cellular protein substrates (1, 2). Although SUMO conjugation or sumoylation is similar to ubiquitination in structure, conjugation process and attachment to target proteins, the biological consequences of these two pathways can be quite distinct. Unlike ubiquitination that normally targets proteins for degradation through proteasome pathways, sumoylation has been implicated in regulation of protein stability, protein-protein interactions, transcriptional activity and subcellular localization (3).
Ubc9 is an E2 conjugating enzyme essential for sumoylation and it transfers the activated SUMO to protein substrates (4, 5). In particular, Ubc9 has been shown to play a key role in nuclear trafficking (6, 7), transcriptional regulation (8–11) and protein stability (12–15) through regulation of sumoylation machinery. In addition, recent evidence indicates that Ubc9 is a multi-functional protein that can exert its functions independent of sumoylation (16–18). In the past years, we have learned that many important proteins, including tumor suppressors and oncoproteins as well as the cell cycle and proliferation-related proteins, are targets for sumoylation or interact with Ubc9; their expression or their activity is regulated by Ubc9 (9, 19). Thus, alterations of Ubc9 could ultimately have an impact on cell growth and cancer development. Indeed, our previous studies indicate that Ubc9 plays a role in tumorigenesis and drug responsiveness (20, 21).
Ubc9 is a single copy gene and is ubiquitously expressed in all human organs and tissues. However, levels of Ubc9 vary in different organs or tissues (22). In tumors Ubc9 is frequently upregulated. We and several other groups have reported upregulation of Ubc9 in various tumors. For example, Ubc9 is upregulated in lung adenocarcinoma, as detected by microarray analysis (23). By semi-quantitative RT-PCR analysis we detected overexpression of Ubc9 in ovarian carcinoma compared to the matched normal ovarian epithelium (20). Moreover, Ubc9 is the most highly expressed protein in protein extracts from melanoma infiltrated lymph nodes identified by antibody array technology (24). However, little is known about the molecular mechanism of Ubc9 upregulation in cancer. In this study, we examine Ubc9 regulation and present evidence that Ubc9 expression is subjected to microRNA regulation at the post-transcriptional level, where miR-30e negatively regulates Ubc9 expression by translation repression. Therefore, these findings might provide a molecular explanation as to why Ubc9 is frequently overexpressed in tumors.
Materials and methods
Reagents
Primary monoclonal Ubc9 antibody from BD Biosciences (San Jose, CA) or custom made polyclonal Ubc9 antibody was used in both Western blot and immunofluorescence microscopy. Anti-SUMO-1 antibody for Western blot and secondary antibodies conjugated with Alex 566 used for immunofluorescence staining were obtained from Invitrogen (Carlsbad, CA). Secondary antibodies conjugated with IRDye 800CW were purchased from LI-COR Biosciences (Lincoln, NE). PCR primers were purchased from Sigma-Genosys (Woodland, TX).
Cell culture
All cell lines were purchased from ATCC (Manassas, VA). Both HeLa and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Cambrex) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO). All media contained 2 mM glutamine, 100 units of penicillin/ml, and 100 µg of streptomycin/ml. Cells were incubated at 37 °C and supplemented with 5% CO2 in the humidified chamber.
Transfection
HeLa cells were transfected using DNAfectin reagent (Applied Biological Materials, British Columbia, Canada) following the manufacturer’s protocol. In brief, cells were seeded at 40% confluence in a 12 or 6-well plate and then transfected with 1 or 3 µg of microRNA expression vectors in serum free medium the following day when the cells reached about 70% confluence. The serum free media was replaced by normal growth media after 15 h of transfection. 293T cells were transfected using the calcium phosphate method, as described previously (25). The transfected cells were grown overnight before they were harvested and lyzed for luciferase assay or extraction of protein or RNA.
Plasmids
pCMV-Ubc9 was described previously (26). To construct pre-microRNA expression vectors, we first amplified ~0.5 kb DNA fragment covering a pre-microRNA, using genomic DNA from a healthy blood donor as a template. PCR reactions were performed using the high fidelity Phusion enzyme (New England Biolabs Ipswich, MA) and corresponding specific primers:
miR-30e-5.1 (sense) 5’-AAAGCTGTGCCTTGTTCTGC
miR-30e-Not1-3.1 (antisense) 5’-GCGGCCGCAGCCCACAGAAAACAAGGAG
miR-30c-5.1 (sense) 5’-TTGGGGAGTTGGAGGCAATC
miR-30c-Not1-3.1 (antisense) 5’-GCGGCCGCAGGTTAATGGGAAACAGGGC
miR-188-5.1 (sense) 5’-CTTCCCTCTCCAGTGCATAG
miR-188-Not1-3.1 (antisense)5’-GCGGCCGCTCCTGCAGGATCCATGTAAG
miR-200c-5.1 (sense) 5’- TAAATCGGTGTGTGTCGCGG
miR-200c-Not1-3.1 (antisense) 5’-GCGGCCGCAAGGTCGACTGTGGGTTCTG
The amplified fragment was first cloned into a PCR cloning vector and subsequently cloned a pCMV vector or lentiviral vector (pCDH-CMV-MCS-EF1-copGFP from System Biosciences, Mountain View, CA) at EcoR1 and Not1 sites. Expression of the mature microRNAs was verified by TaqMan real-time PCR kit (Applied Biosystems) or QuantiMir kit (System Biosciences).
The luciferase-UTR reporter plasmid (pLuc-Ubc9-3’-UTR) was constructed by introducing the Ubc9 3’-UTR carrying putative microRNA binding sites into pGL3 control vector (Promega, Madison, WI). Thus, we amplified the Ubc9 3′-UTR sequence from MCF10A cDNA by PCR using the following primers:
Ubc9-UTR-5.1 (sense), 5’-GCAGCGACCTTGTGGCATCGT
Ubc9-UTR-Not1-3.1 (antisense) 5’-GCGGCCGCGCAGCGACCTTGTGGCATCGT
For construction of deletion mutant pLuc-Ubc9-3’-UTR clones, we used primers Ubc9-UTR-5.2 (see below) and Ubc9-UTR-Not1-3.1, resulting in pLuc-Ubc9-3’-UTR-d1 where the first putative miR-30e binding site was deleted. We then used primers Ubc9-UTR-5.1 and Ubc9-UTR-Not1-3.6 (see below) to generate pLuc-Ubc9-3’-UTR-d2 where the second putative miR-30e binding site was eliminated. Finally, to delete both sites, we used primers Ubc9-UTR-5.2 and Ubc9-UTR-Not1-3.6 to generate pLuc-Ubc9-3’-UTR-d1-d2.
Ubc9-UTR-5.2 (sense), 5’-ACATTTTTGCAAATCTAAAGT
Ubc9-UTR-Not1-3.6 (antisense), 5’-GCGGCCGCAGACAAAACGCCATATAAACAC
The PCR product was also first cloned into a PCR cloning vector and then subcloned into a modified pGL3 control vector where an EcoR1 and Not1 sites were introduced into the Xba1 site so that an insert can be unidirectionally cloned downstream of the luciferase gene. All the amplified products were verified by DNA sequencing before cloning into the final destination vector.
Luciferase Assay
Luciferase assays were carried out in 293T cells to determine the effect of microRNAs on the activity of Luc-Ubc9-3’-UTR and the deletion mutant constructs. First, cells were transfected with appropriate plasmids in 12-well plates. Then, the cells were harvested and lysed for luciferase assay 24 h after transfection. Luciferase activity was determined by using a luciferase assay kit (Promega) according to the manufacturer’s protocol. β-galactosidase was used for normalization.
PCR/RT-PCR and real-time RT-PCR
PCR was performed to amplify pre-microRNA sequences or the Ubc9 3’-UTR sequence according to the standard three-step procedure. Annealing temperature varied depending on the primers used. For RT-PCR, we isolated total RNA using Trizol reagent (Invitrogen) per the manufacturer protocol and used 1µg RNA to synthesize cDNA by SuperScriptase III (Invitrogen) with random primers. Finally, the resultant cDNA was used in regular PCR or real-time PCR reactions. To detect Ubc9 mRNA levels, we used the SYBR Green method with primers Ubc9-5.10 and Ubc9-3.10.
Ubc9-5.10 (sense) 5’-CAGGAGAGGAAAGCATGGAG
Ubc9-3.10 (antisense) 5’-TCGGGTGAAATAATGGTGGT
To detect mature microRNA expression, we also used Trizol reagent to isolate total RNA, which was then amplified by QuantiMir method (System Biosciences) or TaqMan stem-loop RT-PCR method (27, 28) using specific primer sets and TaqMan probe from Applied Biosystems. Real-time PCR reactions were performed in ABI 7500 HT thermal cycler according to the manufacturer’s protocol. Average levels of U6, 5s RNA and β-actin were used as an internal control. The fold-change between vector control and pre-microRNA expression vector was calculated with the 2−ΔΔCt method (27, 28).
Cell growth assay
Cell growth assays were carried out by MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] as described previously (29). In brief, cells were seeded in 96-well plates and incubated for various days before adding MTT. Absorbance at 570 nm was measured in the multi well plate reader (Thermo Scientific, Waltham, MA). The relative values were calculated by expressing the first day data as 1.
Western Blot
Cells were harvested and protein was extracted 2 days after transfection as previously described (30). Protein concentration was determined by protein assay kit (Bio-Rad, Hercules, CA) and samples were separated in 12% SDS polyacrylamide gels. Signals were revealed by a secondary antibody labeled with IRDye 800CW and the signal intensity was determined by Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence microscopy
To detect miR-30e-mediated suppression of Ubc9 by immunofluorescence staining, HeLa cells were first transfected with vector control or miR-30e expression vector and then transferred to coated coverslips in a 12-well plate. After overnight growth, the cells were fixed with 3% paraformaldehyde (Sigma-Aldrich) and permeabilized by 80% cold methanol, followed by washing with PBS (phosphate buffered saline). Coverslips were then incubated with 3%BSA in PBS for 10 min at room temperature. Primary antibodies against Ubc9 in PBST (PBS plus 0.1% Tween 20) were then added and incubated for 1 h at room temperature. After 3 washes with PBS, the cells were incubated with a fluorescence-conjugated secondary antibody in the dark for 1 h. For nuclear staining, the cells were subsequently stained in 0.5 µg/ml Hoechst dye (Sigma-Aldrich) for 5 min before examinations under a fluorescence microscope.
Immunohistochemistry (IHC)
Paraffin-embedded tissue was pretreated at 65°C for 2 h, followed by deparaffinization using standard procedures. Antigen retrieval was carried out in antigen retrieval solution (10 mM Tris, 1 mM EDTA, pH9.0) before applying the primary Ubc9 antibody. Thereafter, slides were incubated for 2 h at room temperature followed by extensive washes with PBST and further incubated for 1 h at room temperature with the secondary antibody conjugated with horse radish peroxidase (HRP). HRP activity was detected using Histostain Plus kit (Invitrogen) according to the manufacturer’s instruction. Finally, sections were counterstained with hematoxylin and mounted.
Patient specimens
Matched breast, head and neck, and lung tumor specimens were obtained from Cooperative Human Tissue Network (CHTN) Midwestern Division (Columbus, OH) or SIU SimmonsCooper Cancer Institute Tissue Bank. The use of these specimens in this study was approved by the Institutional Review Board of Southern Illinois University School of Medicine. Where it is necessary, total protein was isolated in protein extraction buffer using a tissue homogenizer as described previously (30) and protein concentration was determined by protein assays kit (Bio-Rad).
Statistical analysis
Statistical analysis of data was performed using the Student’s t test. Differences with p values less than 0.05 are considered significant.
Results
Ubc9 is upregulated in tumor specimens
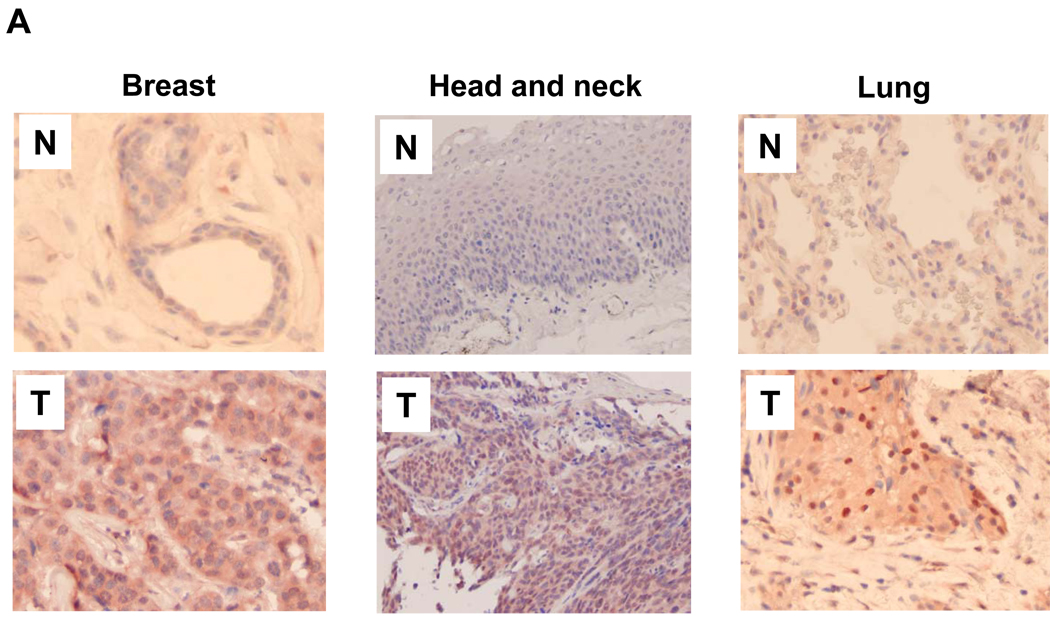
We have previously shown that overexpression of Ubc9 enhances tumor growth in the xenograft mouse model (20). To determine the clinical relevance of this finding, we examined expression levels of Ubc9 in the matched patient specimens including breast, head and neck, and lung by IHC. From 4 cases for each of three types of cancer, we found that the Ubc9 level was higher in tumor than the matched normal tissues. Shown in Fig. 1A were representative fields for each of three cases where the tumor specimens revealed intensive Ubc9 staining, concentrated in the nucleus. However, the matched normal tissues displayed very weak staining, suggesting that Ubc9 is overexpressed in tumors.
Fig. 1. Expression of Ubc9 in the matched tumor specimens.
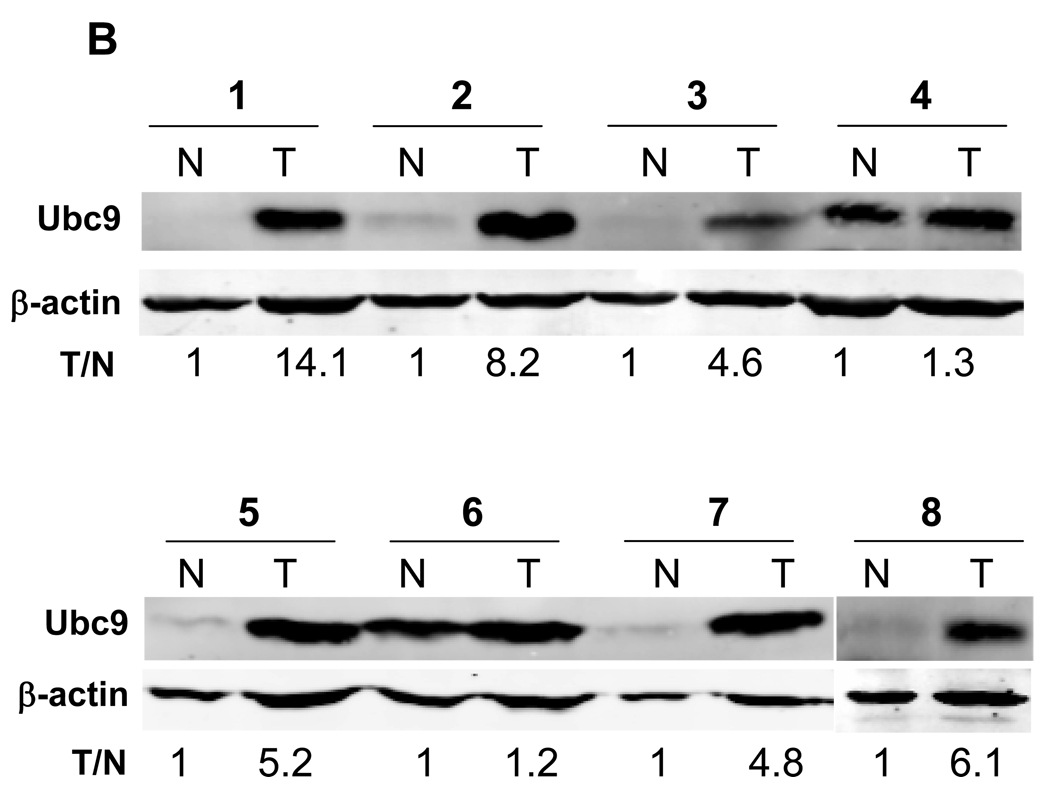
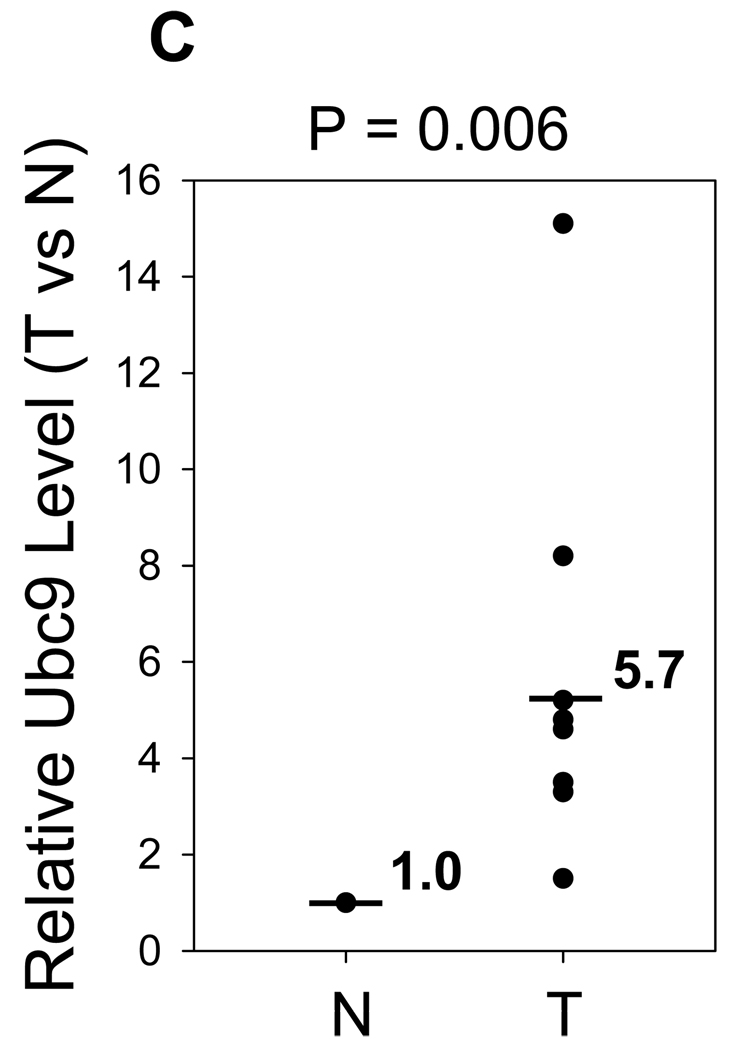
A, Paraffin-embedded specimens were stained by IHC using anti-Ubc9 antibody as described in Materials and Methods. Shown here are representatives of 3 cases for each type. Note strong Ubc9 signals in tumors compared to the matched normal tissues. B, Representative gels for Ubc9 levels in freshly frozen samples of matched breast tumor tissue, as detected by Western blot. Also shown are Ubc9 levels in tumor (T) vs normal tissue (N) after normalization with β-actin. C, Relative expression levels of Ubc9 between tumors and matched normal breast tissues (n = 8) derived from means of two experiments. The Ubc9 level was first normalized with β-actin and was then compared each other; the relative value of normal tissues was set at 1.
To better quantitate the Ubc9 expression in tumor specimens, we examined 8 pairs of frozen samples from the matched breast tumors by Western blot analysis. We found that Ubc9 was upregulated in all 8 cases (Fig. 1B). On average, breast tumors expressed a 5.7-fold higher than the matched normal tissues (Fig. 1C), which is consistent with the IHC data from paraffin-embedded samples (Fig. 1A).
Suppression of Ubc9 by miR-30
To better understand the upregulation of Ubc9 in tumors, we first examined the potential transcriptional regulation. Therefore, we cloned the putative Ubc9 promoter into a luciferase reporter plasmid and then introduced into several cell lines which expressed different levels of Ubc9. However, no significant difference in luciferase activity was seen, suggesting that transcriptional regulation may not be important for the observed difference of Ubc9 expression. Furthermore, we found that epigenetic factors such as methylation and acetylation did not appear to play a significant role in Ubc9 expression because the de-methylation agents such as 5-Aza-deoxycytidine or histone deacetylase inhibitors such as trichostatin A (TSA) had only a marginal effect on Ubc9 expression (not shown).
Therefore, we investigated the post-transcriptional regulation of Ubc9. Newly discovered small non-coding RNAs, microRNAs, have been shown to silence protein-coding genes in a variety of organisms including mammals by translation repression or mRNA degradation (31–33). MicroRNAs are believed to target mRNAs by partial sequence homology to the 3’-untranslated region (3’-UTR) of the target gene. Thus, we searched for potential microRNAs that might play a role in regulation of Ubc9 using several commonly cited microRNA target prediction programs such as TargetScan4 (34), miRBase Target51, PicTar (35) and miRanda (36)2. These four prediction programs all identified 7 putative microRNAs (miR-30a-e, miR-188 and miR-200c) (Table 1). In addition, some other microRNAs were identified by either two or three of these programs.
Table 1.
Putative microRNAs targeting Ubc9
| Name | BS# | Predicted by* |
|---|---|---|
| miR-30e | 2 | T, M, P, R |
| miR-30c | 2 | T, M, P, R |
| miR-30a | 1 | T, M, P, R |
| miR-30b | 1 | T, M, P, R |
| miR-30d | 1 | T, M, P, R |
| miR-188 | 1 | T, M, P, R |
| miR-200c | 1 | T, M, P, R |
| miR-195 | 1 | M, P, R |
| miR-548a | 1 | M, R |
| miR-450b | 1 | M, R |
| miR-361 | 1 | M, R |
| miR-10b | 1 | M, R |
| miR-376c | 1 | M, R |
| miR-200b | 1 | M, R |
| miR-877 | 1 | M, R |
| miR-802 | 1 | M, R |
| miR-49 | 1 | M, R |
| miR-652 | 1 | M, R |
BS, binding site
T, Targetscan4; M, miRBase Target5; P, PicTar; R, miRanda
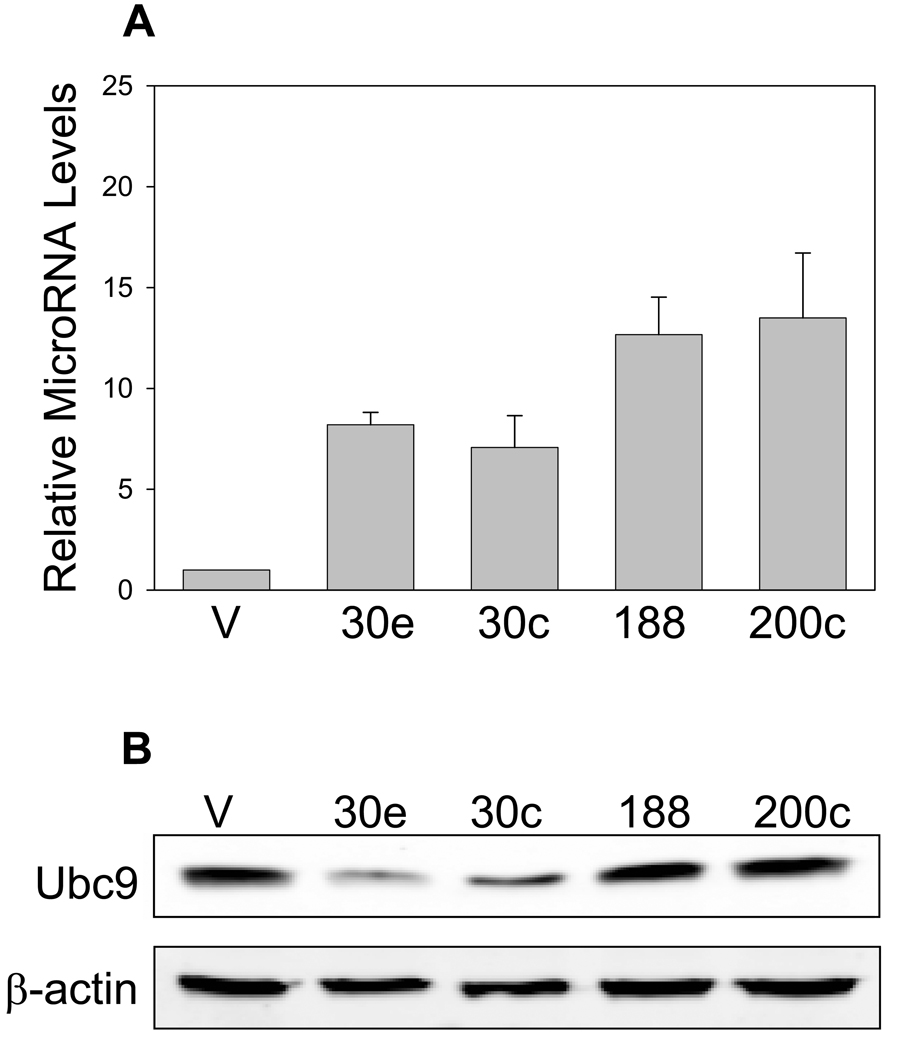
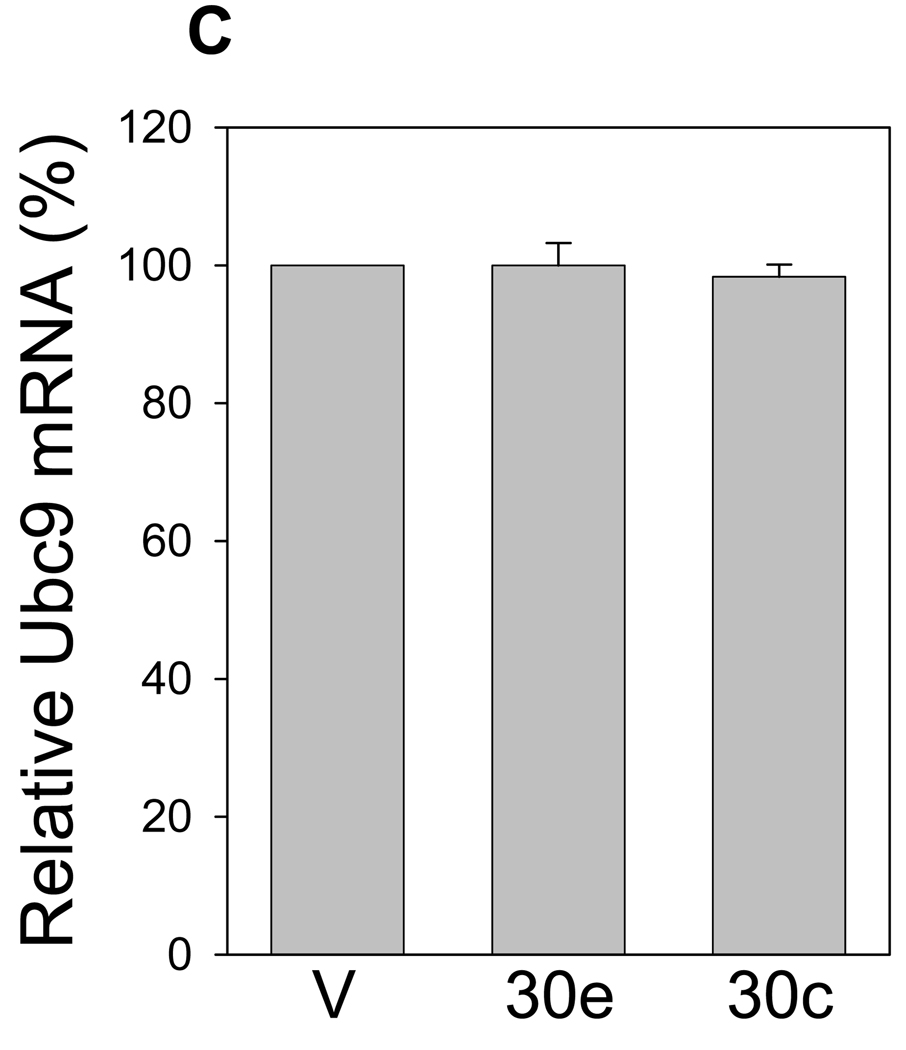
We focused on the miR-30 family and other two microRNAs, miR-188 and miR-200c. Among the miR-30 family, while both miR-30e and miR-30c target Ubc9 at two potential binding sides (Table 1) and the rest of the family have only one site. Thus, we chose miR-30e and miR-30c to represent the miR-30 family. We first confirmed that ectopic expression generated mature microRNAs by real-time RT-PCR (Fig. 2A) and then determined the effect of each microRNA on Ubc9 expression. Western blot analysis revealed that both miR-30e and miR-30c suppressed Ubc9 expression at the protein level (Fig. 2B). In contrast, we detected no significant effect on Ubc9 expression for miR-188 and miR-200c (Fig. 2B), highlighting the specificity of this suppression even though both miR-188 and miR-200c are also predicted to target Ubc9. To determine whether miR-30e and miR-30c affect the Ubc9 mRNA level, we performed real-time RT-PCR analysis for the same cells transfected with miR-30e and miR-30c, and found that these two microRNAs had no effect on the Ubc9 mRNA level (Fig. 2C), suggesting that they regulate Ubc9 expression mainly through translation repression.
Fig. 2. Ubc9 is specifically suppressed by miR-30e and miR-30c.
A, Ectopic expression of microRNAs in 293T cells. Cells were first transfected with microRNA expression vectors as detailed in Materials and Methods and then harvested for extraction of total RNA 2 days later. The mature microRNA levels were determined using QuantiMir real-time PCR method. In addition, we determined the miR-30e levels by the TaqMan real-time PCR method and found that the miR-30e levels were similar, as detected by both methods (not shown). B, A representative Western blot showing specific suppression of Ubc9 by miR-30e and miR-30c. C, Effect of miR-30e and miR-30c on Ubc9 mRNA. Values in (A) and (C) are average of three separate experiments ± SE. V, vector.
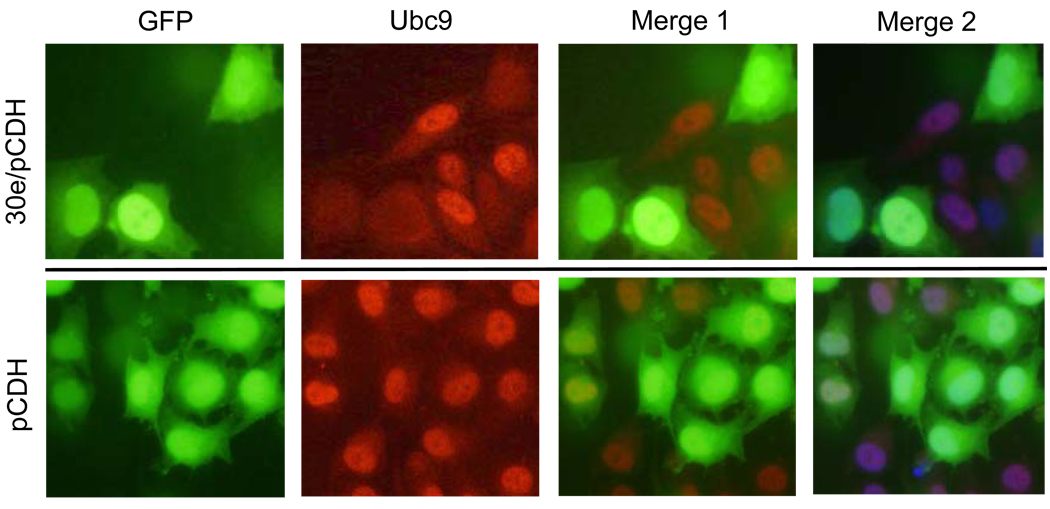
To further confirm the suppressive effect of miR-30e on Ubc9 expression, we introduced the miR-30e expression vector into HeLa cells and then immunostained with Ubc9 specific antibody. As shown in Fig. 3, ectopic expression of miR-30e remarkably suppressed Ubc9 expression because the red signal was clearly reduced (upper panels). In contrast, the vector control (pCDH) had no effect on Ubc9 (Fig. 3, bottom panels), further supporting the notion that Ubc9 is a target for miR-30e.
Fig. 3. Suppression of Ubc9 by miR-30e by immunofluroscence microscopy.
HeLa cells were transfected with miR-30e or vector (pCDH). One day later, the transfected cells were harvested and seeded on glass cover slips in a 12-well plate, and grown for an additional day before immunostaining with anti-Ubc9 antibody (red). Note reduction of Ubc9 expression (red signal) in miR-30e-transfected cells (green cells in upper panels). Merge 1, green plus red; Merge 2, green plus blue (nuclear staining).
Effect of miR-30e on cell growth
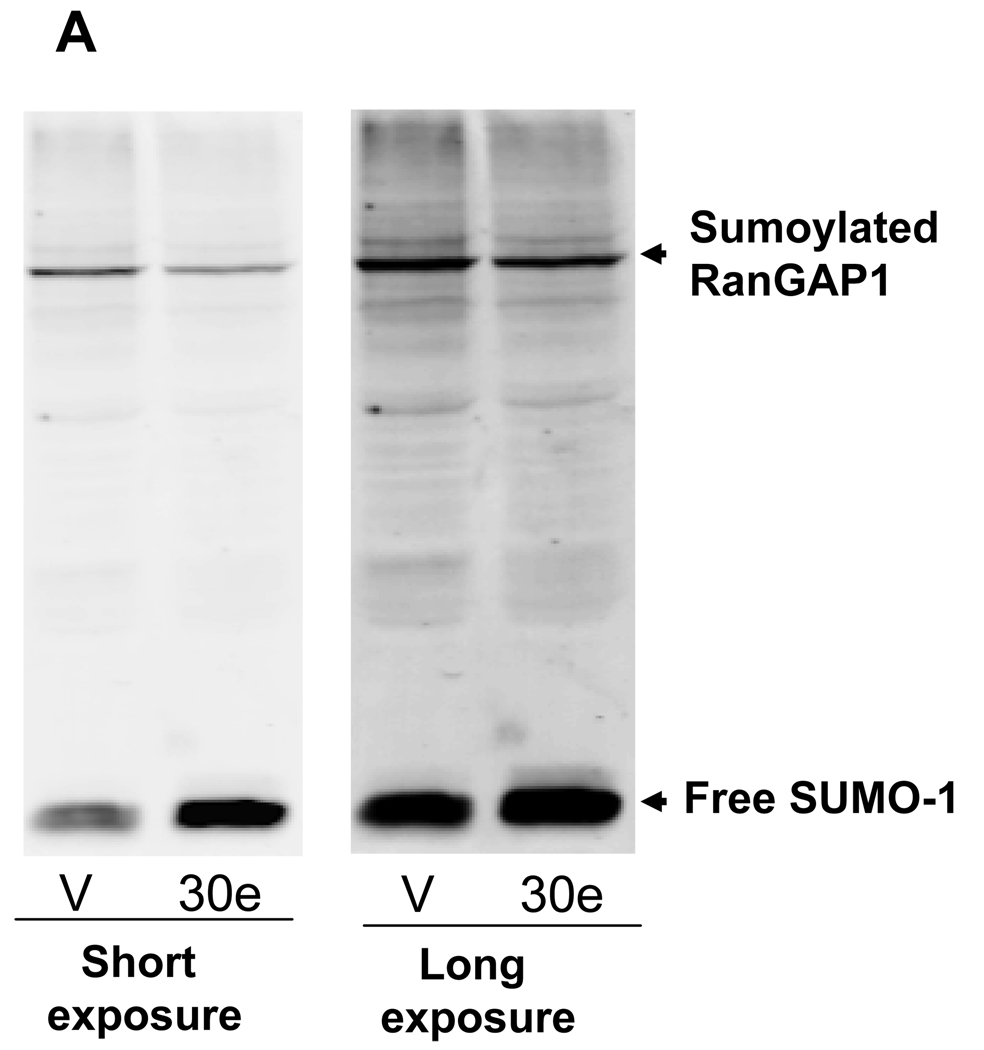
Given that Ubc9 is an E2 enzyme for sumoylation, suppression of Ubc9 by miR-30e would inhibit sumoylation. To test this hypothesis, we determined the effect of miR-30e on the overall levels of protein sumoylation using SUMO-1 antibody. As expected, miR-30e reduced total protein sumoylation (Fig. 4A) as compared to vector control. In particular, we found that miR-30e suppressed the level of sumoylated RanGAP1 (Fig. 4A) because RanGAP1 is a major SUMO substrate (37). In agreement with this, the free SUMO-1 level was higher in miR-30e-transfected cells than in vector control (Fig. 4A), presumably because reduction of overall sumoylation leads to the accumulation of the free SUMO-1.
Fig. 4. Suppression of sumoylation and cell growth by miR-30e.
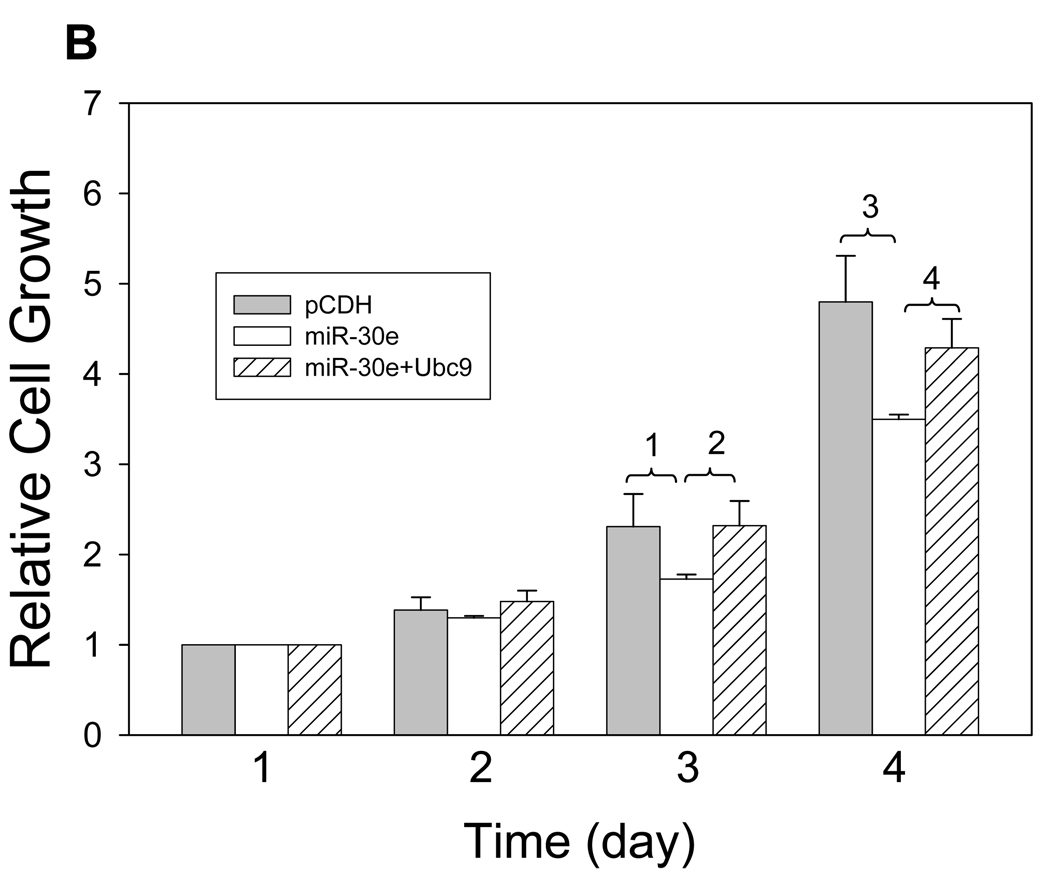
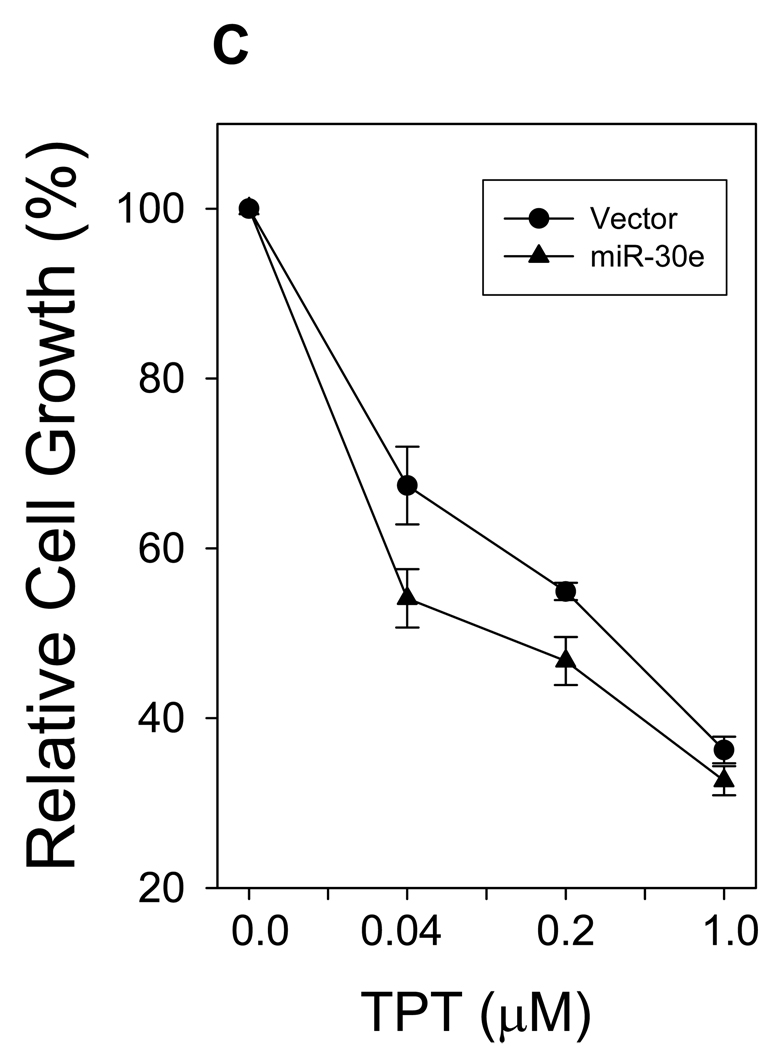
A, Detection of total protein sumoylation. 293T cells were first transfected with miR-30e or vector control. The same number of cells was then directly lyzed in 4% hot SDS, followed by sonication. Signals were detected by SUMO-1 antibody. B, Cells were first transfected with miR-30e, or vector; miR-30e plus pCMV-Ubc9 or miR-30e plus pCMV. There was virtually no difference in cell growth between miR-30e and miR-30e plus pCMV so the data set for miR-30e plus pCMV was not shown here. 1 and 2, 3 = 0.00002; 3, p = 0.0003; 4, p = 0.0002. C, miR-30e sensitizes cells to topotecan (TPT). Cells were first transfected with miR-30e or vector control and then treated with TPT at the indicated concentrations for 4 days. Cells growth was determined by MTT assays for both B and C, as described in Materials and Methods. Values are average of three separate experiments ± SE.
To further determine the effect of suppression of Ubc9 by miR-30e on cellular processes, we examined the cell growth for miR-30e-transfected cells because we have previously reported that suppression of Ubc9 causes cell growth inhibition (29). As expected, we found that miR-30e caused growth inhibition at a time-dependent manner. For example, at the first 2 days, there was no significant difference between vector and miR-30e. However, at days 3 and 4, miR-30e inhibited cell growth by almost 30% compared to the vector control (Fig. 4B). Of interest, this growth inhibition was partially reversed by overexpression of Ubc9 (Fig. 4B), suggesting that Ubc9 is an important target for miR-30e. In addition, miR-30e was able to sensitize cells to the anticancer agent topotecan (Fig. 4C), which is consistent with our previous finding that suppression of Ubc9 by Gam1 can increase the sensitivity to this agent (29). These results suggest that as a negative regulator of Ubc9, miR-30e plays a role in cell growth and drug response, in part through suppression of Ubc9 expression.
Ubc9 is a direct target for miR-30e
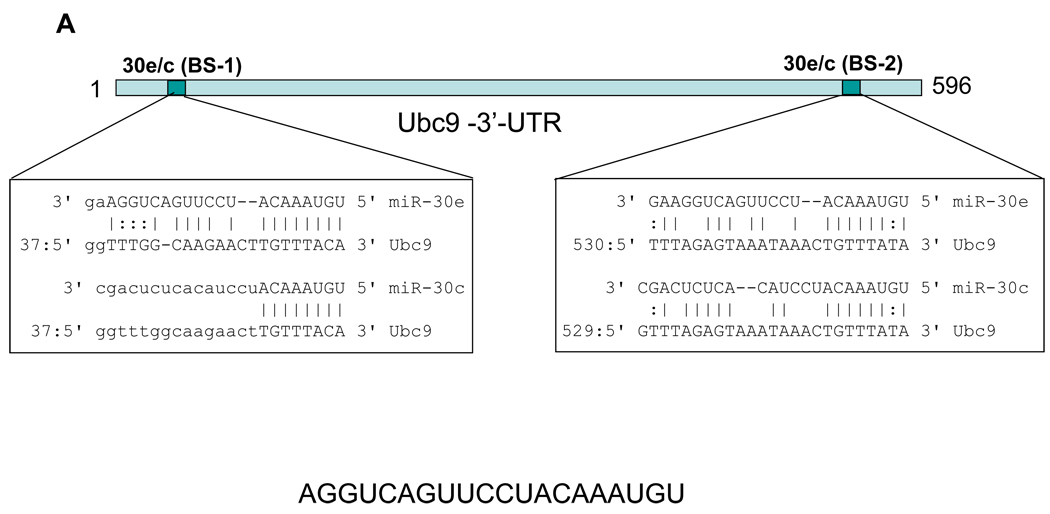
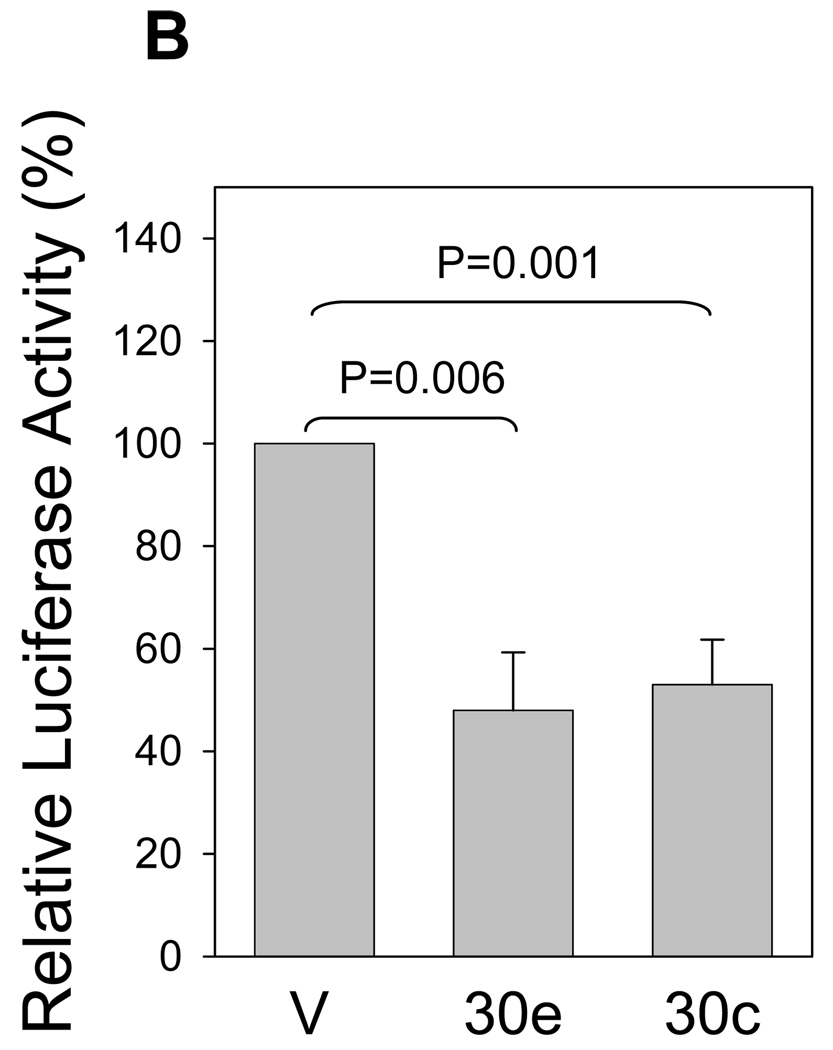
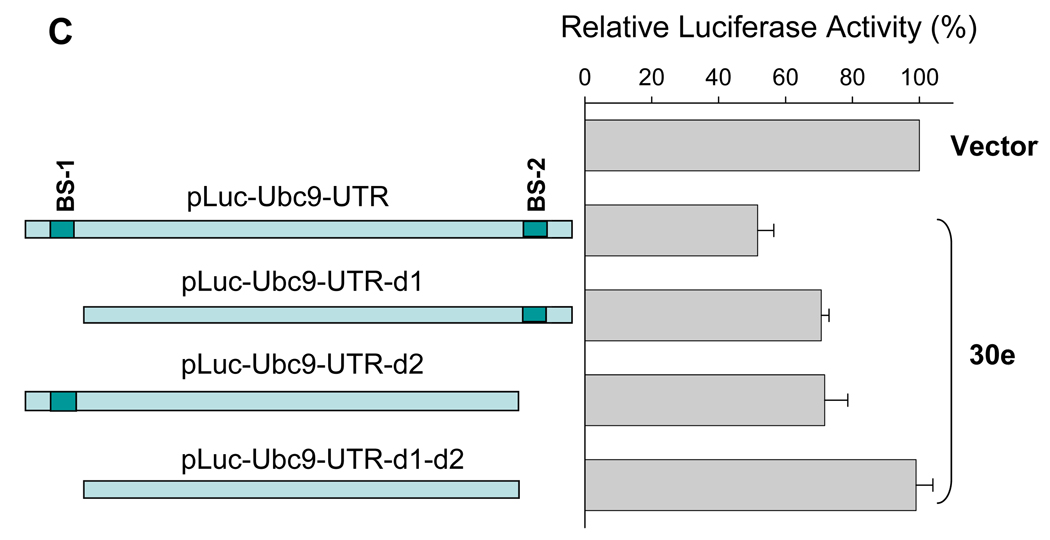
To determine whether miR-30e directly targets Ubc9, we cloned the Ubc9-3’-UTR (Fig. 5A) into pGL3 control vector, resulting in pLuc-Ubc9-3’-UTR (Fig. 5B). After transfection of 293T cells with this reporter construct along with miR-30e or miR-30c, we found that both miR-30e and miR-30c suppressed the luciferase activity by about 50% compared to the vector control (Fig. 5B), suggesting that Ubc9 is a direct target for these two microRNAs. As shown Fig. 5A, there are two potential microRNA binding sites in the 3’-UTR of Ubc9. To determine whether any of these two binding sites is important for microRNA suppression, we deleted the first (pLuc-Ubc9-3’-UTR-d1) or second binding site (pLuc-Ubc9-3’-UTR-d2) or both (pLuc-Ubc9-3’-UTR-d1-d2). As shown in Fig. 5C, deletion of the first binding site impaired the suppression of luciferase activity, but we still detected about 30% suppression; deletion of the second binding site had a similar effect. However, when both sites were deleted, miR-30e-mediated suppression of luciferase activity as abolished. These results suggest that both binding sites are critical for microRNA regulation.
Fig. 5. Ubc9 is a direct target for miR-30e.
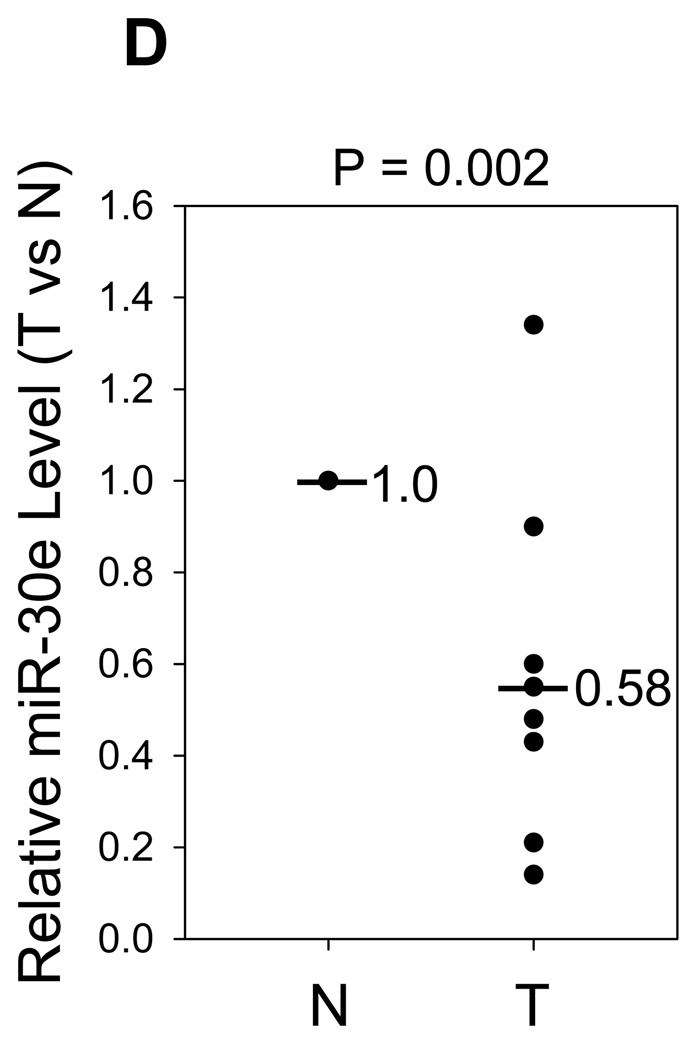
293T cells were transfected with pLuc-Ubc9-3’-UTR or its deletion constructs. At the same time, the cells were co-transfected with pCDH (V) or miR-30e/pCDH (30e) or miR-30c/pCDH (30c) and then harvested for luciferase assays 24 h later, as detailed in Materials and Methods. A, Schematic description of pUbc9 3’-UTR with putative binding sites for miR-30e (30e), and miR-30c (30c). B, Suppression of pLuc-Ubc9-3’-UTR luciferase activity by miR-30e and miR-30c. C, Deletion analysis of the pLuc-Ubc9-3’-UTR. The luciferase activity for each deletion construct was compared between pCDH (100%) and miR-30e. Values are averages of three separate experiments ±SE. D and E, Expression of miR-30e in matched normal breast tissue (N) and breast tumor (T) as CT value (D) or as relative expression of miR-30e (E) by normalizing normal tissue as 1. Each pair of samples is connected by a line in D.
Finally, to investigate the clinical relevance of miR-30e-mediated regulation of Ubc9, we amplified the entire UTR region from 8 pairs of tumors by RT-PCR so that we were able to determine whether there is any mutation or deletion in the Ubc9-3’-UTR, which could be responsible for the observed Ubc9 overexpression. DNA sequencing analysis of the PCR products indicated that both normal and tumor cells expressed wild-type sequences in all of 8 cases. Then we examined the level of miR-30e in these samples. Of interest, we found that the level of miR-30e was lower in tumor than normal tissue in 7 of them (Fig.5D and E), suggesting that Ubc9 overexpresssion in tumor could be in part due to downregulation of miR-30e.
Discussion
Despite its ubiquitous expression in normal tissues, we report here that Ubc9 is deregulated in several types of cancers including breast cancer, head and neck, and lung cancer. In particular, in breast cancer the Ubc9 level is over a 5-fold higher than the matched normal tissues. However, little was known regarding the basis of Ubc9 deregulation in tumors. Although we cannot exclude the possibility of transcriptional regulation of Ubc9, our study indicates that miR-30 family, particularly miR-30e, plays a regulatory role in Ubc9 expression and Ubc9 is a direct target for miR-30e.
Upregulation of Ubc9 expression in tumor specimens, as reported in this study as well as other studies (20, 23, 24), has clinical implications. We have previously shown that while suppression of Ubc9 function by the dominant negative Ubc9 inhibits, ectopic expression of Ubc9 enhances tumor growth in the animal model (20), suggesting that Ubc9 plays a causal role in tumorigenesis. This is likely due to the fact that Ubc9 is an essential enzyme for sumoylation and numerous important proteins, such as tumor suppressors or oncoproteins, are substrates for sumoylation. Thus deregulation of Ubc9 could lead to alterations of sumoylation pathways, ultimately impacting cell growth and cancer development. In this regard, Ubc9-mediated sumoylation is similar to ubiquitination. It is well known that deregulation of ubiquitination pathways could play a key role in cancer development (38, 39) because the timely and irreversible degradation of critical regulators is essential for normal cellular function and turnover of several regulatory proteins resulting from targeted destruction via ubiquitination. Similarly, Ubc9-mediated sumoylation has been shown to play a role in diverse cellular pathways (3), Therefore, cancer cells may have evolved mechanisms to target the basic functions of these protein modification pathways. One such mechanism could involve microRNA regulation of Ubc9 at the post-transcriptional level, leading to its upregulation in tumors.
Four lines of evidence support the notion that Ubc9 is a direct target for miR-30e. First, miR-30e specifically suppresses Ubc9 expression, as demonstrated by both Western blot and immunofluorescence staining. Second, ectopic expression of miR-30e inhibits overall protein sumoylation. Third, miR-30e also causes cell growth inhibition which can be attenuated by overexpression of Ubc9. Fourth, analyses of the luciferase reporter carrying the Ubc9 3’-UTR indicate that miR-30e directly interacts with this sequence and the putative miR-30e binding sites are essential for miR-30e regulation.
MicroRNAs are endogenous small non-coding RNAs that are known to post-transcriptionally regulate gene expression (31, 32). Aberrant expression of microRNAs has been reported in many types of tumors because they may function as oncogenes or tumor suppressor genes. While oncogenic microRNAs are often upregulated (40–42), tumor suppressive microRNAs are often downregulated in cancer (43–45). Little is known regarding the role of miR-30e. Since ectopic expression of miR-30e causes cell growth inhibition, we suggest that miR-30e is a tumor suppressor gene possibly by suppression of tumor promoting factors such as Ubc9. Downregulation of miR-30e in the matched breast tumor specimens supports this notion. Therefore, understanding how miR-30e is expressed in cancer will provide further insight into Ubc9 regulation. As a result, this knowledge will aid in the identification of novel targets in sumoylation-mediated cellular pathways.
Statement of Translational Relevance
As an essential E2 conjugating enzyme for sumoylation, Ubc9 plays a central role in sumoylation-mediated cellular pathways. Available evidence suggests that Ubc9 is a tumor promoting factor. However, little is known about the regulation of Ubc9. In this study, we first show that Ubc9 is overexpressed in several types of cancers, highlighting its clinical significance. We then investigate the underlying mechanism of Ubc9 upregulation and demonstrate that miRNAs such as miR-30e are able to specifically silence Ubc9, thus, providing new insight into Ubc9 regulation. Therefore, our study suggests that Ubc9 may serve as a potential biomarker for diagnosis or prognosis as well as therapeutic target for cancer intervention.
Acknowledgement
This study was supported in part by grants CA102630 (to YM) and CA40570 (to WTB) from NCI.
Abbreviations
- 3’-UTR
3’-untranslated region
- IHC
Immunohistochemistry
- PCR
polymerase chain reaction
- RT
reverse transcription
- qRT-PCR
quantitative RT-PCR
- TPT
topotecan
Footnotes
References
- 1.Muller S, Hoege C, Pyrowolakis G, et al. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 2.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 3.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 4.Tashiro K, Pando MP, Kanegae Y, et al. Direct involvement of the ubiquitin-conjugating enzyme Ubc9/Hus5 in the degradation of IkappaBalpha. Proc Natl Acad Sci U S A. 1997;94:7862–7867. doi: 10.1073/pnas.94.15.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 6.Mahajan R, Delphin C, Guan T, et al. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 7.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong Y, Rogers R, Matunis MJ, et al. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 9.Gostissa M, Hengstermann A, Fogal V, et al. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. Embo J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-del Arco P, Koipally J, Georgopoulos K. Ikaros SUMOylation: switching out of repression. Mol Cell Biol. 2005;25:2688–2697. doi: 10.1128/MCB.25.7.2688-2697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 12.Lin X, Liang M, Liang YY, et al. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J Biol Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- 13.Steffan JS, Agrawal N, Pallos J, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 14.Shao R, Zhang FP, Tian F, et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett. 2004;569:293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 15.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 16.Kurtzman AL, Schechter N. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc Natl Acad Sci U S A. 2001;98:5602–5607. doi: 10.1073/pnas.101129698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu LB, Omata W, Kojima I, et al. The SUMO conjugating enzyme Ubc9 is a regulator of GLUT4 turnover and targeting to the insulin-responsive storage compartment in 3T3-L1 adipocytes. Diabetes. 2007;56:1977–1985. doi: 10.2337/db06-1100. [DOI] [PubMed] [Google Scholar]
- 18.Kaul S, Blackford JA, Jr, Cho S, et al. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J Biol Chem. 2002;277:12541–12549. doi: 10.1074/jbc.M112330200. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez MS, Desterro JM, Lain S, et al. SUMO-1 modification activates the transcriptional response of p53. Embo J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo YY, Yu Y, Theodosiou E, et al. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–2683. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 21.Mo YY, Yu Y, Ee PL, et al. Overexpression of a dominant-negative mutant Ubc9 is associated with increased sensitivity to anticancer drugs. Cancer Res. 2004;64:2793–2798. doi: 10.1158/0008-5472.can-03-2410. [DOI] [PubMed] [Google Scholar]
- 22.Kovalenko OV, Plug AW, Haaf T, et al. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc Natl Acad Sci U S A. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDoniels-Silvers AL, Nimri CF, Stoner GD, et al. Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clin Cancer Res. 2002;8:1127–1138. [PubMed] [Google Scholar]
- 24.Moschos SJ, Smith AP, Mandic M, et al. SAGE and antibody array analysis of melanoma-infiltrated lymph nodes: identification of Ubc9 as an important molecule in advanced-stage melanomas. Oncogene. 2007;26:4216–4225. doi: 10.1038/sj.onc.1210216. [DOI] [PubMed] [Google Scholar]
- 25.Mo YY, Beck WT. Association of human DNA topoisomerase IIalpha with mitotic chromosomes in mammalian cells is independent of its catalytic activity. Exp Cell Res. 1999;252:50–62. doi: 10.1006/excr.1999.4616. [DOI] [PubMed] [Google Scholar]
- 26.Mo YY, Yu Y, Shen Z, et al. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem. 2002;277:2958–2964. doi: 10.1074/jbc.M108263200. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lao K, Xu NL, Yeung V, et al. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem Biophys Res Commun. 2006;343:85–89. doi: 10.1016/j.bbrc.2006.02.106. [DOI] [PubMed] [Google Scholar]
- 29.Wu F, Chiocca S, Beck WT, et al. Gam1-associated alterations of drug responsiveness through activation of apoptosis. Mol Cancer Ther. 2007;6:1823–1830. doi: 10.1158/1535-7163.MCT-06-0771. [DOI] [PubMed] [Google Scholar]
- 30.Zhu S, Si ML, Wu H, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 31.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? Rna. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 36.John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi S, Loda M. The role of the ubiquitination-proteasome pathway in breast cancer: use of mouse models for analyzing ubiquitination processes. Breast Cancer Res. 2003;5:16–22. doi: 10.1186/bcr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipkowitz S. The role of the ubiquitination-proteasome pathway in breast cancer: ubiquitin mediated degradation of growth factor receptors in the pathogenesis and treatment of cancer. Breast Cancer Res. 2003;5:8–15. doi: 10.1186/bcr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 42.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 43.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 44.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]