SUMMARY
The Igf2r imprinted cluster is an epigenetic silencing model in which expression of a ncRNA silences multiple genes in cis. Here, we map a 250 kb region in mouse embryonic fibroblast cells to show that histone modifications associated with expressed and silent genes are mutually exclusive and localized to discrete regions. Expressed genes were modified at promoter regions by H3K4me3 + H3K4me2 + H3K9Ac and on putative regulatory elements flanking active promoters by H3K4me2 + H3K9Ac. Silent genes showed two types of nonoverlapping profile. One type spread over large domains of tissue-specific silent genes and contained H3K27me3 alone. A second type formed localized foci on silent imprinted gene promoters and a nonexpressed pseudogene and contained H3K9me3 + H4K20me3 ± HP1. Thus, mammalian chromosome arms contain active chromatin interspersed with repressive chromatin resembling the type of heterochromatin previously considered a feature of centromeres, telomeres, and the inactive X chromosome.
INTRODUCTION
Genomic imprinting is a cis-acting epigenetic mechanism that results in identical gene sequences being treated differently by the transcription machinery simply because of their inheritance from a maternal or paternal parent. Imprinted expression is restricted to a few hundred genes in the mammalian genome, most of which are found in small clusters. Imprinting of a gene cluster is regulated by an imprint control element that is inactivated on one parental chromosome by a DNA methylation imprint (Solter, 2006). The majority of the genes in an imprinted cluster are mRNA genes; however, at least one is always a noncoding RNA (ncRNA). The function of the ncRNA has been tested in three imprinted clusters. In the Igf2 cluster, the H19 ncRNA has no direct silencing role; instead, silencing operates via an insulator mechanism (Lewis and Reik, 2006). However, in the Igf2r and Kcnq1 imprinted clusters, the Air and Kcnq1ot1 ncRNAs have both been shown to play a direct silencing role (Mancini-Dinardo et al., 2006; Sleutels et al., 2002).
In the two imprinted clusters with functional ncRNAs, one chromosome expresses the ncRNA and silences flanking clustered mRNA genes, while the other chromosome shows the reciprocal expression pattern. Moreover, in these two cases the expressed ncRNA promoter is located in an intron of one of the silenced genes in the cluster. This type of expression pattern, in which closely spaced genes are silent or expressed, indicates that repressive chromatin would not spread throughout an imprinted cluster. Historically, chromatin has been classified by its appearance in interphase nuclei stained with basic dyes as either heterochromatin or euchromatin, which are considered to represent, respectively, the repressed and active regions of the genome (Huisinga et al., 2006). In mammals, heterochromatin-associated histone modifications have only been characterized at pericentric and telomeric regions and the inactive X chromosome (Chadwick and Willard, 2004; Garcia-Cao et al., 2004; Schotta et al., 2004). This has identified H3K9me3, H4K20me3, H3K27me1, and HP1 proteins as markers of pericentric and telomeric regions and two distinct nonoverlapping patterns comprising either H3K9me3 + H4K20me3 + HP1 or Xist ncRNA + macroH2A + H3K27me3 on the inactive X chromosome. It is not yet known if short genomic regions containing silent genes that lack visible staining by basic dyes can also be defined as heterochromatin or if they represent another form of repressed chromatin. Imprinted clusters represent an ideal model system to investigate this, as known imprinted clusters are located in short domains spanning 100–3000 kb and, in addition to imprinted genes, are interspersed with genes that show biparental expression or tissue-specific silencing (http://www.mgu.har.mrc.ac.uk/research/imprinting/imprin-intro.html).
The mouse Igf2r imprinted gene cluster spans 500 kb (Figure 1) and contains the paternally expressed Air ncRNA and the maternally expressed Igf2r mRNA that show widespread imprinted expression in fetal, placental, and adult tissue, plus the maternally expressed Slc22a2 and Slc22a3 mRNAs that are not expressed in fetal tissue but show imprinted expression in placenta (Regha et al., 2006). The Air promoter is located in Igf2r intron 2 and generates an antisense transcript across the 5′ part of the Igf2r gene. Expression of the Air ncRNA has been shown to silence Igf2r, Slc22a2, and Slc22a3 on the paternal chromosome (Sleutels et al., 2002). Surprisingly, however, transcriptional overlap between Air and Igf2r is not necessary for silencing flanking genes in the cluster (Sleutels et al., 2003). A second type of silencing is present on the maternal chromosome, which lacks Air expression because the Air promoter is directly repressed by a DNA methylation imprint acquired during female gametogenesis (Seidl et al., 2006). A methylation imprint acquired in late development that is a consequence, not a cause, of silencing is also found on the silenced paternal Igf2r promoter. However, widespread differential DNA methylation does not mark the three genes silenced by the Air ncRNA (Stoger et al., 1993; Zwart et al., 2001). Allele-specific histone lysine methylation marks have also been identified on the silent and active Air and Igf2r promoters; however, their distribution throughout the region is not known (Fournier et al., 2002; Vu et al., 2004).
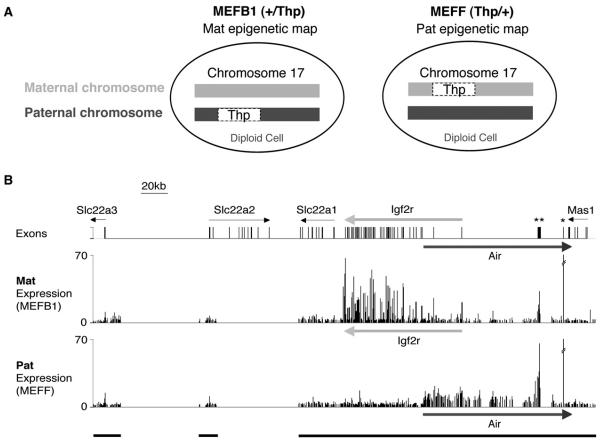
Figure 1. Expression Profile of the Igf2r Imprinted Cluster.
(A) Mouse embryo fibroblast cells (MEFB1 and MEFF) used to detect parental-specific expression and epigenetic modifications were prepared from 13.5 dpc +/Thp and Thp/+ embryos. +, wild-type chromosome 17. Thp, T hairpin chromosome 17 variant contains a 6 Mbp deletion including the 500 kb Igf2r imprinted cluster. The maternal chromosome is written on the left side.
(B) Top row, 380 kb map showing the Slc22a3, Slc22a2, Slc22a1, Igf2r, and Mas1 genes. The Air promoter lies in Igf2r intron 2. Air is a 108 kb single-exon ncRNA that overlaps Igf2r and Mas1 in antisense orientation. Bars, exons or pseudogenes. Thin arrows, silent genes in MEF cells. Thick arrows, maternally expressed gene (light gray), paternally expressed gene (dark gray). *L41 pseudogene, **Au76 pseudogene. The Mat Expression (MEFB1) and Pat Expression (MEFF) rows show cDNA hybridization to a custom PCR tiling array spanning three regions: 19 kb containing the Slc22a3 promoter, 13 kb containing the Slc22a2 promoter, and 220 kb from Slc22a1-3′ end to Mas1-3′ end (indicated by black bars underneath). Each column is the mean ratio of medians of eight replicate spots.
We use here chromatin immunoprecipitation (ChIP) interrogated by PCR or genome tiling arrays (ChIP-Chip) to map the chromatin profile of the Igf2r cluster in mouse embryonic fibroblast (MEF) cells. Surprisingly, silencing of the paternal Igf2r promoter by the Air ncRNA, and silencing of the maternal Air promoter by DNA methylation, does not involve spreading of repressive chromatin. Instead, we show that these silent imprinted promoters and a nonexpressed pseudogene are marked by focal repressive histone modifications resembling those identified at centromeres and telomeres. Furthermore, as these heterochromatic foci are embedded inside active transcription units, they do not block transcription elongation. In contrast, a cluster of flanking tissue-specific silenced genes was modified by a broad domain marked by one type of repressive histone modification. These data show that mammalian chromosome arms contain discrete regions of different types of repressive chromatin previously considered a feature of centromeres, telomeres, and the inactive X chromosome.
RESULTS
Expression Profile of the Igf2r Imprinted Cluster in Thp MEF Cells
We used here MEF cells carrying either a paternal (MEFB1, +/Thp) or maternal (MEFF, Thp/+) Thp deletion, which includes the 500 kb Igf2r imprinted cluster (Figure 1A). Expression of genes in this region in MEFs was determined by cDNA hybridization to a custom PCR tiling array. Figure 1B shows that only Igf2r is expressed from the maternal allele (middle line) and only Air from the paternal allele (bottom line). Igf2r signals correspond to the 48 gene exons while Air ncRNA signals were detected on all PCR fragments throughout the length of unspliced 108 kb Air transcript. Note that the Air promoter lies in Igf2r intron 2, and thus on both parental chromosomes a silent promoter is overlapped by an expressed transcript. Expression of the remaining four tissue-specific silenced genes in this region (Slc22a3, Slc22a2, Slc22a1, and Mas1) was not detected from either chromosome in MEF cells. Hybridization signals were detected on both parental chromosomes at the Au76 and L41 pseudogenes (psg) contained within the Air gene body that represent crosshybridization from expressed genomic copies (Figure 1B, see Figure S1 in the Supplemental Data available with this article online). In MEFB1 and MEFF cells, the DNA methylation status of the Igf2r, Air, Slc22a2, and Slc22a3 promoters was as previously described (Zwart et al., 2001), and Au76-psg was equally methylated on both parental alleles (Figure S1, Table S3).
These MEF cells represent a homogenous population of differentiated cells that allows examination of parental-specific chromatin profiles of three categories of silenced genes: (1) DNA methylation silencing of the maternal Air promoter (Seidl et al., 2006), (2) Air ncRNA silencing of the paternal Igf2r promoter (Sleutels et al., 2002), and (3) tissue-specific biparental silencing of the flanking Slc22a1, a2, and a3 genes (Zwart et al., 2001).
Chromatin Modifications at the Maternal Air Promoter Silenced by DNA Methylation
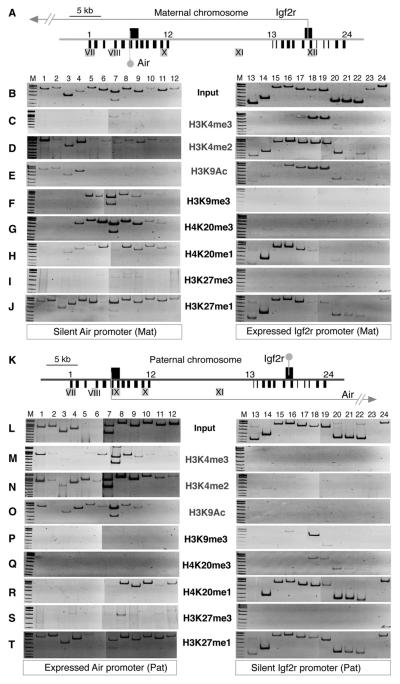
ChIP-PCR was used to test the expressed Igf2r and silent Air promoter regions on the maternal chromosome for active (Figures 2C–2E) and repressive (Figures 2F–2J) histone modifications. Immunoprecipitated ChIP and Input DNA were used as templates for PCR with 24 primer pairs spanning 12 kb around the Air and Igf2r transcription starts at approximately 1 kb intervals. Note that the silent Air promoter is overlapped by transcription from the expressed Igf2r promoter 28 kb away and that four DNaseI hypersensitive (DHS) sites lie in these two regions, of which only DHSXII is maternal specific (Pauler et al., 2005).
Figure 2. Parental-Specific Histone Modifications.
(A–J) Maternal chromosome.
(A) Map (40 kb) showing maternal expression of Igf2r and Air. Arrow, Igf2r transcription; gray lollipop, silent Air transcription start 28 kb downstream of Igf2r. Black boxes above line, CpG islands at the Igf2r and Air promoters. Black bars below line, single-copy regions analyzed by ChIP-PCR. 1–12, primer pairs to assay 11.6 kb of the Air promoter region. 13–24, primer pairs to assay 12.7 kb of the Igf2r promoter region (Table S4, see note on primer pair 7). Gray boxes with roman numerals, DHS sites. DHSIX is absent from the maternal chromosome, DHSXII is unique to the maternal chromosome. (B) Typical Input-PCR in MEFB1 cells containing only the maternal Igf2r cluster.
(C–E) ChIP-PCR using antibodies against H3K4me3, H3K4me2, and H3K9Ac in MEFB1 cells. For H3K4me3, weak reproducible bands were amplified by primer 7–8 downstream of the Air transcription start marked by primer pair 6.
(F–J) ChIP-PCR using antibodies against H3K9me3, H4K20me3, H4K20me1, H3K27me3, and H3K27me1 in MEFB1 cells. One microliter of 1:100 dilution of Input or 1 μl ChIP DNA was amplified per PCR, two to four biological replicas of each ChIP-PCR were performed, and representative images are shown. M, size marker.
(K–T) Paternal chromosome.
(K) Map (40 kb) showing paternal expression of Igf2r and Air. Details in Figure 2A. Arrow, Air transcription; gray lollipop, silent Igf2r transcription start 28 kb downstream of Air. DHSIX is unique to the paternal chromosome, DHSXII is absent from the paternal chromosome.
(L) Typical Input-PCR in MEFF cells containing only the paternal Igf2r cluster. Primer pairs 5 and 6 corresponding to DHSIX generally showed reduced Input signal on the paternal chromosome.
(M–O) ChIP-PCR in MEFF cells using antibodies against H3K4me3, H3K4me2, H3K9Ac.
(P–T) ChIP-PCR in MEFF cells using antibodies against H3K9me3, H4K20me3, H4K20me1, H3K27me3, H3K27me1.
Figures 2C–2E(right panels) show that the expressed Igf2r promoter region is enriched for H3K4me3, H3K4me2, and H3K9Ac. The downstream silent Air promoter region (left panels) was generally depleted for these modifications except for primer pairs 1–4 linked to DHSVII and VIII. On the expressed Igf2r promoter, H3K4me3 showed a focal distribution being most strongly enriched on primers that flank the transcription start. H3K4me2 showed a broader asymmetric distribution being enriched over the transcription start and in the direction of Igf2r transcription. H3K9Ac modifications resembled those of H3K4me2. ChIP-PCR using antibodies to multiple acetylation sites confirmed depletion of H3K9 and H3K14 acetylation on the silent Air promoter and enrichment on DHS sites. However, abundant H4 (K5 + K8 + K12 + K16) acetylation was detected throughout the silent Air and expressed Igf2r promoter regions (Figure S2).
Figures 2F and 2G (left panels) show strong H3K9me3 and H4K20 me3 (spanning 4–6 kb) at the silent Air promoter that were absent from the expressed Igf2r promoter (right panels). H4K20me1 and H3K27me1 were generally present throughout the silent Air promoter region and also found downstream, but not upstream of the expressed Igf2r transcription start (Figures 2H and 2J). H3K27me3 (Figure 2I), H3K27me2, H4K20me2, H3K9me1, and K3K9me2 (Figure S3) were absent from both the silent Air and the expressed Igf2r promoter regions. These data show the Air promoter (silenced by DNA methylation) and the expressed Igf2r promoter and are marked by focal repressive and active histone modifications that show limited spreading.
Chromatin Modifications at the Paternal Igf2r Promoter Silenced by the Air ncRNA
Figures 2K–2T show a parallel analysis of Air ncRNA-mediated silencing of the Igf2r promoter, which shows a similar pattern of focal modifications. On the paternal chromosome, the expressed Air promoter (left panel, Figures 2M–2O) is modified by focal H3K4me3, H3K4me2, and H3K9Ac, while the downstream silent Igf2r promoter (right panel) lacks these modifications. H3K4me2 modifications spread further in the direction of Air transcription but do not reach the silent Igf2r promoter. H3K9Ac was specific to the expressed Air promoter region; however, antibodies to multiple acetylation sites showed more widespread distribution of H3 and H4 acetylation throughout both silent and expressed promoter regions on the paternal chromosome (Figure S2). The modifications on nonpromoter DHS on the paternal chromosome were similar to that described above for the maternal chromosome. DHSX was modified by H3K4me2 alone, and DHSVII and VIII (primer pairs 1–4) were modified by H3K4me2 + H3K9Ac. DHSVII was additionally modified on the paternal chromosome by H3K4me3 (Figure 2M, primer pair 1). All DHS lacked specific repressive modifications, but some were covered by H4K20me1 and H3K27me1 (Figures 2R and 2T).
Figures 2P and 2Q show that H3K9me3 and H4K20me3 modifications were present on the silent Igf2r promoter, albeit to a lesser extent than at the silent Air promoter, but were absent from the expressed Air promoter. Similar to the maternal chromosome, H4K20me1 and H3K27me1 were found at both active and silent promoters (Figures 2R and 2T); however, only H4K20me1 was depleted upstream of the active Air promoter on the paternal chromosome. H3K27me3 (Figure 2S), H3K27me2, H4K20me2, H3K9me1, and H3K9me2 (Figure S3) were absent from both the expressed Air and silent Igf2r promoter regions on the paternal chromosome.
Quantitative PCR of H3K4me3, H3K4me2, and H3K9me3 modifications at the active and silent Air promoters supports the interpretation that expressed and silent promoters show mutually exclusive active and repressive histone modifications (Table S1). The enrichment of the expressed/silent Air promoter at primer pairs 1, 4, 5/6, and 6 is, respectively, 5.4, 0.8, 11.3, 234.2 (for H3K4me3), and 3.0, 54.5, 59.7, and 330.8 (for H3K4me2 for primer pairs 4, 5/6, 6, and 7). H3K9me3 at primer pairs 5/6 and 6 shows, respectively, 11.8- and 17.5-fold enrichment of the silent/expressed Air promoters.
Focal Active and Repressive Histone Modifications Intersperse without Spreading
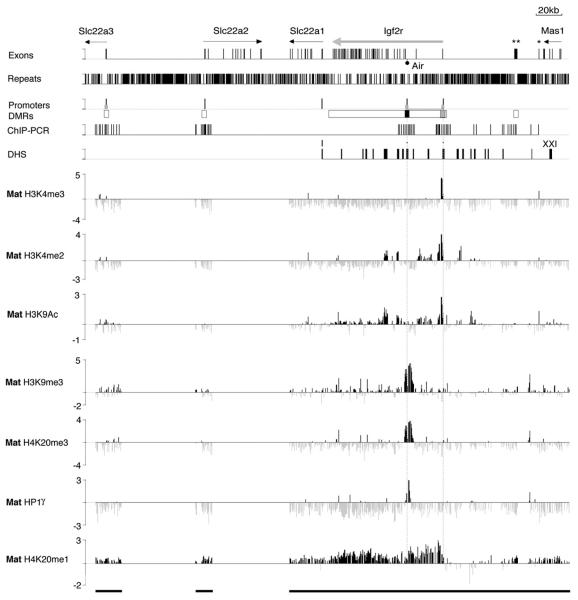
It is notable from the ChIP-PCR of 12 kb at the Air and Igf2r promoter regions that the tested histone modifications do not spread through the body of the expressed or silent gene. To further examine the distribution of active and repressive histone modifications within the Igf2r cluster, we performed ChIP-Chip using the same ChIP/Input samples analyzed in Figures 2 and 3 and the 250 kb custom PCR tiling array shown in Figure 1B. Figure 3 and Figure S4 show a ChIP-Chip analysis of three active (H3K4me3, H3K4me2, and H3K9Ac) and four repressive (H3K9me3, H4K20me3, H4K20me1, and HP1γ) modifications on the maternal chromosome in which Air is silenced by DNA methylation. H3K4me3 was detectable as a single peak at the expressed Igf2r promoter but was absent from the remainder of the tiling array that included silent CpG island promoters (e.g., Slc22a3) and silent CG poor promoters (e.g., Slc22a2, Slc22a1). H3K4me2 showed a broader peak at the expressed Igf2r promoter and was also absent from the four silent promoters on the tiling array. H3K4me2 modifications did not show widespread spreading but were found outside promoter regions including a large domain in Igf2r intron 1. H3K9Ac was detected with a profile broadly similar to H3K4me2. ChIP-Chip with antibodies recognizing multiple acetylation sites (H3K9+K14 or H4K5+K8+K12+K16) confirmed the general pattern for H3 acetylation, but H4 acetylation was more widespread on silent and expressed genes (Figure S5). ChIP-Chip also showed that H3K9me3, H4K20me3, and HP1γ were all localized to the silent Air promoter but were absent from promoters of the four tissue-specific silent genes in the cluster. H4K20me1, in contrast, was generally present throughout the 250 kb tiling array on silent and expressed genes with the exception of 20 kb immediately upstream of the expressed Igf2r promoter. This 20 kb depleted region coincides with four DHS modified by H3K4me2 and H3K9Ac. The results in Figure 3 clearly illustrate that active and repressive modifications show limited spreading and are interspersed in the Igf2r/Air region.
Figure 3. Focal Heterochromatin on the Silent Air Promoter.
ChIP-Chip profile of active (H3K4me3, H3K4me2, H3K9Ac) and repressive (H3K9me3, H4K20me3, and H4K20me1) histone modifications and the heterochromatin protein HP1γ on the maternal (Mat) Igf2r imprinted cluster in MEFB1 cells on the custom PCR tiling array shown in Figure 1B. X axis, 380 kb containing the Igf2r imprinted gene cluster that lacked two regions of 58 kb and 61 kb (gaps in the black bars underneath). Exon track, see Figure 1B. Repeats track, bars show total interspersed repeats (simple repeats, MIR, SINE, LINE1, LINE2, LTRs). Promoters track, bars represent transcription start sites (black) and CpG islands (gray). DMRs track, black and gray boxes; respectively, maternal- and paternal-specific DNA methylated regions; white boxes, regions showing no parental differences in DNA methylation (Stoger et al., 1993). ChIP-PCR track, bars represent 61 sites analyzed by PCR around the Slc22a3, Slc22a2, Igf2r, and Air promoters and Au76-psg. DHS track, bars indicate 21 mapped DHS sites (Pauler et al., 2005). In the seven tracks underneath, each PCR product on the custom tiling array is represented as a single column that is the log2 signal ratio (ChIP/ Input) of the average ratio of medians of eight replicate spots. Vertical dotted lines passing through all tracks mark Air and Igf2r transcription start sites. Only peaks reproducible in biological replicas (Figure S4) were considered positive.
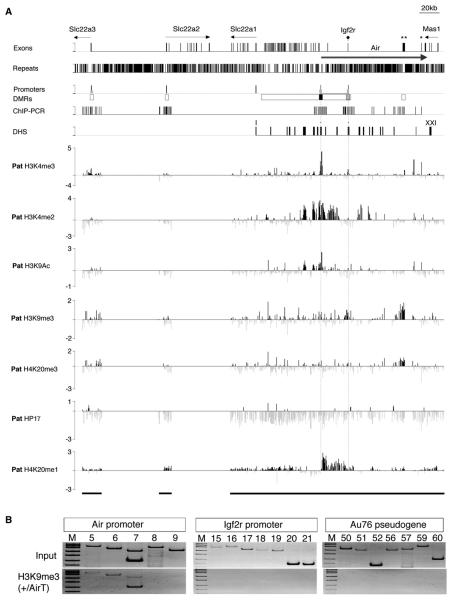
Figure 4A and Figure S6 show a ChIP-Chip analysis of the same seven modifications on the paternal chromosome on which the Igf2r promoter is silenced by the Air ncRNA. Active and repressive modifications on the paternal chromosome show the same interspersed, nonspreading patterns as described above for the maternal chromosome. Thus, on the paternal chromosome, H3K4me3 marks are found only on the expressed Air promoter and the L41-psg, while H3K4me2 and H3K9Ac were found on the expressed Air promoter and also on flanking discrete regions. H3K4me2 was also found extensively in Igf2r intron 1 that was also marked on the maternal chromosome expressing Igf2r (Figure 3). The silent Igf2r promoter was modified by a small but reproducible peak of H3K9me3. A peak of H4K20me3 was not detected on the silent Igf2r promoter despite positive signals in ChIP-PCR assays in Figure 2Q. This likely reflects different sensitivities of these techniques. HP1γ was also not detected. H4K20me1 showed a widespread distribution with a depletion spanning 20 kb upstream of the expressed Air promoter coincident with a region containing DHS modified by H3K4me2 and H3K9Ac.
Figure 4. Focal Heterochromatin on the Silent Igf2r Promoter.
(A) ChIP-Chip profile of active and repressive epigenetic marks on the paternal (Pat) Igf2r imprinted cluster in MEFF cells. Details as in Figure 3. Only peaks reproducible in biological replicas (Figure S6) were considered positive.
(B) Chip-PCR of the Air and Igf2r promoters and the Au76-psg in cells carrying an Air ncRNA truncated from 108 to 3 kb on the paternal allele (Sleutels et al., 2002). This shows a loss of repressive H3K9me3 on Igf2r and Au76-psg when Air is truncated to 3 kb. H4K20me3 was also lost on these two elements (data not shown).
The paternal chromosome also displayed a broad peak of H3K9me3 and H4K20me3 covering the Au76-psg that is contained within the Air transcriptional unit but not expressed in MEF cells (Figure S1). This H3K9me3/H4K20me3 peak is not associated with paternal-specific DNA methylation and is found only in differentiated cells and thus is not a germline imprint (Table S3, Figure S7). Figure 4B shows that the paternal-specific H3K9me3 (and H4K20me3, data not shown) signals are lost in cells that contain a 3 kb truncation of the Air ncRNA that fails to silence the Igf2r promoter (Sleutels et al., 2002).
Of the 21 DHS sites in the profiled region, 11 DHS (VI, VII, VIII, IX, X, XI, XII, XIII, XIV, XV, and XVI) that lie within 20 kb of the expressed Igf2r and Air promoters showed H3K4me2 modifications (DHS sites are positioned with a resolution of ± 0.5–1.0 kb [Pauler et al., 2005]). In contrast, ten DHS (I, II, III, IV, V, XVII, XVIII, XIX, XX, and XXI) located more than 20 kb away from active promoters lacked H3K4me2 (Figures 3 and 4, Figures S4–S6). Three DHS, VII (6.5 kb upstream to the Air promoter), IX (Air transcription start), and XII (Igf2r transcriptional start), were modified by H3K4me3 + H3K4me2 + H3K9Ac. The remaining modified DHS (VI, VIII, X, XI, XIII, XIV, XV, and XVI) were marked either by H3K4me2 + H3K9Ac or by H3K4me2 and H3K9Ac alone. In two cases, the histone modification appeared to span the region between the neighboring DHS (X + XI spanning 13 kb and XIII+XIV spanning 5.2 kb). Paternal-specific histone modifications were present at DHSIX (Air transcription start) and at DHSVII, VIII, XV, and XVI. Maternal-specific histone modifications were found at DHSXII (Igf2r transcription start) and at DHSXV and XVI (Table S2).
The Silent Air and Igf2r Promoter Regions Resemble Heterochromatin
The above Chip-Chip analysis identified three H3K9me3/H4K20me3 peaks; on the silent maternal Air promoter, the silent paternal Igf2r promoter and the paternal copy of the nonexpressed Au76-psg. However, HP1γ was only found on the silent Air promoter. Since HP1 is a classic heterochromatin feature, we tested more extensively for the presence of other HP1 isoforms at these regions. Figure S8 shows that the silent maternal Air promoter is exclusively marked by HP1α, -β, and -γ. The Igf2r promoter lacked HP1α and carried background levels of HP1γ on both silent and active promoters. HP1β, however, exclusively marked the silent Igf2r promoter. The paternal copy of the Au76-psg lacks specific HP1 binding, demonstrating that H4K20me3 can arise independently of HP1 at this region.
H3K9me3 has been shown to be required for induction of H4K20me3 in pericentric DNA (Schotta et al., 2004). We therefore tested H3K9me3 stability in MEF cells lacking the SUV4-20H methylases. Figure S9 shows that, in these cells, H4K20me3 is reduced, but not lost, over the silent Air promoter but is lost over the Au76-psg. However, in both cases, H3K9me3 is unaffected. Thus, H3K9me3 modifications are independent of H4K20me3.
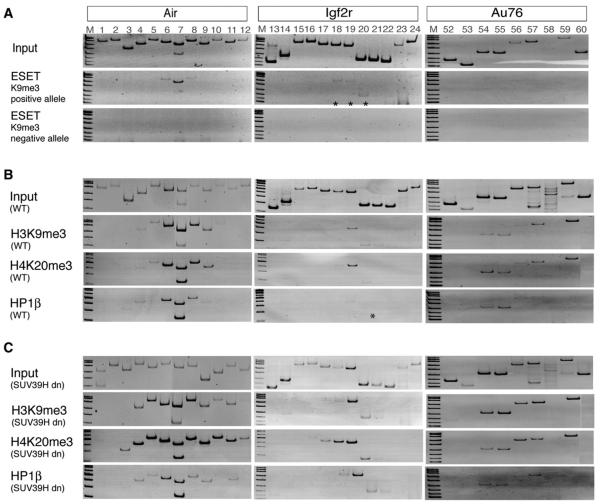
To identify which histone methylase mediates H3K9me3 modifications at the silent Air and Igf2r promoter regions, we performed ChIP-PCR with antibodies to the ESET/SETDB1 H3K9 trimethylase. The silent Air promoter showed ESET signals distributed over three primer pairs; ESET signals were weakly present over a similar sized region on the silent Igf2r promoter and absent from the Au76-psg (Figure 5A). The specificity of the antibody was confirmed by showing absence of a signal on pericentric DNA that carries a SUV39H-dependent H3K9me3 mark (data not shown). To test if the SUV39H methylases contribute to H3K9me3 on Air, Igf2r, and Au76, we examined MEFs lacking these enzymes (Rea et al., 2000). Figures 5B and 5C show that H3K9me3 is retained at the silent Air and Igf2r promoters and at the Au76-psg in cells lacking SUV39H methylases that lack pericentric H3K9me3 (data not shown). Moreover, H3K9me3 plus HP1β and H4K20me3 all increase in SUV39H null cells at the silent Air and Igf2r promoter. This increase was minimal or absent at the Au76-psg.
Figure 5. The Histone H3K9 Trimethylase ESET/SETDB1 Binds the Air and Igf2r HC Peaks.
(A) ESET ChIP-PCR at maternal and paternal alleles of the Air promoter (left), Igf2r promoter (middle), and Au76-psg (right), showing that ESET is present at Air and Igf2r alleles marked by H3K9me3. ESET is absent from Au76-psg and pericentric DNA (data not shown) assayed from the same ChIP preparation.
(B and C) (B) ChIP-PCR of the Air and Igf2r promoters and Au76-psg in wild-type control and (C) SUV39H null (SUV39H dn) diploid MEF cells (Peters et al., 2001) using antibodies for H3K9me3, H4K20me3, and HP1β. Increased signals were seen for HC peaks at the Air and Igf2r promoters, but only minimal changes were seen at Au76-psg. SUV39H dn MEFs maintain DNA methylation over the silent Air and Igf2r promoter regions (data not shown). H3K9me3 is absent from pericentric DNA assayed from the same SUV39H dn ChIP preparation (data not shown) as described (Schotta et al., 2004). *, weak reproducible signals.
H3K27me3 Spreads over Large Regions Containing Tissue-Specific Silenced Genes in MEF Cells
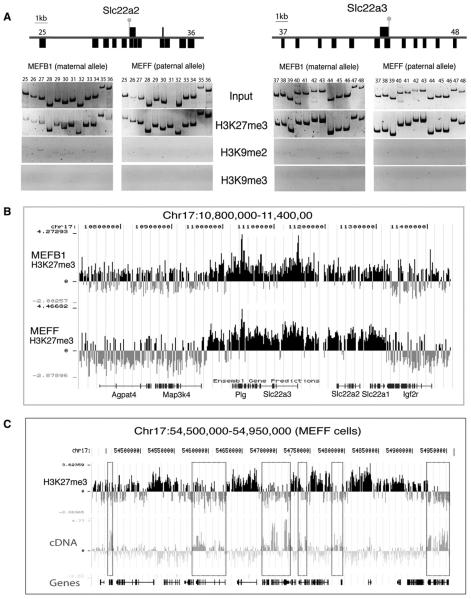
The silent Air and Igf2r genes lack H3K27me3 as determined by ChIP-PCR (Figure 2) and ChIP-Chip (data not shown). However, H3K27me3 (but not H3K9me2 or H3K9me3) was broadly distributed over the promoter regions of the nonexpressed Slc22a2 and Slc22a3 genes on both parental chromosomes (Figure 6A). As the custom PCR tiling array lacked complete coverage of the Slc22a2 and Slc22a3 genes, we hybridized the same H3K27me3 ChIP/Input samples to a NimbleGen oligonucleotide tiling array. This identified a 300 kb H3K27me3 domain on both the maternal and paternal chromosome that included four genes (Plg, Slc22a3, Slc22a2, and Slc22a1) and intergenic regions (Figure 6B). The H3K27me3 domain appeared to have abrupt borders marked by the 5′ end of Map3k4 and the 3′ end of Igf2r. The four genes inside the H3K27me3 domain are repressed in MEF cells, while genes outside this domain are expressed in MEFs (data not shown). Similar H3K27me3 modifications that negatively correlate with gene expression were found on nonimprinted genes lying 43 Mbp distant from Igf2r on chromosome 17 (Figure 6C).
Figure 6. H3K27me3 Modifies Large Domains in MEF Cells.
(A) ChIP-PCR showing the distribution of H3K27me3, H3K9me2, and H3K9me3 in a 13 kb and 19 kb region containing the Slc22a2 (primer pairs 25–36) and Slc22a3 (primer pairs 37–48) promoters on the maternal and paternal allele in MEFB1 and MEFF cells. Details as in Figure 2A. Slc22a2 and Slc22a3 are silent on both parental chromosomes in MEF cells (gray lollipops).
(B and C) NimbleGen mouse chromosome 17 tiling arrays with one 50-mer per 100 bp of single-copy sequence showing the distribution of H3K27me3 over 600 kb containing Igf2r on the maternal (MEFB1) and paternal (MEFF) chromosome and a 450 kb region containing nonimprinted genes in MEFF cells. A transcript profile prepared from hybridizing total cell RNA to the tiling array shows that only some of the genes in this 450 kb region are expressed. Expressed genes are marked by dotted boxes that show expression correlates with H3K27me3 depletion. Nonexpressed genes in this region are marked by widespread H3K27me3, which covers the entire transcription unit and intergenic regions. Bars indicate the log2 signal ratio (ChIP/Input enrichment) of each oligonucleotide on the array. Ensembl genes (http://genome.ucsc.edu) are shown below each array.
DISCUSSION
We have used an imprinted cluster as a model system to examine the type of repressive chromatin associated with silent genes overlapped by antisense transcripts and flanking genes showing tissue-specific silencing. Imprinted genes and a nonexpressed pseudogene overlapped by antisense transcripts were modified by a pattern of focal active chromatin interspersed with focal repressive chromatin that, although resembling pericentric heterochromatin, does not block transcription elongation. However, neighboring groups of tissue-specific silenced genes are contained in a widespread repressed domain devoid of active chromatin marks.
Discrete Foci of Repressive Chromatin Show Limited Spreading
Discrete foci of repressive chromatin were identified on three elements, the silent Air promoter, the silent Igf2r promoter, and the nonexpressed Au76-psg (Table S3). Although the Air and Igf2r promoters are silenced by different initial events, DNA methylation and Air ncRNA expression, respectively (Seidl et al., 2006; Sleutels et al., 2002), both are modified by similar combinatorial marks of H3K9me3, HP1, and H4K20me3, while only H3K9me3 and H4K20me3 modify the nonexpressed Au76-psg. As these modifications are typical of heterochromatin at pericentric and telomeric regions and the inactive X chromosome (Chadwick and Willard, 2004; Garcia-Cao et al., 2004; Schotta et al., 2004), we have named them heterochromatin (HC) peaks. Self-propagation or spreading is considered to be a feature of heterochromatin (Grewal and Jia, 2007). In mammals, the model of heterochromatin spreading at centromeres is based on SUV39H methylase inducing H3K9me3 to generate a binding site for HP1 proteins, which then recruit more SUV39H and also SUV4-20H that induces H4K20me3 (Schotta et al., 2004). While on the mammalian inactive X chromosome, the model is based on initial spreading of the Xist ncRNA that recruits Polycomb group proteins to induce H3K27me3 and H2A ubiquitylation (Chadwick and Willard, 2004). The HC peaks on the silenced Air and Igf2r promoters and Au76-psg are, however, limited to 2–6 kb regions and do not spread through the body of the silent gene. Similar short peaks of H3K9me3 are also present upstream of the imprinted Gtl2 ncRNA, indicating they can occur in other genomic regions (Figure S10). Nonspreading repressive modifications containing H3K9me3 and HP1 limited to a few nucleosomes have also been described for retinoblastoma-mediated and hormone-mediated gene silencing (Ayyanathan et al., 2003; Nielsen et al., 2001). Together, this shows that repressive modifications can be targeted to specific short domains in imprinted and nonimprinted genes by mechanisms that limit spreading.
Similarities between HC Peaks and Pericentric Heterochromatin
Pericentric heterochromatin has been shown to depend on H3K9me3 for subsequent H4K20me3 modification in a pathway suggested to use HP1 to recruit SUV4-20H methylases (Schotta et al., 2004). The H4K20me3 mark in the HC peaks identified here was similarly shown to be dependent on SUV4-20H enzymes. However, HP1 was only associated with HC peaks at Air and Igf2r, but not at the HC peak on the Au76-psg. Thus, H4K20me3 can occur independently of HP1. In addition, reduced H4K20me3 modifications remain on the silent Air promoter in the absence of SUV4-20H methylases, indicating the action of other uncharacterized H4K20 trimethylases. It is notable that H3K9me3 was unchanged at HC peaks in SUV4-20H null cells. This indicates that, similar to repressive pathways at pericentric heterochromatin, H3K9me3 modifications occur prior to SUV4-20H induction of H4K20me3.
The initial event at pericentric heterochromatin is SUV39H-mediated induction of H3K9me3. We have shown that HC peaks at the silent Air and Igf2r promoters, but not that at the Au76-psg, bind ESET, an H3K9 trimethylase also known as SETDB1 that is associated with euchromatic regions (Wang et al., 2003). All three HC peaks retain and even intensify H3K9me3 modifications in cells lacking the SUV39H methylases. Thus, ESET is most likely responsible for H3K9me3 at Air and Igf2r and neither ESET nor SUV39H act on the Au76-psg peak. The absence of viable cells lacking ESET (Dodge et al., 2004) and of antibodies suitable for SUV39H ChIP (Table S4) does not allow us to identify ESET as the sole responsible enzyme for H3K9me3 at HC peaks.
Mutually Exclusive Active and Repressed Chromatin States
In MEF cells, the expressed Igf2r and Air promoters are marked by peaks of H3K4me3 + H3K4me2 + H3K9Ac, while silent promoters lack these modifications. In contrast, silent promoters were marked by peaks of H3K9me3 + H4K20me3 ± HP1 or by broad domains of H3K27me3, while expressed promoters lack these modifications. Previous analyses identified active and repressive histone modifications on the Air and Igf2r promoters that were not mutually exclusive but enriched on one parental allele compared to the other (Fournier et al., 2002; Vu et al., 2004). The reason for this difference is not clear but could arise from mixtures of cell types, only some of which express or show imprinted expression of the tested gene (e.g., Igf2r in mouse brain is imprinted in glial cells but not in neurons [Yamasaki et al., 2005]). In addition to the Air and Igf2r promoters, the ChIP-Chip tiling array contains 19 DHS that are present on both parental chromosomes and may be shared by both promoters (Pauler et al., 2005). It is notable that all these DHS lacked specific repressive marks and only DHS within 20 kb of the expressed Igf2r and Air promoters showed active modifications, either by H3K4me2 + H3K9Ac or by H3K4me2 or H3K9Ac alone (Table S2). We do not yet have an explanation for this pattern of DHS-associated active histone modifications. It is possible that only DHS sites marked by active histone modifications are involved in activation of Igf2r and Air promoters in MEF cells, but this interpretation needs to be tested further. ChIP-Chip tiling array studies in the human and mouse genome have previously shown that focal active modifications composed of H3K4me1, H3K4me2, and H3 acetylation, but not H3K4me3, mark putative regulatory elements of nonimprinted expressed genes (Heintzman et al., 2007; Bernstein et al., 2005). However, these studies did not examine repressive modifications or characterize DHS sites.
Focal Heterochromatin Does Not Block or Impede Transcript Elongation
The ChIP-Chip maps show that the Air and Igf2r genes contain interspersed peaks of active and repressive chromatin. Notably, the three identified HC peaks in this region lie within actively transcribed gene bodies (Figure 7). On the maternal chromosome, the expressed Igf2r transcript runs over the silent Air promoter HC peak. On the paternal chromosome, the expressed Air transcript runs over HC peaks at the silent Igf2r promoter and the nonexpressed Au76-psg. The Air HC peak does not impede Igf2r transcription elongation, as maternal chromosomes carrying the silent modified Air promoter or an Air promoter deletion express similar levels of Igf2r (Wutz et al., 2001). In addition, HC peaks do not arise simply from transcription overlap, as Air promoter silencing is independent of Igf2r expression (Sleutels et al., 2002, 2003). The Air ncRNA has been shown to silence Igf2r (Sleutels et al., 2003); here we also show that Air ncRNA expression is needed for formation of HC peaks over both the silent Igf2r promoter and the Au76-psg, reinforcing its specific role as a cis-acting silencer. The significance of repressive modifications on the paternal Au76-psg allele is not known. It is unlikely to play a role in silencing Igf2r, as it is not conserved in the rat genome that shows imprinted Igf2r expression (Mills et al., 1998).
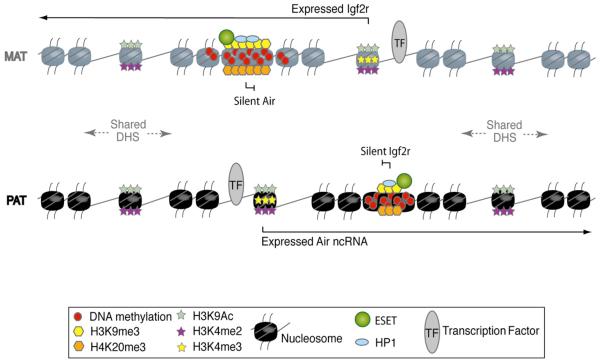
Figure 7. Active and Repressive Chromatin States Intersperse without Overlap.
Mutually exclusive active and repressive chromatin modifications that mark the expressed and silent Air and Igf2r genes that lie 28 kb apart are illustrated for the maternal (MAT) and paternal (PAT) chromosomes. The expressed Air and Igf2r promoters lack repressive modifications and are marked by H3K4me3, H3K4me2, and H3K9Ac. DHS sites are present on both parental chromosomes, but only those that lie within 20 kb of the expressed promoters are marked by active modifications (H3K4me2 and H3K9Ac, but not by H3K4me3). The silent Air and Igf2r promoters lack active modifications and are marked by focal HC peaks of H3K9me3, HP1, and H4K20me3 that resemble heterochromatin defined at centromeres, telomeres, and the inactive X chromosome. Note that these HC peaks do not impede transcript elongation from the expressed Igf2r and Air promoters.
Imprinted Gene Silencing and Tissue-Specific Gene Silencing
In contrast to the focal HC peaks found on the imprinted genes in this study, we show that tissue-specific silent genes from two regions on mouse chromosome 17 are marked by a different type of widespread repressive chromatin comprising only H3K27me3. This modification was present on both parental chromosomes, covered the silent gene body and intergenic regions, and was reduced or absent on expressed genes. We also observed that silent genes marked by H3K27me3 lacked H3K9me3 modifications and vice versa. Although our data show that different repressive modifications mark silent imprinted genes and silent tissue-specific genes, we consider it unlikely that imprinted genes carry unique silencing marks. Instead, it is probable that silent nonimprinted genes in the mammalian genome are modified by the same two types of nonoverlapping repressive chromatin described here, one based on HC peaks and one based on spreading of H3K27me3, that may index permanent or reversible silent states. In support of this argument, large H3K27me3 domains have been identified on silent genes showing cell-type-specific expression in many mouse and human cell types and in large regions of the inactive X chromosome (Bernstein et al., 2006; Chadwick and Willard, 2004; Squazzo et al., 2006). These reports also described a negative correlation between H3K27me3 and H3K9me3.
Models of Epigenetic Gene Silencing in Mammals
We show here (Figure 7) that discrete foci of repressed chromatin resembling classic pericentric heterochromatin can intersperse with focal regions of active chromatin in silent genes overlapped by antisense transcripts. Although we do not yet know the extent of HC peaks in the mammalian genome, the existence of interspersed active and repressive chromatin states questions our current understanding of how chromatin regulates genes in two ways. First, this finding contrasts with the classic view of chromosome chromatin organization in which silent heterochromatin is considered to be restricted to large domains at centromeres, telomeres, and the inactive X chromosome, while active euchromatin is found on chromosome arms that contain the genomes genes (Huisinga et al., 2006). Second, the demonstration that focal heterochromatin exists in the body of actively transcribed genes not only challenges current models of transcription-coupled remodeling processes but also raises questions about the exact role of heterochromatin in gene silencing. Chromosome-wide tiling array maps of repressive chromatin marks will be important tools to resolve these questions.
EXPERIMENTAL PROCEDURES
Chromatin Immunoprecipitation
ChIP was performed as described (Umlauf et al., 2004) with three modifications to minimize nonspecific chromatin binding to protein A Sepharose (PAS): (1) MNase-digested chromatin was precleared with swollen PAS, (2) PAS was pretreated overnight with blocking buffer (10 mM Tris [pH 8.0], 10 mM EDTA [pH 8.0], 1% Triton X-100, 0.01% SDS, 50 mM NaCl, 0.5 mg/ml BSA, 0.25 mg/ml salmon sperm DNA) before binding antigen-antibody complexes, and (3) the PAS-antigen-antibody complex was stringently washed with solutions containing 125, 250, and 500 mM NaCl. Antibodies are listed in Table S4. The resulting ChIP material was analyzed by ChIP-PCR, ChIP-Chip, and/or QPCR. Two to four biological replicas were performed.
Nonquantitative and Quantitative PCR
Nonquantitative PCR was performed with 61 primer pairs (Table S5). PCR mix contained the following: 1 × buffer, 2.5 mM MgCl2, 200 μM dNTPs, 0.5 U Taq polymerase (Promega), and 0.8 M betaine (Sigma). PCR cycles were the following: 94°C/3 min, 30 cycles of 96°C/10 s (12 cycles for pericentric primers), 94°C/30 s, 58°C/1 min, 72°C/1 min, and final extension 72°C/5 min. PCR template was the following: 1 μl of 1:100 diluted Input DNA, 1 μl of ChIP DNA, and 1 μl of mock precipitate. Quantitative real-time PCR assays are listed in Table S1.
Custom PCR Genome Tiling Array Design and Preparation
A custom PCR tiling array spanning 250 kb contained continuous single-copy sequences from bp 12342011–12362277, bp 12420070–12433039, and bp 12493360–12703950 (UCSC Mouse, February 2006, chromosome 17). Repeats were excluded by RepeatMasker (www.repeatmasker.org). PCR products were overlapping with an average size of 430 bp. Control PCR fragments were also spotted on the custom PCR tiling array (Figure S11). PCR reactions were as described above, with the exception of 1.5 M betaine. The PCR template was 2 ng of cosmid (cosOT1, cosMS4, cosMS6, cosMS1, or cos3LA3) or BAC DNA (BACs RP23-84H13, RP23-367L3, or RP23-81B3), or 10 ng of genomic DNA. Products were gel checked, Speedvac concentrated to 10–20 μl, and SSC added to a final 3× concentration. Samples were spotted in eight positions on Nexterion slides (Schott) pretreated with 1-methyl 2-pyrrolidone in blocking solution, with a Promedia Associates Inc. printer (Model PA-MP2000) using SMP3 printing tips (Telechem).
Custom PCR Genome Tiling Array Probe Amplification and Hybridization
ChIP DNA and a 1:100 dilution of Input DNA was amplified using two rounds of a T7 in vitro transcription protocol modified from Liu et al. (2003) (full protocol available upon request) labeled with Alexa 555 or Alexa 647 dyes (Molecular Probes), and ChIP and Input DNA were hybridized together on the tiling array for 18 hr at 42°C. Two biological replicates of MEFB1 and MEFF cells (including a dye swap) were hybridized to the custom PCR tiling array for each mapping experiment. For the high-resolution Oligo tiling arrays, amplified DNA/cDNA was sent for hybridization to NimbleGen Systems or Agilent Technologies Inc.
Custom PCR Genome Tiling Array Scanning and Analysis
Custom PCR tiling array slides were scanned with a GenePix 4000B scanner (Axon Instruments). Microarrays were gridded and the average ratio of medians (with background subtraction) of the eight replicate spots calculated for each PCR product. Individual spots with a ratio greater than or less than two standard deviations from the mean were excluded from the analysis. The average ratio of medians was then log2 transformed and plotted. Data were displayed with Signal Map (NimbleGen).
Custom PCR Genome Tiling Array Expression Analysis
PolyA+ RNA was isolated from MEFB1 and MEFF cells (Micro Poly[A] Purist, Ambion) and cDNA prepared and labeled and used as described above. The Input sample was as used for ChIP analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Feil for the ChIP protocol, Peter Steinlein for tiling array advice, the GEN-AU Epigenetics team for support, and Barlow group members for comments on the manuscript. Project support was from GEN-AU Epigenetic Plasticity of the Mammalian Genome (GZ200.141/1-VI/2006) and the EU-FW6 IP “HEROIC” (LSHG-CT-2005-018883) and NoE “The Epigenome” (LSHG-CT-2004-053433).
Footnotes
Supplemental Data
Supplemental Data include 11 figures, 5 tables, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/27/3/353/DC1/.
The authors declare no conflict of interest.
Accession Numbers
The Gene Expression Omnibus (GEO) accession number for the microarray data reported in this paper is GSE5834.
REFERENCES
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, Feil R. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- Lewis A, Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4:1–11. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JJ, Falls JG, De Souza AT, Jirtle RL. Imprinted M6p/Igf2 receptor is mutated in rat liver tumors. Oncogene. 1998;16:2797–2802. doi: 10.1038/sj.onc.1201801. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Pauler FM, Stricker SH, Warczok KE, Barlow DP. Long-range DNase I hypersensitivity mapping reveals the imprinted Igf2r and Air promoters share cis-regulatory elements. Genome Res. 2005;15:1379–1387. doi: 10.1101/gr.3783805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Regha K, Latos PA, Spahn L. The imprinted mouse Igf2r/Air cluster—a model maternal imprinting system. Cytogenet. Genome Res. 2006;113:165–177. doi: 10.1159/000090829. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Tjon G, Ludwig T, Barlow DP. Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J. 2003;22:3696–3704. doi: 10.1093/emboj/cdg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter D. Imprinting today: end of the beginning or beginning of the end? Cytogenet. Genome Res. 2006;113:12–16. doi: 10.1159/000090809. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Vu TH, Li T, Hoffman AR. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum. Mol. Genet. 2004;13:2233–2245. doi: 10.1093/hmg/ddh244. [DOI] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Wutz A, Theussl HC, Dausman J, Jaenisch R, Barlow DP, Wagner EF. Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development. 2001;128:1881–1887. doi: 10.1242/dev.128.10.1881. [DOI] [PubMed] [Google Scholar]
- Yamasaki Y, Kayashima T, Soejima H, Kinoshita A, Yoshiura K, Matsumoto N, Ohta T, Urano T, Masuzaki H, Ishimaru T, et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum. Mol. Genet. 2005;14:2511–2520. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001;15:2361–2366. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.