Abstract
Prostate cancer (PrCa) is the most frequently diagnosed male cancer in developed countries. To identify common PrCa susceptibility alleles, we have previously conducted a genome-wide association study in which 541, 129 SNPs were genotyped in 1,854 PrCa cases with clinically detected disease and 1,894 controls. We have now evaluated promising associations in a second stage, in which we genotyped 43,671 SNPs in 3,650 PrCa cases and 3,940 controls, and a third stage, involving an additional 16,229 cases and 14,821 controls from 21 studies. In addition to previously identified loci, we identified a further seven new prostate cancer susceptibility loci on chromosomes 2, 4, 8, 11, and 22 (P=1.6×10−8 to P=2.7×10−33).
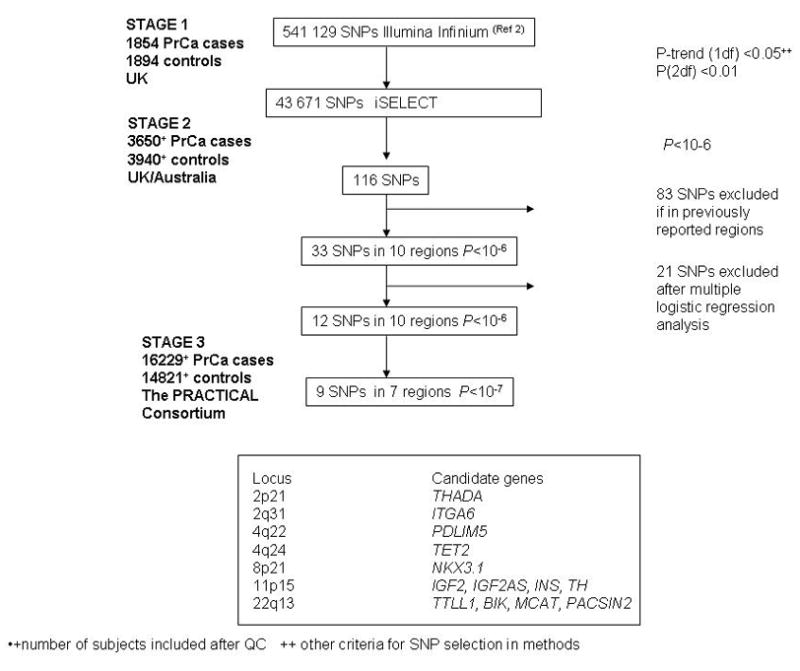
Genome-wide association studies (GWAS) provide a powerful approach to identify common disease alleles. We previously conducted a GWAS1, based on genotyping of 541, 129 SNPs in 1,854 clinically detected PrCa cases and 1,894 controls (see Figure 1, stage 1). Follow-up genotyping of SNPs exhibiting strong evidence of association (P<10−6), in a further 3,268 cases and 3,366 controls, allowed us to identify SNPs at 7 susceptibility loci associated with the disease at genome-wide levels of significance1. Other studies have identified an additional 8 loci2–9. These loci, however, explain only a small fraction of the familial risk of PrCa. Moreover, the strength of the associations that have been detected are generally small (per-allele odds ratios, OR, 1.1–1.2), and the power of the existing studies to detect many of the susceptibility alleles has been limited. It is highly likely, therefore, that other PrCa predisposition loci exist, and that such loci should be detectable by studies with larger sample sizes.
Figure 1.
Summary of experimental design and results. Numbers quoted are after QC (see Online Methods).
In an attempt to identify further susceptibility loci, we conducted a more extensive follow-up of SNPs showing evidence of association in stage 1 of our GWAS. We designed a panel of 47,120 SNPs, aiming to include all SNPs with a significant association in stage 1 at P-trend (1df)<.05 or P(2df)<.01 (see Online Methods). These SNPs were genotyped using the Illumina iSELECT platform in 3,894 PrCa cases and 4,055 controls from the United Kingdom (UK) and Australia (Figure 1, stage 2). After quality control (QC) exclusions (as described in Online Methods), we utilised data from 43,671 SNPs in 3,650 PrCa cases and 3,940 controls.
Genotype frequencies in cases and controls were compared using a 1 degree of freedom (df) Cochran-Armitage trend test (for QQ plots see Supplementary Figure 1). There was little evidence of inflation in the test statistics in the UK samples (estimated inflation factor λ=1.08), but there was more marked inflation in those from Australia (λ=1.23; λ=1.19 for stage 2 overall), suggestive of some population substructure. The Australian samples were selected from three studies (MCCS, RFPCS and EOPCS; see Supplementary Note for cohort descriptions), and further analysis revealed that adjustment for sub-study substantially reduced the inflation (λ=1.08 for Australia, λ=1.14 overall). This inflation may reflect oversampling of MCCS for individuals of South European ancestry. Principal components analysis identified a distinct cluster that is overrepresented in the MCCS study, consistent with admixture in this population (see Online Methods and Supplementary Figure 2). Adjustment for the first two principal components in addition to stratification by sub-study did not, however, reduce the inflation further. The residual inflation could reflect weak population substructure, or may reflect the combined effects of weak susceptibility alleles.
There was a clear excess of nominally significant associations in stage 2, with 132 SNPs significant at P<.0001 compared with ~4 that would be expected by chance (Supplementary Table 1). After combining with the stage 1 data, 116 SNPs were significant at P <10−6 (Supplementary Table 2). Of these, twenty-six of the SNPs were on chromosome 8q24, a region known to harbour multiple PrCa susceptibility loci 1, 3, 4, 6, 7 In addition, 42 SNPs were in the 7 regions we identified in our previous analysis1 and 13 were in two regions on 17q identified by Gudmundsson et al5. We also found strong evidence for an association with two SNPs on 2p15 (rs2710647, P=7.1×10−8; rs6545977, P=4.5×10−7), within the EHBP1 gene, close to that recently reported by Gudmundsson et al8. rs2710647 is however, only weakly correlated with the previously reported SNP rs721048, r2=0.19, and might reflect an independent association. Two additional susceptibility loci on chromosomes 7 and 10 were identified in the GWAS by Thomas et al9. We found supporting evidence for an association with rs10486567 on chromosome 7 (JAZF1; stage 2 per-allele OR 1.05, 95%CI 0.92, 95%CI 0.85–1.00, combined P=.00008), but only limited evidence for an association with rs4962416 on chromosome 10 (CTBP2; per-allele OR 1.05, 95%CI 0.98–1.13; P=.04).
The remaining 33 SNPs significant at P<10−6 were in 10 regions not previously associated with PrCa. Multiple logistic regression analysis was used to define a minimal subset of 12 independently significant SNPs, such that the remaining SNPs were not significant after adjustment for these SNPs. The strength of these associations in stage 2 was not substantially affected by principal components adjustment (Supplementary Table 3). These 12 SNPs were then subjected to further replication analysis in a third stage, involving 16,229 cases and 14,821 controls from 21 studies participating in the PRACTICAL Consortium (Supplementary Table 4).
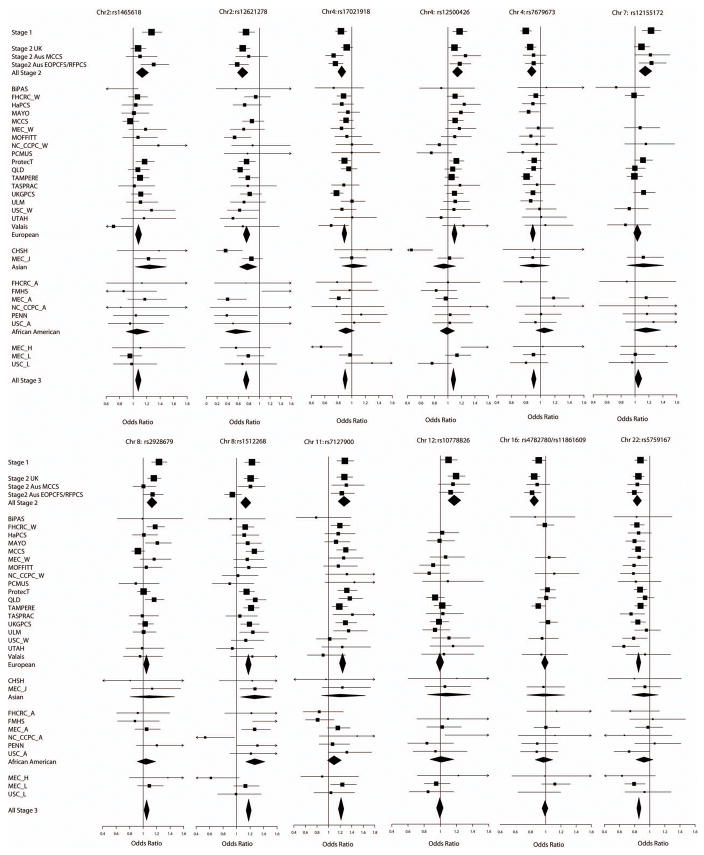
Eight SNPs in 7 regions showed clear evidence of replication in stage 3 (P=.0002 or lower, in each case in the same direction as in stages 1 and 2). In each case the combined P-trend over all 3 stages reached P<10−7, with a range of P=1.6×10−8 to P=2.7×10−33. Two SNPs on chromosome 4, rs17021918 and rs12500426, are correlated (r2 =0.5) but both showed independent association after multiple logistic regression analyses (P=0.014 and P=0.0003 for the effect of rs12500426 after adjustment for rs17021918 in stage 3 and overall, respectively; P=.0002 and P=3.6×10−7 for the effects of rs17021918 after adjustment for rs12500426; Supplementary Table 5). In addition to the above SNPs, an additional SNP on chromosome 8 showed more limited evidence of replication (P=.007 in stage 3, P=7.1×10−8 overall). It is ~90kb from rs1512268, which showed very clear evidence of association (P=3.4×10−30), but the SNPs are in neighbouring linkage disequilibrium (LD) blocks and are only weakly correlated (r2=0.03). rs12155172 on chromosome 7 showed weak evidence of association in stage 3 (P=0.06, with an effect in the same direction as stages 1 and 2), but did not reach genome-wide significance in the combined dataset (P=8.8×10−6). Thus, this locus may harbour a susceptibility allele, but further large case-control studies will be required to confirm or refute this finding. For the remaining 2 loci, on chromosomes 12 and 16, there was no evidence of association in stage 3. We conclude that these two loci were probably false positive associations in stages 1 and 2.
We are able to compare our results with those from from the Cancer Genetic Markers of Susceptibility (CGEMS) PrCa study, a GWAS of 1,117 PrCa cases and 1,105 controls that utilized the same genotyping platform as our GWAS. Of the 9 SNPs that reached genome-wide significance in our study, 8 were typed in CGEMS, and all had an estimated OR in the same direction as our study (Supplementary Table 6). Only two of the SNPs were nominally significant in CGEMS (rs5759167; P=.0035, and rs7679673; P=.014). However, in all cases the estimated the 95% confidence interval for per-allele OR from the CGEMS study contained our estimate. This suggests that the failure to replicate association of some of these loci in CGEMS may be related to the relatively smaller size of the CGEMS stage 1.
All but two of the SNPs associated with PrCa risk exhibit an association with allele dose consistent with a log-additive model, as observed for most common cancer susceptibility alleles. rs12621278 on chromosome 2 exhibits a strong dose response, with an odds ratio (OR) in homozygotes (0.35, 95%CI 0.24–0.52) being smaller than would be expected under a log-additive model (P=.0076). rs17021918 on chromosome 4 showed no difference in risk between heterozygotes and homozygotes (P=.023 compared with a log-additive model).
There was no evidence for a difference in the per-allele ORs among European, Asian and African-American populations (Figure 2), with the exception of rs12500426, on chromosome 4, which exhibited an association in Europeans but not in Asian or African-American studies (P=.046 for heterogeneity in the OR by population); and rs7679673, on chromosome 4, for which the association in European and Asian populations was not observed in African-Americans (P=.032). These differences might reflect differences in the LD structure and the frequency of the causal variant(s) in different populations. There was no evidence of heterogeneity in the per-allele ORs in European and African-American studies for any SNP and only weak evidence of heterogeneity for two SNPs in Asian studies (Supplementary Table 7). We also found no marked differences in the per-allele ORs between studies based on populations where PSA screening was prevalent (studies in the US, and the ProtecT study in the UK) and those in which screening was less common (Supplementary Table 8).
Figure 2.
Forrest plots for the 12 SNPs replicated in stage 3. Squares represent the estimated per-allele odds ratio (OR) for each study. Lines indicate the 95% confidence interval. Diamonds represent the OR estimates and confidence limits for the subgroups indicated. The SNPs rs4782780 and rs11861609 are highly correlated and only one was typed in any study. The OR is shown for the SNP typed in each study (see Online Methods).
The controls in stage 1 of our GWAS were selected for low Prostate Specific Antigen (PSA) levels, and this may have led to the preferential selection of SNPs associated with PSA levels10, 11. We were able to examine the associations between genotypes and serum PSA levels in 1,585 control samples from the ProtecT study in stage 2 of our scan (Supplementary Table 9). Two SNPs, rs17021918 (chromosome 4) and rs1512268 (chromosome 8) showed weak association with PSA levels, in the same direction as the PrCa association (P=.043 and P=.037 respectively). Both SNPs, however, showed very strong evidence of association in all three stages, and we conclude that none of the associations are likely to be mediated simply through associations with PSA level.
Data on Gleason score were available for 7,855 PrCa cases from 14 studies. There was no difference in the OR for cases with high/intermediate grade disease (Gleason score ≥7) versus low grade disease (Gleason score <7), for any of the associated SNPs (Supplementary Table 10). This consistency (also seen for the previously identified loci1) suggests that most susceptibility loci identified to date modulate the early stages of disease development rather than progression.
For 2 of the SNPs, rs12621278 on chromosome 2 (P= 1.1×10−5) and rs7127900 on chromosome 11 (P=.006), the per-allele OR varied with age, with a higher OR at younger ages (Supplementary Table 11). One SNP, rs7679673, also exhibited a stronger association when analyses were restricted to cases with a family history of PrCa (P=.02; Supplementary Table 12).
The associated SNPs identified in the second stage of our GWAS lie in LD blocks that include several plausible causative genes (see Figure 1 for candidate gene list and Supplementary Note for detail). Particularly notable are rs12621278 on 2q31, which is in intron 1 of ITGA6, the gene encoding integrin alpha 6, and rs2928679 and rs1512268 (90kb apart on 8p21). rs2928679 lies 10kb downstream of NKX3.1, which codes for an androgen-regulated homeobox protein NKX3.1 which is in the HDAC1 pathway.
Most of the per-allele ORs estimated for these variants in this study population were modest, ranging from 1.08–1.28 fold. We found no further loci with associations as strong as the SNPs on 8q or the MSMB locus, as would be expected since the power to detect these loci in our first analysis was already high. Nevertheless, there are now more than 20 loci conferring ORs >1.1 for PrCa, more than for all other cancer types. We estimate that the power to detect these associations in this experiment varied from >80% (for rs1512268 and rs7127900) to <1% (for rs1465618, rs12155172 and rs2928679; Supplementary Table 13). This strongly suggests that additional loci of similar magnitude remain to be identified. Mapping of such loci will require the synthesis and follow-up of larger GWAS datasets. We have demonstrated that we have power to confirm such loci using the PRACTICAL Consortium.
Based on an overall two-fold familial relative risk to first-degree relatives of PrCa cases, and on the assumption that the SNPs combine multiplicatively, the new loci reported here together explain approximately 4.3% of the familial risk of PrCa. Including the previously reported loci, approximately 21.5% of familial risk in PrCa may now be explained. Under this model, the top 10% of the population at highest risk has a relative risk approximately 2.3-fold greater than the average risk in the general population, while the top 1% has an estimated 3-fold increased relative risk. In contrast, the individuals classified at the bottom 1% of genetic risk according to this model are estimated to have a relative risk of about one-fifth of the population average. Such risk prediction may have implications for targeted screening and prevention. Moreover, the associations we have found using tagSNPs may represent stronger associations with the causal variants. If so, the overall contribution of the causal variants will be greater. Resequencing of these regions, further genotyping and functional analyses will be required to identify the genetic variants responsible for each risk locus.
Methods
Samples
PrCa cases and controls used in stage 1 of the GWAS have been described previously1. PrCa cases and controls for stage 2 (Figure 1) were selected from studies in the UK and Australia. UK cases were drawn from the UK Genetic Prostate Cancer Study (UKGPCS). UKGPCS includes cases PrCa cases that were either diagnosed at age ≤60 years (n=341) and/or those that had a first or second degree family history of prostate cancer (n=220), recruited from urologists throughout the UK, and a series of cases recruited from PrCa clinics in the Urology Unit at The Royal Marsden NHS Foundation Trust over a 14 year period. UK controls were identified through two sources. Six hundred and fifty-six controls were drawn from the UKGPCS study (Prostate Cancer Research Foundation Study component) and were geographically, ethnically and age matched to the UKGPCS young onset cases. They had no family or personal history of PrCa. The remaining controls (n=1636) were selected from men in the ProtecT (Prostate testing for cancer and Treatment) study27. ProtecT is a national study of community-based PSA testing and a randomised trial of subsequent PrCa treatment. Approximately 110,000 men between the ages of 50 and 69 years, (with a small set of men aged 45–49 years from one centre), were ascertained through general practices in nine regions in the UK. For this study we selected as controls men who had a PSA of <10ng/ml and negative prostate biopsies. Men with PSA ≥3ng/ml were excluded if they had a positive prostatic biopsy. We excluded, from both cases and controls, men who reported to be non-white. The majority of men in the UK are diagnosed via a clinical presentation; amongst the cases in this study 100% of those from the ProtecT study were diagnosed through asymptomatic PSA screening.
The Australian cases were ascertained from three studies28–30: (i) a population-based series of PrCa cases identified from the Victorian Cancer Registry since 1999, diagnosed at <56 years (Early Onset Prostate Cancer Study, EOPCFS; n=631); (ii) a population-based case-control study consisting of cases diagnosed in Melbourne and Perth (Risk Factors for Prostate Cancer Study, RFPCS; n=702). Cases were identified from the population cancer registries, with histopathologically confirmed PrCa, excluding tumors with Gleason scores of < 5, and diagnosed at < 70 years with sampling stratified by age at diagnosis and (iii) a prospective cohort study of 17,154 men aged 40 to 69 years at recruitment in 1990–4 (Melbourne Collaborative Cohort Study, MCCS; n=378). Controls were selected from the RFPCS study, in which they were identified through government electoral rolls and frequency matched to the age distribution of the RFPCS cases (n=667), together with a random sample from the MCCS cohort (n=981).
Stage 3 included samples from 21 PrCa case-control studies from groups in the PRACTICAL Consortium (Supplementary Table 4).
All studies were approved by the appropriate ethics committees.
Genotyping
Stage 2 genotypes were generated using an Illumina iSELECT array. SNPs were selected on the basis of the stage 1 results to include those with (a) a 1df P-trend <.059 (n= 34,484) and (b) a 2df genotype test P<.01 (n=2,202). We also included (c) all SNPs from the 1M array in LD blocks defined around “hits” from stage 1, defined as a SNP with P-trend <.0001; (d) all SNPs from the Illumina 1M array on 8q24; (e) all SNPs from the 550k array in the HLA region and (f) all SNPs significant in the CGEMS GWAS with P-trend <.01. We also included a further set of SNPs of interest in collaboration with CGEMS group (these were not considered in this paper; results to be reported separately). For analysis, we utilized samples on which genotypes could be called for at least 97% of SNPs at a confidence score of ≥0.25. Data were generated on 43,671 of 47,120 SNPs.
To identify close relatives we computed identity-by-state (IBS) probabilities for all pairs. We identified 93 cryptic duplicate samples (or monozygotic twins) or probable brothers (IBS >0.86). In each case we excluded the individual with the lower call rate. By computing IBS scores between participants and individuals in HapMap and using multi-dimensional scaling, we identified 252 individuals who appeared to have significant Asian or African ancestry (approximately >10% non-European ancestry). We removed 14 cases with a significant level of heterozygosity on X (16–39%; including 3 known cases of Klinefelter’s syndrome). After these exclusions, 3,650 cases and 3,940 controls were used in the final analysis of stage 2.
We filtered out all SNPs with a call rate <95%, a minor allele frequency in controls of <1%, or whose genotype frequency in controls departed from Hardy-Weinberg equilibrium at p<.00001. After these exclusions, we analyzed 43,671 SNPs, of which 42,186 were genotyped in both stage 1 and stage 2. Duplicate concordance was 99.998%.
In stage 3, genotyping of samples from all but one study site was performed by 5′ exonuclease assay (Taqman™) using the ABI Prism 7900HT sequence detection system according to the manufacturer’s instructions. Primers and probes were supplied directly by Applied Biosystems as Assays-By-Design™. The Queensland site used the iPlex Sequenom MassArray® system. Ten of the 12 SNPs worked well with the initial assay; one (rs12155172) had to be redesigned and one (rs4782780) had to be replaced by a proxy SNP (rs11861609, r2=0.93) for all groups except Queensland. These latter two SNPs were therefore only run by 8 groups (Supplementary Table 4).
Assays at all sites included at least two negative controls and 2–5% duplicates per plate. Quality control guidelines were followed by all the participating groups as previously described31. We excluded individuals that were not typed for at least 80% of the SNPs attempted. Data on a given SNP for a given site were also excluded if they failed QC criteria, which necessitated: a call rate >95%, no deviation from Hardy-Weinberg equilibrium in controls at P<.00001 and a <2% discordance between genotypes in duplicate samples. Cluster plots were re-examined centrally where necessary. Overall, 12 site/SNP combinations were excluded.
Statistical methods
We assessed associations between each SNP and PrCa at stage 2 using a 1df Cochran-Armitage trend test and a general 2df chi-squared test. Inflation in the chi-squared statistic was assessed using the genomic control approach; we derived an inflation factor (λ) by dividing the median of the lowest 90% of the 1df statistics by the 45% percentile of a 1df chi-squared distribution (0.357). This cutoff was used to avoid inclusion of SNPs likely to be associated with risk. Analyses were stratified by country and, within Australia, by study (MCCS and EOPCS/RFPCS). This stratification was made because the MCCS were known to be oversampled for individuals of Southern Europe ancestry, and because this stratification materially reduced the overdispersion. To further assess population structure, we performed principal components analysis using 15,363 uncorrelated SNPs (r2<0.1). The first component was strongly related to stratum (MCCS vs. EOPCS/RFPCS vs. UK; Supplementary Figure 2). Addition of up to five principal components as covariates made no difference to the inflation, after adjustment for stratum, and we therefore chose not to use the principal components in the primary analysis, to preserve consistency with stage 3. However, subsequent adjustment for the SNPs reaching genome-wide significance made no material difference to the strength of the associations or significance levels. SNPs were selected for evaluation in stage 3 on the basis of a significance level of P<10−6 based on a 1df trend test. Multiple logistic regression was used to define the minimal set of SNPs that showed evidence of association at P<.05, after adjustment for other SNPs.
In stage 3, we stratified analyses by study and racial/ethnic group (white, African-American, south-east Asian, Latino and Hawaiian). Where <100 individuals were recorded in a minority ethnic group, these individuals were excluded. The Mayo Clinic study genotyped multiple cases for the same family; we included only one case per family at random in the analysis. After exclusions, analyses were based on 16,229 PrCa cases and 14,821 controls from stage 3.
ORs and confidence limits were estimated using unconditional logistic regression, stratified by study and racial/ethnic group. In the text we have reported the combined tests of association over all three stages, but have emphasized the OR estimates from stage 3, to minimize the effect of “winner’s curse”. Tests of homogeneity of the ORs across strata were assessed using likelihood ratio tests. The associations between genotype and family history and Gleason score were assessed using a case-only analysis; Gleason score was analysed both using the binary endpoint of Gleason score ≥ 7 and as an ordinal variable, by polytomous regression. Modification of the ORs by age was assessed using a case-only analysis, assessing the association between age and SNP genotype in the cases using polytomous regression. The associations between SNP genotypes and PSA level were assessed using linear regression, after log-transformation of PSA level to correct for skewness. Analyses were performed in R (principally using SNPMatrix32) and Stata.
Supplementary Material
Table 1.
Summary results for SNPs selected for genotyping in Stage 3.
| Marker | Position | Stage | MAF | Per-allele OR (95%CI) | Heterozygous OR (95%CI) | Homozygous OR (95%CI) | P-value | |
|---|---|---|---|---|---|---|---|---|
| Stage | Combined | |||||||
| rs1465618 G/A |
2 43407453 |
Stage 1 | .18 | 1.27 (1.14–1.43) | 1.20 (1.04–1.39) | 1.89 (1.36–2.61) | 3.3×10−5 | |
| Stage 2 | .21 | 1.13 (1.05–1.23) | 1.16 (1.05–1.28) | 1.20 (.95–1.50) | .002 | |||
| Stage 3 | .23 | 1.08 (1.03–1.12) | 1.08 (1.03–1.14) | 1.15 (1.04–1.26) | .0002 | 1.6×10−8 | ||
| rs12621278 A/G |
2 173019799 |
Stage 1 | .06 | .74 (.61–.91) | .76 (.61–.94) | .45 (.16–1.30) | .004 | |
| Stage 2 | .06 | .67 (.58–.78) | .68 (.58–.80) | .37 (.16–.89) | 1.5×10−7 | |||
| Stage 3 | .06 | .75 (.70–.80) | .78 (.72–.84) | .35 (.24–.52) | 4.9×10−15 | 8.7×10−23 | ||
| rs17021918 C/T |
4 95781900 |
Stage 1 | .37 | .84 (.76–.93) | .89 (.77–1.02) | .67 (.54–.83) | .0005 | |
| Stage 2 | .35 | .85 (.79–.91) | .80 (.72–.88) | .77 (.66–.89) | 4.7×10−6 | |||
| Stage 3 | .34 | .90 (.87–.93) | .86 (.82–.91) | .85 (.78–.92) | 1.8×10−8 | 4.2×10−15 | ||
| rs12500426 C/A |
4 95733632 |
Stage 1 | .44 | 1.17 (1.07–1.28 | 1.29 (1.11–1.50) | 1.35 (1.13–1.62) | .001 | |
| Stage 2 | .44 | 1.14 (1.07–1.22) | 1.11 (1.00–1.24) | 1.31 (1.14–1.49) | 9.6×10−5 | |||
| Stage 3 | .46 | 1.08 (1.05–1.12) | 1.08 (1.03–1.14) | 1.17 (1.10–1.25) | 1.8×10−6 | 1.3×10−11 | ||
| rs7679673 C/A |
4 106280983 |
Stage 1 | .38 | .80 (.73–.88) | .73 (.63–.83) | .69 (.56–.85) | 3.2×10−6 | |
| Stage 2 | .39 | .88 (.82–.94) | .87 (.79–.97) | .78 (.67–.90) | .0002 | |||
| Stage 3 | .45 | .91 (.88–.94) | .89 (.84–.94) | .83 (.77–.89) | 6.7×10−8 | 2.6×10−14 | ||
| rs12155172 G/A |
7 20961016 |
Stage 1 | .20 | 1.23 (1.10–1.37) | 1.26 (1.10–1.45) | 1.42 (1.05–1.92) | .0002 | |
| Stage 2 | .21 | 1.15 (1.06–1.24) | 1.13 (1.03–1.25) | 1.35 (1.09–1.68) | .001 | |||
| Stage 3 | .20 | 1.05 (.998–1.10) | 1.04 (.98–1.10) | 1.11 (.97–1.27) | .06 | 8.8×10−6 | ||
| rs2928679 C/T |
8 23494920 |
Stage 1 | .45 | 1.24 (1.13–1.36) | 1.26 (1.08–1.47) | 1.53 (1.27–1.83) | 6.5×10−6 | |
| Stage 2 | .46 | 1.13 (1.06–1.21) | 1.16 (1.04–1.30) | 1.27 (1.11–1.45) | .0003 | |||
| Stage 3 | .42 | 1.05 (1.01–1.09) | 1.04 (.98–1.10) | 1.11 (1.03–1.19) | .007 | 7.1×10−8 | ||
| rs1512268 G/A |
8 23582408 |
Stage 1 | .40 | 1.23 (1.12–1.35) | 1.28 (1.11–1.48) | 1.49 (1.23–1.80) | 1.0×10−5 | |
| Stage 2 | .42 | 1.13 (1.06–1.21) | 1.13 (1.02–1.26) | 1.29 (1.12–1.47) | .0002 | |||
| Stage 3 | .45 | 1.18 (1.14–1.22) | 1.19 (1.13–1.26) | 1.39 (1.30–1.48) | 2.5×10−23 | 3.4×10−30 | ||
| rs7127900 G/A |
11 2190150 |
Stage 1 | .18 | 1.28 (1.14–1.44) | 1.40 (1.22–1.61) | 1.27 (.92–1.76) | 1.7×10−5 | |
| Stage 2 | .18 | 1.27 (1.17–1.38) | 1.29 (1.16–1.42) | 1.52 (1.19–1.95) | 2.4×10−8 | |||
| Stage 3 | .20 | 1.22 (1.17–1.27) | 1.24 (1.18–1.30) | 1.42 (1.27–1.57) | 1.2×10−22 | 2.7×10−33 | ||
| rs10778826 G/A |
12 80626985 |
Stage 1 | .41 | 1.10 (1.00–1.21) | 1.05 (.91–1.21) | 1.24 (1.02–1.50) | .04 | |
| Stage 2 | .39 | 1.17 (1.10–1.25) | 1.12 (1.01–1.24) | 1.40 (1.22–1.61) | 3.5×10−6 | |||
| Stage 3 | .35 | .99 (.95–1.04) | .99 (.93–1.05) | .99 (.91–1.09) | .8 | .006 | ||
| rs4782780 T/C |
16 81960548 |
Stage 1 | .40 | .91 (.83–1.00) | .88 (.77–1.02) | .84 (.69–1.02) | .05 | |
| Stage 2 | .43 | .85 (.79–.90) | .91 (.82–1.01) | .69 (.60–.80) | 1.1×10−6 | |||
| Stage 3 (One study) | .41 | 1.01 (.89–1.13) | 1.17 (.98–1.42) | .95 (.74–1.21) | .92 | 3.7×10−6 | ||
| rs11861609 C/G |
16 81942167 |
Stage 3 (Nine studies) | .40 | .99 (.95–1.03) | .98 (.92–1.05) | .98 (.90–1.07) | .58 | N/A |
| Stage 3 (Ten studies) | .40 | .99 (.95–1.03) | 1.00 (.94–1.07) | .98 (.90–1.06) | .63 | .0007 | ||
| rs5759167 G/T |
22 41830156 |
Stage 1 | .49 | .88 (.80–.96) | .84 (.72–.98) | .77 (.64–.93) | .005 | |
| Stage 2 | .50 | .83 (.78–.89) | .80 (.72–.90) | .70 (.61–.80) | 6.3×10−8 | |||
| Stage 3 | .47 | .86 (.83–.88) | .84 (.80–.88) | .74 (.69–.79) | 6.0×10−21 | 5.9×10−29 | ||
Acknowledgments
These are detailed in the Supplementary Note
Footnotes
Competing Financial Interests
None
Author contribution
RAE and DFE designed the study, wrote the paper and are joint PIs on the GWAS. RAE is PI of the UKGPCS and project managed this study. ZKJ coordinated the PRACTICAL genotyping, MG coordinated the administration.
ZKJ, SME, DAL, MT, EJS, CSC coordinated sample collation, provided molecular advice and performed molecular work on UK samples.
AAAO and DE performed the analyses; JM collated the dataset.
GGG, JH and DRE are PIs of the Australian studies; MCS led the Australian genotyping GS coordinated the Australian studies.
KM and RE are joint PIs of the PCRF study; AL and JFL coordinated sample collection BEH and CH lead the MEC consortium.
JS is PI of the Tampere study; TW and TLT coordinated sample collation, provided molecular advice and performed molecular work.
FCH, DN and JD are joint PIs of ProtecT; SJL helped with control matching for the UK.
JLS is PI of the Fred Hutchinson study and EAO is PI of the NHGRI genotyping for PROGRESS; LMF and JSK coordinated data collation.
SAI is PI of the USC study, EMJ is PI of the NC_CCPC study, SNT and DS are PIs of the Mayo clinic study; SKD coordinated data collation.
JYP is PI of the Moffit study, JC is PI of the Queensland study; AS led the Queensland genotyping, JLD is PI of the Tasprac study, WV is PI of the Ulm study; CM led the genotyping. TD is PI of the Hannover study, TRR is PI of the UPenn study, KAC is PI of the FMHS study, LCS is PI of the Utah study, POC and PH are PIs of the Valais study; WDF led the molecular work. MZ is PI of the BiPAS study, RK is PI of the PCMUS study GC is PI of the CHSH; HWZ led the genotyping and Y-JL provided study set up advice to the CHSH. ATAJ, ALH, LTO’B, RAW, ECP, EJS, DPD, AH, RAH, VSK, CCP, NVA, CJW, AT, TC, CO, LNK, LLM, AA, AC, DMK, EMK, MCS, RC, ADJ, AS, TAS,, JPS, SC, JA, RAG, JB, MAK, FL, AP, BP, JS, AM, ML, KL, AMR, EML, JF, HK, CS, and AM identified and collected clinical material/processed samples/undertook genotyping/collated data. Other members of The UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, The UK ProtecT Study Collaborators, and The PRACTICAL Consortium(membership lists provided in the Supplementary Note) collected clinical samples and provided data management.
Reference List
- 1.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 2.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 6.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J, et al. Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat Genet. 2008;40:1032–1034. doi: 10.1038/ng0908-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eeles R, Giles G, Neal D, Muir K, Easton DF. Reply to “Variation in KLK genes, prostate-specific antigen and risk of prostate cancer”. Nat Genet. 2008;40:1035–1036. doi: 10.1038/ng0908-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drieschner N, et al. A domain of the thyroid adenoma associated gene (THADA) conserved in vertebrates becomes destroyed by chromosomal rearrangements observed in thyroid adenomas. Gene. 2007;403:110–117. doi: 10.1016/j.gene.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Rippe V, et al. Identification of a gene rearranged by 2p21 aberrations in thyroid adenomas. Oncogene. 2003;22:6111–6114. doi: 10.1038/sj.onc.1206867. [DOI] [PubMed] [Google Scholar]
- 14.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King TE, et al. The role of alpha 6 integrin in prostate cancer migration and bone pain in a novel xenograft model. PLoS ONE. 2008;3:e3535. doi: 10.1371/journal.pone.0003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, et al. Cloning and identification of a novel human gene PDLIM5, a homolog of AD-associated neuronal thread protein (AD7c-NTP) DNA Seq. 2004;15:144–147. doi: 10.1080/10425170310001656756. [DOI] [PubMed] [Google Scholar]
- 17.Jankowska AM, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009 doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.bate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008;76:717–727. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei Q, et al. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 20.Lee MJ, Kim YS, Kummar S, Giaccone G, Trepel JB. Histone deacetylase inhibitors in cancer therapy. Curr Opin Oncol. 2008;20:639–649. doi: 10.1097/CCO.0b013e3283127095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Nandi AK, Li X, Bieberich CJ. NKX-3.1 interacts with prostate-derived Ets factor and regulates the activity of the PSA promoter. Cancer Res. 2002;62:338–340. [PubMed] [Google Scholar]
- 22.Fu VX, et al. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 2008;68:6797–6802. doi: 10.1158/0008-5472.CAN-08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camp NJ, Farnham JM, Cannon-Albright LA. Localization of a prostate cancer predisposition gene to an 880-kb region on chromosome 22q12.3 in Utah high-risk pedigrees. Cancer Res. 2006;66:10205–10212. doi: 10.1158/0008-5472.CAN-06-1233. [DOI] [PubMed] [Google Scholar]
- 25.Johanneson B, et al. Fine mapping of familial prostate cancer families narrows the interval for a susceptibility locus on chromosome 22q12.3 to 1.36 Mb. Hum Genet. 2008;123:65–75. doi: 10.1007/s00439-007-0451-y. [DOI] [PubMed] [Google Scholar]
- 26.Camp NJ, et al. Compelling evidence for a prostate cancer gene at 22q12.3 by the International Consortium for Prostate Cancer Genetics. Hum Mol Genet. 2007;16:1271–1278. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donovan J, et al. Prostate Testing for Cancer and Treatment (ProtecT) feasibility study. Health Technol Assess. 2003;7:1–88. doi: 10.3310/hta7140. [DOI] [PubMed] [Google Scholar]
- 28.Giles GG, et al. Smoking and prostate cancer: findings from an Australian case-control study. Ann Oncol. 2001;12:761–765. doi: 10.1023/a:1011131105617. [DOI] [PubMed] [Google Scholar]
- 29.Giles GG, et al. Androgenetic alopecia and prostate cancer: findings from an Australian case-control study. Cancer Epidemiol Biomarkers Prev. 2002;11:549–553. [PubMed] [Google Scholar]
- 30.Severi G, et al. ELAC2/HPC2 polymorphisms, prostate-specific antigen levels, and prostate cancer. J Natl Cancer Inst. 2003;95:818–824. doi: 10.1093/jnci/95.11.818. [DOI] [PubMed] [Google Scholar]
- 31.Kote-Jarai Z, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clayton D, Leung HT. An R package for analysis of whole-genome association studies. Hum Hered. 2007;64:45–51. doi: 10.1159/000101422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.