Abstract
There is strong clinical evidence that implicates tenofovir in the loss of bone mineral density during treatment of human immunodeficiency virus infection. In this study, we sought to test the hypothesis that tenofovir treatment of osteoblasts causes changes in the gene expression profile that would impact osteoblast function during bone formation. Primary osteoblasts were isolated and then treated with the tenofovir prodrug, tenofovir disoproxil fumarate (TDF). Total RNA from TDF-treated and untreated osteoblasts were extracted and used for microarray analysis to assess TDF-associated changes in the gene expression profile. Strikingly, the changes in gene expression profiles involved in cell signaling, cell cycle and amino acid metabolism, which would likely impact osteoblast function in bone formation. Our findings demonstrate for the first time that tenofovir treatment of primary osteoblasts results in gene expression changes that implicate loss of osteoblast function in tenofovir-associated bone mineral density loss.
Keywords: bone, nucleotide, metabolism, HIV, osteoclast, cell cycle, signaling
Introduction
There are currently over 30 million people infected with human immunodeficiency virus (HIV). Highly active antiretroviral therapy (HAART) has been very successful in the clinical management of HIV infection. Nucleoside reverse transcriptase inhibitors (NRTIs) are used in virtually all HAART regimens. The NRTIs target HIV reverse transcriptase and prevent the synthesis of viral cDNA from the genomic RNA. NRTI's act as chain terminators of HIV DNA synthesis since NRTIs lack a 3′-hydroxyl group on the deoxyribose and following incorporation the next incoming deoxynucleotide cannot form a new 5′-3′ phosphodiester bond that is required to extend the synthesis of the DNA chain. HIV drug resistance and associated drug metabolic disorders have driven the need for the identification of new drug targets and the development of new anti-HIV drugs.
Tenofovir, 9-(R)-{2-(phosphonomethoxy)propyl}adenine or PMPA, is a nucleotide analog that was approved in 2001 for use in treating HIV infection and more recently for chronic hepatitis B infection. Tenofovir disoproxil fumarate (TDF) is the prodrug form. Tenofovir is popular and well prescribed for several reasons. First, tenofovir has improved potency as it is a nucleotide and has an abbreviated intracellular activation pathway to allow a more rapid and complete conversion to the active drug. Second, TDF has a labile lipophilic group to facilitate penetration through target cell membranes. Third, tenofovir is not known to be a substrate, inducer or inhibitor of human cytochrome P450 enzymes in vitro or in vivo. Fourth, tenofovir has high potency and an unusually durable response when used in trials of single-agent therapy intensification in highly treatment-experienced individuals. Fifth, the active metabolite, tenofovir diphosphate, exhibits a long intracellular half-life in both resting and activated peripheral blood mononuclear cells, which allows for single daily dosing.
HIV infection [1; 2; 3; 4] as well as tenofovir treatment of adults [5] have been implicated in causing osteopenia. Tenofovir-associated loss of bone mineral density has also been observed with children and adolescents [6; 7; 8]. Similar findings of reduced bone mineral density have been reported with use of tenofovir during infection of macaques with simian immunodeficiency virus [9; 10; 11].
Osteoblasts, which are derived from meschenchymal stem cells, synthesize collagen and glycoproteins that forms osteoid [12]. Osteoclasts, which are derived from hematopoietic stem cells, on the other hand, resorb bone. Bone is a dynamic tissue characterized by cycles of bone resorption and bone formation. Osteoblast differentiation is regulated by many cytokines including bone morphogenetic proteins (BMPs), which are necessary for inducing osteoblast differentiation and maintaining the differentiated state of committed osteoblasts [13; 14]; transforming growth factor beta (TGF-beta), which is a regulator of osteoblast differentiation [15]; and fibroblast growth factor (FGFs), which antagonizes the positive osteoblast differentiation effects of the Wnt signaling pathway [16]. Extensive cell signaling between osteoblasts and osteoclasts is required for maintaining a balance in the activities of osteoblasts and osteoclasts. Changes in osteoblast function can lead to reduced bone and either osteopenia or osteoporosis.
Based upon the observations of tenofovir-associated reduced bone mineral density, we sought to explore the basis for these clinical observations. In this study, we investigated whether tenofovir could perturb osteoblast gene expression profiles in a manner that could impact osteoblast function in bone formation. To do this, we initially harvested primary murine osteoblasts and exposed them to various TDF concentrations. TDF concentrations that were physiologically relevant to the clinical dosing in humans were then further used for treating primary osteoblasts followed by isolation of total RNA for use in microarray analysis. We observed a wide spectrum of gene expression changes in osteoblast genes, including many associated with osteoblast function in bone formation. Our findings show for the first time that tenofovir can result in the perturbation of osteoblast gene expression and that these changes implicate loss of osteoblast function that could lead to reduced bone mineral density.
Materials and Methods
Primary osteoblast cultures
Primary osteoblasts were isolated as previously described [17]. Calvaria from 7-10 day old C57/BL6 mice were dissected and subjected to sequential digestions with 2 mg/mL of collagenase A (Roche Molecular Biomedicals) in MEM solution containing 0.25% trypsin (Gibco) for 20, 40 and 90 min. The third digestion was plated at 1.5 × 104 cells/cm2. Cells were maintained at 37 C in a humidified atmosphere of 5% CO2, in MEM media containing 10% FBS, 1% penicillin/streptomycin, and 1% non-essential amino acids. Upon confluency, cells were differentiated with ascorbic acid (50 μg/mL)-containing media (Sigma-Aldrich).
Cell viability
A serial dilution series (i.e., 500 uM, 250 uM, 50 uM 5 uM, 500 nM, or 50 nM) of TDF was added to the cell culture media of primary murine osteoblast cultures for 3 days, refreshing the cells with fresh media with TDF every 24h. Cell viability was determined by measurement of cellular ATP using the CellTiter-Glo Luminescent Cell Viability Assay using the manufacturer's instructions (Promega, Madison, WI).
Microarray analysis
Primary murine osteoblasts were prepared and treated with 500 nM TDF for 10 days as described above. Total RNA from primary osteoblasts was then extracted using the RNeasy mini plus kit (Qiagen, Valencia, CA). Four independent replicates from TDF-treated and untreated primary osteoblasts were analyzed. Two gene chips were used in each replicate experiment. The purity and concentration of total RNA was determined based on OD 260/280 readings. Assistance in RNA quality control, labeling, hybridization, and initial data analysis was provided by Genome Explorations Inc. (Memphis, TN). The integrity of the RNA was examined by capillary electrophoresis using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) with RNA 6000 Neno Lab-in-a-Chip Kit (Agilent Technologies) following manufacturer's instructions. Total RNA was used for cDNA synthesis with the reverse transcription-in vitro transcription (RT-IVT) method [18] using the GeneChip WT cDNA Synthesis and Amplification kit (Affymetrix, Santa Clara, CA) according to manufacturer's instructions.
Fragmented and labeled cDNA was hybridized for 17 h at 45 C to GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix). The mouse Gene 1.0 ST array is an expression array featuring whole genome-transcript coverage with 750,000 unique oligonucleotide probes representing a total number of 28,853 mouse genes. There were approximately 27 probes covering the full length of each gene. Arrays were washed and then stained using a Fluidics Station 450 reagent (Affymetrix) followed by a second staining step with phycoerythrin-conjugated streptavidin (Invitrogen) according to recommended procedures from the manufacturer. The fluorescence intensities were the measured using a GCS 3000 7G high-resolution confocol laser scanner and analyzed with Affymetrix GeneChip Expression Console™ software (Affymetrix). Background correction, normalization, and signal summarization (per probe set) were calculated accordingly [19]. For each transcript, an independent t-test (5% confidence) was applied to assess significance of expression level based on RMA absolute signal log ratios ≥ 1.0. The following resources were used to compose gene annotation, gene ontology, and biochemical pathway: National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), NetAffx (http://www.affymetrix.com), Gene Ontology Consortium (http://amigo.geneontology.org), Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg), and WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt).
Quantitative real-time PCR
Primary murine osteoblasts were prepared and treated with 500 nM, 1 uM, and 2 uM TDF as described earlier. Ten days post-treatment, cells were washed in 1× phosphate-buffered saline (PBS) twice prior to RNA extraction. The RNeasy mini plus kit (Qiagen, Valencia, CA) was used to obtain total RNA and to remove traces of genomic DNA contamination from cells based on manufacturer's instructions. Immediately following RNA extraction, 500 ug of total RNA from each sample was used to generate first stand cDNA as templates for quantitative PCR (qPCR) with the Transcriptor High Fidelity cDNA kit (Roche Applied Science, Indianapolis, IN). SYBR green qPCR reagents from Invitrogen (Carlsbad, CA) were used to detect real-time gene expression with an initial denaturation at 95 C for 3 minutes and 35 cycles of 15 seconds at 94 C, 30 seconds at 60 C, and 30 seconds at 72 C.
Results
The goal of this study was to determine whether in vitro treatment of primary osteoblasts with TDF, the prodrug of tenofovir (Fig. 1A, B), would alter gene expression as determined by microarray analysis, and provide insights into how TDF exposure may influence osteoblast function. We first investigated the effect of TDF exposure on cell viability. We sought to determine whether physiologically relevant TDF concentrations corresponding to the dosing of TDF in antiretroviral therapy had any effects on cell viability. A range of TDF concentrations (50 nM to 500 uM) were analyzed for their effects on osteoblast viability. The highest TDF concentrations analyzed, i.e., 50 uM, 250 uM and 500 uM, were found to significantly reduce cell viability (Fig. 1C). The lower TDF concentrations, i.e., 50 nM, 500 nM and 5 uM, had no effect on cell viability. Tenofovir has been shown to cause very little cytotoxicity in human HepG2 and skeletal muscle cell lines, with average CC50 of 399 and 870 uM, respectively [20]. It is to be expected that primary culture is more sensitive to drug treatment in comparison to immortalized cell lines. As shown in our cytotoxicity assay, primary osteoblast cultures are sensitive to much lower concentrations of TDF (Fig. 1C). Based upon these results, we chose the TDF concentration of 500 nM for analyzing the impact of TDF exposure on osteoblast gene expression in primary cells. This TDF concentration is physiologically relevant based upon the serum concentrations and dosing regimens used in antiretroviral therapy of HIV infected individuals [21; 22].
Fig. 1.
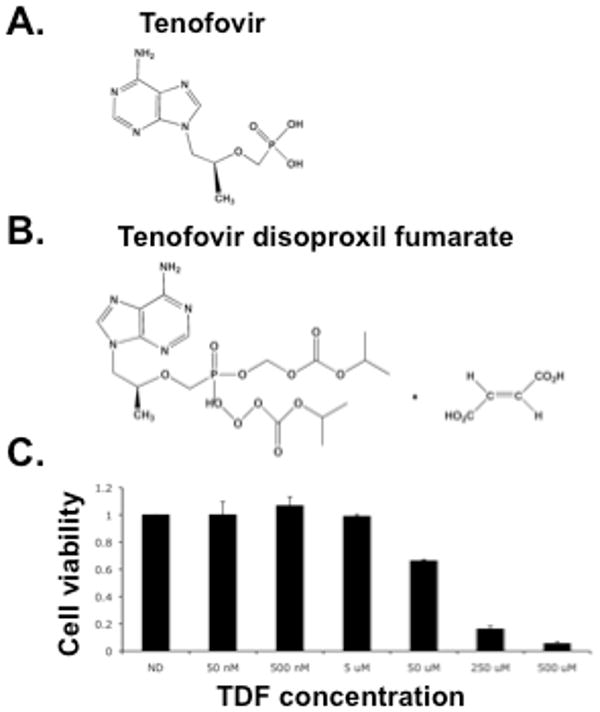
Tenofovir structure and primary osteoblast cell viability following drug exposure. The structure of tenofovir (A) and tenofovir disoproxil fumarate (TDF, the prodrug of tenofovir) (B) is shown. C. Viability of primary osteoblasts following exposure to TDF. A TDF dilution series was added to primary osteoblast cultures for 3 days, refreshing every 24h. Cell viability was determined by ATP detection as described in the Materials and Methods.
Primary osteoblasts were treated with 500 nM TDF, and then total RNA was extracted and analyzed for yield and integrity by capillary electrophoresis (data not shown). Samples were used as templates for cDNA synthesis and used to generate cDNAs that were then prepared for use in microarray analysis. Two gene chips were used in each replicate experiment and a total of four independent replicate experiments were performed. A student t-test (p-value ≤ 0.05) was used to filter for probe sets that exhibit an absolute fold-change ≥ 1.5. The microarray analysis was intensive and allowed for the thorough analysis of osteoblast gene expression profiles. As described below, this intensive approach helped focus the gene expression profile on a relatively limited number of transcripts (i.e., under 80). Another likely variable that was responsible for the focused gene expression profile was the replicate-to-replicate variability of TDF treatment of primary osteoblasts. Finally, limited uptake of TDF by osteoblasts in tissue culture and in the absence of the bone microenvironment could also explain the focused gene expression profile.
A heat map was generated from the gene expression profile obtained from the microarray data (Fig. 2). From this heat map, a total of seventy-nine transcripts were identified to have significantly altered gene expression profiles in groups treated with TDF. Hierarchical clustering analysis was done for the gene expression data. The dendrograms were generated based on average linkage hierarchical clustering of expression data. Supplemental Table 1 lists the resulting gene expression profile of transcripts with significant expression changes as identified from intensive microarray analysis of TDF-treated primary osteoblasts. The Log2 fold change was determined as well as the t-test p-values for TDF treatment versus that of the control. Twenty-six of the 79 (i.e., 33%) transcripts were upregulated, with the largest increases observed with Serpina3n, Ass1, and Anxa8, (1.266939, 1.134408,1.128678, respectively). Fifty-three of the 79 (i.e., 67%) transcripts were downregulated, with Rspo2, Snora44, and Pgm5 (-1.039561, -1.000873, and -0.993176, respectively) having the greatest reduction in transcript levels.
Fig. 2.
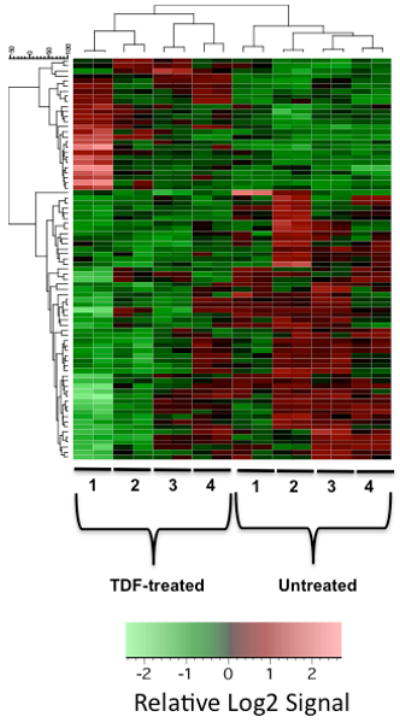
Heat map of gene expression profile from TDF-treated and untreated primary osteoblasts as determined from microarray analysis. Two gene chips were used in each replicate experiment and a total of four independent replicate experiments were performed. Hierarchical clustering analysis of gene expression data from primary osteoblasts exposed to TDF was also performed. The dendrograms were generated based on average linkage hierarchical clustering of expression data from 79 probe sets. Significant change was defined as having an absolute fold change >= 1.5 and a t-test p-value of <= 0.05. Signal values were log2 transformed, row mean centered and then subjected to unsupervised hierarchical clustering by the UPGMA algorithm using Pearson Correlation as the similarity metric.
The KEGG (i.e., Kyoto Encyclopedia of Genes and Genomes) database was used for analyzing the pathways associated with the identified genes (Table 1). The use of KEGG allowed us to identify networks of molecular interactions involved in the response of primary osteoblasts to TDF treatment. Notably, several signaling pathways (i.e., Wnt, TGF-beta, Hedgehog, VEGF and MAPK) were identified as well as several involved in amino acid metabolism (i.e., arginine and proline, glycine, serine and threonine, and alanine and aspartate) and energy metabolism. The regulation of cell cycle was also implicated by the identification of Cdkn1a, cyclin-dependent kinase inhibitor 1A (p21), which is a negative regulator of CDK and the cell cycle.
Table 1.
Organization of osteoblast genes altered by TDF treatment by KEGG Biochemical Pathways.
| KEGG pathway | Gene number | Entrez IDs | Enrichment a |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | 3 | 11480 16324 20296 | O=3;E=0.4497; R=6.6711;P=1.05e-2 |
| Wnt signaling pathway | 2 | 14366 170758 | O=2;E=0.2784; R=7.1839;P=3.18e-2 |
| Colorectal cancer | 2 | 14366 170758 | O=2;E=0.1596;R=12.5313;P=1.12e-2 |
| Arginine and proline metabolism | 2 | 11898 209027 | O=2;E=0.1012;R=19.7628;P=4.66e-3 b |
| Urea cycle and metabolism of amino groups | 2 | 11898 209027 | O=2;E=0.0467;R=42.8266;P=1.00e-3 b |
| Axon guidance | 2 | 108151 170758 | O=2;E=0.2434; R=8.2169;P=2.49e-2 |
| TGF-beta signaling pathway | 2 | 11480 16324 | O=2;E=0.1596;R=12.5313;P=1.12e-2 |
| Cell cycle | 1 | 12575 | O=1;E=0.2064;R=4.845;P=1.87e-1 |
| SNARE interactions in vesicular transport | 1 | 53611 | O=1;E=0.0623;R=16.0514;P=6.05e-2 |
| Hedgehog signaling pathway | 1 | 15245 | O=1;E=0.1032; R=9.6899;P=9.82e-2 |
| VEGF signaling pathway | 1 | 170758 | O=1;E=0.1382;R=7.2359;P=1.29e-1 |
| Focal adhesion | 1 | 170758 | O=1;E=0.3582; R=2.7917;P=3.02e-1 |
| Cell adhesion molecules (CAMs) | 1 | 14961 | O=1;E=0.2492; R=4.0128;P=2.21e-1 |
| Adherens junction | 1 | 170758 | O=1;E=0.1421;R=7.0373;P=1.33e-1 |
| Gap junction | 1 | 19091 | O=1;E=0.1635;R=6.1162;P=1.51e-1 |
| Complement and coagulation cascades | 1 | 14962 | O=1;E=0.1227;R=8.15;P=1.16e-1 |
| Antigen processing and presentation | 1 | 14961 | O=1;E=0.1402;R=7.1327;P=1.31e-1 |
| Natural killer cell mediated cytotoxicity | 1 | 170758 | O=1;E=0.2142; R=4.6685;P=1.93e-1 |
| B cell receptor signaling pathway | 1 | 170758 | O=1;E=0.1246;R=8.0257;P=1.17e-1 |
| Fc epsilon RI signaling pathway | 1 | 170758 | O=1;E=0.146;R=6.8493;P=1.36e-1 |
| Long-term depression | 1 | 19091 | O=1;E=0.1441;R=6.9396;P=1.34e-1 |
| Olfactory transduction | 1 | 19091 | O=1;E=0.0584;R=17.1233;P=5.68e-2 |
| Regulation of actin cytoskeleton | 1 | 170758 | O=1;E=0.3777;R=2.6476;P=3.15e-1 |
| Type I diabetes mellitus | 1 | 14961 | O=1;E=0.0934;R=10.7066;P=8.93e-2 |
| Pancreatic cancer | 1 | 170758 | O=1;E=0.1402;R=7.1327;P=1.31e-1 |
| Glioma | 1 | 12575 | O=1;E=0.1207; R=8.285;P=1.14e-1 |
| MAPK signaling pathway | 1 | 170758 | O=1;E=0.5179; R=1.9309;P=4.05e-1 |
| Folate biosynthesis | 1 | 14528 | O=1;E=0.0662;R=15.1057;P=6.41e-2 |
| Glycine, serine and threonine metabolism | 1 | 236539 | O=1;E=0.0915; R=10.929;P=8.76e-2 |
| Alanine and aspartate metabolism | 1 | 11898 | O=1;E=0.0584;R=17.1233;P=5.68e-2 |
| Purine metabolism | 1 | 14544 | O=1;E=0.2609; R=3.8329;P=2.30e-1 |
| Chronic myeloid leukemia | 1 | 12575 | O=1;E=0.1441;R=6.9396;P=1.34e-1 |
| Pentose and glucuronate interconversions | 1 | 67880 | O=1;E=0.0234;R=42.735;P=2.31e-2 |
| Oxidative phosphorylation | 1 | 333182 | O=1;E=0.2142; R=4.6685;P=1.93e-1 |
O = the observed gene number in the KEGG pathway; E = is the expected gene number in the KEGG pathway (expected number of genes in a specific KEGG pathway for an interestings gene set=total number of genes in the KEGG pathway for the reference set * Total number of genes in the interesting set/Total number of genes in the reference set).; R = is the ration of enrichment for the KEGG pathway (R=O/E); P = the p value indicating the significance of enrichment calculated from Hypergeometric test. (it is given for the KEGG pathways with R>1.
KEGG pathways with at least 2 genes with p<0.01
Gene Ontology (GO) analysis was done on the transcripts showing either up-regulation or down-regulation following TDF-exposure of primary osteoblasts (Supplemental Fig. 1). A couple of particularly notable observations can be made from this analysis. First, a large number of the transcripts encode for extracellular proteins, which include molecules involved in signaling pathways as well as proteins involved in molecule transporters. Second, many types of proteins involved in amino acid metabolism were identified as having their gene expression altered by TDF exposure.
To further assess the results from the microarray analysis, a limited number of transcripts were selected (i.e., Rac3, Acvr2a, Hhip, and Ecm1) for qPCR analysis. These gene transcripts were selected based upon their general relevance to cell signaling pathways or processes that could be associated with osteoblast function or differentiation. Three independent experiments were performed for this assay and the 18S rRNA was used as an internal control for determining the relative level of gene expression. We were able to conclude general verification on the up- or down-regulation of transcript expression as determined by the microarray results at a success rate of 83% (Table 2). The differences for Rac3 and Ecm1 were statistically significant. The observed discrepancies with the microarray data may be due to the SYBR green method used for qPCR, which is a relatively non-quantitative method for qPCR compared to other methodologies (e.g, TaqMan). Also, more than 1 housekeeping gene is likely required in order to establish accurate normalization for the verification of the small fold differences observed in the microarray analysis. Finally, alternative splicing and the differences in detection between the microarray probes versus that of the qPCR probes could result in discrepancies.
Table 2.
Real-time PCR analysis of selected genes from TDF-treated primary osteoblasts.
| Relative Gene Expression a | |||||
|---|---|---|---|---|---|
| Replicate | Rac3 | Acvr2a | Hhip | Ecm1 | |
| 1 | 1.230 | 1.619 | 1.212 | 1.250 | |
| 2 | 1.472 | 0.764 | 0.570 | 1.000 | |
| 3 | 1.159 | 0.724 | 0.651 | 1.247 | |
| Avg. SD | 1.287 | 1.036 | 0.811 | 1.166 | |
| 0.164 | 0.246 | 0.350 | 0.127 | ||
Gene expression in TDF-treated primary osteoblasts compared to untreated osteoblasts. Gene expression was normalized to 18S rRNA.
Discussion
Tenofovir is a successful anti-HIV drug used in HAART regimens that has been associated with reduced bone density [23]. In this study, we investigated whether the exposure of primary osteoblasts in vitro led to significant changes in gene expression profiles. We chose primary murine osteoblasts for analysis as they provide a readily tractable model system to assess the mechanism(s) involved in tenofovir-mediated reduction in bone density that can be readily translated into mouse models of HIV infection. Such models would allow for the in vivo analysis of TDF administration on bone morphology in the context of HIV infection.
Microarray analysis was used for the identification of genes whose gene expression was altered due to TDF exposure. The gene expression of several genes involved in amino acid biosynthesis and metabolism were down regulated in response to TDF exposure (Table 1, Supplemental Table 1). The general impact of this downregulation is reduced osteoblast growth, activity and differentiation. We also identified genes with altered gene expression that are involved in six different signaling pathways. These pathways include the Wnt (frizzled homolog 4, frizzled-related protein, Rac3), transforming growth factor-beta (TGF-beta) (activin receptor IIA, inhibin beta-B), Hedgehog (hedgehog-interacting protein), MAPK (Rac3), VEGF (Rac3), B cell receptor (Rac3), and the Fc epsilon RI signaling pathway (Rac3). The Wnt proteins are secreted morphogens required in many species and organs for many basic developmental processes, such as cell-fate specification, control of asymmetric cell division, and progenitor-cell proliferation. There are minimally three distinct Wnt pathways: the canonical pathway, the planar cell polarity (PCP) pathway and the Wnt/Ca2+ pathway. There were 3 Wnt-associated genes identified from our microarray analysis whose gene expression was perturbed by exposure to TDF: frizzled homolog 4, frizzled-related protein, and Rac3. The secreted frizzled-related proteins have been reported to inhibit osteoblast differentiation [24] and their observed downregulation of frizzled homolog 4 and frizzled-related protein by TDF (Supplemental Table 1) could result in reduction of bone density.
We recently evaluated the effect of tenofovir exposure on the gene expression profile of primary osteoclasts [25]. Specific downregulation of Gnas, Got2 and Snord32a was observed. The downregulation of Gnas gene expression may result in less MAPK/ERK signaling and ultimately a reduction in osteoclast proliferation and actin filament formation – resulting in decreased bone resorption. In context of TDF-mediated reduced bone density, this supports the model that the downregulation of bone formation by osteoblasts is associated with bone density loss. Got2 is a mitochondrial enzyme involved in energy transduction, specifically amino acid metabolism as well as the urea and tricarboxylic acid cycles. It is of particular interest that in this study, we also observed altered gene expression of Ass1 and Pycr1, which are also both involved in amino acid metabolism (and the urea cycle in the liver of mammals). KEGG pathway analysis indicated that the enrichment of these particular genes was highly significant (Table 2). Ass1 is a cellular enzyme while Pycr1 is a mitochondrial enzyme. Perturbation of amino acid metabolism following exposure to TDF in both osteoblasts and osteoclasts suggests alteration in bone homeostasis. Like Snord32a, SnorA44 is also a small nucleolar RNA (snoRNA). In particular, SnorA44 belongs to the H/ACA box class of snoRNAs as it has the predicted hairpin-hinge-hairpin-tail structure, and is predicted to guide the pseudouridylation of U822 and U686 of 18S ribosomal RNA (rRNA). The snoRNAs are essential for regulating biological functions such as RNA processing and modifications, gene expression, protein trafficking, and genome stability. The reduction in SnorA44 expression, like that of Snord32a, could result in a decrease of fully functional ribosomes, which would lead to a reduction in gene expression.
In summary, our observations indicate that TDF exposure of primary osteoblasts results in the distinct alteration of gene expression including signal transduction pathways, the cell cycle, as well as amino acid and energy metabolism. Our findings show for the first time that tenofovir can result in the perturbation of osteoblast gene expression and that these changes implicate defective osteoblast function leading to decreased bone formation that could result in reduced bone mineral density.
Supplementary Material
Acknowledgments
The following reagent was obtained through the NIH AIDS Research and Reference Reagent program, Division of AIDS, NIAID, NIH: Tenofovir disoproxil fumarate, Catalog Number 10198. We thank Andy Kaplan for stimulating discussions, and David Largaespada and Raha Allaei for assistance with mice. Supported by NIH grants AR53946 (to K.C.M.), DE16093 (to R.G.), and GM56615 (to L.M.M.). I.F.G. and L.P. were supported by T32DE07288.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arpadi SM, Horlick M, Thornton J, Cuff PA, Wang J, Kotler DP. Bone mineral content is lower in prepubertal HIV-infected children. J Acquir Immune Defic Syndr. 2002;29:450–4. doi: 10.1097/00126334-200204150-00004. [DOI] [PubMed] [Google Scholar]
- 2.Tebas P, Powderly WG, Claxton S, Marin D, Tantisiriwat W, Teitelbaum SL, Yarasheski KE. Accelerated bone mineral loss in HIV-infected patients receiving potent antiretroviral therapy. AIDS. 2000;14:F63–7. doi: 10.1097/00002030-200003100-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigano A, Mora S. Adverse effects of antiretroviral therapy: focus on bone density. Expert Opin Drug Saf. 2004;3:199–208. doi: 10.1517/14740338.3.3.199. [DOI] [PubMed] [Google Scholar]
- 4.Madeddu G, Spanu A, Solinas P, Calia GM, Lovigu C, Chessa F, Mannazzu M, Falchi A, Mura MS. Bone mass loss and vitamin D metabolism impairment in HIV patients receiving highly active antiretroviral therapy. Q J Nucl Med Mol Imaging. 2004;48:39–48. [PubMed] [Google Scholar]
- 5.Gallant JE, Staszewski S, Pozniak AL, DeJesus E, Suleiman JM, Miller MD, Coakley DF, Lu B, Toole JJ, Cheng AK. Efficacy and safety of tenofovir DF vs stavudine in combination therapy in antiretroviral-naive patients: a 3-year randomized trial. JAMA. 2004;292:191–201. doi: 10.1001/jama.292.2.191. [DOI] [PubMed] [Google Scholar]
- 6.Hazra R, Gafni RI, Maldarelli F, Balis FM, Tullio AN, DeCarlo E, Worrell CJ, Steinberg SM, Flaherty J, Yale K, Kearney BP, Zeichner SL. Tenofovir disoproxil fumarate and an optimized background regimen of antiretroviral agents as salvage therapy for pediatric HIV infection. Pediatrics. 2005;116:e846–54. doi: 10.1542/peds.2005-0975. [DOI] [PubMed] [Google Scholar]
- 7.Gafni RI, Hazra R, Reynolds JC, Maldarelli F, Tullio AN, DeCarlo E, Worrell CJ, Flaherty JF, Yale K, Kearney BP, Zeichner SL. Tenofovir disoproxil fumarate and an optimized background regimen of antiretroviral agents as salvage therapy: impact on bone mineral density in HIV-infected children. Pediatrics. 2006;118:e711–8. doi: 10.1542/peds.2005-2525. [DOI] [PubMed] [Google Scholar]
- 8.Purdy JB, Gafni RI, Reynolds JC, Zeichner S, Hazra R. Decreased bone mineral density with off-label use of tenofovir in children and adolescents infected with human immunodeficiency virus. J Pediatr. 2008;152:582–4. doi: 10.1016/j.jpeds.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo AB, Tarantal AF, Watnik MR, Martin RB. Tenofovir treatment at 30 mg/kg/day can inhibit cortical bone mineralization in growing rhesus monkeys (Macaca mulatta) J Orthop Res. 2002;20:1185–9. doi: 10.1016/S0736-0266(02)00074-8. [DOI] [PubMed] [Google Scholar]
- 10.Tarantal AF, Marthas ML, Shaw JP, Cundy K, Bischofberger N. Administration of 9-[2-(R)-(phosphonomethoxy)propyl]adenine (PMPA) to gravid and infant rhesus macaques (Macaca mulatta): safety and efficacy studies. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:323–33. doi: 10.1097/00042560-199904010-00001. [DOI] [PubMed] [Google Scholar]
- 11.Van Rompay KK, Brignolo LL, Meyer DJ, Jerome C, Tarara R, Spinner A, Hamilton M, Hirst LL, Bennett DR, Canfield DR, Dearman TG, Von Morgenland W, Allen PC, Valverde C, Castillo AB, Martin RB, Samii VF, Bendele R, Desjardins J, Marthas ML, Pedersen NC, Bischofberger N. Biological effects of short-term or prolonged administration of 9-[2-(phosphonomethoxy)propyl]adenine (tenofovir) to newborn and infant rhesus macaques. Antimicrob Agents Chemother. 2004;48:1469–87. doi: 10.1128/AAC.48.5.1469-1487.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis JE, Merriam A, Awadallah A, Yoo JU, Johnstone B, Caplan AI. A quadripotential mesenchymal progenitor cell isolated from the marrow of an adult mouse. J Bone Miner Res. 1999;14:700–9. doi: 10.1359/jbmr.1999.14.5.700. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi A, Katagiri T, Ikeda T, Wozney JM, Rosen V, Wang EA, Kahn AJ, Suda T, Yoshiki S. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J Cell Biol. 1991;113:681–7. doi: 10.1083/jcb.113.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzawa M, Takeuchi Y, Fukumoto S, Kato S, Ueno N, Miyazono K, Matsumoto T, Fujita T. Extracellular matrix-associated bone morphogenetic proteins are essential for differentiation of murine osteoblastic cells in vitro. Endocrinology. 1999;140:2125–33. doi: 10.1210/endo.140.5.6704. [DOI] [PubMed] [Google Scholar]
- 15.Erlebacher A, Filvaroff EH, Ye JQ, Derynck R. Osteoblastic responses to TGF-beta during bone remodeling. Mol Biol Cell. 1998;9:1903–18. doi: 10.1091/mbc.9.7.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrosetti D, Holmes G, Mansukhani A, Basilico C. Fibroblast growth factor signaling uses multiple mechanisms to inhibit Wnt-induced transcription in osteoblasts. Mol Cell Biol. 2008;28:4759–71. doi: 10.1128/MCB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCauley LK, Koh AJ, Beecher CA, Cui Y, Rosol TJ, Franceschi RT. PTH/PTHrP receptor is temporally regulated during osteoblast differentiation and is associated with collagen synthesis. J Cell Biochem. 1996;61:638–47. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C638::AID-JCB18%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 18.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–7. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 20.Cihlar T, Birkus G, Greenwalt DE, Hitchcock MJ. Tenofovir exhibits low cytotoxicity in various human cell types: comparison with other nucleoside reverse transcriptase inhibitors. Antiviral Res. 2002;54:37–45. doi: 10.1016/s0166-3542(01)00210-8. [DOI] [PubMed] [Google Scholar]
- 21.Barditch-Crovo P, Deeks SG, Collier A, Safrin S, Coakley DF, Miller M, Kearney BP, Coleman RL, Lamy PD, Kahn JO, McGowan I, Lietman PS. Phase i/ii trial of the pharmacokinetics, safety, and antiretroviral activity of tenofovir disoproxil fumarate in human immunodeficiency virus-infected adults. Antimicrob Agents Chemother. 2001;45:2733–9. doi: 10.1128/AAC.45.10.2733-2739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boffito M, Pozniak A, Kearney BP, Higgs C, Mathias A, Zhong L, Shah J. Lack of pharmacokinetic drug interaction between tenofovir disoproxil fumarate and nelfinavir mesylate. Antimicrob Agents Chemother. 2005;49:4386–9. doi: 10.1128/AAC.49.10.4386-4389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigsby IF, Pham L, Mansky LM, Gopalakrishnan R, Mansky KC. Tenofovir-associated bone density loss. Ther Clin Risk Manag. 2009 In press. [PMC free article] [PubMed] [Google Scholar]
- 24.Bodine PV, Stauffer B, Ponce-de-Leon H, Bhat RA, Mangine A, Seestaller-Wehr LM, Moran RA, Billiard J, Fukayama S, Komm BS, Pitts K, Krishnamurthy G, Gopalsamy A, Shi M, Kern JC, Commons TJ, Woodworth RP, Wilson MA, Welmaker GS, Trybulski EJ, Moore WJ. A small molecule inhibitor of the Wnt antagonist secreted frizzled-related protein-1 stimulates bone formation. Bone. 2009;44:1063–8. doi: 10.1016/j.bone.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Grigsby IF, Pham L, Gopalakrishnan R, Mansky LM, Mansky KC. Downregulation of Gnas, Got2 and Snord32a following tenofovir exposure of primary osteoclasts. Biochem Biophys Res Comm. 2010 doi: 10.1016/j.bbrc.2009.12.039. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


