Abstract
Neuroimaging centers and pharmaceutical companies are working together to evaluate treatments that might slow the progression of Alzheimer’s disease (AD), a common but devastating late-life neuropathology. Recently, automated brain mapping methods, such as tensor-based morphometry (TBM) of structural MRI, have outperformed cognitive measures in their precision and power to track disease progression, greatly reducing sample size estimates for drug trials. In the largest TBM study to date, we studied how sample size estimates for tracking structural brain changes depend on the time interval between the scans (6–24 months). We analyzed 1309 brain scans from 91 probable AD patients (age at baseline: 75.4±7.5 years) and 189 individuals with mild cognitive impairment (MCI; 74.6±7.1 years), scanned at baseline, 6, 12, 18, and 24 months. Statistical maps revealed 3D patterns of brain atrophy at each follow-up scan relative to the baseline; numerical summaries were used to quantify temporal lobe atrophy within a statistically-defined region-of-interest. Power analyses revealed superior sample size estimates over traditional clinical measures. Only 80, 46, and 39 AD patients were required for a hypothetical clinical trial, at 6, 12, and 24 months respectively, to detect a 25% reduction in average change using a two-sided test (α=0.05, power=80%). Correspondingly, 106, 79, and 67 subjects were needed for an equivalent MCI trial aiming for earlier intervention. A 24-month trial provides most power, except when patient attrition exceeds 15–16%/year, in which case a 12-month trial is optimal. These statistics may facilitate clinical trial design using voxel-based brain mapping methods such as TBM.
Introduction
Alzheimer’s disease (AD) is a pathological condition associated with aging. It affects over 24 million individuals worldwide according to a Delphi consensus study in 2005 (Ferri et al., 2005), and the incidence doubles every 5 years after age 60 years (Brookmeyer et al., 1998; Jellinger, 2006; Jorm et al., 1987). AD has been described as the most feared illness in the elderly population (Harris and Interactive, 2006), even more so than cancer and cardiovascular diseases, mainly due to the lack of effective treatment or prevention methods. The optimal treatment window is in the non-symptomatic stage, during which misfolded proteins begin to aggregate into extracellular senile plaques and intracellular neurofibrillary tangles (Selkoe, 2004), followed by inflammatory damage and cell death in the central nervous system (Frank et al., 2003; Shaw et al., 2007). Mild cognitive impairment (MCI), especially the amnestic type (Petersen, 2003b; Petersen et al., 2001), is a transitional stage between normal aging and fully-developed AD. With an annual conversion rate of 10–25% to AD, MCI has attracted much attention for treatment trials targeting minimally symptomatic individuals (Grundman et al., 2004; Petersen, 2003a, 2007). Development of disease-modifying therapies is mainly hindered by the lack of robust biomarkers to aid early diagnosis and assess treatment efficacy (Shaw et al., 2007).
Longitudinal, structural magnetic resonance imaging (MRI) has substantial power to track subtle brain changes over time, and there has been great interest in which neuroimaging-derived measures, either used separately or jointly, offer greatest power to track AD. Several automated methods have been proposed to measure hippocampal atrophy (Morra et al., 2009a,b; Schuff et al., 2009), ventricular enlargement (Carmichael et al., 2006; Chou et al., 2008, 2009a,b; Nestor et al., 2008; Thompson et al., 2004), and whole brain atrophy assessed using methods such as the brain boundary shift integral (BBSI) (Fox et al., 2000) or SIENA (Ho et al., in press; Smith et al., 2002, 2004). These neuroimaging measures have several advantages as outcome measures as they can differentiate patients from controls (Davatzikos et al., 2008; Fox and Freeborough, 1997), correlate with clinical and cognitive decline (Fox et al., 1999; Hua et al., 2008b; Jack et al., 2009; Leow et al., 2009), correlate with pathologically confirmed neuronal loss (Vemuri et al., 2008; Whitwell et al., 2008), predict future conversion from preclinical to symptomatic AD (Apostolova et al., 2006; Hua et al., 2008b; Misra et al., 2009), and have a high test-retest reliability (Leow et al., 2006). Voxel-based morphometry studies, including the ones using TBM, have depicted the characteristic pattern of structural degeneration in AD, providing an automated and convenient platform for image-based disease characterization (Apostolova et al., 2007; Apostolova and Thompson, 2008; Busatto et al., 2008; Chetelat et al., 2005, 2008a,b; Davatzikos et al., 2001; Dickerson et al., 2009; Frisoni et al., 2007; Hua et al., 2008b; Ishii et al., 2005; Leow et al., 2009).
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a large-scale multi-site study to define optimal neuroimaging methods for clinical trials. In prior studies, we found that tensor-based morphometry gave better sample size estimates than standard clinical scores for tracking disease-related changes over a one-year interval (Hua et al., 2009a); sample sizes were comparable for subjects scanned 3 versus 1.5 Tesla (Ho et al., in press). As those analyses focused on people scanned with a 1-year follow-up interval, it is of great interest to know whether a shorter interval (6 months) or a longer one (24 months) would provide better power. In choosing a follow-up interval, clinical trial designers must consider the risk of waiting a long time for the disease to progress. This must be traded off against the more limited sensitivity to disease progression at shorter intervals, when biological changes have had less time to accumulate. To investigate these trade-offs, we analyzed the serial brain scans of 91 patients diagnosed with probable AD, followed up at 6, 12, and 24 months, and 189 MCI subjects followed up at 6, 12, 18, and 24 months. 3D maps of atrophy (i.e., volume loss - a widely-used neuroimaging measure of AD progression) were created, based on nonlinearly warping the follow-up scan to baseline scan. These maps charted disease progression in great spatial detail for each individual. To boost power, a numerical summary was derived from a statistically-defined region-of-interest (Stat-ROI (Hua et al., 2009a; Chen et al., 2009a; Ho et al., in press) inside the temporal lobes to quantify the rate of atrophy. Using the TBM-derived neuroimaging measure, we computed sample size estimates for hypothetical AD and MCI clinical trials. We compared sample size requirements for trials with different scan intervals of 6, 12, 18, and 24 months, and explored the influence of using atlas-based versus statistically pre-defined ROIs.
We hypothesized that, at all time intervals examined, TBM-derived neuroimaging measures would lead to greatly reduced sample size estimates for clinical trials compared to traditional cognitive measures. We also expected that longitudinal studies with longer follow-up periods would require smaller sample sizes to detect treatment effects. Finally, we assessed whether the use of a statistically pre-defined ROI boosted power at each follow-up interval; in exploratory analyses, we determined whether the power estimates depended on how the statistically pre-defined ROI was generated (i.e., which diagnostic group and which time-points were used to generate it).
Materials and methods
Subjects
Longitudinal brain MRI scans and associated clinical data were downloaded from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) public database (http://www.loni.ucla.edu/ADNI/Data/). ADNI is a large five-year study launched in 2004 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million public-private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments acquired at multiple sites (as in a typical clinical trial), can replicate results from smaller single site studies measuring the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to monitor the effectiveness of new treatments, and lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California, San Francisco.
Serial brain MRI scans were analyzed from 91 probable AD patients (age at baseline: 75.4±7.5 years) scanned at baseline and followed up at 6, 12, and 24 months, and 189 individuals with amnestic MCI (74.6±7.1) followed up at 6, 12, 18, and 24 months, amounting to a total of 1309 scans. These subjects are from the same cohort as those in our prior TBM studies, which examined a subset of 676 of these individuals at baseline, and 515 of them at 12-month follow-up, but not at other time-points (Hua et al., 2008a,b, 2009a). All subjects underwent thorough clinical and cognitive assessment at the time of scan acquisition. Cognitive tests examined here included the Alzheimer’s Disease Assessment Scale-cognitive subscale (ADAS-Cog), a 70-point scale designed to measure the severity of cognitive impairment, which is currently the most widely used cognitive measure in AD trials (Rosen et al., 1984). It consists of 11 tasks assessing learning and memory, language production and comprehension, constructional and ideational praxis, and orientation. The sum-of-boxes Clinical Dementia Rating (CDR-SB), ranging from 0 to 18, measures dementia severity by evaluating patients’ performance in six domains: memory, orientation, judgment and problem solving, community affairs, home and hobbies, and personal care (Berg, 1988; Hughes et al., 1982; Morris, 1993). The Mini-Mental State Examination (MMSE) provides a global measure of mental status, evaluating five cognitive domains: orientation, registration, attention and calculation, recall, and language (Cockrell and Folstein, 1988; Folstein et al., 1975). The maximum MMSE score is 30; scores of 24 or lower are generally consistent with dementia. For both ADAS-Cog and CDR-SB, higher scores indicate poorer cognitive function, whereas for MMSE, higher scores denote better cognitive function. All AD patients met NINCDS/ADRDA criteria for probable AD (McKhann et al., 1984). The ADNI protocol lists more detailed inclusion and exclusion criteria (Mueller et al.), 2005a,b which can also be found online at http://www.alzheimers.org/clinicaltrials/fullrec.asp?PrimaryKey=208.
These data sets were downloaded on or before June 1, 2009, and reflect the status of the database at that point; as data collection is ongoing, we focused on analyzing all available baseline and follow-up scans up to 2 years, together with associated clinical and cognitive scores. The study was conducted according to the Good Clinical Practice guidelines, the Declaration of Helsinki and U.S. 21 CFR Part 50-Protection of Human Subjects, and Part 56-Institutional Review Boards. Written informed consent was obtained from all participants before experimental procedures, including cognitive tests, were performed.
MRI acquisition and image correction
All subjects were scanned with a standardized MRI protocol developed for ADNI (Jack et al., 2008). Briefly, high-resolution structural brain MRI scans were acquired at 59 ADNI sites using 1.5 Tesla MRI scanners (GE Healthcare, Philips Medical Systems, or Siemens). Using a sagittal 3D MP-RAGE scanning protocol, the typical 1.5T acquisition parameters were repetition time (TR) of 2400 ms, minimum full TE, inversion time (TI) of 1000 ms, flip angle of 8°, 24 cm field of view, 192 × 192 × 166 acquisition matrix in the x-, y-, and z- dimensions, yielding a voxel size of 1.25 × 1.25 × 1.2 mm3, later reconstructed to 1 mm isotropic voxels. The scan quality was evaluated by the ADNI MRI quality control center at the Mayo Clinic following standardized criteria.
Image corrections were applied using a processing pipeline at the Mayo Clinic, consisting of: (1) correction of geometric distortion due to gradient non-linearity (Jovicich et al., 2006), i.e., “gradwarp” (2) “B1-correction” for adjustment of image intensity inhomogeneity due to B1 non-uniformity (Jack et al., 2008), (3) “N3” bias field correction for reducing residual intensity inhomogeneity (Sled et al., 1998), and (4) geometrical scaling for removing scanner- and potential session-specific calibration errors using a phantom scan acquired for each subject (Gunter et al., 2009). All original image files as well as images with all of these corrections are available to the general scientific community at http://www.loni.ucla.edu/ADNI/Data/.
Image pre-processing
To adjust for linear drifts in position and scale within the same subject, the follow-up scan (6-, 12-, 18-, or 24-month) was linearly registered to its matching baseline scan using a 9-parameter (9P) registration, driven by a mutual information (MI) cost function (Collins et al., 1994). 9P linear registration was chosen over 6P rigid-body registration as previous studies have shown that 9P registration can correct for scanner voxel size variations in large longitudinal studies involving multiple sites, scanners and acquisition sequences, and consistently outperforms 6P registration (Hua et al., 2009a; Paling et al., 2004). Using the ADNI data, 9P linear registration was shown to achieve similar level of scaling correction to phantom-based image correction (Clarkson et al., 2009), and to correct for any remaining scanner voxel size variation that was not accounted for by image correction algorithms (Hua et al., 2009a). To account for global differences in brain scale across subjects, the mutually aligned time-series of scans was then linearly registered to the International Consortium for Brain Mapping template (ICBM-53) (Mazziotta et al., 2001), applying the same 9P transformation to both mutually aligned scans. Globally aligned images were re-sampled in an isotropic space of 220 voxels along x-, y- and z-dimensions with a final voxel size of 1 mm3.
Tensor-based morphometry (TBM) and 3D Jacobian maps
Individual Jacobian maps illustrating local expansion or compression were also created to estimate 3D patterns of structural brain change over time, by warping the follow-up scan to match the baseline scan. The nonlinear registration algorithm was driven by a mutual information cost function, and as a regularizing term, we used the symmetrized Kullback-Leibler (sKL-MI) distance, with the registration parameters of sigma=6 and lambda=8, chosen as the best parameter set from an earlier optimization study (Hua et al., 2009a). The parameters sigma and lambda control the Jacobian field smoothness and weighting of the regularization, respectively (Yanovsky et al., 2008, 2009) (see Hua et al., 2009a, for a detailed discussion of the sKL-MI registration parameters and their implications for detecting change). Color-coded maps of the Jacobian determinants were created to illustrate regions of volume expansion (i.e., with detJ (r)>1), or contraction (i.e., with detJ (r)>1) (Ashburner and Friston, 2003; Chung et al., 2001; Freeborough and Fox, 1998; Riddle et al., 2004; Thompson et al., 2000; Toga, 1999) over time. These maps of tissue change were also spatially normalized across subjects by nonlinearly aligning all individual Jacobian maps to a minimal deformation template (MDT), for regional comparisons and group statistical analyses. The MDT was constructed based on images from 40 normal controls as detailed elsewhere (Hua et al., 2008a,b).
Group averages and statistical comparisons
To illustrate the average degree of atrophy, relative to the baseline image, at each follow-up time in AD and MCI groups, we constructed voxel-wise mean maps by taking the average at each voxel of the Jacobian maps, across subjects. These maps were color-coded to show the percentage of regional brain tissue loss and ventricular enlargement. The Jacobian maps of AD and MCI groups were compared at each time point using permutation-based two sample t tests, to assess the overall significance of group differences inside the whole brain as well as inside the temporal lobes, corrected for multiple comparisons (Bullmore et al., 1999; Chiang et al., 2007; Hua et al., 2009b; Nichols and Holmes, 2002; Thompson et al., 2003). In brief, a null distribution for the group differences in Jacobian determinant (atrophic rate) at each voxel was constructed using 10,000 random permutations of the data. For each test, the subjects’ diagnosis (AD vs. MCI) was randomly permuted and voxel-wise t tests were conducted to identify voxels more significant than p=0.05. The volume of voxels inside a mask (whole brain or temporal lobes) more significant than p=0.05 was computed for the real experiment and for the random assignments. Finally, a ratio, describing the fraction of the time the suprathreshold volume was greater in the randomized maps than the real effect (the original labeling), was calculated to give an overall p-value for the significance of the map (corrected for multiple comparisons by permutation). The correction is for the number of tests, so it quantifies the level of surprise in seeing the overall map within the mask or search region.
Regions of interest (ROIs) and a numerical summary for temporal lobe atrophy
To give a summary of the 3D map of brain atrophy for each subject, a single numerical measure was derived by computing an average within an ROI. Both anatomically and statistically-defined ROIs were used in this study. First, a temporal lobe ROI (Temp-ROI), including the temporal lobes of both brain hemispheres, was manually delineated on the MDT template by a trained anatomist using the Brainsuite software program (Shattuck and Leahy, 2002) (Fig. 2). Secondly, a statistically-defined ROI (Stat-ROI) was defined based on voxels with significant atrophic rates (p<0.001) within the temporal lobes, in a non-overlapping training set of 20 AD patients (age at baseline: 74.8±6.3 years; 7 men and 13 women) scanned at baseline and 12 months (Fig. 2). The method is detailed fully in two prior reports, Hua et al. (2009a) and Ho et al. (in press). The training set used to define the ROI was deliberately based on scans that were independent of (and not overlapping with) the testing set consisting of 91 AD and 189 MCI subjects, in order to maximize the available testing set (from which maps and sample sizes were computed) at all the time points. Next, we aimed to answer the following question: does a “customized” Stat-ROI—specifically made for each diagnostic group and time point—further improve the statistical power? We created separate Stat-ROI based on 20 AD patients (age at baseline: 75.4±7.5 years; 12 men and 8 women) scanned at baseline and followed up at 6, 12, and 24 months. And, we made a corresponding set of separate Stat-ROIs based on 20 MCI subjects (75.4±7.4 years; 12 men and 8 women) scanned at baseline and followed up at 6, 12, 18, and 24 months, selected from the common set consisting of 91 AD and 189 MCI subjects across all time points. To ensure that the training and testing sets did not overlap, all subjects chosen for the training set were subsequently removed from the testing set. For example, when 20 AD patients were selected to compute the customized Stat-ROI at 6-month, the testing set at 6-months for AD was reduced to 71 subjects (91–20=71). This was done to comply with the rule in machine learning that evaluations should be made on data sets independent of those used to select the regions of interest.
Fig. 2.
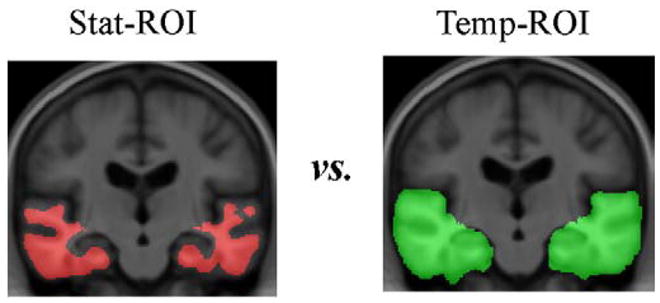
Statistically defined regions of interest (Stat-ROI) and temporal lobe regions of interest (Temp-ROI) are shown. The Stat-ROI (left, shown in red) was defined based on voxels that had showed significant atrophic rates over time (p < 0.001) within the temporal lobes, in a non-overlapping training set of 20 AD patients (age at baseline: 74.8±6.3 years; 7 men and 13 women) scanned at baseline and 12-month, chosen outside the independent evaluation set. The Temp-ROI (right, shown in green), including the temporal lobes of both brain hemispheres, was manually delineated on the MDT template by a trained anatomist using the Brainsuite software program (Shattuck and Leahy, 2002). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To allow a simple statistical analysis, a numeric summary—the mean atrophy rate for all voxels within an ROI—was also computed for each follow-up scan, to summarize the overall amount of temporal lobe atrophy detected during an observation time of 6, 12, 18, and 24 months, respectively.
Sample size calculations and power analysis
A power analysis was defined by the ADNI Biostatistics Core to estimate the sample size required to detect a 25% reduction in the mean annual rate of atrophy, using a two-sided test and standard significance level (α=0.05) for a hypothetical two-arm study (treatment versus placebo). The estimated minimum sample size for each arm is computed from the formula below. Briefly, β̂ denotes the estimated annual change (average of the group) and σD refers to the standard deviation of the rate of atrophy across subjects.
Here zα is the value of the standard normal distribution for which P[Z<zα]=α and in this case we set α to its conventional value of 0.05 (Rosner, 1990). The sample sizes required to achieve 80% and 90% power were computed in this study, subsequently referred to as n80 and n90. As the observation time ranged from 6 to 24 months, instead of converting the overall amount of change to an annual rate of atrophy, we computed the number of subjects required to detect a 25% reduction in the overall atrophy occurring over the interval (which gives the same result). These sample size estimates define how many patients would need to be recruited for clinical trials with the duration of 6, 12, 18, and 24 months respectively. We used the individual atrophy measures computed within the Stat-ROI or Temp-ROI for sample size calculations.
The 95% confidence interval for the n80 estimate was computed from 10,000 bootstrapped samples. At each bootstrapped resample, a subset of the numerical summaries (n=91 for AD and n=189 for MCI) was randomly drawn from the overall pool of numerical summaries within each group (AD or MCI). This was performed separately for the data at each follow-up interval. The confidence intervals were estimated with a bias corrected and accelerated percentile method (Davison and Hinkley, 1997; Efron and Tibshirani, 1993).
Exploratory analysis
As an exploratory analysis, we further computed the n80s derived from a Stat-ROI within the anatomical regions of the frontal, parietal, occipital lobes, CSF, whole brain, hippocampus, cerebral white matter, and cerebral gray matter (see Table 2). The hippocampus was delineated on the MDT template by investigators at University College, London using MIDAS (Medical Image Display and Analysis System) software (Freeborough et al., 1997). This delineation included the hippocampus proper, dentate gyrus, subiculum, and alveus (Fox et al., 1996; Scahill et al., 2003). The cerebral gray and white matter was classified using the partial volume classifier (PVC) in the Brainsuite software package (Shattuck and Leahy, 2002), after removing the brain stem and cerebellum from the whole-brain mask.
Table 2.
The n80s and 95% bootstrap confidence intervals (c) derived from the Stat-ROIs within the anatomical regions of the temporal, frontal, parietal, occipital lobes, CSF, whole brain, cerebral white matter and cerebral gray matter, performed here as an exploratory analysis.
| 6 months |
12 months |
18 months |
24 months |
||||||
|---|---|---|---|---|---|---|---|---|---|
| n80 | c | n80 | c | n80 | c | n80 | c | ||
| AD | Temporal | 80 | [55,120] | 46 | [36,65] | N/A | N/A | 39 | [30,53] |
| Frontal | 100 | [69,160] | 82 | [60,117] | 70 | [48,114] | |||
| Parietal | 102 | [72,154] | 66 | [49,95] | 63 | [47,85] | |||
| Occipital | 109 | [77,161] | 53 | [40,75] | 56 | [44,74] | |||
| CSF | 90 | [66,138] | 59 | [44,85] | 40 | [33,53] | |||
| Whole brain | 88 | [61,133] | 58 | [44,82] | 47 | [36,67] | |||
| Hippocampus | 114 | [78,189] | 68 | [49,100] | 84 | [58,140] | |||
| White matter | 88 | [63,131] | 67 | [51,95] | 59 | [43,84] | |||
| Gray matter | 100 | [71,167] | 45 | [33,62] | 38 | [29,52] | |||
| MCI | Temporal | 106 | [81,148] | 79 | [64,105] | 81 | [66,106] | 67 | [55,86] |
| Frontal | 151 | [111,226] | 130 | [98,183] | 127 | [99,173] | 107 | [82,149] | |
| Parietal | 135 | [100,196] | 121 | [96,161] | 132 | [102,185] | 115 | [91,155] | |
| Occipital | 123 | [95,176] | 94 | [75,120] | 87 | [70,116] | 84 | [69,108] | |
| CSF | 146 | [113,209] | 85 | [66,115] | 93 | [75,124] | 78 | [64,102] | |
| Whole brain | 112 | [85,159] | 92 | [73,123] | 93 | [74,124] | 80 | [63,103] | |
| Hippocampus | 143 | [108,202] | 125 | [97,172] | 117 | [92,160] | 103 | [82,137] | |
| White matter | 135 | [99,206] | 117 | [89,164] | 125 | [96,176] | 129 | [81,140] | |
| Gray matter | 115 | [89,155] | 84 | [66,108] | 73 | [69,94] | 66 | [53,82] | |
Two-sample t tests and correction for multiple comparisons
To determine if the sample size estimate was affected by the choice of ROI (Temp-ROI versus Stat-ROI, and various Stat-ROIs based on specific time point and diagnostic group), we conducted serial pairwise Student t tests. Because we were testing a large number of methodological alternatives to compare the effects of varying follow-up periods and Stat-ROIs (29 two-sample t tests: including 10 tests to compare rate of atrophy at different follow-up periods with Stat- and Temp-ROI, respectively, 7 tests to compare the Stat- versus Temp-ROI, and 12 tests to compare the effects of customized ROI based on diagnostic group and time point), we took the necessary precautions to avoid inflating the false positive rate due to multiple comparisons. We used Bonferroni correction to adjust each p-value, i.e., p=b·n, where b stands for the uncorrected p-value from each t test and n is the total number of tests (n=29). All the p-values Results in the section are corrected p-values after this Bonferroni correction.
Sample size estimates accounting for attrition
In reality, participants have the right to drop out of a clinical trial or research study at any time, or they may be unable to return due deteriorating health, disability, or death, or because they have moved out of the area or have lost interest in the study. Attrition rate for ADNI is around 7% per year, but in other studies with repeated assessments, attrition rates may be as high as 50% per additional visit (Thompson et al., 2009). Because of this, it is reasonable to build in estimates of the attrition rate in sample size estimation because larger samples may need to be recruited at the outset to ensure that an adequate sample remains later. In a simplified model, assuming that the participants who drop out are a random sub-sample, we adjusted the sample size estimates for attrition, by using the formula: Adjusted n80=(n80 from power estimate)/[(1 − attrition rate)T]. Here T denotes the inter-scan interval or trial interval in years, e.g., 1.5 for an 18-month interval. Although the assumption of random drop-out might not hold up in real clinical trials, it allows approximate adjustments to be made for attrition.
Results
Average atrophy maps visualizing patterns of disease progression measured at 6, 12, 18, and 24 months
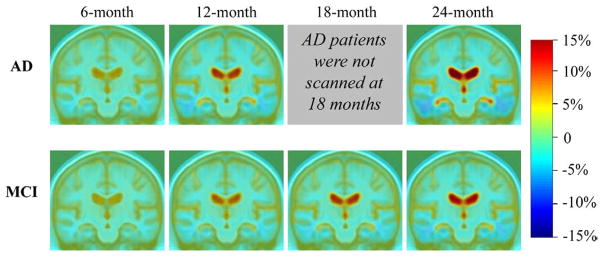
Individual Jacobian maps were averaged in AD and MCI groups to demonstrate the average amount of brain tissue loss (in blue colors) and ventricular enlargement (in red colors), characteristic of disease progression in AD, at a follow-up period of 6, 12, 18, and 24 months (Fig. 1). These tissue changes are shown as percentages, relative to the baseline scan, and are computed within each individual, before averaging across subjects in the group. Small but visible changes are noted at 6 months, the shortest follow-up period, in AD and MCI. As expected, greater progressive change, evidenced by ventricular expansion and temporal lobe atrophy, was detected at longer observation times. AD patients had progressed faster than MCI at all time points, as expected. The corrected p-values for comparing AD and MCI groups within a search region encompassing the entire brain were p=0.07, p=0.002, and p=0.004 at 6-, 12- and 24-month respectively; and the corrected p-values within the temporal lobes were p=0.02, p<0.0001, and p<0.0001 at 6, 12 and 24 months, respectively. This is in line with the intuition that AD versus MCI differences in atrophic rates are hardest to detect at the shortest follow-up interval (6 months), but can be detected so long as a temporal lobe region of interest is used.
Fig. 1.
Mean level of progressive atrophy in AD and MCI groups after intervals of 6, 12, 18, and 24 months, respectively. Per the study design, AD patients were not scanned at the 18-month follow-up interval. Clearly, the profile of atrophy, and group differences, are easier to distinguish at later time points. Also, as expected, and as previously reported for 1-year follow-up only (Hua et al., 2009a; Ho et al., in press; Leow et al., 2009), the MCI mean atrophic rate is lower than that seen in AD, at every time-point.
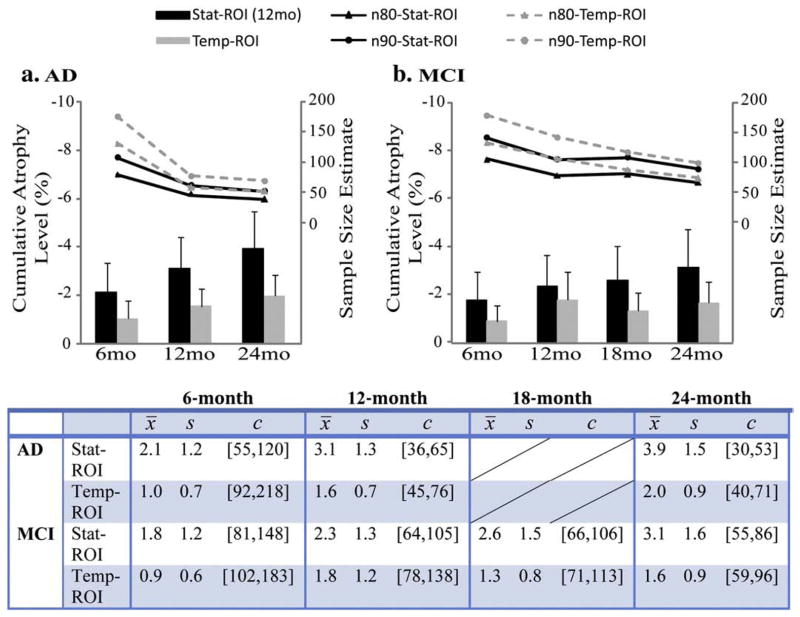
Sample size estimates using numerical summaries derived from the Stat-ROI versus Temp-ROI
As detailed in the Materials and methods section, the Temp-ROI was defined based on the average anatomy of the MDT (Fig. 2). The Stat-ROI was defined based on a non-overlapping training set of AD patients followed up for a 12-month period (Fig. 2). Since the training set was chosen to be independent of the evaluation set, sample size estimates (n80 and n90) were computed from the full sample of 91 AD patients and 189 MCI subjects. The same Stat-ROI was applied to compute all sample size estimates in Fig. 3. Across the same set of patients in AD, longer intervals led to greater effect sizes for the measurement of temporal lobe atrophy, resulting in smaller sample size estimates (n80 and n90 in Fig. 3a). MCI subjects showed the same trend as AD, with greater cumulative atrophy at longer follow-up intervals. The MCI subjects were examined at an additional time point (18 months), but this time-point, when used on its own, offered little extra benefit relative to a 12-month follow-up, showing comparable n80 and n90 at 12 and 18 months. Sample size estimates based on numeric summaries derived from the Stat-ROI consistently out-performed the ones derived from the Temp-ROI (Fig. 3) (all pair-wise comparisons, of the Stat-ROI versus the Temp-ROI, had corrected p-values less than 0.05). Additionally, all TBM-derived neuroimaging markers demonstrated drastic sample size reductions relative to standard clinical measures (ADAS-Cog, MMSE, and CDR-SB) at all follow-up periods (Table 1).
Fig. 3.
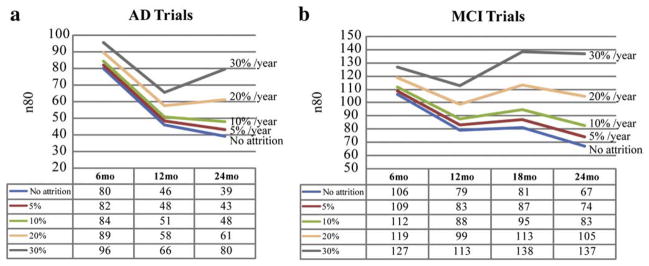
Average level of cumulative temporal lobe atrophy (as a % of the baseline tissue volume) and sample size estimates required to detect a 25% slowing of degeneration with 80% (n80) and 90% (n90) power, with an observation time of 6, 12, and 24 months for AD (a) and 6, 12, 18, and 24 months for MCI (b). The table shows numerical values of the cumulative atrophy, in percent, as a mean loss (x̄, the sign is omitted here) and standard deviation (s). The 95% confidence interval (c) for the n80 sample size measure was estimated from 10,000 bootstrapped samples. At 6 months, the required sample sizes are much higher than those that would be needed at later time points. Sample size estimates are generally better (i.e., lower) for numeric summaries derived from a statistically pre-defined region of interest (solid black lines) versus an atlas-defined region of interest that covers the entire temporal lobes (dashed gray lines). Also, the degree to which a 6-month interval inflates the sample size estimate is not so extreme for the statistical region of interest, as it focuses on regions where changes are likely to be occurring at this very short follow-up interval.
Table 1.
Sample size estimates for TBM-derived measures of atrophy are compared with traditionally used clinical measures.
| Stat-ROI | Temp-ROI | ADAS-Cog | MMSE | CDR | |||
|---|---|---|---|---|---|---|---|
| AD | 6 months | n80 | 80 | 131 | 1,371 | 2,224 | 1,709 |
| n90 | 107 | 176 | 1,834 | 2,975 | 2,285 | ||
| 12 months | n80 | 46 | 58 | 483 | 809 | 577 | |
| n90 | 62 | 77 | 647 | 1,082 | 771 | ||
| 24 months | n80 | 39 | 52 | 215 | 365 | 215 | |
| n90 | 52 | 69 | 287 | 488 | 288 | ||
| MCI | 6 months | n80 | 106 | 134 | 16,645 | 2,382 | 3,372 |
| n90 | 142 | 179 | 22,265 | 3,788 | 4,511 | ||
| 12 months | n80 | 79 | 106 | 8,212 | 2,767 | 836 | |
| n90 | 106 | 142 | 10,985 | 3,701 | 1,118 | ||
| 18 months | n80 | 81 | 89 | 1,381 | 1,217 | 791 | |
| n90 | 109 | 119 | 1,847 | 1,628 | 1059 | ||
| 24 months | n80 | 67 | 75 | 1,013 | 879 | 626 | |
| n90 | 90 | 100 | 1,355 | 1,176 | 838 |
Clinical scores were obtained at the time of each scan acquisition, so they have intervals that match those of the MRI scans. The sample size estimates for clinical measures were computed using the power analysis formula defined in the Materials and methods section. Smaller numbers are better—they mean that fewer subjects are needed to demonstrate an effect size of fixed magnitude (a 25% slowing).
The exploratory analysis (Table 2) suggested that a statistically defined ROI generated within the temporal lobes is the best search region in terms of yielding the lowest n80 s (smaller n80s are better), although several anatomical ROIs performed almost equally well. Summaries of atrophic rates derived within the cortical gray matter showed comparable statistical power to the ones derived from the temporal lobes. The performance of the statistical ROIs was somewhat insensitive to the choice of the search region, with the temporal region giving marginally better results, as would be predicted from the sequence of AD progression.
Choices of statistical ROI
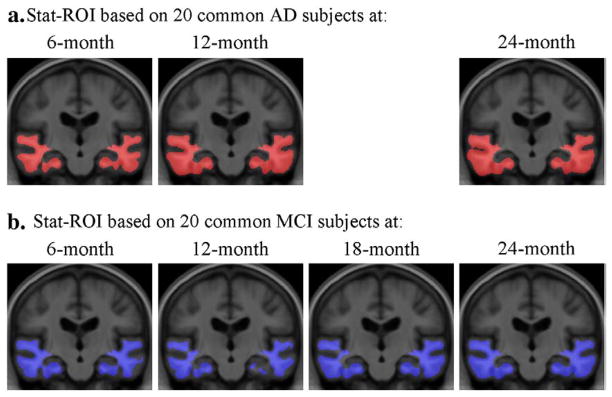
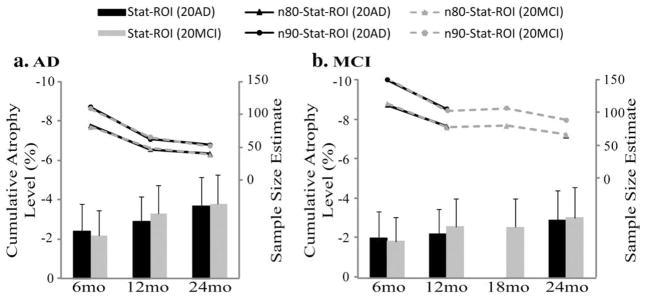
In theory, the statistically pre-defined region of interest could be based on any chosen set of AD or MCI subjects, even from a separate study, so long as they do not overlap with the testing (or evaluation) set. Apart from the Stat-ROI created from the non-common set (Figs. 2 and 3), we created 7 additional Stat-ROIs based on 20 AD patients and 20 MCI subjects chosen from the common set consisting of 91 AD patients and 189 MCI subjects, at each follow-up time. As shown in Fig. 4, the Stat-ROIs were anatomically very similar regardless of the diagnostic group and follow-up period that they were based on, indicating a consistent pattern of longitudinal change detected in both AD and MCI. Moreover, the choice of Stat-ROI did not significantly affect the sample size estimates in AD (Fig. 5a) and MCI (Fig. 5b) (all pair-wise comparisons had corrected p-values greater than 0.05).
Fig. 4.
Statistically pre-defined ROIs based on 20 AD patients (a) and 20 MCI subjects (b) chosen from the common set, at a follow-up period of 6, 12, 18, and 24 months. AD patients were not scanned at 18 months so no Stat-ROI was created at that time-point.
Fig. 5.
Average level of cumulative atrophy in the temporal lobes (as a % of baseline tissue volume) and sample size estimates required to detect a 25% slowing of degeneration with 80% (n80) and 90% (n90) power, with an observation time of 6, 12, and 24 months for AD (a) and 6, 12, 18, and 24 months for MCI (b), using various Stat-ROIs. The Stat-ROIs were either based on 20 AD patients (“Stat-ROI (20AD)”) or 20 MCI subjects (“Stat-ROI (20MCI)”) chosen from the common set, at each follow-up interval. In AD, sample size estimates using AD-specific ROIs (data points connected with solid black lines) were almost identical to the ones using an MCI-based ROI (data points connected with dashed gray lines). The same was true for the MCI group, as the gray (MCI-based ROI) and black (AD-based ROI) points mostly overlapped. In other words, it does not matter greatly which group the statistical ROI is generated from.
Sample size estimates accounting for attrition rate
In practice, the attrition rate would have to be considered in making sample size estimates. ADNI, as a research project, has a low attrition rate of 5–7%. Real clinical trials usually have a much higher attrition rate ranging from 20% to 30% per year. To illustrate this effect, we computed sample size estimates using TBM-derived neuroimaging measures with attrition rates built into the power analysis, assuming that the people who drop out are a random sub-sample of the overall sample (Figs. 6 and 7). With little to no attrition, longer trials are better in terms of sample size estimates. As the attrition rate goes up, the longer trials, i.e., 18- and 24-month, begin to fall behind the shorter trials, as the benefit of waiting longer for the disease to progress is outweighed by the number of people who fail to return for follow-up evaluation. The 12-month interval was the optimal trial duration (among those considered here) when the attrition rate was greater than 20% per year for both AD and MCI (Fig. 6). To determine the cut-off point, the adjusted n80s were plotted against the attrition rate (Fig. 7). We found that the 24-month trial gave the best sample size estimate so long as the attrition rate was below 15–16% per year. If the attrition rate rose further, the 12-month trial became the optimal choice, outperforming all other trial durations (Fig. 7). More complex models could be fitted if the attrition rate is itself variable over time, but prior studies have shown that a somewhat consistent proportion of subjects tends to drop out, with each successive follow-up (Thompson et al., 2008).
Fig. 6.
Sample size estimates after adjusting for attrition rate in AD (a) and MCI (b) trials. The sample size estimates with attrition rates of 0%, 5%, 10%, 20% and 30% per year are plotted and summarized in tables. When all patients remain in a trial, i.e., no attrition, longer trials give better power or lower sample size estimates compared to shorter trials. As the attrition rate goes up, shorter trials, particularly the 12-month trial, become the optimal trial duration.
Fig. 7.
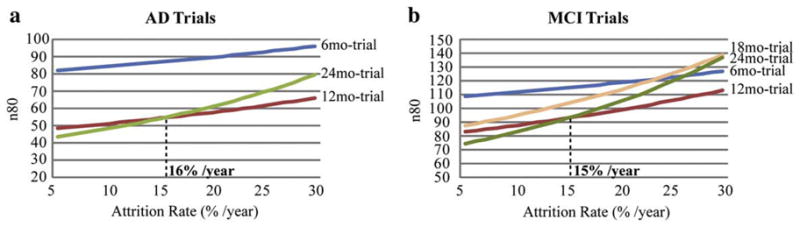
Finding the optimal trial duration by plotting the adjusted power estimates (i.e., minimal sample sizes; n80s) against attrition rate in AD (a) and MCI (trials). When the attrition rate is below 16% in AD trials and 15% in MCI trials, a 24-month trial is best; if the attrition rate falls below the cut-off points mentioned above, then a 12-month trial is best, as demonstrated by smaller sample size estimates.
Discussion
Serial structural brain changes in Alzheimer’s disease were visualized in patients with probable AD (N = 91) and MCI (N=189), scanned at baseline and after intervals of 6, 12, 18, and 24 months. 3D maps showed the regional distribution of cumulative atrophy across the brain, relative to the baseline scan, and the resulting patterns, at all time-points, are consistent with many past reports that pinpoint areas most affected by AD (Frisoni et al., 2009; Scahill and Fox, 2007; Scahill et al., 2003; Thompson and Apostolova, 2007; Whitwell et al., 2007). The most prominent features detected by longitudinal MRI and TBM were ventricular expansion and temporal lobe atrophy (Fig. 1), consistent with earlier findings.
To improve the utility of voxel-based neuroimaging for clinical trials, which tend to deal with “single-number” outcome measures, we also derived a numeric summary to quantify the overall amount of temporal lobe atrophy in regions consistently affected by AD. After reducing a 3D map into this single atrophy score, we evaluated its statistical power versus standard clinical measures for detecting disease progression in AD and MCI. Sample size estimates were far smaller for neuroimaging measures than for clinical scores, suggesting that fewer patients or shorter observation times would still offer sufficient power in clinical trials using neuroimaging surrogate markers (Chen et al., 2009a,b; Jack et al., 2003; Reiman et al., 2008b; Schuff et al., 2009).
Probable AD patients, followed up after 6, 12, and 24-month intervals, demonstrated successively greater cumulative brain atrophy, and progressively smaller sample sizes to detect a 25% reduction in the average change in a hypothetical clinical trial. MCI subjects also showed a gradual increase in the cumulative level of brain atrophy (versus baseline) in maps created at 6, 12, 18, and 24 month intervals. The 18-month follow-up for MCI, when used on its own, did not exhibit a detectably improved sample size estimate compared to a 12-month trial. Brain atrophy is detectable on MRI as early as 6 months in both AD and MCI groups. Even so, changes had greater effect sizes at longer intervals, illustrating the trade-off between recruitment requirements (and therefore costs) and the required observation time. More patients are needed for shorter trials, but patient enrollment may be reduced if a longer observation time is acceptable. And regardless of the scanning interval, TBM is clearly a useful neuroimaging marker that may help reduce costs for clinical trials.
Theoretical and practical considerations
Minimal sample sizes directly relate to effect sizes, which depend upon both the mean and standard deviation (SD) of the atrophy measures. For TBM measures derived from the statistically pre-defined region of interest, both the mean level of cumulative atrophy and its SD increased monotonically with longer inter-scan intervals (Fig. 3); as the mean rose faster than the variance, incrementally greater effect sizes (and smaller sample size estimates) were observed. According to one interpretation, most of the sources of measurement error in estimating the atrophic rate from serial MRI scans (e.g., scanner calibration, RF bias fields) may be roughly the same regardless of the scan interval, but the interval must always be long enough for sufficient systematic atrophy to accumulate and be detectable above the noise that is inevitable whenever measurements are made.
Even so, it is not logically necessary that long intervals must always give best effect sizes: they may not if the population variability increases drastically. Attrition effects are also relevant in practice. A long study may be useful to maximize the theoretical effect size, but only if patients are willing and able to stay in the trial to allow a longer follow-up. With attrition rates greater than 15–16% per year, shorter trials (e.g., 12-month) became a better choice (Fig. 7), probably because the added effect sizes from cumulative change over a longer period were outweighed by the exponentially growing attrition. Additionally, shorter trials may also be favored due to cost concerns, as the cost generally rises with longer follow-up periods. Even when considering the attrition rate, the 12-month trial is consistently better than the 6-month trial. Although a reasonable amount of change can be detected at 6 months (and differences between MCI and AD group rates of atrophy were detectable at 6 months), a 12-month trial might be the optimal trial duration in practice, assuming a typical attrition rate (20–30% per year) for clinical trials. Clearly, the optimal follow-up interval depends not only on the effect size, but also on the attrition rate, cost and other potentially unmodeled factors.
In one recent clinical trial, we used 3D cortical mapping methods to assess cortical thickness at 3-, 6-, and 12-month follow-up intervals in patients with first-episode schizophrenia, randomized to two different medications (Thompson et al., 2008). Intriguingly, medication effects—favoring olanzapine over haloperidol treatment—were significant as early as 3 and 6 months, but were no longer significant at 12 months. This may be because, in that case, medications are most effective soon after disease onset, and less so later, or it may be because, in schizophrenia, atrophic rates are greater soon after disease onset, and less so later (the opposite appears to be the case in AD and MCI, where atrophic rates are thought to accelerate over time as the disease progresses).
In another MRI study measuring atrophy in AD (Schott, 2005), 38 AD patients and 19 controls were scanned at 0, 6, and 12 months, and the boundary shift integral (BSI) method was used to measure whole brain volume loss over time. Consistent with our findings, smaller sample sizes were needed to detect the same effect size at longer intervals. In Schott et al.’s study, only 154 subjects per arm were needed to detect a 20% reduction in the rate of atrophy when measurements were made after 1 year, versus 410 for a 6-month interval. Schott et al. also noted that smaller samples were needed when measuring rates of ventricular enlargement versus whole brain atrophy, if the trial duration was 6 months, but not at 12 months. They found that the variance in the estimated rate of whole brain atrophy was almost twice as high at 6 versus 12 months, but the variance in the ventricular atrophy rate—which can be measured more consistently—was about the same at 6 versus 12 months.
We also found that the refining the search region can improve sample size estimates. Use of a statistically pre-defined ROI gave better power estimates at all time-points, compared to an anatomically defined ROI that included the entire temporal lobes (Fig. 3 and Table 1). The use of a statistically-defined ROI based on an independent training sample was first proposed for positron emission tomography (PET) images (Chen et al., 2009a,b; Reiman et al., 2008a). We recently applied the same idea to MRI image analysis, and found consistently higher power to detect disease-related brain atrophy (Hua et al., 2009a; Ho et al., in press). A Stat-ROI reduces the search regions to areas showing the most robust changes in AD or MCI, eliminating regions with greater variance or smaller effect sizes across subjects, and avoiding regions with changes in the opposite direction (e.g., ventricular enlargement or intra-sulcal CSF). The Stat-ROI is slightly smaller in size than an atlas-based definition of the temporal lobes (Fig. 2) and, relative to the full temporal lobe ROI, it largely removes partial volumed voxels (containing mixtures of different tissues) at gray matter/CSF interfaces. At the interface of brain tissue and CSF, voxels with progressive tissue loss (in the gray matter) are partial volumed with expanding voxels (in the ventricular space) (Hua et al., 2008a, 2009b), which can give high variance across subjects and can reduce statistical power. The Stat-ROI retains regions changing the most in AD, and boosts power substantially.
We set the temporal lobes as the default search region to compute the Stat-ROI because it is one of the earliest regions to become atrophic in AD (Braak and Braak, 1991; Braskie et al., 2008; Jack et al., 1997, 1998; Killiany et al., 1993; Thompson et al., 2003), showing most differentiation between patients and healthy elderly (Hua et al., 2008a). The exploratory analysis in multiple anatomical regions (Table 2) further confirmed that the temporal lobes were the best search region to use, when this TBM method is used to identify AD-associated brain degeneration. Several other regions performed similarly in terms of n80. The hippocampus did not show especially high power with TBM, perhaps because it is too small to be modeled accurately with TBM due to partial volume effects (Hua et al., 2009a). The Stat-ROI within the temporal lobe includes several regions with early pathological changes in AD such as the medial temporal lobe and entorhinal cortex (Fig. 2), but largely avoids the hippocampus due to the limitation of TBM in picking up changes with high effects sizes from small structures. In other work (Protas et al., 2009), we have combined data from multiple ROIs for diagnostic classification in AD and MCI, and a similar approach could be taken here. Even so, the temporal lobe statistical ROI almost always gave best results when used alone, so we report data from that region here for simplicity of implementation and interpretation.
Stat-ROIs based on different diagnostic groups (AD vs. MCI) and based on different time points were very similar. They covered similar regions within the temporal lobes (Fig. 4), and power analysis was insensitive to these different choices of Stat-ROIs (Fig. 5). This suggests that a Stat-ROI derived from one time point may be reasonably applied to data from other time-points, making it easy to implement in clinical trials or other longitudinal studies. As picking a training set at each time point or for each diagnostic group is unnecessary, the number of scans in the testing set is preserved to maximize the sample size for power analysis.
Based on the power formula described in the Materials and methods section, the estimated minimum sample size for each arm is computed assuming a 25% slowing of the atrophy rate, indicated by a multiplier of 0.25 in the denominator. In reality, treatments may slow atrophy to different degrees, which may be denoted by k%, for different k. The sample size estimates required to detect a k% slowing of atrophy can be easily derived by multiplying the sample size estimates (n80 or n90) in this paper by (25/k)2, as the numbers follow an inverse-square law. For example, 4 times as many subjects would be needed to detect a 12.5% slowing of atrophy (half of 25%), versus a 25% slowing of atrophy (Ho et al., in press). The quadratic relationship between the sample size estimates and the percentage atrophic rate is illustrated in Fig. 8, using the n80s for a 12-month trial derived from the Stat-ROI (Table 1), as an example. The results of this paper can be easily translated to studies aiming to detect a different level of treatment effect, and our findings remain unaffected as multiplying the variables by a constant (25/k)2 does not alter the ranking of the effect sizes in the statistical tests (it is a monotone transformation, i.e., it preserves the rank order).
Fig. 8.
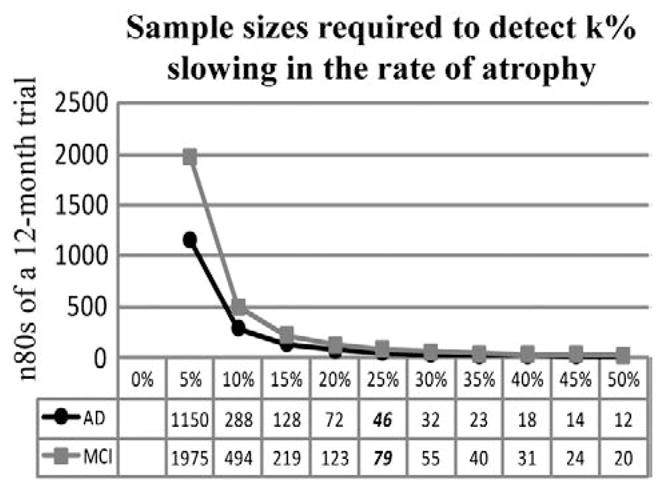
Sample sizes required to detect different degrees (k%) of slowing in the rate of atrophy show a quadratic dependency, for a hypothetical 12-month clinical trial using numerical summaries derived from a Stat-ROI. Our sample size estimates (n80AD =46; n80MCI =79 at 12 months) are based on assuming a 25% slowing of the rate of atrophy, whereas in reality, treatments may slow atrophy to different degrees. Even so, the sample size estimates required to detect a k% slowing of atrophy can be easily derived by multiplying the numbers in this paper by (25/k)2.
Localization of Changes with TBM
It may seem paradoxical that the brain changes with greatest effect sizes in TBM were generally found in large homogeneous regions of the white matter, when AD is widely accepted to be a predominantly hippocampal and cortical gray matter pathology. The predominant site of plaque and tangle accumulation in AD is the hippocampus and cortex, and the molecular hallmarks of AD spread through the cortex in a characteristic trajectory (Braak and Braak, 1991; Braskie et al., 2008). Volumetric atrophy is widespread in the cortical gray matter (GM). Our group, along with many others, has mapped the spatial-temporal trajectory of GM loss in AD, and it largely mirrors the Braak and Braak sequence of AD pathology (Dickerson et al., 2009; Thompson et al., 2003), and the cortical trajectory of plaque and tangle build-up tracked with [18F]FDDNP-PET (Braskie et al., 2008).
For some time, white matter (WM) changes in AD were relatively difficult to quantify in conventional MRI because of the lack of visible anatomical boundaries that would be required to parcellate white matter. Other than white matter hyperintensities, which are hallmark lesions of cerebral vascular disease (Brickman et al., 2009), the nature of WM degeneration in AD has not been well characterized until recently. Diffusion tensor imaging (DTI), relaxometry, and functional connectivity studies have now provided substantial evidence for diffuse WM abnormalities in AD (Buckner et al., 2009; Wozniak and Lim, 2006). Myelin breakdown and Wallerian degeneration both lead to WM atrophy, perhaps secondary to the effect of cortical neuronal loss in AD (Bartzokis, 2009; Bartzokis et al., 2006, 2007; Spires-Jones et al., 2009). Disease-related WM degeneration is detectable in the form of reduced fractional anisotropy on DTI (Huang et al., 2007; Naggara et al., 2006; Rose et al., 2000; Sandson et al., 1999; Wang et al., 2009), lowered relaxation rates in T2-based MRI relaxometry (Bartzokis et al., 2003, 2004), and disrupted connectivity observed using resting-state functional MRI (Agosta et al., 2009; Buckner et al., 2009; Hedden et al., 2009; Supekar et al., 2008; Wang et al., 2007; Zhou et al., 2008).
As both GM and WM changes are occurring, a key question is which of the MRI-derived measures is the most reliable for detecting group differences or dynamic changes over time, and which can resolve them with greatest effect sizes and accuracy in longitudinal studies. As a percentage, more cortical and hippocampal gray matter may be lost over time than white matter. Even so, the effect sizes for the changes in gray matter may be lower than expected, as these structures are convoluted and difficult to measure accurately. The cortex is thin and the hippocampus is narrow, often only a few voxels thick, and automated measures of the same structures using different algorithms can disagree substantially (Morra et al., 2009b), leading to calls for better harmonization of hippocampal measurement methods across studies (Frisoni et al., 2010).
This is especially the case in voxel-based maps, where changes may be greatest, as a percentage of their volume, in the cortex and hippocampus, but when pooling data across subjects voxel-by-voxel, the interiors of large white matter structures still tend to be better registered than the cortical and hippocampal boundaries once all the data are aligned. In the interiors of structures, such as the white matter, coherent patterns (such as atrophy) are more likely to be reinforced across all members of a group than at boundary voxels where loss patterns may be less well registered, even after nonlinear registration. This effect can be partially overcome using voxel-based morphometry (VBM; Ashburner and Friston, 2000) but that method multiplies gray matter “density” measures by registration-based estimates of changes, whereas TBM uses the registration field only. Cortical thickness measures also tend to have relatively poor reproducibility; in fact, different algorithms give mean cortical thickness values for normal subjects that differ by a factor of two (Aganj et al., 2009).
Relation to Prior Work
Our earlier TBM paper on the 12-month ADNI follow-up data (Hua et al., 2009) was mainly focused on optimizing the TBM processing pipeline for statistical power. We studied the effects on the results of critical parameters such as the linear registration steps, and the regularization parameters of the deformation model. Because so many parameter options and analysis choices could be made, we compared TBM designs with different linear and nonlinear registration parameters (including different regularizing functions) and found the set of best-performing parameters from the standpoint of maximizing effect sizes. A secondary focus of that prior paper was to motivate and evaluate the use of a data-driven statistical ROI to compute power estimates. We found that the statistical power of tracking brain degeneration could be further enhanced by using a statistically-defined ROI within the temporal lobes, based on voxels found to change the most in an independent sample. That study served as a foundation for the current study, in which we used the best TBM design and parameters from the prior study.
The current study asks a different question about the optimal length of a clinical trial, with the intent of understanding factors that might influence power in a clinical trial. Expanding the temporal sampling significantly relative to the prior study, we analyzed brain scans collected at baseline, 6, 12, 18, and 24 months, for both AD patients and MCI subjects. Unlike our prior report, the current study used scans at 5 time-points, and from many more subjects, so we were able to gauge the tradeoff between trial duration and recruitment requirements, both with and without considering likely attrition rates. The resulting information offers an evaluation of a simple and readily implementable image analysis method as well as a guideline for trials involving a neuroimaging component.
Limitations and caveats
The current study has some limitations, and some qualifications are needed. As noted before, a 25% reduction in the atrophic rate may have a different functional significance for a patient than a 25% reduction in the rate of decline for clinical or cognitive test scores (and may in reality be either better or worse for the patient). As such, a head-to-head comparison of clinical and neuroimaging measures is useful, but in reality both measures are informative and will continue to be widely used. Secondly, longitudinal studies with more than two time-points can employ more advanced statistical designs that use all the data at once, such as random effects or mixed effects models, to estimate intra-subject variance and take advantage of the repeated measures (Fitzmaurice et al., 2004; Frost et al., 2004). In recent ADNI analyses, using all scans in the time-series has been shown to boost power estimates (Schuff et al., 2009). Some advocate the use of two scans taken on the same day from each subject, showing a net beneficial effect on sample size estimates, especially when the follow-up interval is short (Schott et al., 2006). Finally, it is reasonable that multiple complementary neuroimaging measures, from MRI, PET and other modalities (arterial spin labeling, diffusion imaging, and resting-state fMRI) may be combined in the future, with genetic and CSF biomarker data, to give more robust metrics of disease progression, and to obtain better predictive value in longitudinal models.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., and Wyeth, as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation. Algorithm development and image analysis for this study was funded by grants to P.T. from the NIBIB (R01 EB007813, R01 EB008281, R01 EB008432), NICHD (R01 HD050735), and NIA (R01 AG020098), and National Institutes of Health through the NIH Roadmap for Medical Research, Grants U54-RR021813 (CCB) (to AWT and PT).
Footnotes
Author contributions were as follows: XH, SL, DH, IY, AL, AT, and PT performed the image analyses; CJ, MB, ER, DH, JK, NS, GA, and MW contributed substantially to the image and data acquisition, study design, quality control, calibration and pre-processing, databasing and image analysis. Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. ADNI investigators include (complete listing available at: http://www.loni.ucla.edu/ADNI/Collaboration/ADNI_Manuscript_Citations.pdf). We thank Anders Dale for his contributions to the image pre-processing and the ADNI project.
References
- Aganj I, Sapiro G, Parikshak N, Madsen SK, Thompson PM. Measurement of cortical thickness from MRI by minimum line integrals on soft-classified tissue. Hum Brain Mapp. 2009;30:3188–3199. doi: 10.1002/hbm.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F, Rocca MA, Pagani E, Absinta M, Magnani G, Marcone A, Falautano M, Comi G, Gorno-Tempini ML, Filippi M. Sensorimotor network rewiring in mild cognitive impairment and Alzheimer’s disease. Hum Brain Mapp. 2009 doi: 10.1002/hbm.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM. Mapping progressive brain structural changes in early Alzheimer’s disease and mild cognitive impairment. Neuropsychologia. 2008;46:1597–1612. doi: 10.1016/j.neuropsychologia.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006;63:693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Steiner CA, Akopyan GG, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Three-dimensional gray matter atrophy mapping in mild cognitive impairment and mild Alzheimer disease. Arch Neurol. 2007;64:1489–1495. doi: 10.1001/archneur.64.10.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Human Brain Function. Academic Press; 2003. Morphometry. [Google Scholar]
- Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G, Cummings JL, Sultzer D, Henderson VW, Nuechterlein KH, Mintz J. White matter structural integrity in healthy aging adults and patients with Alzheimer disease: a magnetic resonance imaging study. Arch Neurol. 2003;60:393–398. doi: 10.1001/archneur.60.3.393. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Sultzer D, Lu PH, Nuechterlein KH, Mintz J, Cummings JL. Heterogeneous age-related breakdown of white matter structural integrity: implications for cortical “disconnection” in aging and Alzheimer’s disease. Neurobiol Aging. 2004;25:843–851. doi: 10.1016/j.neurobiolaging.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Geschwind DH, Edwards N, Mintz J, Cummings JL. Apolipoprotein E genotype and age-related myelin breakdown in healthy individuals: implications for cognitive decline and dementia. Arch Gen Psychiatry. 2006;63:63–72. doi: 10.1001/archpsyc.63.1.63. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Geschwind DH, Tingus K, Huang D, Mendez MF, Edwards N, Mintz J. Apolipoprotein E affects both myelin breakdown and cognition: implications for age-related trajectories of decline into dementia. Biol Psychiatry. 2007;62:1380–1387. doi: 10.1016/j.biopsych.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Berg L. Clinical dementia rating (CDR) Psychopharmacol Bull. 1988;24:637–639. [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Klunder AD, Hayashi KM, Protas H, Kepe V, Miller KJ, Huang SC, Barrio JR, Ercoli LM, Siddarth P, Satyamurthy N, Liu J, Toga AW, Bookheimer SY, Small GW, Thompson PM. Plaque and tangle imaging and cognition in normal aging and Alzheimer’s disease. Neurobiol Aging. 2008 Nov 10; doi: 10.1016/j.neurobiolaging.2008.09.012. [Electronic publication ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman AM, Siedlecki KL, Muraskin J, Manly JJ, Luchsinger JA, Yeung LK, Brown TR, Decarli C, Stern Y. White matter hyperintensities and cognition: testing the reserve hypothesis. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.10.013. [Electronic publication ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88:1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore ET, Suckling J, Overmeyer S, Rabe-Hesketh S, Taylor E, Brammer MJ. Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural MR images of the brain. IEEE Trans Med Imaging. 1999;18:32–42. doi: 10.1109/42.750253. [DOI] [PubMed] [Google Scholar]
- Busatto GF, Diniz BS, Zanetti MV. Voxel-based morphometry in Alzheimer’s disease. Expert Rev Neurother. 2008;8:1691–1702. doi: 10.1586/14737175.8.11.1691. [DOI] [PubMed] [Google Scholar]
- Carmichael OT, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Kuller LH, Lopez OL, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Mapping ventricular changes related to dementia and mild cognitive impairment in a large community-based cohort. IEEE ISBI. 2006:315–318. [Google Scholar]
- Chetelat G, Landeau B, Eustache F, Mezenge F, Viader F, de la Sayette V, Desgranges B, Baron JC. Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. Neuroimage. 2005;27:934–946. doi: 10.1016/j.neuroimage.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Desgranges B, Landeau B, Mezenge F, Poline JB, de la Sayette V, Viader F, Eustache F, Baron JC. Direct voxel-based comparison between grey matter hypometabolism and atrophy in Alzheimer’s disease. Brain. 2008a;131:60–71. doi: 10.1093/brain/awm288. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Fouquet M, Kalpouzos G, Denghien I, De la Sayette V, Viader F, Mezenge F, Landeau B, Baron JC, Eustache F, Desgranges B. Three-dimensional surface mapping of hippocampal atrophy progression from MCI to AD and over normal aging as assessed using voxel-based morphometry. Neuropsychologia. 2008b;46:1721–1731. doi: 10.1016/j.neuropsychologia.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Chen K, Langbaum J, Fleisher AS, Reschke C, Lee W, Liu X, Ayutyanont N, Bandy D, Alexander GE, Thompson PM, Foster NL, Harvey DJ, De Leon MJ, Koeppe RA, Jagust WJ, Weiner MW, Reiman EM the Alzheimer’s Disease Neuroimaging Initiative. Twelve-month metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment assessed using an empirically pre-defined statistical region-of-interest: findings from the Alzheimer’s Disease Neuroimaging Initiative (submitted) 2009a doi: 10.1016/j.neuroimage.2010.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reschke C, Lee W, Napatkamon A, Liu X, Bandy D, Langbaum J, Alexander GE, Foster NL, Koeppe RA, Jagust WJ, Weiner MW, Reiman EM. Cross-sectional and longitudinal analyses of fluorodeoxyglucose positron emission tomography images from the Alzheimer’s disease neuroimaging initiative. ADNI Data Presentations Meeting; Seattle, WA. 2009b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. 3D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. Neuroimage. 2007;34:44–60. doi: 10.1016/j.neuroimage.2006.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YY, Lepore N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. Neuroimage. 2008;40:615–630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YY, Leporé N, Avedissian C, Madsen SK, Hua X, Jack CR, Weiner MW, Toga AW, Thompson PM. Mapping ventricular expansion and its clinical correlates in Alzheimer’s disease and mild cognitive impairment using multi-atlas fluid image alignment. SPIE Medical Imaging; February 7–12 2009; Lake Buena Vista, Florida. 2009a. [Google Scholar]
- Chou YY, Lepore N, Avedissian C, Madsen SK, Parikshak N, Hua X, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. Neuroimage. 2009b;46:394–410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. Neuroimage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- Clarkson MJ, Ourselin S, Nielsen C, Leung KK, Barnes J, Whitwell JL, Gunter JL, Hill DL, Weiner MW, Jack CR, Jr, Fox NC. Comparison of phantom and registration scaling corrections using the ADNI cohort. Neuroimage. 2009;47:1506–1513. doi: 10.1016/j.neuroimage.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell JR, Folstein MF. Mini-Mental State Examination (MMSE) Psychopharmacol Bull. 1988;24:689–692. [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the RAVENS maps: methods and validation using simulated longitudinal atrophy. Neuroimage. 2001;14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AC, Hinkley DV. Bootstrap Methods and their Application. Cambridge University Press; 1997. [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, Sperling RA, Atri A, Growdon JH, Hyman BT, Morris JC, Fischl B, Buckner RL. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman & Hall; New York: 1993. [Google Scholar]
- Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, Jorm A, Mathers C, Menezes PR, Rimmer E, Scazufca M. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. Wiley-Interscience; 2004. [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fox NC, Freeborough PA. Brain atrophy progression measured from registered serial MRI: validation and application to Alzheimer’s disease. J Magn Reson Imaging. 1997;7:1069–1075. doi: 10.1002/jmri.1880070620. [DOI] [PubMed] [Google Scholar]
- Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, Rossor MN. Presymptomatic hippocampal atrophy in Alzheimer’s disease. A longitudinal MRI study. Brain. 1996;119(Pt 6):2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- Fox NC, Scahill RI, Crum WR, Rossor MN. Correlation between rates of brain atrophy and cognitive decline in AD. Neurology. 1999;52:1687–1689. doi: 10.1212/wnl.52.8.1687. [DOI] [PubMed] [Google Scholar]
- Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- Frank RA, Galasko D, Hampel H, Hardy J, de Leon MJ, Mehta PD, Rogers J, Siemers E, Trojanowski JQ. Biological markers for therapeutic trials in Alzheimer’s disease. Proceedings of the biological markers working group; NIA initiative on neuroimaging in Alzheimer’s disease. Neurobiol Aging. 2003;24:521–536. doi: 10.1016/s0197-4580(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC. Modeling brain deformations in Alzheimer disease by fluid registration of serial 3D MR images. J Comput Assist Tomogr. 1998;22:838–843. doi: 10.1097/00004728-199809000-00031. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC, Kitney RI. Interactive algorithms for the segmentation and quantitation of 3-D MRI brain scans. Comput Methods Programs Biomed. 1997;53:15–25. doi: 10.1016/s0169-2607(97)01803-8. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Pievani M, Testa C, Sabattoli F, Bresciani L, Bonetti M, Beltramello A, Hayashi KM, Toga AW, Thompson PM. The topography of grey matter involvement in early and late onset Alzheimer’s disease. Brain. 2007;130:720–730. doi: 10.1093/brain/awl377. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Prestia A, Rasser PE, Bonetti M, Thompson PM. In vivo mapping of incremental cortical atrophy from incipient to overt Alzheimer’s disease. J Neurol. 2009;256:916–924. doi: 10.1007/s00415-009-5040-7. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Rev Neurol. 2010;6:1–11. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost C, Kenward MG, Fox NC. The analysis of repeated ‘direct’ measures of change illustrated with an application in longitudinal imaging. Stat Med. 2004;23:3275–3286. doi: 10.1002/sim.1909. [DOI] [PubMed] [Google Scholar]
- Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA, Foster NL, Jack CR, Jr, Galasko DR, Doody R, Kaye J, Sano M, Mohs R, Gauthier S, Kim HT, Jin S, Schultz AN, Schafer K, Mulnard R, van Dyck CH, Mintzer J, Zamrini EY, Cahn-Weiner D, Thal LJ. Mild cognitive impairment can be distinguished from Alzheimer disease and normal aging for clinical trials. Arch Neurol. 2004;61:59–66. doi: 10.1001/archneur.61.1.59. [DOI] [PubMed] [Google Scholar]
- Gunter JL, Bernstein MA, Borowski BJ, Ward CP, Britson PJ, Felmlee JP, Schuff N, Weiner M, Jack CR. Measurement of MRI scanner performance with the ADNI phantom. Med Phys. 2009;36(6):2193–2205. doi: 10.1118/1.3116776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, Interactive. MetLife Foundation Alzheimer’s Survey: What America Thinks. 2006. [Google Scholar]
- Hedden T, Van Dijk KR, Becker JA, Mehta A, Sperling RA, Johnson KA, Buckner RL. Disruption of functional connectivity in clinically normal older adults harboring amyloid burden. J Neurosci. 2009;29:12686–12694. doi: 10.1523/JNEUROSCI.3189-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AJ, Hua X, Lee S, Yanovsky I, Leow AD, Gutman B, Dinov ID, Toga AW, Jack CR, Jr, Bernstein MA, Reiman EM, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Comparing 3T and 1.5T MRI for tracking AD progression with tensor-based morphometry. NeuroImage. in press. [Electronic publication ahead of print] [Google Scholar]
- Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou YY, Brun C, Chiang MC, Barysheva M, Jack CR, Jr, Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes RG, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM the Alzheimer’s Disease Neuroimaging Initiative. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008a;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008b;43:458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Lee S, Yanovsky I, Leow AD, Chou YY, Ho AJ, Gutman B, Toga AW, Jack CR, Jr, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009a;48:668–681. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Levitt JG, Caplan R, Thompson PM, Toga AW. Detecting brain growth patterns in normal children using tensor-based morphometry. Hum Brain Mapp. 2009b;30:209–219. doi: 10.1002/hbm.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Friedland RP, Auchus AP. Diffusion tensor imaging of normal-appearing white matter in mild cognitive impairment and early Alzheimer disease: preliminary evidence of axonal degeneration in the temporal lobe. AJNR Am J Neuroradiol. 2007;28:1943–1948. doi: 10.3174/ajnr.A0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- Ishii K, Kawachi T, Sasaki H, Kono AK, Fukuda T, Kojima Y, Mori E. Voxel-based morphometric comparison between early- and late-onset mild Alzheimer’s disease and assessment of diagnostic performance of z score images. AJNR Am J Neuroradiol. 2005;26:333–340. [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, Smith GE, Ivnik RJ, Kokmen E. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology. 1997;49:786–794. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer’s disease. Neurology. 1998;51:993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, Xu Y, Shiung M, O’Brien PC, Cha R, Knopman D, Petersen RC. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003;60:253–260. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, Shiung MM, Gunter JL, Boeve BF, Kemp BJ, Weiner M, Petersen RC. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain. 2009;132:1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger KA. Alzheimer 100—highlights in the history of Alzheimer research. J Neural Transm. 2006;113:1603–1623. doi: 10.1007/s00702-006-0578-3. [DOI] [PubMed] [Google Scholar]
- Jorm AF, Korten AE, Henderson AS. The prevalence of dementia: a quantitative integration of the literature. Acta Psychiatr Scand. 1987;76:465–479. doi: 10.1111/j.1600-0447.1987.tb02906.x. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Killiany RJ, Moss MB, Albert MS, Sandor T, Tieman J, Jolesz F. Temporal lobe regions on magnetic resonance imaging identify patients with early Alzheimer’s disease. Arch Neurol. 1993;50:949–954. doi: 10.1001/archneur.1993.00540090052010. [DOI] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Jr, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009;45:645–655. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp. 2009a;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009b;45:S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005a;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005b;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggara O, Oppenheim C, Rieu D, Raoux N, Rodrigo S, Dalla Barba G, Meder JF. Diffusion tensor imaging in early Alzheimer’s disease. Psychiatry Res. 2006;146:243–249. doi: 10.1016/j.pscychresns.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Nestor SM, Rupsingh R, Borrie M, Smith M, Accomazzi V, Wells JL, Fogarty J, Bartha R. Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain. 2008;131:2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paling SM, Williams ED, Barber R, Burton EJ, Crum WR, Fox NC, O’Brien JT. The application of serial MRI analysis techniques to the study of cerebral atrophy in late-onset dementia. Med Image Anal. 2004;8:69–79. doi: 10.1016/j.media.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment clinical trials. Nat Rev Drug Discov. 2003a;2:646–653. doi: 10.1038/nrd1155. [DOI] [PubMed] [Google Scholar]
- Petersen RC. Mild Cognitive Impairment: Aging to Alzheimer’s Disease. Oxford University Press; New York: 2003b. [Google Scholar]
- Petersen RC. Mild cognitive impairment: current research and clinical implications. Semin Neurol. 2007;27:22–31. doi: 10.1055/s-2006-956752. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Protas HD, Huang SC, Kepe V, Hayashi K, Klunder A, Braskie MN, Ercoli L, Bookheimer S, Thompson PM, Small GW, Barrio JR. FDDNP binding using MR derived cortical surface maps. Neuroimage. 2009;49:240–248. doi: 10.1016/j.neuroimage.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Ayutyanont N, Bandy D, Lee W, Reschke C, Alexander GE, Weiner MW, Koeppe RA, Jagust WJ. Twelve-month cerebral metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment: preliminary findings from the Alzheimer’s disease neuroimaging initiative. ICAD; Chicago, Illinois: 2008a. [Google Scholar]
- Reiman EM, Chen K, Ayutyanont N, Lee W, Bandy D, Reschke C, Alexander GE, Weiner MW, Koeppe RA, Foster NL, Jagust WJ. Twelve-month cerebral metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment: Preliminary findings from the Alzheimer’s disease neuroimaging initiative (ADNI) Alzheimer’s and Dementia. 2008b;4:T110–T111. [Google Scholar]
- Riddle WR, Li R, Fitzpatrick JM, DonLevy SC, Dawant BM, Price RR. Characterizing changes in MR images with color-coded Jacobians. Magn Reson Imaging. 2004;22:769–777. doi: 10.1016/j.mri.2004.01.078. [DOI] [PubMed] [Google Scholar]
- Rose SE, Chen F, Chalk JB, Zelaya FO, Strugnell WE, Benson M, Semple J, Doddrell DM. Loss of connectivity in Alzheimer’s disease: an evaluation of white matter tract integrity with colour coded MR diffusion tensor imaging. J Neurol Neurosurg Psychiatry. 2000;69:528–530. doi: 10.1136/jnnp.69.4.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer’s disease. Am J Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- Rosner B. Fundamentals of Biostatistics. PWS-Kent Publishing Company; Boston: 1990. [Google Scholar]
- Sandson TA, Felician O, Edelman RR, Warach S. Diffusion-weighted magnetic resonance imaging in Alzheimer’s disease. Dement Geriatr Cogn Disord. 1999;10:166–171. doi: 10.1159/000017099. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Fox NC. Longitudinal imaging in dementia. Br J Radiol. 2007;2(80 Spec No):S92–98. doi: 10.1259/bjr/78981552. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch Neurol. 2003;60:989–994. doi: 10.1001/archneur.60.7.989. [DOI] [PubMed] [Google Scholar]
- Schott JM, Frost C, Whitwell JL, Macmanus DG, Boyes RG, Rossor MN, Fox NC. Combining short interval MRI in Alzheimer’s disease: implications for therapeutic trials. J Neurol. 2006;253:1147–1153. doi: 10.1007/s00415-006-0173-4. [DOI] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132 (4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Korecka M, Clark CM, Lee VM, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat Rev Drug Discov. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23 (Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, De Stefano N. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Spires-Jones TL, Stoothoff WH, de Calignon A, Jones PB, Hyman BT. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci. 2009;32:150–159. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Apostolova LG. Computational anatomical methods as applied to aging and dementia. Br J Radiol. 2007;80(Spec No 2):S78–91. doi: 10.1259/BJR/20005470. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Bartzokis G, Hayashi KM, Klunder AD, Lu PH, Edwards N, Hong MS, Yu M, Geaga JA, Toga AW, Charles C, Perkins DO, McEvoy J, Hamer RM, Tohen M, Tollefson GD, Lieberman JA. Time-lapse mapping of cortical changes in schizophrenia with different treatments. Cereb Cortex. 2009;19:1107–1123. doi: 10.1093/cercor/bhn152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW. Brain Warping. 1. Academic Press; San Diego: 1999. [Google Scholar]
- Vemuri P, Whitwell JL, Kantarci K, Josephs KA, Parisi JE, Shiung MS, Knopman DS, Boeve BF, Petersen RC, Dickson DW, Jack CR., Jr Antemortem MRI based STructural Abnormality iNDex (STAND)-scores correlate with postmortem Braak neurofibrillary tangle stage. Neuroimage. 2008;42:559–567. doi: 10.1016/j.neuroimage.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liang M, Wang L, Tian L, Zhang X, Li K, Jiang T. Altered functional connectivity in early Alzheimer’s disease: a resting-state fMRI study. Hum Brain Mapp. 2007;28:967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Goldstein FC, Veledar E, Levey AI, Lah JJ, Meltzer CC, Holder CA, Mao H. Alterations in cortical thickness and white matter integrity in mild cognitive impairment measured by whole-brain cortical thickness mapping and diffusion tensor imaging. AJNR Am J Neuroradiol. 2009;30:893–899. doi: 10.3174/ajnr.A1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain. 2007;130:1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Josephs KA, Murray ME, Kantarci K, Przybelski SA, Weigand SD, Vemuri P, Senjem ML, Parisi JE, Knopman DS, Boeve BF, Petersen RC, Dickson DW, Jack CR., Jr MRI correlates of neurofibrillary tangle pathology at autopsy: a voxel-based morphometry study. Neurology. 2008;71:743–749. doi: 10.1212/01.wnl.0000324924.91351.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak JR, Lim KO. Advances in white matter imaging: a review of in vivo magnetic resonance methodologies and their applicability to the study of development and aging. Neurosci Biobehav Rev. 2006;30:762–774. doi: 10.1016/j.neubiorev.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky I, Thompson P, Osher S, Leow AD. Asymmetric and symmetric unbiased image registration: statistical assessment of performance. IEEE Comput Soc Workshop Math Methods Biomed Image Anal. 2008;1:1–8. doi: 10.1109/CVPRW.2008.4562988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky I, Leow AD, Lee S, Osher SJ, Thompson PM. Comparing registration methods for mapping brain change using tensor-based morphometry. Medical Image Analysis. 2009;13:679–700. doi: 10.1016/j.media.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Dougherty JH, Jr, Hubner KF, Bai B, Cannon RL, Hutson RK. Abnormal connectivity in the posterior cingulate and hippocampus in early Alzheimer’s disease and mild cognitive impairment. Alzheimers Dement. 2008;4:265–270. doi: 10.1016/j.jalz.2008.04.006. [DOI] [PubMed] [Google Scholar]