Abstract
Although the JAK/STAT pathway regulates numerous processes in vertebrates and invertebrates through modulating transcription, its functionally-relevant transcriptional targets remain largely unknown. With one jak and one stat (stat92E), Drosophila provides a powerful system for finding new JAK/STAT target genes. Genome-wide expression profiling on eye discs in which Stat92E is hyperactivated, revealed 584 differentially-regulated genes, including known targets domeless, socs36E and wingless. Other differentially-regulated genes (chinmo, lama, Mo25, Imp-L2, Serrate, Delta) were validated and may represent new Stat92E targets. Genetic experiments revealed that Stat92E cell-autonomously represses Serrate, which encodes a Notch ligand. Loss of Stat92E led to de-repression of Serrate in the dorsal eye, resulting in ectopic Notch signaling and aberrant eye growth there. Thus, our micro-array documents a new Stat92E target gene and a previously-unidentified inhibitory action of Stat92E on Notch signaling. These data suggest that this study will be a useful resource for the identification of additional Stat92E targets.
Keywords: Unpaired, JAK, STAT, micro-array, target genes, Drosophila, eye disc, Chinmo, Serrate, Delta, Notch, Imp-L2, Lama, Wingless, Pannier, Mo25, Pointed
INTRODUCTION
During development, extracellular cues activate conserved signal transduction pathways, which trigger changes in gene expression and ultimately lead to pleiotropic effects, including growth and differentiation. Frequently dys-regulation of these pathways leads to human diseases like cancer. One such pathway, Janus kinase/signal transducer and activator of transcription (JAK/STAT) was first identified as a key regulator of interferon and cytokine signaling in mammals (Schindler et al., 1992; Shuai et al., 1992; Velazquez et al., 1992; Watling et al., 1993; Shuai et al., 1994). These studies showed that JAKs are an unusual class of tyrosine kinases that are activated by IFN binding to its receptor. STATs are a unique family of latent cytoplasmic transcription factors that are recruited to phosphorylated IFN receptors and are then activated by JAKs. STAT dimers transit to the nucleus to modulate target gene transcription (Bach et al., 1997; Darnell, 1997).
Gene ablation experiments revealed that the four jak and seven stat genes regulate numerous processes in mammals, including growth and immunity (Russell et al., 1995; Thomis et al., 1995; Takeda et al., 1997; Parganas et al., 1998; Teglund et al., 1998; Metcalf et al., 2000; Laron, 2002). Other studies subsequently showed that sustained activation of the JAK/STAT pathway is a causal event in human leukemia and myeloproliferative disorders and that persistent activation of Stat3 is associated with a dozen other types of human cancer, including all classes of carcinoma (Lacronique et al., 1997; Baxter et al., 2005; Darnell, 2005; James et al., 2005; Levine et al., 2005; Jones et al., 2009; Kilpivaara et al., 2009; Olcaydu et al., 2009). Moreover, a dominant-active form of Stat3 called Stat3-c is oncogenic, transforms fibroblasts and causes tumors in nude mice (Bromberg et al., 1999). Inhibition of Stat3 function by siRNA knock-down or by small molecules arrests the growth of primary human cancer cells, which makes Stat3 an attractive target for cancer therapy (Blaskovich et al., 2003; Chiarle et al., 2005; Song et al., 2005; Sun et al., 2005). However, the functionally-relevant transcriptional targets of this pathway remain largely unidentified.
Drosophila serves as an excellent model for studying this pathway as it has a single jak and a single stat gene (Zeidler et al., 2000). In Drosophila, three related cytokines, Unpaired (Upd), Upd2 and Upd3, activate the receptor Domeless (Dome), which leads to the activation of the JAK called Hopscotch (Hop) and the STAT called Stat92E. Activated Stat92E induces expression of target genes dome and socs36E, the latter of which encodes a negative regulator (see Fig. 1E and (Binari and Perrimon, 1994; Hou et al., 1996; Yan et al., 1996; Harrison et al., 1998; Sefton et al., 2000; Brown et al., 2001; Callus and Mathey-Prevot, 2002; Chen et al., 2002; Ghiglione et al., 2002; Karsten et al., 2002; Agaisse et al., 2003; Bach et al., 2003; Rawlings et al., 2004; Gilbert et al., 2005; Hombria et al., 2005)). Work from numerous labs has shown that this pathway plays important roles in many aspects of Drosophila development, including growth and immunity (reviewed in (Arbouzova and Zeidler, 2006)). Importantly, two gain-of-function hop mutations were the first to link the JAK/STAT pathway to hyper-proliferation and cancer. These hop alleles result in hyperactive kinases and lead to a profound over-proliferation of blood cells, ultimately causing a fly “leukemia” and subsequent lethality (Harrison et al., 1995; Luo et al., 1995; Luo et al., 1997).
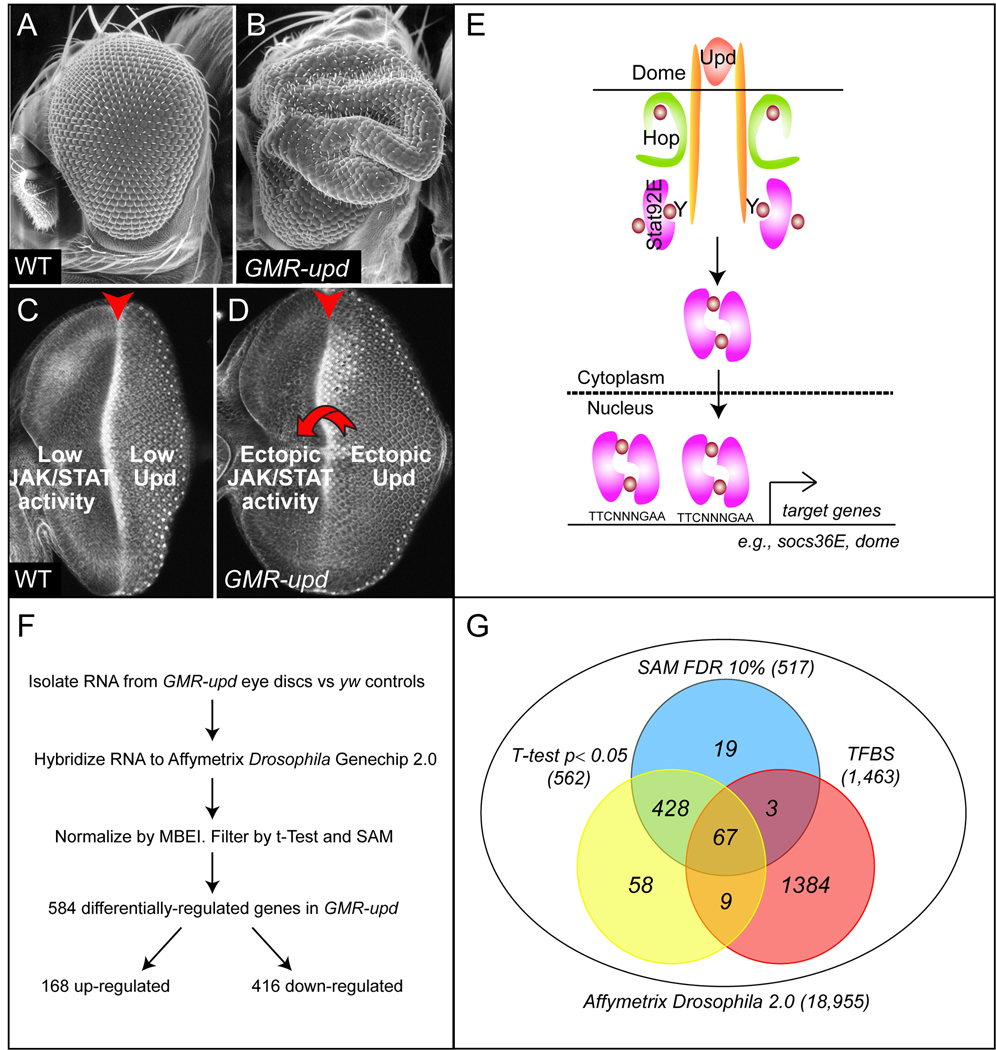
Figure 1. Over-expression of upd driven by the GMR promoter generates an enlarged-eye phenotype.
(A,B) Scanning electron micrographs (100x). Wild type (WT) adult eye (A). GMR-upd adult eye (B). (C,D) Immuno-fluorescence image of a WT (C) or GMR-upd (D) third instar eye imaginal disc dissected at 110 hours AED and stained with phalloidin. In wild type third instar eye discs, JAK/STAT signaling is low or not detectable because the ligand Upd stops being synthesized in posterior midline cells at the onset of third instar (Bach et al., 2003; Ekas et al., 2006; Bach et al., 2007). In contrast, in GMR-upd eye discs, Upd is ectopically produced throughout third instar by cells posterior to the morphogenetic furrow. This ectopic Upd diffuses anteriorly and induces activation of the JAK/STAT pathway in undifferentiated anterior cells (D, arrow). (E) The Drosophila JAK/STAT pathway. Upd is a secreted cytokine that activates a transmembrane cytokine receptor Dome, which in turn activates the JAK tyrosine kinase Hop and the STAT transcription factor Stat92E. Activated Stat92E dimers translocate from the cytoplasm to the nucleus, where they induce growth by promoting the transcription of targets, only a few of which like dome and socs36E have been identified. (F) Micro-array design and outcome (see also text for details). Biological quintuples of individual eye discs were analyzed by Affymetrix Drosophila 2.0 GeneChip arrays to identify sets of mRNAs significantly modulated between yw and GMR-upd samples. (G) The statistical filtering of array data (SAM or T-test) was performed as described in Materials and Methods, and overlap with a set of genes harboring Stat92E binding sites (TFBS) is shown.
We and others have previously shown that the JAK/STAT pathway plays important roles in growth and patterning of the Drosophila eye. The adult eye is derived from an epithelial imaginal disc, which arises from an embryonic primordium of ~50 progenitor cells (reviewed in (Cohen, 1993; Wolff and Ready, 1993; Dominguez and Casares, 2005)). These progenitors undergo exponential rates of growth during the first two of three larval stages or instars. During the third larval instar, this high rate of growth is curbed by signals to differentiate originating from the morphogenetic furrow as it moves across the eye disc in the anterior direction. Cells posterior to the furrow begin to differentiate into photoreceptors and their support cells, while cells anterior to it remain undifferentiated. The differentiated eye disc everts in the pupa to become functional in the adult.
In wild type eye discs, Upd synthesis is restricted to only a few cells at the posterior midline during the first and second larval instar, and its expression is extinguished in early third instar (Bach et al., 2003; Tsai and Sun, 2004; Ekas et al., 2006; Bach et al., 2007). Conversely, in the GMR-upd transgenic line, Upd is broadly mis-expressed in the eye disc at later larval stages in cells posterior to the furrow. Although the GMR promoter is active only in posterior eye cells, Upd is a secreted protein that diffuses away from the producing cells and, for reasons that are not completely clear, activates the JAK/STAT pathway only in undifferentiated eye cells located anterior to the morphogenetic furrow (Fig. 1C,D and (Bach et al., 2003; Ekas et al., 2006; Bach et al., 2007)). Activated Stat92E in anterior cells results in additional mitoses and increased cellular growth. These additional anterior cells are patterned normally by the furrow, ultimately leading to an adult eye that is 2 times larger than wild type (Fig. 1A,B and (Bach et al., 2003)). In contrast, loss of Stat92E activity leads to an adult eye that is both reduced in size and aberrantly patterned. We also previously reported that eye discs with large stat92E clones in the dorsal domain frequently exhibit large overgrowth in this region (Ekas et al., 2006).
Work from numerous labs has established that proliferative growth in the eye disc is continuous from late first instar to late second/early third instar (Dominguez and Casares, 2005). A well-known proliferative signal in the developing eye disc is provided by the Notch pathway. Although the Notch receptor is ubiquitously expressed in the eye disc, it is activated only at the D–V midline by the apposition of Notch ligands Delta (Dl) and Serrate (Ser) expression domains there (Kopczynski et al., 1988; Thomas et al., 1991). This D–V boundary acts as an organizing center for the growth of the disc (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998). Numerous genes are thought to act sequentially in early larval development to establish this localized Notch signaling (see Fig. 6J). During second instar, Wingless (Wg) and Hedgehog (Hh) are dorsally restricted and activate expression of the Iroquois complex (Iro-C) genes in the dorsal half of the eye disc (Rijsewijk et al., 1987; Lee et al., 1992; Cavodeassi et al., 1999). Iro-C gene products act redundantly to repress the expression of fringe (fng), which encodes a glycosyltransferase, to the ventral half of the eye primordium (Irvine and Wieschaus, 1994; Cho and Choi, 1998; Dominguez and de Celis, 1998; Cavodeassi et al., 1999; Yang et al., 1999; Haines and Irvine, 2003). Fng has been shown to potentiate the ability of Dl to activate Notch and to inhibit the ability of Ser to do so in the eye and wing disc, as well as in other tissues (Fleming et al., 1997; Panin et al., 1997; Klein and Arias, 1998; Haines and Irvine, 2003). It is currently postulated that asymmetric expression of fng, which generates a border of fng-expressing and fng-nonexpressing cells, is one of the most important steps in establishing local Notch activation at the D–V boundary, which results in global eye disc growth (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998). Once the Notch receptor is activated at the D–V boundary, it stimulates eye growth by induction of its target eyegone (eyg), which encodes a Pax6-like protein (Jun et al., 1998; Jang et al., 2003; Chao et al., 2004; Dominguez et al., 2004). eyg is expressed in a wedge along D–V boundary from second instar; this expression pattern depends upon Notch receptor activity and is required downstream of Notch for eye growth (Chao et al., 2004; Dominguez et al., 2004). Consistent with this model of Notch activation, eyg is only ectopically expressed in clones over-expressing Dl that reside in the ventral domain of the eye disc, where fng is normally expressed. Conversely, eyg expression is only induced by Ser-mis-expressing clones that reside in the dorsal region of the eye disc, where fng is normally not expressed (see Fig. 1j–m in (Dominguez et al., 2004) for examples).
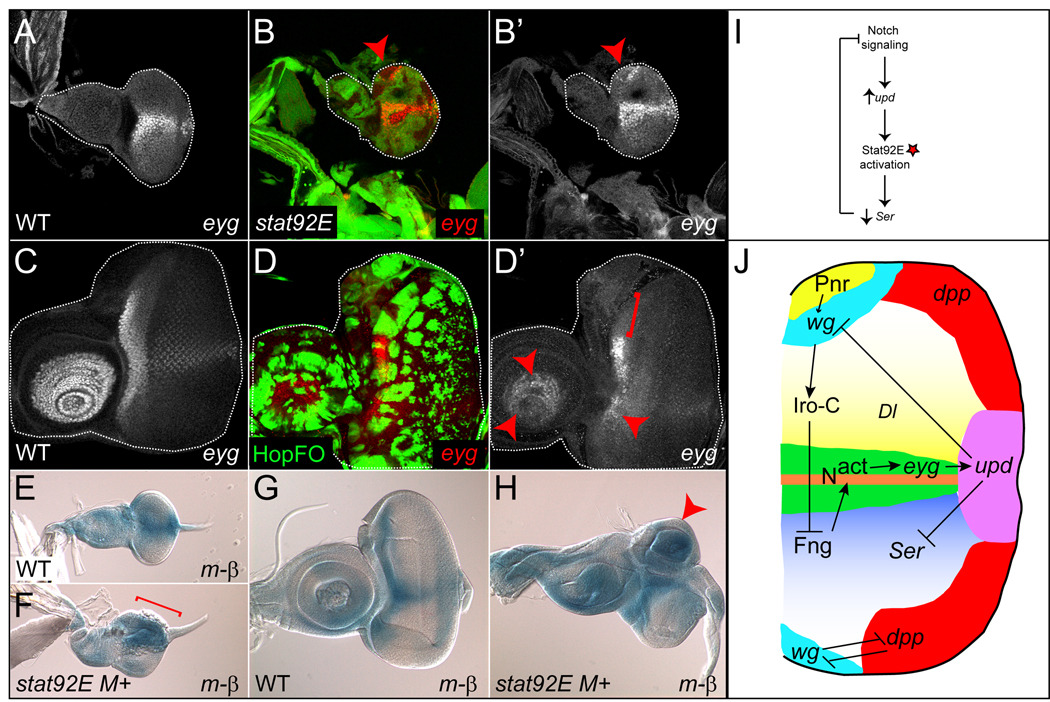
Figure 6. Notch signaling is ectopically activated when JAK/STAT signaling is reduced.
(A) eyg-lacZ (white) is expressed at the D–V midline in a wildtype (WT) late second instar eye disc. (B) eyg-LacZ (red) is ectopically expressed within a stat92E mosaic clone at the dorsal pole of the a second instar eye disc (B,B’, arrowhead). (C) In WT third instar, eyg-lacZ (white) is expressed at the anterior margin of the eye disc and in the distal antenna. (D) hop-expressing clones (green) cause repression of eyg-lacZ in a cell autonomous manner (D’, arrowheads and bracket). (E) The m-β reporter is normally expressed at the D–V boundary in a WT second instar eye disc. (F) However, in a second instar disc containing large stat92E M+ clones, this reporter is ectopically expressed throughout the dorsal eye disc (F, bracket), precisely in the region where Ser is ectopically expressed (see Fig. 4E). (G) In a WT third instar eye disc, the m-β reporter is normally expressed at the D–V boundary and at the anterior margin. (H) However, in a third instar disc containing large stat92E M+ clones, a circular, independent growth domain is observed in the dorsal eye that contains high levels of m-β reporter activity (arrowhead). (I) Model of negative feedback loop between Notch and JAK/STAT pathways. Notch regulates upd autonomously in the eye disc. Upd then acts on neighboring cells to activate Stat92E (red star). Stat92E represses Ser expression in a cell-autonomous manner. This results in reduction in Notch ligand levels and a decrease in Notch receptor activity. (J) Model of negative feedback loop between Notch and JAK/STAT pathways in the context of the developing eye disc. See text for details. eyg-LacZ was detected by anti-β-gal. Single channel for eyg-LacZ is (A,B’,C,D’). hop-expressing clones are GFP+ (D). m-β was detected by Xgal staining.
In the last few years, work from several laboratories have shown that Notch regulates growth of the eye disc, at least in part through cell-autonomous of induction of the upd gene, most likely directly via Eyg (see Fig. 6I,J and (Chao et al., 2004; Dominguez et al., 2004; Reynolds-Kenneally and Mlodzik, 2005)). The critical role of JAK/STAT pathway signaling in growth of the eye disc is highlighted by the fact that upd expression and Stat92E activity are highest from first to early third larval instar, the proliferative growth phase of the eye disc (Dominguez and Casares, 2005; Ekas et al., 2006; Bach et al., 2007). Moreover, hyper-activation of Notch in clones, either by over-expressing an activated form of Notch or by trapping activated Notch receptors in the endocytic pathway by loss-of-function mutations in ESCRT genes, leads to dramatic cell-autonomous increases in upd expression. This, in turn, triggers non-autonomous activation of Stat92E in neighboring cells and results in tissue overgrowth (Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005; Herz et al., 2006). Furthermore, additional molecules have been shown to increase Stat92E activity and cause over-growth of the eye. Most notably, a mutation in the Drosophila C-terminal src kinase gene leads to ectopic activation of Src and then of Stat92E, which results in overgrowth of the eye (Read et al., 2004).
Mammalian STAT binding elements share a similar overall sequence of TT(N5)AA (Seidel et al., 1995). Studies of in vitro selection of oligonucleotides bound to Stat92E revealed that it binds to a consensus sequence similar to the mammalian one: TTC(N)3GAA (Yan et al., 1996). Stat92E can function as a transcriptional co-activator and induce expression of several in vivo and in vitro reporters (Baeg et al., 2005; Gilbert et al., 2005; Muller et al., 2005; Bach et al., 2007; Tsai et al., 2007). However, only a handful of Stat92E target genes are currently known. dome, socs36E, even-skipped (eve) stripe 3 enhancer, D-eIF1A, Turandot A, thiolester-containing protein 1–4 (tep 1–4), ptp61F, apontic and potentially c-raf appear to be positively regulated by JAK/STAT signaling (Yan et al., 1996; Kwon et al., 2000; Lagueux et al., 2000; Myrick and Dearolf, 2000; Ghiglione et al., 2002; Karsten et al., 2002; Agaisse et al., 2003; Bach et al., 2003; Baeg et al., 2005; Muller et al., 2005; Starz-Gaiano et al., 2008). Of these genes, only dome and socs36E have been shown by clonal analysis to be both positively- and cell-autonomously regulated by Stat92E (Ghiglione et al., 2002; Bach et al., 2003; Bach et al., 2007). Furthermore, only the Stat92E binding sites in eve stripe 3 have been proven by mutational analysis to be critical for Stat92E–dependent transcriptional regulation (Yan et al., 1996). Stat92E has also been shown to negatively regulate the wg gene in an cell-autonomous manner in the eye, antenna and leg discs, as well as in the presumptive notum of the wing disc (Ekas et al., 2006; Ayala-Camargo et al., 2007; Tsai et al., 2007). However, it is not known whether Stat92E can act as a repressor to inhibit wg transcription or whether Stat92E’s regulation of wg is indirect, for example by Stat92E inducing a direct target gene that encodes a wg repressor. Taken together, these pioneering studies highlight the need to identify and characterize more target genes that are autonomously regulated by the JAK/STAT pathway, especially those that have roles in growth control.
To identify new JAK/STAT target genes, we performed rigorous genome-wide expression profiling using RNA from GMR-upd eye discs, in which the JAK/STAT is hyper-activated, compared to control yw eye discs. This analysis led to the identification of 584 differentially-regulated genes, three of which are known targets: socs36E, dome, and wg. We validated in vivo in GMR-upd eye imaginal discs the differential expression of 19 up-regulated genes, including chronologically inappropriate morphogenesis (chinmo), lamina ancestor (lama), Mo25 and pointed (pnt) and 9 down-regulated genes, including pannier (pnr), ecdysone-inducible gene L2 (Imp-L2), dachsous (ds), Serrate (Ser) and Delta (Dl). In total, we validated by at least one method 28 differentially-regulated genes in this micro-array. We then showed that Ser and Dl are ectopically expressed within stat92E loss-of-function clones. Furthermore, we found that Ser is robustly repressed in a cell-autonomous manner by activated Stat92E. Most notably, we determined the functional consequence of Stat92E–mediated repression of Ser: loss of JAK/STAT pathway actvity in clones leads to inappropriate activation of Notch signaling in the dorsal domain of the eye by ectopic expression of Ser there in the absence of Fng. This results in the generation of ectopic growth organizing centers and leads to over-growth of the dorsal domain of the eye disc. These data have defined a new and unexpected role for the JAK/STAT pathway in regulating growth of the eye disc through restricting Notch activity by repressing Notch ligand expression. Lastly, these data indicate that a negative feedback loop exists between Notch and JAK/STAT pathways in the developing eye.
RESULTS
We previously reported that Upd is expressed by a few cells at the posterior margin of the eye disc beginning in the first larval instar and ending in early third instar (Bach et al., 2003; Ekas et al., 2006; Bach et al., 2007). We took advantage of this temporally and spatially restricted expression pattern to generate the GMR-upd transgenic line, in which Upd is mis-expressed throughout third instar by being placed directly under the regulatory elements of the Glass multiple repeat (GMR) promoter (Hay et al., 1994). We previously reported that GMR-upd animals have a dramatically enlarged adult eye (Fig. 1A,B and (Bach et al., 2003)). As mentioned above, the GMR promoter is active only in posterior eye cells, but the mis-expressed Upd diffuses away from the cells that secreted it and activates Stat92E only in undifferentiated eye cells located anterior to the morphogenetic furrow (Fig. 1C,D and (Bach et al., 2003; Ekas et al., 2006; Bach et al., 2007)). In early third instar, GMR-upd eye discs are the same size as yw controls (data not shown and (Bach et al., 2003)). However, later at ~110 hours after egg deposition (AED) (i.e., the middle of the third larval instar), GMR-upd eye discs become larger than controls, as a result of Upd over-expression (Fig. 1C,D and (Bach et al., 2003)). The sensitivity of undifferentiated eye cells to Upd is exemplified by the up-regulation of target genes socs36E and dome only in cells anterior to the furrow, as well as the increased proliferation of these anterior cells in GMR-upd eye discs (Fig. 2A,B,D,E and (Karsten et al., 2002; Bach et al., 2003; Bach et al., 2007)). We previously reported that the additional anterior progenitor cells in GMR-upd eye discs differentiate in an appropriate manner and give rise to an enlarged, but normally patterned, adult eye that has substantially increased numbers of ommatidia (Fig. 1A,B and (Bach et al., 2003)).
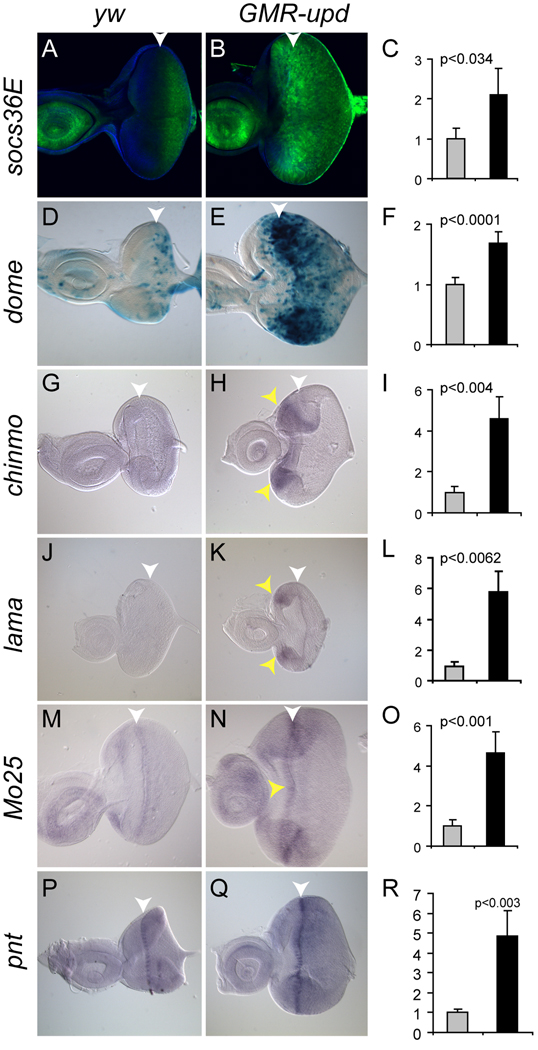
Figure 2. Up-regulated genes anterior to the furrow in GMR-upd discs.
The expression of all genes was analyzed in mid-third instar eye discs. The morphogenetic furrow (MF) is marked by a white arrowhead. The Y-axis in the bar graphs represents relative mRNA abundance. (A,B) socs36E expression as assessed by the 10xSTAT92E-GFP transcriptional reporter (green) in yw control discs (A). socs36E is up-regulated anterior to the MF in GMR-upd discs (B). (C) socs36E is increased 2.10 fold in the GMR-upd micro-array. (D,E) dome-Gal4, UAS-lacZ (abbreviated dome) expression (blue) in control eye discs (D). dome is up-regulated in cells anterior to the MF in GMR-upd discs (E). (F) dome is increased 1.68 fold in the GMR-upd micro-array. (G,H) chinmo-RC (labeled chinmo) mRNA, as assessed by in situ hybridization, is absent in control discs (G) and is increased in cells anterior to the MF in GMR-upd discs (H, yellow arrowheads). (I) chinmo is increased 4.6 fold in the GMR-upd micro-array. (J,K) lama mRNA, as assessed by in situ hybridization, is not present in wild type discs (J). However, it is up-regulated in cells anterior to the MF in GMR-upd discs (K, yellow arrowheads). (L) lama is increased 5.44 fold in the GMR-upd micro-array. (M,N) Mo25 mRNA, as assessed by in situ hybridization, is present faintly in cells within and immediately surrounding the furrow in wild type discs (M). In GMR-upd discs Mo25 has a broader expression domain as well as increased expression levels (N, yellow arrowheads). (O) Mo25 is increased 4.65 fold in the GMR-upd micro-array. (P,Q) pnt mRNA, as assessed by in situ hybridization, is present in cells within the furrow in wild type discs (P). In GMR-upd discs, pnt is expressed at a significantly higher level in cells within the furrow, as well as at higher levels in cells adjacent to the furrow (Q). (R) pnt is increased 4.8 fold in the GMR-upd micro-array. Gray bar is yw and black bar is GMR-upd (C,F,I,L,O,R).
To identify Stat92E target genes, we performed a genome-wide micro-array analysis using GMR-upd eye discs as compared to controls from identically-aged animals. We isolated single larval eye discs from GMR-upd and yw controls at the 110-hour AED time point and performed five independent replicates of both samples (Fig. 1C,D,F see Materials and Methods). The micro-array data was normalized using MBEI, and analyzed using two different statistical methods, T-test and SAM (Fig. 1F and Materials and Methods). We identified 584 statistically significant, differentially-regulated genes, out of which 495 (or 84%) were identified by both statistical methods, suggesting that the expression values are robust, while 23 and 67, respectively, were identified by either SAM or T-test alone (Fig. 1G, Suppl. Table 1). For this 584 transcript list, the overall measurement reproducibility and limited variance within each tested genotype and the simultaneous magnitude of differential expression between the two genotypes is summarized by box-plot analysis (Suppl. Fig. 1). We compared these 584 genes to the list of those identified in a whole-genome bio-informatics search for clusters of Stat92E binding sites using Target Explorer, the web-based search engine designed for Drosophila genomes (Suppl. Table 2 and see Materials and Methods and (Sosinsky et al., 2003)). 79 (13.5%) of these genes had at least one cluster of Stat92E binding sites, increasing the possibility that they could be direct Stat92E targets (Fig. 1G and Suppl. Table 2).
We used the NIH DAVID suite to functionally annotate the lists of differentially-modulated genes extracted from our micro-array data (Huang da et al., 2009). From the 584 differentially-regulated genes, this platform was able to identify dome, socs36E, ken and barbie (ken), and Fps oncogene analog (Fps85D) as JAK/STAT pathway components, indicating that this program has a high probability of assigning correct function to the genes in the GMR-upd micro-array (Suppl. Table 3 and (Jiang et al., 2001; Bach et al., 2003; Arbouzova et al., 2006)). We also identified many genes involved in the regulation of processes in which the JAK/STAT pathway has well-established roles, including oogenesis, cell migration, embryogenesis, proximal-distal pattern formation, immune response, hemocyte differentiation and hindgut development (Suppl. Table 3 and (Hou et al., 1996; Yan et al., 1996; Silver and Montell, 2001; Baksa et al., 2002; Beccari et al., 2002; Agaisse et al., 2003; Johansen et al., 2003; Ayala-Camargo et al., 2007; Krzemien et al., 2007)). These data suggest that the GMR-upd micro-array accurately identified genes that are differentially-regulated by JAK/STAT signaling.
Genes up-regulated in the GMR-upd micro-array
168 (28.8%) of the 584 differentially-regulated genes in the GMR-upd micro-array were up-regulated (Suppl. Table 1). The white (w) gene served as an internal control for this study. The GMR-upd transgene contains a copy of the w cDNA and is maintained in a Drosophila stock that was homozygous for a null mutation in the endogenous w gene. Since the control RNA samples were derived from flies that were also homozygous mutant for the w null allele, w mRNA should be up-regulated in GMR-upd eye discs. Indeed, w is increased 6.4 fold in the micro-array and 20 fold by Q-PCR (Suppl. Table 1, Suppl. Fig. S2). As an additional control, upd was not expected to be up-regulated in this analysis because the GMR-upd transgene contains only the upd coding sequence (and not sequences from the 5’ and 3’ untranslated regions (UTRs) (Bach et al., 2003)), while the upd Affymetrix probes are designed for the 3’ UTR of this transcript. Indeed, upd is not a regulated transcript in this micro-array (not shown). Importantly, we found that the expected target genes dome and socs36E are significantly increased 1.68 and 2.10 fold, respectively, in GMR-upd samples versus controls (Suppl. Table 1, Fig. 2C,F). We validated these results in vitro and in vivo. Q-PCR revealed that dome was increased 3.3 fold, while socs36E was increased 2.4 fold in GMR-upd samples as compared with controls (Suppl. Fig. S2). More importantly, in GMR-upd eye discs both genes exhibited significantly increased expression in cells anterior to the morphogenetic furrow, the region of this disc where Stat92E transcriptional activity is the highest (compare Fig. 2A to 2B and 2D to 2E and (Bach et al., 2007)). The fact that our analysis revealed the two best characterized Stat92E targets (dome and socs36E) as up-regulated transcripts further supports the validity of our results.
We were also able to demonstrate that four other potential Stat92E target genes are specifically increased in cells anterior to the furrow in GMR-upd eye discs as compared to yw controls: chinmo, lama, Mo25 and pnt. Flybase predicts the chinmo transcription unit to have four splice-variants: chinmo-RA, -RB, -RC, -RD. We found that the -RC isoform is increased 4.6 fold while the -RD variant is increased 2.73 fold as compared to controls (Fig. 2I and Suppl. Table 1). Q-PCR using primers for a region of chinmo shared by all isoforms revealed that chinmo mRNA is increased 2 fold in GMR-upd samples (Suppl. Fig. S2). Furthermore, in situ hybridization with chinmo-RC and –RD specific ribo-probes showed that both chinmo isoforms are absent in mid-third instar yw control eye discs (Fig. 2G and not shown), while both are strongly up-regulated in cells anterior to the furrow in GMR-upd eye discs (Fig. 2H, yellow arrowheads and not shown). Target Explorer identified one cluster of Stat92E binding sites in putative regulatory regions of the chinmo gene, raising the possibility that it is directly regulated by Stat92E activity (Suppl. Table 2).
lama is up-regulated 5.44 fold in the GMR-upd micro-array (Fig. 2L, Suppl. Table 1). Consistent with this finding, Q-PCR revealed that it is increased 3 fold in GMR-upd samples (Suppl. Fig. S2). lama encodes a Phospholipase B protein that is expressed in neural and glial precursors prior to differentiation (Perez and Steller, 1996). in situ hybridization showed that lama is not expressed in control third instar eye discs (Fig. 2J). However, it is up-regulated in cells anterior to the furrow in GMR-upd eye discs, particularly at the dorsal and ventral poles (Fig. 2K, yellow arrowheads). Target Explorer identified two clusters of Stat92E binding sites in non-coding, putative regulatory regions of the lama gene, raising the possibility that lama is directly regulated by Stat92E (Suppl. Table 2).
Mo25 was increased 4.65 fold in the GMR-upd micro-array (Fig. 2O, Suppl. Table 1). Although the specific function of Drosophila Mo25 is not currently known, Mo25 family members are widely conserved in eukaryotes, and there is growing evidence that they play important roles in regulating growth and cell polarity in yeast, worms and humans (Watts et al., 2000; Milburn et al., 2004; Mendoza et al., 2005). Mo25 mRNA can be detected at low levels in cells surrounding the furrow in yw control eye discs (Fig. 2M). However, we observed an increase in Mo25 expression in a broader swath of cell surrounding the furrow in GMR-upd eye discs (Fig. 2N, yellow arrowheads). These results suggest that the ectopic JAK/STAT signaling in GMR-upd discs can up-regulate the Mo25 gene. However, the lack of any clusters of Stat92E binding sites in the Mo25 gene suggests that Stat92E may regulate it indirectly or through the three single Stat92E binding sites present in this gene (not shown).
Lastly, pnt, which encodes an ETS family transcription factor that is directly induced upon activation of the Epidermal growth factor receptor, is increased 4.8 fold in the GMR-upd micro-array (Fig. 2R, Suppl. Table 1 and (Klambt, 1993; Gabay et al., 1996)). In wild-type eye discs, pnt mRNA is strongly expressed in groups of cells within the morphogenetic furrow (Fig. 2P). Consistent with the micro-array results, we observed an increase in pnt expression within cells in the furrow in GMR-upd eye discs (Fig. 2Q). Furthermore, Target Explorer identified two clusters of Stat92E binding sites in the pnt gene, raising the possibility that Stat92E may directly regulate pnt expression (Suppl. Table 2).
Additionally, we validated 13 genes up-regulated in the GMR-upd micro-array by Q-PCR: w, ken, CG11784, Fps85D, atypical Protein Kinase C (aPKC), PAR-domain protein 1 (pdp1), escargot (esg), terribly reduced optic lobes (trol), Signal recognition particle receptor β (SrpRβ), brain tumor (brat), domino (dom), tep-2 and polychaetoid (pyd) (Suppl. Table 1 and Suppl. Fig. 2). Of these, one gene (tep-2) is highly homologous to a complement-like gene tep-1 that is strongly induced in hopTum-l animals (Lagueux et al., 2000). Five others (pdp1, esg, trol, SrpRβ and pyd) all have one cluster of Stat92E binding sites in putative regulatory regions, raising the possibility that they may be direct Stat92E target genes (Suppl. Table 2). Furthermore, deficiencies that removed ken, aPKC, trol, tep-2 and pyd dominantly modified the GMR-upd enlarged eye phenotype in an F1 modifier genetic screen (Bach et al., 2003). c-fes oncogene, a Src-related fps protein tyrosine kinase member and the mammalian Fps85D ortholog, acts downstream of Jak1 in proliferation of B lymphocytes (Jiang et al., 2001). The remaining genes have not previously been linked to JAK/STAT pathway signaling. In sum, we successfully validated 19 genes up-regulated in the GMR-upd micro-array by at least one method.
Genes down-regulated in the GMR-upd micro-array
416 genes (71.2%) were down-regulated in GMR-upd samples. We previously reported that in the developing eye disc Stat92E represses both wg and pannier (pnr), which encodes a GATA transcription factor (Maurel-Zaffran and Treisman, 2000; Ekas et al., 2006). Therefore, these genes are predicted to be down-regulated when JAK/STAT signaling is hyper-activated in the eye disc. As expected, pnr and wg were down-regulated 2.13 and 1.61 fold, respectively, in GMR-upd samples (Fig. 3C,F, Suppl. Table 1). Furthermore, Q-PCR revealed that both transcripts are significantly down-regulated, 4.60 and 2.02 fold, respectively, in GMR-upd samples (Suppl. Fig. S3). In the eye imaginal epithelium, pnr is normally expressed dorsally in peripodial cells located “above” undifferentiated cells anterior to the furrow (Fig. 3A). Consistent with previous results, we find that pnr is repressed in dorsal peripodial cells by ectopic expression of Upd (Fig. 3B and (Ekas et al., 2006)). The area of the pnr expression domain is 98 pixel sq. in control eye discs, but this value is reduced by 30% to 60 pixel sq. in GMR-upd eye discs (Fig. 3A,B). In wild type eye discs, wg is expressed in cells at the dorsal and ventral poles anterior to the furrow (Fig. 3D). In GMR-upd discs, wg expression is diminished in these cells anterior (Fig. 3E). Furthermore, as we previously reported, clones that over-express Hop, which autonomously activates Stat92E, cause cell-autonomous repression of wg at both the dorsal and ventral poles of the eye disc (Suppl. Fig. 4E,E’,E” and (Ekas et al., 2006)). Thus, the GMR-upd micro-array identified the only two known genes repressed by Stat92E (wg and pnr) as differentially-regulated in the GMR-upd samples. This observation strongly suggests that our analysis is likely to detect other targets that are negatively regulated by Stat92E.
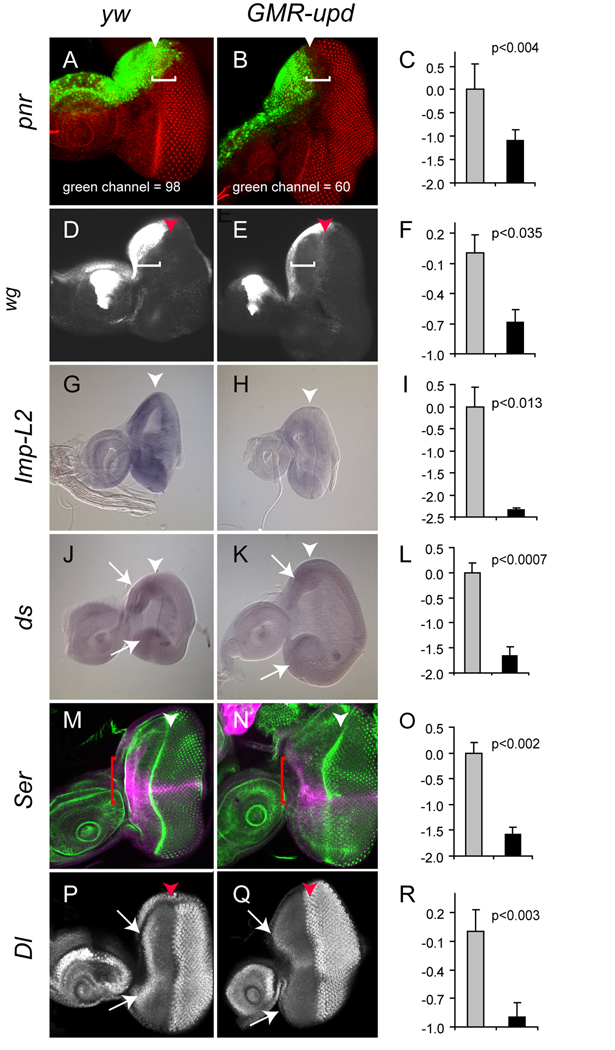
Figure 3. Down-regulated genes in GMR-upd.
The expression of all genes was analyzed in mid-third instar eye discs. The morphogenetic furrow (MF) is marked by an arrowhead. The Y-axis in the bar graphs represents relative mRNA abundance. (A,B) pnr-Gal4, UAS-gfp (abbreviated pnr) (green) is expressed in dorsal peripodial cells in yw control discs (A, bracket). In GMR-upd discs, pnr is repressed in dorsal peripodial cells located “above” cells anterior to the MF (B, bracket). The mean area of pnr expression in wild type eye discs (A) is 98 pixel sq, while in GMR-upd (B) it is reduced to 60 pixels sq. (C) pnr is down-regulated 2.13 fold in the GMR-upd micro-array. (D,E) wg-lacZ (white) is expressed in cells anterior to the furrow at the lateral margins of the dorsal and ventral poles in control discs (D, bracket). In GMR-upd discs, wg expression is decreased in cells anterior to the MF (E, bracket). (F) wg is down-regulated 1.61 fold in the GMR-upd micro-array. (G,H) Imp-L2 mRNA, as assessed by in situ hybridization, is down-regulated in cells anterior to the MF in GMR-upd discs (H) as compared to yw controls (G). (I) Imp-L2 is decreased 5.08 fold in the GMR-upd micro-array. (J,K) ds is expressed in cells at the dorsal and ventral poles in controls discs (J, arrows). Its expression at the poles is greatly reduced in GMR-upd discs (K, arrows). (L) ds is decreased 3.14 fold in the GMR-upd micro-array. (M,N) Ser-lacZ (magenta) is expressed at the D–V boundary and in cells at the anterior edge of control discs (M, bracket). Ser is repressed in cells anterior to the MF in GMR-upd discs (N, bracket). (O) Ser is down-regulated 2.98 fold in the GMR-upd micro-array. (P,Q) In a wild type third instar eye disc, Dl-lacZ (white) is expressed in cells at the anterior margin of the disc and in cone cells posterior to the furrow (P, arrowheads). In GMR-upd discs, Dl is repressed in cells anterior to the MF (Q, arrow). Dl is decreased 1.86 fold in the GMR-upd micro-array (R). The brackets in A,B,D,E,M,N indicate the region in which gene expression is repressed in GMR-upd discs. Gray bar is yw and black bar is GMR-upd (C,F,I,L,O,R).
We find that several genes (Imp-L2, ds, Ser and Dl) have significantly reduced expression in GMR-upd eye discs (Fig. 3I,L,O,R and Suppl. Table 1). Imp-L2 was decreased 5.08 fold in the GMR-upd micro-array and 5 fold by Q-PCR analysis of GMR-upd total RNA (Fig. 3I, Suppl. Table 1, Suppl. Fig. S3). Imp-L2 encodes a secreted Ig domain protein that can bind to and inhibit insulin function (Osterbur et al., 1988; Garbe et al., 1993; Sloth Andersen et al., 2000). Imp-L2 transcripts are reduced in GMR-upd discs, most noticeably in undifferentiated cells anterior to the furrow (Fig. 3G,H). Imp-L2 contains two clusters of Stat92E binding sites, suggesting that it may be a direct target of Stat92E (Suppl. Table 2). The ds gene encodes an atypical Cadherin and can be autonomously induced in the eye disc by activation of the Wg signaling pathway (Clark et al., 1995; Yang et al., 2002). As a result, its expression is enriched at the dorsal and ventral poles of the eye disc, where Wg is expressed. Since ds is a target of wg in the eye disc and since wg is autonomously repressed by activated Stat92E, ds expression should be decreased in the GMR-upd eye discs. Indeed, ds is down-regulated 3.14 fold in the GMR-upd micro-array and 2 fold by Q-PCR analysis (Fig. 3L, Suppl. Table 1, Suppl. Fig. 3). Furthermore, ds transcripts are reduced in GMR-upd discs, most strongly in cells anterior to the furrow (Fig. 3J,K). Although we favor the interpretation that ds levels are reduced in GMR-upd eye discs because its inducer (Wg) is reduced, Target Explorer did reveal one cluster of Stat92E binding sites in putative regulatory regions of the ds gene, raising the possibility that it may be regulated by Stat92E (Suppl. Table 2).
Ser and Dl transcripts were decreased 2.98 and 1.86 fold, respectively, in the GMR-upd micro-array (Fig. 3O,R, Suppl. Table 1). In addition, Ser and Dl transcripts were also decreased 1.5 and 3 fold, respectively, by Q-PCR (Suppl. Fig. 3). To confirm the micro-array values, we used a Ser-lacZ reporter and a Dl-lacZ enhancer trap, which mimic expression of these genes in the eye (Bachmann and Knust, 1998; Spradling et al., 1999; Wang and Struhl, 2004). In control third instar eye discs, Ser is expressed at the D–V boundary and along the lateral margin (Fig. 3M, Suppl. Fig 4A and (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998)). In third instar GMR-upd eye discs, we find that Ser is significantly reduced in cells located immediately anterior to the furrow (Fig. 3N, bracket, and Suppl. Fig 4B). In a control third instar eye disc, Dl is expressed at moderate levels in cells anterior to the furrow, and at high levels in cone cells posterior to the furrow (Fig. 3P, arrows; Suppl. Fig. 5A; and (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998)). In contrast, in a third instar GMR-upd eye disc, Dl expression is significantly reduced in cells anterior to the furrow (Fig. 3Q, arrows). This suggests that Ser and Dl are negatively regulated by Stat92E. Target Explorer identified two clusters of Stat92E binding sites in putative regulatory regions of Ser, one cluster at ~5,000 bp upstream of the start site that resides within the 9.5 kb Ser reporter, and also two clusters of Stat92E binding sites in the Dl gene (Suppl. Table 2). In addition, a deficiency that removed Ser modified the GMR-upd enlarged-eye phenotype (Bach et al., 2003). These data raise the possibility that Stat92E may direct negatively regulate these genes.
Additionally, we validated 3 genes down-regulated in the GMR-upd micro-array by Q-PCR: mirror (mirr); gram-positive specific serine protease (grass) and Angiotensin converting enzyme (Ance) (Suppl. Table 1 and Suppl. Fig. 3). Although Target Explorer did not identify clusters of Stat92E binding sites in non-coding regions of these genes, deficiencies that removed grass and Ance modified the GMR-upd enlarged-eye phenotype (Bach et al., 2003). We favor the model that mirr is repressed in GMR-upd eye discs because levels of its inducer (Wg) are reduced in GMR-upd tissue (Fig. 3E). Ance family genes have been best studied for their role in D–V patterning of the Drosophila embryo (Stathopoulos and Levine, 2005). No direct link between Ance and JAK/STAT signaling has as-yet been made, however, both are critical for Drosophila immune function (Agaisse and Perrimon, 2004; Kambris et al., 2006). In sum, we successfully validated 9 genes down-regulated in the GMR-upd micro-array by at least one method.
Ser and Dl are ectopically expressed in cells lacking stat92E
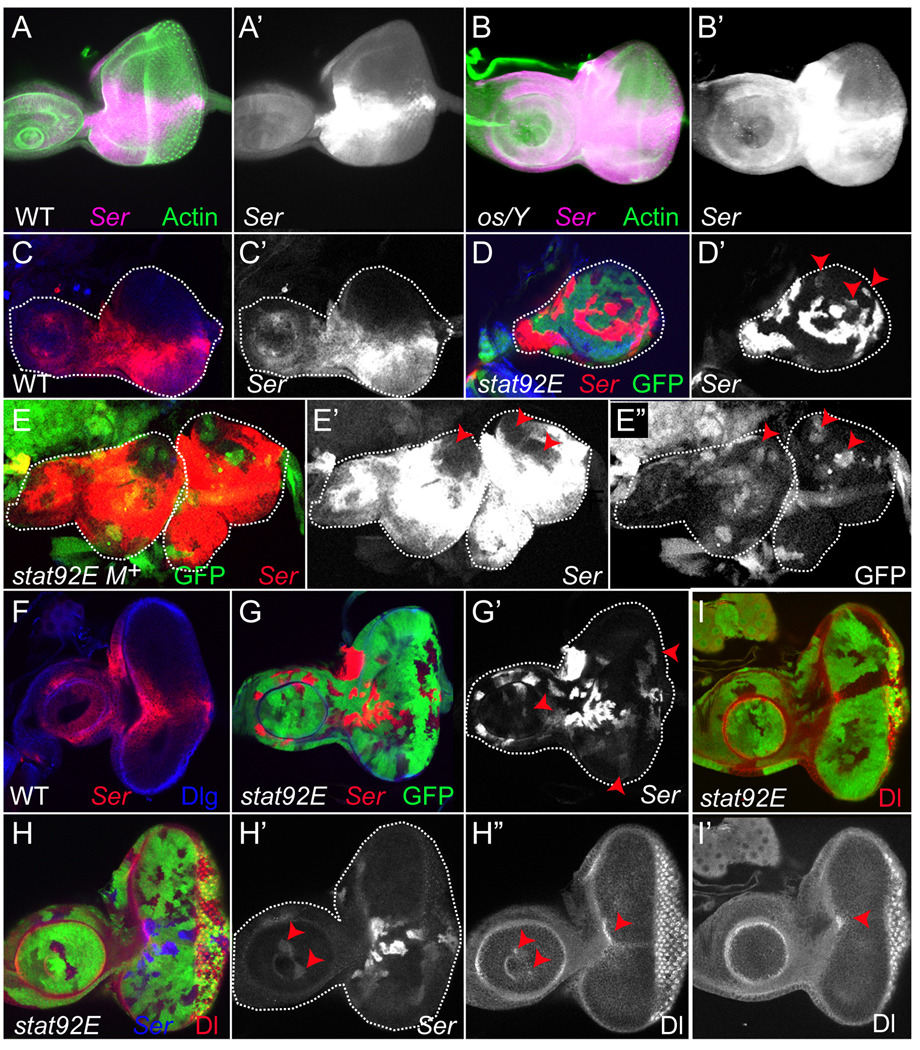
To test the hypothesis that Ser and Dl are negatively regulated by JAK/STAT signaling, we monitored expression of the Ser gene in an upd hypomorphic allele called outstretched (os). Homozygous os flies have small eyes and outstretched wings (Lindsley and Grell, 1968). In os/+ heterozygous control animals, Ser gene expression pattern is identical to wild-type, primarily along the D–V boundary and at the anterior lateral margin (Figs. 4A,A’,F and 5A,A’ and not shown). In contrast, in os/Y hemizygous animals, the Ser expression domain is significantly expanded (Fig. 4B,B’). We next monitored expression of Ser in clones lacking stat92E. We made large patches of eye tissue that are homozygous mutant for stat92E using ey-FLP and Minute techniques (see Materials and Methods). Minutes are mutations in ribosomal genes that are cell lethal when homozygous and confer an autonomous growth disadvantage when heterozygous (Morata and Ripoll, 1975; Lambertsson, 1998). In wild type second instar eye discs, Ser is expressed in the ventral domain (Fig. 4C,C’ and (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998)). In contrast, in a second instar eye disc containing large stat92E clones in a Minute background (labeled stat92E M+), Ser is ectopically expressed at higher intensity and throughout the stat92E M+ clones (Fig. 4E,E’), except in heterozygous tissue which contains one wild type copy of the stat92E gene (Fig. 4E”, arrowheads). A similar observation was made in older discs containing stat92E M+ clones (n=22 second or third instar discs with stat92E M+ clones examined). We also examined Ser expression in mosaic stat92E clones generated by ey-flp in a non-Minute background (n=39 discs). We scored for ectopic Ser in stat92E clones residing outside of the endogenous Ser expression domain at second or third instar. We found that Ser is ectopically expressed in at least one stat92E clone per disc in the dorsal domain in second instar eye discs or in the dorsal and/or ventral domain in third instar eye discs (Fig. 4D,D’G,G’,H,H’ arrowheads) (n=91 stat92E clones in 39 discs with ectopic Ser in the eye field). We observed a similar but weaker effect of loss of stat92E on Dl. When large stat92E clones are induced, Dl protein is ectopically expressed at high levels anterior to the furrow, but its expression in cone cells posterior to the furrow remains unchanged (n= 40 discs examined) (Supp. Fig. 5A,B). In mosaic stat92E clones, Dl protein expression is autonomously increased, with this effect being most pronounced in clones located at the anterior margin of the eye disc (Fig. 4I,I’, arrowheads). Moreover, Ser and Dl are always ectopically expressed within the same stat92E clone when that clone resides within the distal antenna. In wild type antennal discs, Stat92E is activated in the distal antenna (Ayala-Camargo et al., 2007), Ser is not expressed in this region, and Dl is expressed in a ring around it (Fig. 4,H’,H”, Suppl. Fig. 4F, Suppl. Fig. 5A). Ser is ectopically expressed in at least one stat92E clone per disc in the distal antenna (Fig. 4G,G’, H,H’, arrowheads) (n=34 stat92E clones in 39 discs with ectopic Ser in the antennal field). Within these clones, Dl expression becomes concentrated into dots in the center of the clone where Ser is ectopically expressed (n= 8 clones in 8 discs with both the Ser gene and Dl protein ectopically expressed in the same stat92E clone residing in the distal antenna) (Fig. 4H’,H”, arrowheads). We also observed that many stat92E clones did not contain ectopic Ser or Dl. These data suggest that the timing and/or spatial location of stat92E clones is key in determining whether Notch ligands are ectopically expressed.
Figure 4. JAK/STAT signaling negatively regulates Ser and Dl.
(A–B) Heterozygous os/+; Ser-LacZ/+ third instar eye discs show an expression pattern of Ser (magenta) that is indistinguishable from wild type (A,A’, compare Fig. 4A to Fig. 3M). In contrast, hemizygous os/Y; Ser-LacZ:+ third instar eye discs show ectopic expression of Ser in both the eye and antennal disc (B,B’). (C–G) Ser is ectopically expressed in stat92E clones. In a wild type second instar eye disc, Ser (red) is expressed in the ventral domain (C). Ser is ectopically expressed in mosaic stat92E clones, except those residing at the dorsal pole (D,D’, arrowheads). Large stat92E clones generated in a Minute background (labeled stat92E M+) lack expression of GFP. In stat92E M+ clones, Ser is ectopically expressed throughout the entire mutant tissue (E, red and E’, white), except in GFP+ heterozygous cells which contain a wild type copy of the stat92E gene (E, green, E’, arrowheads and E” white and arrowheads). In a wild type third instar disc, Ser (red) is expressed in cells at the D–V boundary and at the anterior lateral margin; it is excluded from the distal antenna (F). Ser (red) is ectopically expressed in stat92E clones residing in both the dorsal and ventral domains of the eye as well as those in the distal antenna (G,G’ arrowheads). (H–I) Ser (blue) and Dl (red) are ectopically expressed in stat92E clones. Dl is ectopically expressed in stat92E clones residing at the anterior margin of the eye disc (H,H”,I,I’, arrowheads in eye disc). In wild type disc, Dl is expressed in ring around A3 (see Suppl Fig 5A). However, in stat92E clones residing in the distal antenna, Dl expression is altered such that it now appears as a “dot” (H”, arrowhead in antennal disc) within the middle of ectopic Ser (H’, arrowheads in antennal disc). Single channel for Ser (A’-E’,G’,H’). Single channel for Dl (H”,I”). Phalloidin is green in A. Dlg is blue in C,D,F. Ser-LacZ was detected by anti-β-gal. stat92E397 clones lack GFP in D,E,G-I.
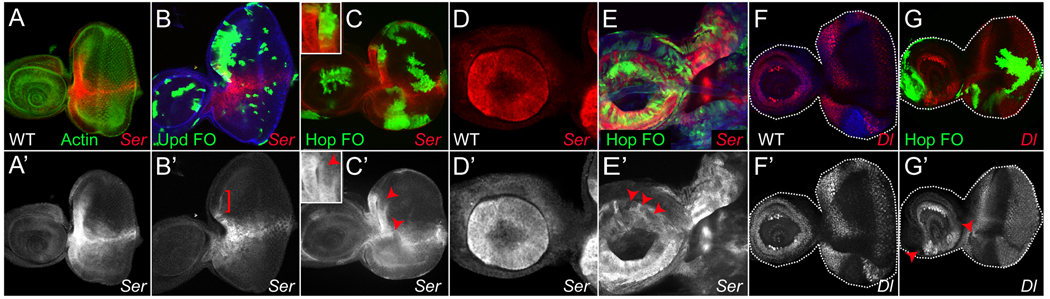
Figure 5. Activated Stat92E represses Ser.
(A) Expression of Ser-LacZ (red) in a mid-third instar eye disc. (B) A large upd-expressing clone (green) located at the dorsal anterior margin of the eye significantly represses endogenous expression of Ser there (B’, bracket). (C) hop-expressing clones located at the dorsal anterior margin (magnified in inset), D–V midline or in the ventral domain significantly repress Ser (red) in a cell-autonomous manner (C’, arrowhead and inset). (D) Ser (red) is expressed in a ring around the distal antenna in a wild type eye-antennal disc. (E) hop-expressing clones repress Ser in a cell-autonomous manner in the antenna (E’, arrowheads). (F) Expression of Dl-lacZ in a wild type third instar eye-antennal disc. (G) a hop-expressing clone represses Dl-lacZ expression at the ventral anterior lateral margin in a cell-autonomous manner (G’, arrowhead in eye disc). Note the re-patterning induced by the hop-expressing clone in the antenna (G’, arrowhead in antennal disc). Single channel for Ser (A’-E’) and for Dl (F’,G”). Phalloidin is green in A. Dlg is blue in B,E. Ser-LacZ and Dl-LacZ were detected by anti-β-gal. upd- or hop-expressing clones are GFP+ in B,C,E,G.
Ser and Dl are repressed cell-autonomously by JAK/STAT pathway activity
To test the prediction that Ser is repressed by JAK/STAT signaling, we examined Ser gene expression in cells that had hyper-activated Stat92E. We generated clones of cells that mis-expressed the ligand Upd, which activate Stat92E non-cell autonomously. In 7/7 discs, we found that large upd-expressing clones strongly repressed endogenous Ser expression at the anterior margin of the eye disc (compare Fig. 5B,B’, bracket, with Fig. 5A,A’). We also hyper-activated the JAK/STAT pathway by inducing clones that mis-express Hop. Indeed, in 11/12 discs examined, we found Hop-expressing clones repressed Ser in a cell-autonomous manner at the D–V boundary or the anterior margin of the eye disc, or in the proximal antenna (compare Fig. 5A,A’,D,D’ with Fig. 5C,C’,E,E’, arrowheads). The fact that low levels of Ser-lacZ are still detectable in some hop-expressing clones is likely due to perdurance of the β-gal protein. Taken together, these data indicate that activation of the JAK/STAT pathway represses Ser cell-autonomously. We also addressed if activation of Stat92E could repress the Dl gene. In 1/5 discs examined, we found Hop-expressing clones could repress a Dl enhancer trap at the anterior margin of the eye disc (Fig. F,F’,G.G’) but not in other regions of this disc. These data suggest that Stat92E activity more strongly impacts the expression of Ser than of Dl. Moreover, when taken together with the loss-of-function experiments, these data suggest that Stat92E represses Ser, possibly directly or via an intermediate (see discussion), and that once Ser is ectopically expressed in the dorsal domain of the eye disc, the expression of Dl is subsequently increased. Our results are consistent with previous reports that Ser and Dl up-regulate each other’s expression when Notch signaling is activated at growth organizers in imaginal discs (Panin et al., 1997). In sum, our data indicate that JAK/STAT pathway activity represses Dl less potently than it does Ser, and they strongly suggest that Ser (and not Dl) is the relevant target of Stat92E.
Stat92E represses Notch activity
To examine the functional consequence of Stat92E–mediated repression of Ser, we monitored Notch pathway activity in eye discs that contained mosaic stat92E clones using two Notch targets that faithfully mirror Notch activity in the eye disc: eyg and Enhancer of split m-β (m-β) (Cooper et al., 2000; Dominguez et al., 2004). In wild type second instar eye discs, eyg is expressed at the D–V boundary of the developing eye (Fig. 6A). We found in 8/22 discs that eyg is ectopically expressed in a cell-autonomous manner in mosaic stat92E clones in the dorsal eye (Fig. 6B,B’, arrowheads). Moreover, in 8/10 discs hyper-activation of Stat92E results in repression of eyg within Hop-expressing clones (Fig. 6D,D’, arrowheads and bracket). This repression of eyg by activated Stat92E occurs at the D–V boundary and at the anterior margin of the eye disc, as well as in the antennal disc. We observe similar results for the m-β reporter. In control second instar eye discs, this reporter is expressed at the D–V midline anterior to the furrow, while in third instar, it is expressed at both the D–V boundary and the anterior margin (Fig. 6E,G and (Cooper et al., 2000)). As expected, in 45/45 eye discs with stat92E M+ clones, m-β expression shifts dorsally (Fig. 6F), precisely where ectopic Ser is also observed (Fig. 4E). Pronounced “blebbing” is also observed, which may be a result of increased growth in the dorsal domain of stat92E mutant eye discs (Fig. 6F, bracket). Later in third instar, independent circular growth organizers with high levels of Notch activity are observed only in the dorsal domain in stat92E M+ mutant discs, presumably as a result of aberrant Notch activation there (Fig. 6H, arrowhead). This is never observed in control discs (Fig. 6G). We were able to rule out abnormal expression of fng as a cause of the ectopic Notch signaling observed in stat92E M+ discs. Consistent with published reports, in 5/5 second instar control eye discs, we found that fng mRNA is expressed in the ventral domain (Suppl. Fig. 5C, bracket, and (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998)). Moreover, in 5/5 second instar stat92E M+ eye discs, fng expression remains confined to the ventral domain (Suppl. Fig. 5D, bracket). Furthermore, fng expression is not altered in third instar GMR-upd discs as compared to controls (Fig. 5E,F). Taken together, these data strongly suggest that JAK/STAT signaling normally acts to restrict Ser. In the absence of stat92E in the dorsal domain of the eye, Ser is ectopically expressed there, and this leads to the induction of growth-regulatory Notch target genes like eyg, and formation of ectopic growth organizing centers and over-growth of the dorsal eye. Thus, in wild-type discs, Notch induces expression of the upd gene in cells at the posterior margin of the eye, but Upd acts at a distance to activate Stat92E, which represses the expression of Ser and, as a result, limits the extent of Notch pathway activity (Fig. 6I,J).
DISCUSSION
The JAK/STAT pathway plays important roles in conserved processes, including growth and patterning during development. However, the transcriptional targets of this signaling system are largely unknown. We have combined three powerful techniques, whole-genome expression profiling, Drosophila genetics, and whole-genome bio-informatics screening, to identify new targets of the JAK/STAT pathway. Our study identified 584 genes with significantly altered expression in GMR-upd eye discs, in which the JAK/STAT pathway is hyper-activated, as compared to controls. 79 of these genes were also found to have a least one cluster of Stat92E binding sites, raising the possibility that they may be direct Stat92E targets. Of the 584 differentially-regulated genes, 168 (28.8%) genes were up-regulated while 416 (71.2%) were down-regulated. The fact that we identified the known target genes socs36E, dome and wg as being differentially-regulated in GMR-upd tissue indicates that our micro-array can data-mined as a source for additional Stat92E target genes.
Up-regulated genes
We were able to validate a total of 19 up-regulated genes in the GMR-upd micro-array. Five (dome, socs36E, chinmo, lama and Mo25) were validated both in vitro by Q-PCR and in vivo by mRNA analysis (Fig. 2, Suppl. Table 1, Suppl. Fig. S2), while one (pnt) was validated only in vivo by in situ hybridization (Fig. 2). Thirteen additional genes (w, ken, CG11784, Fps85D, aPKC, pdp1, esg, trol, SrpRβ, brat, dom, tep-2 and pyd) were also validated by Q-PCR as significantly up-regulated in GMR-upd discs (Suppl. Fig. S2). chinmo and lama are not expressed in control third instar eye discs, while Mo25 and pnt are expressed in cells in the morphogenetic furrow. However, when the JAK/STAT pathway is hyper-activated in GMR-upd discs, all four genes are up-regulated in undifferentiated cells anterior to the furrow. The fact that lama expression is strongly increased only in anterior cells at the poles of the eye disc suggests that not all undifferentiated cells may be competent to express lama following reception of the Upd signal.
chinmo has one cluster of Stat92E binding sites, suggesting that it may be a direct Stat92E target. We previously reported that Stat92E transcriptional activity is highest in first and second instar wild type eye discs (Ekas et al., 2006; Bach et al., 2007). Consistent with these results, chinmo is expressed in early eye development, and may be a target of the Pax 6 homolog Eyeless (Ostrin et al., 2006). Moreover, Stat92E may be able to promote chinmo expression in other Drosophila tissues, since it was identified as a differentially-regulated gene in a micro-array screen for JAK/STAT target genes in the adult testis (Terry et al., 2006). Since we did not validate chinmo expression in vivo in the testis, the ability of Stat92E to induce this gene in other tissues remains unclear. chinmo was identified in 2006 as a gene required for the temporal identity of early-born neurons in the Drosophila mushroom body (Zhu et al., 2006). However, these authors did not report what signals control chinmo expression in this tissue. To the best of our knowledge, we are the first to identify a factor (i.e., Stat92E) that can lead to up-regulation of the chinmo gene. In the future, it will be critical to determine if activated Stat92E also controls chinmo expression in developing neurons, as a role for the JAK/STAT pathway in temporal neuronal identity has as-yet not been reported.
lama encodes a conserved Phospholipase B protein that is expressed in neural and glial precursors prior to differentiation (Perez and Steller, 1996). lama has two clusters of Stat92E binding sites, suggesting that it may be a direct Stat92E target. In support of this hypothesis, lama, like maximal Stat92E transcriptional activity, is strongly detected in young (second instar) eye discs (Klebes et al., 2005). In addition, both upd and lama transcripts are significantly up-regulated during “trans-determination”, a process during which certain Drosophila imaginal disc cells switch fates (or trans-determine) (Klebes et al., 2005). These results suggest that upd and lama are expressed in pluripotent imaginal cells that exhibit developmental plasticity. Although the epistasis between these genes was not established by Klebes and colleagues, our results indicate that JAK/STAT signaling can positively regulate transcription of the lama gene.
JAK/STAT signaling functions to reduce Notch activity by repressing Ser
We showed that the Notch ligands Ser and Dl are significantly down-regulated in GMR-upd discs. Furthermore, we were able to validate this observation by demonstrating the reduced expression of these genes in situ in GMR-upd eye discs. Clonal analysis indicated that Ser and Dl are ectopically expressed in cells lacking stat92E, which suggests that Stat92E either directly or indirectly represses these genes. However, the effect of Stat92E on Ser is more pronounced than it is on Dl. Ser is frequently ectopically expressed in stat92E clones in the dorsal, ventral and anterior portions of the eye disc, as well as in the distal antenna. In contrast, Dl protein is ectopically expressed only in stat92E clones located at the anterior margin of the eye disc or in the distal antenna and only in clones that also have ectopic Ser. These data suggest that Stat92E may in fact negatively regulate Ser, and once Ser is de-repressed, Dl levels are up-regulated in these stat92E clones as a result of increased Ser. This model is supported by the observation that Ser is routinely repressed in a cell-autonomous manner by hyper-activation of the JAK/STAT pathway while Dl is not, and is consistent with a published report that Ser and Dl up-regulate each other’s expression as a result of Notch pathway activation (Panin et al., 1997). In this study, we used a Ser-lacZ reporter gene in which the 9.5 kb of genomic DNA located immediately upstream of the start site drives expression of β-galactosidase (Bachmann and Knust, 1998). This fragment contains one cluster of Stat92E binding sites, which raises the possibility that Stat92E directly represses Ser (see below for a more detailed discussion of STAT proteins as repressors).
We then showed the functional consequence of loss of JAK/STAT pathway activity on Notch signaling. Ectopic Notch activity (as assessed by eyg and m-β) is only observed in dorsal stat92E M+ clones, precisely where high levels of ectopic Ser are also observed. Additionally, independent, circular growth organizing domains that have high levels of Notch activity are only observed in the dorsal eye. fng expression is not altered in second instar eye discs containing large stat92E clones, indicating that aberrant expression of this critical regulator of Notch pathway activation is not the reason for excessive growth in large dorsally-located stat92E clones. Rather de-repression of Ser and subsequent induction of Dl in these clones causes ectopic growth organizing centers in the dorsal eye.
Our study is the first to uncover the negative regulation of Notch signaling by the JAK/STAT pathway. As mentioned in the introduction, the activity of Wg and Hh induce Iro-C genes in the dorsal half of the eye. Iro-C proteins repress fng to the ventral domain, thus established a fng+/fng− interface, where Notch receptor activation occurs (Cho and Choi, 1998; Dominguez and de Celis, 1998; Cavodeassi et al., 1999; Yang et al., 1999). The ability of Fng to promote Dl-dependent activation of Notch, while inhibiting Ser-dependent activation, leads to Notch signaling at the D–V boundary and induction of the eyg gene there. Notch autonomously regulates expression of the upd gene, presumably via Eyg (although Eyg regulation of upd has not as-yet been shown to be autonomous). However, Notch regulates growth of the entire eye disc through both upd-dependent and -independent mechanisms (Chao et al., 2004; Dominguez et al., 2004; Reynolds-Kenneally and Mlodzik, 2005). Our study extends these previous observations by showing that loss of JAK/STAT pathway activity leads to ectopic expression of Ser. In wild type animals, Upd protein is produced by cells at the anterior margin of the eye disc, but it acts as a long-range mitogen and activates Stat92E in most cells in a second instar eye disc (Bach et al., 2003; Tsai and Sun, 2004; Ekas et al., 2006; Bach et al., 2007; Tsai et al., 2007). When Stat92E activity is lacking from cells in the dorsal eye disc, Ser is strongly ectopically expressed there. Since Fng inhibits Ser’s ability to activate Notch and since Fng is excluded from the dorsal domain of the eye, ectopic expression of Ser in dorsal stat92E clones leads to inappropriate activation of the Notch pathway there. This results in excessive growth within independent growth-organizing domains in the dorsal eye (Fig. 6H). Thus, our findings indicate for the first time that there is a negative feedback loop between the Notch and JAK/STAT pathways (Fig. 6I,J).
Other down-regulated genes in the GMR-upd micro-array
The Imp-L2 gene is also significantly down-regulated by JAK/STAT signaling. Imp-L2 was originally reported to be a secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila (Osterbur et al., 1988; Garbe et al., 1993). Biochemical analysis in insect cells indicates that Imp-L2 can bind to human insulin and inhibits it from binding the insulin receptor (InR) (Sloth Andersen et al., 2000). The InR pathway in Drosophila, as well as in other species, is a key positive growth regulator (Brogiolo et al., 2001). This suggests that Imp-L2 may function to negatively regulate insulin action and hence growth in Drosophila. The fact that this gene is decreased in the GMR-upd micro-array suggests that JAK/STAT signaling may repress it either directly or indirectly in order to promote growth in the eye disc. We attempted to test this hypothesis by monitoring in control and GMR-upd third instar eye discs Akt phosphorylated on Ser505 using an antibody from Cell Signaling as a read-out of InR pathway activation. However, this antibody does not work well for immmuno-fluorescence and we were unable to draw any conclusions from these experiments. Thus, the model that JAK/STAT signaling represses a negative regulator of the InR pathway to promote growth in the eye disc remains to be tested
Potential explanations for why so many transcripts in the GMR-upd micro-array are down-regulated
Stat92E may directly downregulate gene expression. Although it is not currently known if Stat92E functions as a transcriptional repressor as well as an activator, the dual property of being able to either induce or arrest gene transcription has been observed for other transcription factors, including the Drosophila proteins Orthodenticle, Dorsal and Hunchback (Schulz and Tautz, 1994; Stathopoulos and Levine, 2002; Cook et al., 2003). Despite the fact that most published reports suggest that mammalian STATs and Stat92E can robustly activate gene transcription, there is precedence for STAT proteins as repressors: the Dictyostelium Dd-STATa protein acts as a repressor by binding to an element in the regulatory region of the ecmA gene (Kawata et al., 1996; Mohanty et al., 1999). This STAT-mediated repression is required for the commitment to stalk cell differentiation and chemotaxis in this organism. Moreover, we and others found that Stat92E can repress transcription of the wg gene in multiple Drosophila tissues (Ekas et al., 2006; Ayala-Camargo et al., 2007; Tsai et al., 2007). In the developing eye, we were able to narrow the Stat92E–responsive element to a small 263 bp enhancer wg2.11Z (Ekas et al., 2006; Pereira et al., 2006). The lack of well-characterized Stat92E binding sites in this enhancer led to the hypothesis that Stat92E represses wg indirectly through another protein (Ekas et al., 2006). The model that Stat92E can directly repress the wg gene through the wg2.11Z enhancer has as-yet not been directly tested, but this will be important to do in future experiments to determine if Stat92E can act as a repressor. This information will also help to clarify whether the large number of down-regulated genes in the GMR-upd micro-array is due to Stat92E’s repressive action directly on chromatin.
It is possible that Stat92E acts to repress transcription through induction of one or more target genes that encode transcriptional repressors. One potential candidate is chinmo, which encodes a novel protein with one N-terminal BTB/POZ domain and two C-terminal C2H2 Zinc (Zn) fingers, that is localized to the nucleus in mushroom body neuroblasts (Zhu et al., 2006; Maurange et al., 2008). However, the molecular function of Chinmo is currently unknown. The presence of the Zn finger domains suggests that it may be bind DNA, as many nuclear hormone receptors possess only two Zn fingers and yet bind DNA (Freedman et al., 1988; Luisi et al., 1991). The BTB/POZ domain in Chinmo suggests that it may function to downregulate expression of specific, as-yet unidentified target genes by recruiting HDACs and/or Polycomb proteins to chromatin as has been shown for the mammalian BTB/POZ, Zn proteins Bcl-6 and PLZF (Deweindt et al., 1995; Huynh and Bardwell, 1998; Melnick et al., 2002). However, recently BTB/POZ-domain proteins, including those that have both BTB/POZ and Zn finger domains, have also been shown to be adaptors for Cullin 3 E3 Ubiquitin ligases, which promote protein degradation (Geyer et al., 2003; Weber et al., 2005; Zhang et al., 2006). Future experiments will be needed to address if Chinmo is a direct Stat92E target gene and elucidate the cellular function of Chinmo.
MATERIALS AND METHODS
Fly stocks
The following stocks are described in Flybase: yellow white (yw); ey-FLP; stat92E397; stat92E85C9; Mo25-lacZ (P(PZ)Mo2500274 ry506); eyg-lacZ (P(lacW)eygM3–12); UAS-hop; UAS-upd; Ser-lacZ (Bachmann and Knust, 1998); pnr-Gal4, UAS-gfp (pnr>gfp) (Singh and Choi, 2003); FM7 ubi-gfp(FM7-gfp). We used Enhancer of split m-β (E(spl)mβ-lacZ) transgenic line (Cooper et al., 2000). We also used GMR-updΔ3’ #19 (referred to as GMR-upd) (Bach et al., 2003) and 10xSTAT92E–GFP (Bach et al., 2007). We generated a dome-Gal4, UAS-lacZ (dome>lacZ) recombinant line (dome-Gal4 was originally described in (Ghiglione et al., 2002)). We also generated a recombinant chromosome FRT82B stat92E397 Ser-lacZ II-9.5, which contains a stat92E allele that is a strong hypomorph and likely acts as an activity null allele (Ekas et al., in preparation) and a Ser gene reporter containing a 9.5 kilobase (kb) region of the Ser gene immediate 5’ of the start site (Bachmann and Knust, 1998). The “patchy” appearance of Ser-lacZ in stat92E clones is due to the fact that stat92E clones have 2 copies of the reporter, whereas the sister clones or twin spots have none. We also generated a recombinant chromosome eyg-lacZ FRT82B stat92E85C9 contains a stat92E allele that behaves as an activity null (Ekas et al., in preparation) and eygM3–12 that behaves as an eyg enhancer trap (Jun et al., 1998; Jang et al., 2003).
Clonal analysis
Clones were generated by ey-FLP using the FLP/FRT technique (Xu and Rubin, 1993; Newsome et al., 2000). Since ey-FLP can induce clones in the eye-antennal disc primordium prior to its segregation into eye and antennal fields, it can induce clones in both the eye and antennal disc. stat92E clones were generated using FRT82B ubi GFP(S65T)nls 3R/TM6B, Tb. Minute clones were generated by FRT82B M(3)96C arm-lacZ. upd or hop-expressing flip-out clones were generated using UAS-upd or UAS-hop and the flip-out cassette stock P(AyGAL4)25 P(UASGFP. S65T)T2; hs-flp MKRS/ TM6B, in which FLP is under the control of the heat-shock promoter (Ito et al., 1997). Flip-out clones express both Upd or Hop and GFP.
Timed collections
yw or GMR-upd/(FM7-gfp) flies were grown in vials at 25°C. For timed collections, we allowed the flies to lay eggs for 2 hours. The embryos were maintained at 25°C until 110 hours after egg deposition (AED), which corresponds to mid-third instar. At this time, we isolated GFP negative larvae, which represent GMR-upd/Y animals. One of the pair of eye discs in a single larva was taken for RNA isolation (see below). The other was fixed in 50% glutaraldehyde, mounted on a microscope slide and visually inspected by brightfield microscopy for the morphogenetic furrow having progressed approximately half-way across the eye disc.
RNA isolation
For each micro-array, total RNA was extracted from a single mid-third instar larval eye disc using the Arcturus Isolation kit (PicoPure™ RNA Isolation Kit). The RNA quality and quantity was assessed using the Agilent 2100 Bioanalyzer and Nanodrop ND-1000, and subsequently amplified using the Arcturus Amplification kit (RiboAmp® RNA Amplification Kit). Labeled anti-sense RNA (aRNA) was synthesized from the resulting cDNA using the ENZO BioArray™ High Yield™ RNA Transcript Labeling Kit. After isolation and amplification, the aRNA was again assayed by the Agilent 2100 Bioanalyzer and Nanodrop ND-1000.
Micro-array data acquisition and analysis
Equal amounts (9 µg) of amplified control and GMR-upd aRNA were separately hybridized onto the GeneChipR Drosophila Genome 2.0 Arrays (Affymetrix). The chip processing and image acquisition were obtained following the recommendations of the array manufacturer. The raw data were normalized using Model Based Expression Index (MBEI) (Li and Wong, 2001) and further filtered using GeneSpring 7.2 (Agilent). To identify the differentially abundant mRNAs between the two groups, the pre-processed data were rigorously statistically filtered by T-test (p<0.05, alpha correction) and also by Significance Analysis of Micro-array (SAM) at False Discovery Rate (FDR) set to 10% (see Suppl. Table 1) (Tusher et al., 2001). (Gene Ontology, KEGG pathways) of the resulting gene lists were performed using a web based tool DAVID bioinformatics resources (Huang da et al., 2009). Primary data from this study has been deposited at NCBI GEO database (under GSE ###, which will be submitted following acceptance for publication of the manuscript).
Quantitative real-time PCR (Q-PCR)
We performed Q-PCR for validation of potential candidate genes using the SYBR Green PCR Mix (Applied Biosystems) protocol and a real-time PCR machine (ABI 7900HT) from Applied Biosystems. We isolated and amplified the RNA using the same kits and protocols as the ones used for the micro-array. We measured the cDNA concentration using a Nanodrop ND-1000. We used 3 ng of cDNA per sample per reaction, 5 uM of each primer and 1X SYBR. We did triplicates per primer per sample. We used six different reference genes: CG1091, CG7424, CG15693, CG2093, CG10728, CG33054, RPL31 using the primer sequences as described (Livak and Schmittgen, 2001). For all other genes, we used the following primers: white F: TATTCTGCAACGAGCGACAC and R: CAAAAGTTCGCCCGGATAG socs36E F: GCTGCCAGTCAGCAATATGT and R: GACTGCGGCAGCAACTGT dome F: CGGACTTTCGGTACTCCATC and R: GATCGATCATCGCCGAGTT ken F: GCCCACAAGTTGGTCCTG and R: CCGGAAAGTATACGGTGGTG lama F: TGATATTGCTGCTTCCTGGAC and R: TGGTTTGGCGATGGTTTTAT CG11784 F: GTTGACTTCGCCAAGAAGGA and R: ATCGCTGTCCACAAACACC Fps85D F: ACCAACTCCAGAGCCAGAGA and R: CTGGAGCATCAGTCGGTACA aPKC F: TACAGTTGACCCCGGATGAT and R: TCCTCCAGAGACATCAGCAA chinmo F: CAGTGCCAATGAGGCTAATG and R: TCAAGTTCTCCAGCTTCACG pdp1 F: GACAAGACCCTGCCCTATGA and R: CAGGCCATCAGGTATGTTGTT esg: F: TCAGCTGCAAGGATTGTGAC and R: CGTAGTTGAGTTCGCTGCTG trol F: CTATGCAGACTGCGAAGACA and R: AGCTATCGCATTCGAACTCAG srpRβ F: TTGTCTTTGTGGTGGACTCG and R: AGGGTTGTGTGGCACTGTCT brat F: GGAAACCAGGAAACGAACTG and R: GGTGGCTCCGTTTACCTTTA dom F: GTGGCTTCACAGGCCAAC and R: ACATGGGTGCGCAGATTT Mo25 F: TAATACGACTCACTATAGGGCGCCTGGTCTCGATCAAGAACATGC and R: AATTAACCCTCACTAAAGGGAAGGAGCGATCCGTATGGAAGTTGG Tep-2 F: TTCGTTCTGCTGGCTTTCTT and R: CTTCGGCCACATAGCGTACT pyd F: AATCGAGAGGCAACTTCTTCC and R: ACCACATCGTCCCAGTTCTC pnr F: TTGGAGGCCATCAAGGAGT and R: TCCGTGTGCAGCTTACTGAG Ser F: CTTTGTGCTCAGCGATCC and R: CATATCCAACGCCTGCAGTA mirr F: GCCAATATCGACGATGACG and R: GTCGTCCGTGGAACCAAC ds F: TACAACGTATCCGTCGCTGA and R: ATGGCATTTACTCCGCAATC Imp-L2 F: GTGAAAGTGCCAACGAAGC and R: GAACAGCAGCAGCGCTAAG grass F: TGCATGACATAGCTCTCCTGA and R: TGCCTTCTCCTTCAGCTCAT wg F: GACGAAATGGACGTCGTCAG and R: TGGCTTGTGCTCGGGATT Dl F: GGCTGTGAACATGGACATTG and R: CATGGATGCAGTTCGGTTC Ance F:GGAGGCGGAGAACATTAAGA and R: GACAAGACCCTGCCCTATGA
Ribo-probe synthesis
RNA probes were designed against the contiguous cDNA sequence of differentially-expressed genes. We used cDNA clones from Drosophila Genomics Resource Center (DGRC). The probes were synthesized using 1–5 µg of linearized plasmid in a 20 µL transcription reaction mix. We used a DIG-labeling kit per the manufacturer’s instructions (Roche). The resulting labeled ribo-probes were ethanol precipitated and re-suspended in 100 µL of HB4.
in situ hybridization
Mid-third instar eye discs were dissected in cold PBS and fixed in 8% paraformaldehyde on ice for 1 hour. They were subsequently washed three times in PBS-T (PBS 0.1% Tween 20) for 10 minutes and pre-hybridized for 1 hour at 65°C in hybridization buffer (HB4) that contains 50% formamide, 5x SSC, 2 mg/µl Heparin, 0.1% Tween-20, 500 mg Tortula Yeast RNA extract and 0.1 mg/ml herring sperm DNA. After pre-hybridization, the discs were hybridized overnight in 100 µL of HB4 and 1 µL of the ribo-probe that had already been denatured at 80°C for 10 min in HB4 and then put on ice. After hybridization, the discs were washed two times for 25 minutes in a buffer containing 50% formamide, 50% 2xSSC with 0.1% Tween-20. They were rinsed in PBS-T at room temperature three times for 10 minutes. Subsequently, they were incubated for 2 hours with anti-Digoxigenin (DIG) (Roche) (diluted 1:2000) and then washed three times for 10 minutes in PBS-T. After this, they were rinsed once and washed for 5 minutes in alkaline phosphate buffer pH=9.5 containing 0.1M NaCl, 0.05M MgCl2, 0.1M Tris (pH=9.5) and 0.1% Tween-20. The reaction was developed by adding 40 µL of NBT/BCIP stock solution to 2 ml of PBS.
Antibody staining
Antibody and X-gal stainings were performed as described in (Ekas et al., 2006). We used the following primary antibodies: rat anti-Elav (1:50), mouse anti-β-galactosidase (1:50), mouse anti-Discs large (Dlg) (1:50), mouse anti-Delta (Dl) mAb C594.9B (Qi et al., 1999) (1:50) (all from the Developmental Studies Hybridoma Bank) and rabbit anti-β-galactosidase (Cappel) (1:100). We used fluorescent secondary antibodies at 1:250 (Jackson Laboratories). We collected fluorescent images (at 25X magnification) using a Zeiss LSM 510 confocal microscope and scanning electron micrographs (at 100X) using a Leo SEM (Zeiss) (Harvard School of Public Health).
Bio-informatics search for Stat92E binding sites
We searched the entire non-coding region of the Drosophila melanogaster genome for two Stat92E binding sites located within 100 base pairs (bp) of each other. For this analysis, we used Target Explorer, which was designed for the Drosophila genome (Sosinsky et al., 2003). This platform generated a matrix using Stat92E binding sites uploaded by the user. We employed known Stat92E binding sites from eve stripe 3 enhancer (Yan et al., 1996), as well as putative Stat92E binding sites found in intron 1 of the socs36E gene (Karsten et al., 2002; Baeg et al., 2005; Bach et al., 2007). We searched for two Stat92E binding sites matching the matrix (using cut off score 6.5) that were located within 100 bp of each other, since work in mammalian systems has shown that two STAT sites located within this distance is sufficient to impart stronger transcriptional regulation (Xu et al., 1996). We then searched for genes with one, two or three pairs of Stat92E binding sites. This platform identified the three clusters of Stat92E binding sites in socs36E intron 1, indicating that it can accurately identify known Stat92E target genes. Taken together, we identified 1,463 genes that contained at least one pair (or cluster) of Stat92E binding sites within 100 bp of each other (see Fig. 1G and Suppl. Table 2).
Supplementary Material
Box plot analysis shows a five-number summary (the smallest observation, lower quartile, median, upper quartile, and largest observation) for the 584 significantly modulated mRNAs found either by SAM or T-test and harboring Stat92E TFBS’s, based on five individual eye discs per group (R1 through R5).
See Materials and Methods for procedures. In all figures, gray bar is yw and black bar is GMR-upd. The Y-axis in the bar graphs represents relative mRNA abundance.
See Materials and Methods for procedures. In all figures, gray bar is yw and black bar is GMR-upd. The Y-axis in the bar graphs represents relative mRNA abundance.
(A,B) single channel for Ser-lacZ expression (white) for Fig. 3M and N. Brackets indicate region where Ser expression is reduced in GMR-upd eye discs (B) as compared to yw controls (A). (C,D) merge of wg-lacZ (magenta) and Phalloidin (actin) for panels in Fig. 3D,E. Brackets indicate region where wg expression is reduced in GMR-upd eye discs (D) as compared to yw controls (C). (E-E”). Hop-expressing clones (green in E, white in E”) autonomously repress wg-lacZ (E’, arrowheads). wg-lacZ is magenta in E and white in E’. Furrow is marked by arrow in A–D. Ser-lacZ and wg-lacZ were detected with β–gal antibody.
(A,B) In wild type (WT) third instar eye discs, Dl (brown) is expressed at low levels anterior to the furrow and in cone cells posterior to the furrow (A). In eye discs containing stat92E M+ clones, Dl expression is significantly increased in cells anterior to the furrow (B, arrow). (C,D) in situ hybridization reveals that fng is expressed, as previously reported, in the ventral domain of a wild type second instar eye disc (C, bracket). In second instar eye discs containing stat92E M+ clones, fng expression is still restricted to the ventral domain (D, bracket). In C and D, the eye discs are outlined with a dashed black line. (E,F) in situ hybridization reveals that fng expression is not changed in third instar wild type (E) as compared to GMR-upd eye discs (F).
The table lists statistically significant mRNA transcripts detected by 584 Affymetrix probe sets (AFFY ID, column B) as described in Materials and Methods and in Fig 2A,B. The Gene Symbol and Gene Titles (Columns C and D) were updated using Affymetrix NETAFFX™ Analysis Center, release of November 13th, 2007. Column E lists the median value of fold change in GMR-upd samples that was baseline-normalized to median yw measurements set equal to 1. Column F lists either or both statistical algorithms applied to determine significant regulation of each gene (SAM, T-test, see Fig 2A and Materials and Methods). Column G indicates presence of Stat92E binding site in the target gene locus. Columns H through AA list the fold-change abundance (FC), raw intensities normalized by MBEI (Raw) and Present/Absent calls (Flags) of individual quintuple measurements in each genotype.
See Materials and Methods for procedures. Columns A and B list gene name and gene symbol, respectively, which were obtained from Flybase. Column C lists chromosomal location of the gene: X, 2L, 2R, 3L, 3R, 4, U= unmapped, Het = heterochromatin. Columns D and E list information for the first and second Stat92E binding site in the cluster, respectively. Specifically, they list the direction of the Stat92E binding site (for = forward, rev = reverse); the sequence identified by Target Explorer; its position on the chromosome (using Drosophila genome release 3.1, for which Target Explorer was designed (Sosinsky et al., 2003)); its binding site score (i.e., how similar it was to the consensus generated by matrix (minimum cut-off = 6.63)). Column F lists the location of the cluster in gene (i.e., 5’, 3’ or intronic regions). Column G lists the number of base pairs between the Stat92E binding site cluster and the start of the region of the gene (5’, 3’, intronic) in which the cluster was found.
The table summarizes the results of functional annotation and pathway analysis (KEGG and Gene Ontologies). Genes that populate each Term in corresponding GO or KEGG Category are separated into either upregulated or downregulated sections (yellow field/up and blue field/down, respectively). Count = number of genes per Term whose symbols are listed to the right. All molecules with microarray expression profiles validated by independent techniques (see Materials and Methods for details) are shown in red.
ACKNOWLEDGEMENTS
We thank S. Noselli, E. Knust, A. Bachmann, J. Treisman, C. Desplan, H. Sun, S. Bray, the Bloomington Stock Center and the DGRC for flies and cDNA clones. Several monoclonal antibodies used in this study were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. We thank the NYU Cancer Institute Genomics Facility for micro-array data processing and analysis and for assistance with Q-PCR and Rebecca Stearns (Harvard School of Public Health) for use of the Leo SEM. We thank J. Treisman, R. Dasgupta and members of the Bach and Dasgupta labs for helpful comments on the manuscript and discussions throughout the course of this study. We also are grateful to two anonymous reviewers whose comments significantly improved this study. This study was supported in part by a Young Investigator Award from the Breast Cancer Alliance, Inc. and a Basil O’Connor Starter Award from the March of Dimes (E.A.B.), as well as a Pharmacological Sciences Training Grant (GM066704-01) from the NIH awarded to New York University School of Medicine (L.A.E.).
Grant Sponsor: March of Dimes Basil O’Connor Starter Award; Grant number 5-FY06-579
Grant Sponsor: Breast Cancer Alliance, Inc.; Young Investigator Award (2004-6)
Grant Sponsor: Pharmacological Sciences Training Grant; Grant Number GM066704-01
REFERENCES
- Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling Role of Hemocytes in Drosophila JAK/STAT-Dependent Response to Septic Injury. Dev Cell. 2003;5:441–450. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Bach EA, Zeidler MP. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Ayala-Camargo A, Ekas LA, Flaherty MS, Baeg GH, Bach EA. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev Dyn. 2007;236:2721–2730. doi: 10.1002/dvdy.21230. [DOI] [PubMed] [Google Scholar]
- Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A, Knust E. Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev Genes Evol. 1998;208:346–351. doi: 10.1007/s004270050190. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksa K, Parke T, Dobens LL, Dearolf CR. The Drosophila STAT protein, stat92E, regulates follicle cell differentiation during oogenesis. Dev Biol. 2002;243:166–175. doi: 10.1006/dbio.2001.0539. [DOI] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Beccari S, Teixeira L, Rorth P. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech Dev. 2002;111:115–123. doi: 10.1016/s0925-4773(01)00615-3. [DOI] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–1279. [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Diez Del Corral R, Campuzano S, Dominguez M. Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development. 1999;126:4933–4942. doi: 10.1242/dev.126.22.4933. [DOI] [PubMed] [Google Scholar]
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, Karras JG, Levy DE, Inghirami G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- Cho KO, Choi KW. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998;396:272–276. doi: 10.1038/24394. [DOI] [PubMed] [Google Scholar]
- Clark HF, Brentrup D, Schneitz K, Bieber A, Goodman C, Noll M. Dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila. Genes Dev. 1995;9:1530–1542. doi: 10.1101/gad.9.12.1530. [DOI] [PubMed] [Google Scholar]
- Cohen SM. Imaginal disc development. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 747–841. [Google Scholar]
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Tyler DM, Furriols M, Chalkiadaki A, Delidakis C, Bray S. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev Biol. 2000;221:390–403. doi: 10.1006/dbio.2000.9691. [DOI] [PubMed] [Google Scholar]
- Darnell JE. Validating Stat3 in cancer therapy. Nat Med. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Deweindt C, Albagli O, Bernardin F, Dhordain P, Quief S, Lantoine D, Kerckaert JP, Leprince D. The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: a novel function for the BTB/POZ domain as an autonomous repressing domain. Cell Growth Differ. 1995;6:1495–1503. [PubMed] [Google Scholar]
- Dominguez M, Casares F. Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. Dev Dyn. 2005;232:673–684. doi: 10.1002/dvdy.20311. [DOI] [PubMed] [Google Scholar]
- Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Ekas LA, Baeg GH, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133:4721–4729. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- Fleming RJ, Gu Y, Hukriede NA. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- Freedman LP, Luisi BF, Korszun ZR, Basavappa R, Sigler PB, Yamamoto KR. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- Gabay L, Scholz H, Golembo M, Klaes A, Shilo BZ, Klambt C. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development. 1996;122:3355–3362. doi: 10.1242/dev.122.11.3355. [DOI] [PubMed] [Google Scholar]
- Garbe JC, Yang E, Fristrom JW. IMP-L2: an essential secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila. Development. 1993;119:1237–1250. doi: 10.1242/dev.119.4.1237. [DOI] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Georgenthum E, Carballes F, Medioni C, Cerezo D, Noselli S. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129:5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- Gilbert MM, Weaver BK, Gergen JP, Reich NC. A novel functional activator of the Drosophila JAK/STAT pathway, unpaired2, is revealed by an in vivo reporter of pathway activation. Mech Dev. 2005;122:939–948. doi: 10.1016/j.mod.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. Embo J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombria JC, Brown S, Hader S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, Jun S, Desplan C, Beckendorf SK, Sun YH. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jiang H, Foltenyi K, Kashiwada M, Donahue L, Vuong B, Hehn B, Rothman P. Fes mediates the IL-4 activation of insulin receptor substrate-2 and cellular proliferation. J Immunol. 2001;166:2627–2634. doi: 10.4049/jimmunol.166.4.2627. [DOI] [PubMed] [Google Scholar]
- Johansen KA, Iwaki DD, Lengyel JA. Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development. 2003;130:135–145. doi: 10.1242/dev.00202. [DOI] [PubMed] [Google Scholar]
- Jones AV, Chase A, Silver RT, Oscier D, Zoi K, Wang YL, Cario H, Pahl HL, Collins A, Reiter A, Grand F, Cross NC. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet. 2009 doi: 10.1038/ng.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci U S A. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- Kawata T, Early A, Williams J. Evidence that a combined activator-repressor protein regulates Dictyostelium stalk cell differentiation. Embo J. 1996;15:3085–3092. [PMC free article] [PubMed] [Google Scholar]
- Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL, Bass A, Marubayashi S, Heguy A, Garcia-Manero G, Kantarjian H, Offit K, Stone RM, Gilliland DG, Klein RJ, Levine RL. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009 doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt C. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development. 1993;117:163–176. doi: 10.1242/dev.117.1.163. [DOI] [PubMed] [Google Scholar]
- Klebes A, Sustar A, Kechris K, Li H, Schubiger G, Kornberg TB. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132:3753–3765. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- Klein T, Arias AM. Interactions among Delta, Serrate and Fringe modulate Notch activity during Drosophila wing development. Development. 1998;125:2951–2962. doi: 10.1242/dev.125.15.2951. [DOI] [PubMed] [Google Scholar]
- Kopczynski CC, Alton AK, Fechtel K, Kooh PJ, Muskavitch MA. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 1988;2:1723–1735. doi: 10.1101/gad.2.12b.1723. [DOI] [PubMed] [Google Scholar]
- Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- Kwon EJ, Park HS, Kim YS, Oh EJ, Nishida Y, Matsukage A, Yoo MA, Yamaguchi M. Transcriptional regulation of the Drosophila raf proto-oncogene by Drosophila STAT during development and in immune response. J Biol Chem. 2000;275:19824–19830. doi: 10.1074/jbc.M001114200. [DOI] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard OA. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci U S A. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Laron Z. Growth hormone insensitivity (Laron syndrome) Rev Endocr Metab Disord. 2002;3:347–355. doi: 10.1023/a:1020905725012. [DOI] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Grell EH. Genetic variations of Drosophila melanogaster. Washington, DC: Publs Carnegie Instn.; 1968. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. Embo J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Rose P, Barber D, Hanratty WP, Lee S, Roberts TM, D'Andrea AD, Dearolf CR. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17:1562–1571. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Treisman JE. pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development. 2000;127:1007–1016. doi: 10.1242/dev.127.5.1007. [DOI] [PubMed] [Google Scholar]
- Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, Prive GG, Licht JD. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M, Redemann S, Brunner D. The fission yeast MO25 protein functions in polar growth and cell separation. Eur J Cell Biol. 2005;84:915–926. doi: 10.1016/j.ejcb.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- Milburn CC, Boudeau J, Deak M, Alessi DR, van Aalten DM. Crystal structure of MO25 alpha in complex with the C terminus of the pseudo kinase STE20-related adaptor. Nat Struct Mol Biol. 2004;11:193–200. doi: 10.1038/nsmb716. [DOI] [PubMed] [Google Scholar]
- Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Mohanty S, Jermyn KA, Early A, Kawata T, Aubry L, Ceccarelli A, Schaap P, Williams JG, Firtel RA. Evidence that the Dictyostelium Dd-STATa protein is a repressor that regulates commitment to stalk cell differentiation and is also required for efficient chemotaxis. Development. 1999;126:3391–3405. doi: 10.1242/dev.126.15.3391. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Myrick KV, Dearolf CR. Hyperactivation of the Drosophila Hop jak kinase causes the preferential overexpression of eIF1A transcripts in larval blood cells. Gene. 2000;244:119–125. doi: 10.1016/s0378-1119(99)00568-5. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Olcaydu D, Harutyunyan A, Jager R, Berg T, Gisslinger B, Pabinger I, Gisslinger H, Kralovics R. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009 doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- Osterbur DL, Fristrom DK, Natzle JE, Tojo SJ, Fristrom JW. Genes expressed during imaginal discs morphogenesis: IMP-L2, a gene expressed during imaginal disc and imaginal histoblast morphogenesis. Dev Biol. 1988;129:439–448. doi: 10.1016/0012-1606(88)90391-0. [DOI] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281:2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Pereira PS, Pinho S, Johnson K, Couso JP, Casares F. A 3' cis-regulatory region controls wingless expression in the Drosophila eye and leg primordia. Dev Dyn. 2006;235:225–234. doi: 10.1002/dvdy.20606. [DOI] [PubMed] [Google Scholar]
- Perez SE, Steller H. Molecular and genetic analyses of lama, an evolutionarily conserved gene expressed in the precursors of the Drosophila first optic ganglion. Mech Dev. 1996;59:11–27. doi: 10.1016/0925-4773(96)00556-4. [DOI] [PubMed] [Google Scholar]
- Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- Rawlings JS, Rennebeck G, Harrison SM, Xi R, Harrison DA. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 2004;5:38. doi: 10.1186/1471-2121-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Bach EA, Cagan RL. Drosophila C-terminal Src kinase negatively regulates organ growth and cell proliferation through inhibition of the Src, Jun N-terminal kinase, and STAT pathways. Mol Cell Biol. 2004;24:6676–6689. doi: 10.1128/MCB.24.15.6676-6689.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Rijsewijk F, Schuermann M, Wagenaar E, Parren P, Weigel D, Nusse R. The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987;50:649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- Schulz C, Tautz D. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development. 1994;120:3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- Sefton L, Timmer JR, Zhang Y, Beranger F, Cline TW. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405:970–973. doi: 10.1038/35016119. [DOI] [PubMed] [Google Scholar]
- Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci U S A. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Singh A, Choi KW. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development. 2003;130:6351–6360. doi: 10.1242/dev.00864. [DOI] [PubMed] [Google Scholar]
- Sloth Andersen A, Hertz Hansen P, Schaffer L, Kristensen C. A new secreted insect protein belonging to the immunoglobulin superfamily binds insulin and related peptides and inhibits their activities. J Biol Chem. 2000;275:16948–16953. doi: 10.1074/jbc.M001578200. [DOI] [PubMed] [Google Scholar]
- Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky A, Bonin CP, Mann RS, Honig B. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 2003;31:3589–3592. doi: 10.1093/nar/gkg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev Cell. 2008;14:726–738. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Dorsal gradient networks in the Drosophila embryo. Dev Biol. 2002;246:57–67. doi: 10.1006/dbio.2002.0652. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Sun J, Blaskovich MA, Jove R, Livingston SK, Coppola D, Sebti SM. Cucurbitacin Q: a selective STAT3 activation inhibitor with potent antitumor activity. Oncogene. 2005;24:3236–3245. doi: 10.1038/sj.onc.1208470. [DOI] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Terry NA, Tulina N, Matunis E, DiNardo S. Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol. 2006;294:246–257. doi: 10.1016/j.ydbio.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111:749–761. doi: 10.1242/dev.111.3.749. [DOI] [PubMed] [Google Scholar]
- Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Tsai YC, J.G. Y, P.H. C, Posakony JW, Barolo S, Kim J, Henry Sun Y. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev Biol. 2007;306:760–771. doi: 10.1016/j.ydbio.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn BA, Quelle FW, Rogers NC, Schindler C, Stark GR, Ihle JN, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- Watts JL, Morton DG, Bestman J, Kemphues KJ. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467–1475. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
- Weber H, Bernhardt A, Dieterle M, Hano P, Mutlu A, Estelle M, Genschik P, Hellmann H. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 2005;137:83–93. doi: 10.1104/pp.104.052654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DR. Pattern formation in the Drosophila retina. In: Martinez Arias M, Bate M, editors. The Development of Drosophila melanogaster. Plainview, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Yang CH, Axelrod JD, Simon MA. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675–688. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Yang CH, Simon MA, McNeill H. mirror controls planar polarity and equator formation through repression of fringe expression and through control of cell affinities. Development. 1999;126:5857–5866. doi: 10.1242/dev.126.24.5857. [DOI] [PubMed] [Google Scholar]
- Zeidler MP, Bach EA, Perrimon N. The roles of the Drosophila JAK/STAT pathway. Oncogene. 2000;19:2598–2606. doi: 10.1038/sj.onc.1203482. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Box plot analysis shows a five-number summary (the smallest observation, lower quartile, median, upper quartile, and largest observation) for the 584 significantly modulated mRNAs found either by SAM or T-test and harboring Stat92E TFBS’s, based on five individual eye discs per group (R1 through R5).
See Materials and Methods for procedures. In all figures, gray bar is yw and black bar is GMR-upd. The Y-axis in the bar graphs represents relative mRNA abundance.
See Materials and Methods for procedures. In all figures, gray bar is yw and black bar is GMR-upd. The Y-axis in the bar graphs represents relative mRNA abundance.
(A,B) single channel for Ser-lacZ expression (white) for Fig. 3M and N. Brackets indicate region where Ser expression is reduced in GMR-upd eye discs (B) as compared to yw controls (A). (C,D) merge of wg-lacZ (magenta) and Phalloidin (actin) for panels in Fig. 3D,E. Brackets indicate region where wg expression is reduced in GMR-upd eye discs (D) as compared to yw controls (C). (E-E”). Hop-expressing clones (green in E, white in E”) autonomously repress wg-lacZ (E’, arrowheads). wg-lacZ is magenta in E and white in E’. Furrow is marked by arrow in A–D. Ser-lacZ and wg-lacZ were detected with β–gal antibody.
(A,B) In wild type (WT) third instar eye discs, Dl (brown) is expressed at low levels anterior to the furrow and in cone cells posterior to the furrow (A). In eye discs containing stat92E M+ clones, Dl expression is significantly increased in cells anterior to the furrow (B, arrow). (C,D) in situ hybridization reveals that fng is expressed, as previously reported, in the ventral domain of a wild type second instar eye disc (C, bracket). In second instar eye discs containing stat92E M+ clones, fng expression is still restricted to the ventral domain (D, bracket). In C and D, the eye discs are outlined with a dashed black line. (E,F) in situ hybridization reveals that fng expression is not changed in third instar wild type (E) as compared to GMR-upd eye discs (F).
The table lists statistically significant mRNA transcripts detected by 584 Affymetrix probe sets (AFFY ID, column B) as described in Materials and Methods and in Fig 2A,B. The Gene Symbol and Gene Titles (Columns C and D) were updated using Affymetrix NETAFFX™ Analysis Center, release of November 13th, 2007. Column E lists the median value of fold change in GMR-upd samples that was baseline-normalized to median yw measurements set equal to 1. Column F lists either or both statistical algorithms applied to determine significant regulation of each gene (SAM, T-test, see Fig 2A and Materials and Methods). Column G indicates presence of Stat92E binding site in the target gene locus. Columns H through AA list the fold-change abundance (FC), raw intensities normalized by MBEI (Raw) and Present/Absent calls (Flags) of individual quintuple measurements in each genotype.
See Materials and Methods for procedures. Columns A and B list gene name and gene symbol, respectively, which were obtained from Flybase. Column C lists chromosomal location of the gene: X, 2L, 2R, 3L, 3R, 4, U= unmapped, Het = heterochromatin. Columns D and E list information for the first and second Stat92E binding site in the cluster, respectively. Specifically, they list the direction of the Stat92E binding site (for = forward, rev = reverse); the sequence identified by Target Explorer; its position on the chromosome (using Drosophila genome release 3.1, for which Target Explorer was designed (Sosinsky et al., 2003)); its binding site score (i.e., how similar it was to the consensus generated by matrix (minimum cut-off = 6.63)). Column F lists the location of the cluster in gene (i.e., 5’, 3’ or intronic regions). Column G lists the number of base pairs between the Stat92E binding site cluster and the start of the region of the gene (5’, 3’, intronic) in which the cluster was found.
The table summarizes the results of functional annotation and pathway analysis (KEGG and Gene Ontologies). Genes that populate each Term in corresponding GO or KEGG Category are separated into either upregulated or downregulated sections (yellow field/up and blue field/down, respectively). Count = number of genes per Term whose symbols are listed to the right. All molecules with microarray expression profiles validated by independent techniques (see Materials and Methods for details) are shown in red.