Abstract
Mitochondrial respiratory chain defects have been associated with various diseases and normal aging, particularly in tissues with high energy demands including skeletal muscle. Muscle-specific mitochondrial DNA (mtDNA) mutations have also been reported to accumulate with aging. Our understanding of the molecular processes mediating altered mitochondrial gene expression to dysfunction associated with mtDNA mutations in muscle would be greatly enhanced by our ability to transfer muscle mtDNA to established cell lines. Here, we report the successful generation of mouse cybrids carrying skeletal muscle mtDNA. Using this novel approach, we performed bioenergetic analysis of cells bearing mtDNA derived from young and old mouse skeletal muscles. A significant decrease in oxidative phosphorylation coupling and regulation capacity has been observed with cybrids carrying mtDNA from skeletal muscle of old mice. Our results also revealed decrease growth capacity and cell viability associated with the mtDNA derived from muscle of old mice. These findings indicate that a decline in mitochondrial function associated with compromised mtDNA quality during aging leads to a decrease in both the capacity and regulation of oxidative phosphorylation.
INTRODUCTION
Mitochondria are ubiquitous organelles in eukaryotic cells whose primary function is to generate energy through oxidative phosphorylation (1), the step-wise transfer of electrons to oxygen coupled with ATP production. High-energy electrons, which leave the electron transport chain (ETC) prematurely, generate reactive oxygen species (ROS) (2). Disruption of ETC has been suggested to alter the production of ROS (3). Oxidative damage caused by ROS to DNA, proteins and lipids in animal tissues has been reported, and the quantity of such damage increases during aging (4,5).
Skeletal muscles, which contain long fibers formed from a large number of fused cells, support or move the skeleton. The performance of skeletal muscle, speed and strength, deteriorates with aging due to fiber loss and atrophy (6), a condition known as sarcopenia. Cytochrome c oxidase (COX), the terminal component of ETC, -negative muscle fibers have been detected in human, monkey, mouse and Drosophila, and their quantity increases with age (7). In addition, studies carried out with different ages of rhesus monkeys indicate regions of muscle fibers exhibiting high levels of ETC defects also contain a large accumulation of mtDNA deletions (8). Further, mtDNA deletions in old animals could focally accumulate to high levels in certain muscle fibers, and that could directly contribute to sarcopenia (9,10). Interestingly, in calorie-restricted mice, a consistently proven animal model of extended mean and maximum life span and attenuated development of age-related diseases (11,12), both the accumulation of deleted mtDNAs and the number of COX-negative fibers, were decreased (13).
Mitochondrial function is under dual genome control. Although the majority of structural genes and all of the regulatory genes are encoded by the nuclear genome, the mitochondrial genome contains the sequences for 37 genes, including 13 for essential polypeptides of the oxidative phosphorylation apparatus, 22 tRNAs and two rRNAs for mitochondrial protein synthesis (14,15). It is believed that since mtDNA is close to the ROS generation site, it undergoes relatively more cycles of replication (especially in postmitotic cells such as muscles), and is subject to a reduced breadth of DNA repair activities. Consistent with this hypothesis, mtDNA exhibits a relatively high mutation frequency (16).
The successful development of mitochondrial-mediated transformation has opened the way to elucidate the role of mtDNA (17). However, in-depth studies of the contributions of mtDNA mutations to tissue-specific pathogenesis are limited as the transfer of mitochondria from tissues to cell lines has only successfully been carried out with platelets (18) and synaptosomes (19,20). A noticeable missing technique is a method to establish cybrids with mtDNA derived from muscle. In the present study, we developed a new approach to generate cybrids containing mtDNA from mouse skeletal muscle. Using this method, we further analyzed respiratory activity and growth capacity in glucose and galactose media of cybrids with mtDNA from old and young mice.
MATERIALS AND METHODS
Animals and sample preparation
C57BL/6 and DBA/2 mice were purchased from Charles River Laboratories. Among them, six each of C57BL/6 were aged to 6 and 26 months at the UTHSCSA animal facility. Skeletal muscles were dissected from the hind legs. Each muscle was cut into small pieces on a 10-mm dish on ice, and rinsed several times with Hank’s Balanced Salt Solution (HBSS, Invitrogen). The muscle samples were resuspended with 6 vol hypotonic buffer (RSB) (10.0 mM Tris,10.0 mM KCl, 0.15 mM MgCl2, pH 6.7) and further processed with a Dounce A type homogenizer for about 30 strokes. The processed samples were then centrifuged at 2300 r.p.m. for 3 min and the supernatant was then subjected to another centrifugation at 1000 r.p.m. for 10 min. Finally, muscle mitochondria enclosed by plasma membrane were collected by centrifugation at 6500 r.p.m. for 10 min.
Cell lines and media
All cell lines used in the present work were grown in monolayer culture. The mtDNA-less ρ0 LL/2-m21 cell line was a derivative of mouse cell line LL/2 as described previously (21). Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
mtDNA analysis
For mtDNA sequencing, total DNA was extracted with the Easy-DNATM kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and then subjected to polymerase chain reaction (PCR) amplification of the fragments containing part of the tRNAArg and ND3 genes. Amplified mtDNA fragments were purified using the AllStar PCR Clean Up kit (AllStar Sci. Inc., Sunnyvale, CA, USA), and purified PCR products were sequenced.
The sequences of primers used were:
ND3-5 (9312-9331): TGAAGCCGCAGCATGATACT
ND3-3: (9900-9881): GTTGAAGAAGGTAGATGGCA
O2 consumption measurement
The measurements were carried out in two chambers of a YSI Model 5300 Biological Oxygen Monitor as described earlier (22). Briefly, determination of the O2 consumption rate was carried out in about 5×106 cells in Tris-based, Mg2+, Ca2+-deficient (TD) buffer (0.137 M NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 25 mM Tris–HCl, pH 7.4). After recording the base respiration rate, 2.5 µg/ml oligomycin was added to measure uncoupled respiration, and then 0.5 µM FCCP was added to determine maximal respiration.
Cell growth and viability analysis
Multiple identical samples of 5×104 cells were grown for 4 days on six-well plates in the appropriate medium (DMEM, which contains 4.5 mg/ml glucose and 0.11 mg/ml pyruvate, or DMEM lacking glucose and containing 0.9 mg/ml galactose and 0.11 mg/ml pyruvate, both supplemented with 10% dialyzed FBS), and counted on a daily basis. Doubling time = 72 h×Ln (2)/[Ln (total cell number at 96 h)-Ln (total cell number at 24 h)].
Cell viability was analyzed with a Vi-CellTM XR cell viability analyzer (Beckman Coulter).
mtDNA content analysis
The relative mtDNA levels were measured by a real-time PCR and normalized by simultaneous measurement of the nuclear DNA. Primers and probes for quantitative PCR (qPCR) were designed using Primer Express (Applied Biosystems). QPCR was carried out using an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) in 20-µl reaction in different tubes containing 0.5 µM each of the forward and reverse primers, 0.1 µM for each probe (ND3 and β-actin genes) and 50 ng DNA sample. The PCR conditions were 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 60 s. The threshold cycle number (Ct) values of the β-actin gene and the mitochondrial ND3 gene were determined. Each measurement was carried out four times and normalized against a serial dilution of a control DNA sample. Ct values can be used as a measure of the input DNA contents and Ct value differences were used to quantify mtDNA levels relative to the β-actin gene with the following equation: Relative copy number (Rc) = 2ΔCt, where ΔCt is the Ctβ-actin − CtND3
The primer and probe information is as follows:
For mtDNA ND3: forward, TCTGACTCCCCCAAATAAATCTG; reverse, GGGTCGAATCCGCATTCA.
For nuclear β-actin: forward, AAATCGTGCGTGACATCAAAGA; reverse, GGCCATCTCCCTGCTCGAA.
Probes: Cox III, FAM- TCAGAAAAAGCAAATCC; β-actin, FAM- AGCTGTGCTATGTTGCT.
Statistical analysis
P, probability, values were calculated by using the unpaired Student’s t-test contained in the Microsoft Excel program. The difference is considered as statistically significant when P < 0.05. r, Pearson product-moment correlation coefficient, is a measure of the strength of linear dependence between two variables. It ranges from −1 to 1. A value of 1 implies that a linear equation describes the relationship perfectly; a value of −1 implies a perfect negative correlation; a value of 0 implies that there is no linear correlation between the variables.
RESULTS
Generation of cybrids with mitochondria from mouse skeletal muscle
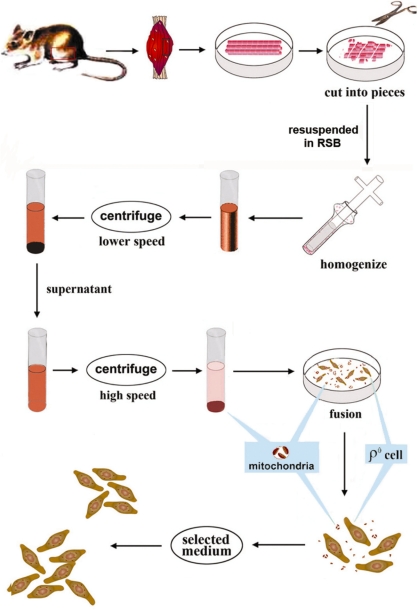
As shown in Figure 1, muscle samples were obtained from hind legs of healthy mice right after the animals were euthanized. In particular, muscles were dissected out; fatty and nerve cells were removed. After being rinsed with HBSS, the muscle samples were cut into small pieces on ice. Fine muscle pieces were subjected to treatment with hypotonic buffer and then homogenized. The processed muscle was constantly checked under the microscope after 20 strokes with the homogenizer until the nuclei were released and plasma membrane-enclosed mitochondria formed. The processed samples were then centrifuged at 2300 r.p.m. for 3 min and the supernatant was then subjected to another centrifugation at 1000 r.p.m. for 10 min. Finally, muscle mitochondria enclosed by plasma membrane were collected by centrifugation at 6500 r.p.m. for 10 min.
Figure 1.
Generation of cybrids containing mtDNA from skeletal muscle. Flowchart of how mtDNA from the skeletal muscle was transferred to ρ0 cells to produce cybrids.
Fusion solution was prepared as 9 g of polyethylene glycol (PEG) (Sigma P-5402; MW: 1450) was autoclaved with slow exhaust for 15 min, and then 2 ml DMSO and 9 ml DMEM were added when it was cooled to about 65°C. The solution was mixed gently and then kept in 37°C incubator. The mouse mtDNA-less LL/2-m21 ρ0 cell line was established previously (23). Approximately 106 ρ0 cells were mixed with freshly isolated membrane enclosed muscle mitochondria prepared from about 1 g of muscle sample from two hind legs. The mixture was centrifuged for 10 min at 2000g, and the pellet was resuspended with 45% PEG by mild and brief pipetting while avoiding the creation of air bubbles. After one-min treatment with PEG, the fusion process was stopped by diluting with 10 volumes of DMEM. The fusion products were incubated with DMEM supplemented with 50 µg/ml uridine at 37°C for 24 h. On Day 2, the medium was replaced by semi-selection media with the amount of uridine reduced to10 µg/ml. Five days later, the medium was changed again to selection medium, i.e. the same medium without uridine. Transformants were selected based on the pyrimidine auxotrophy of the ρ0 cells (24) as the cybrids containing exogenous mitochondria from muscle were able to survive in the selective medium.
Validation of the transfer of mtDNA from muscle to cybrids
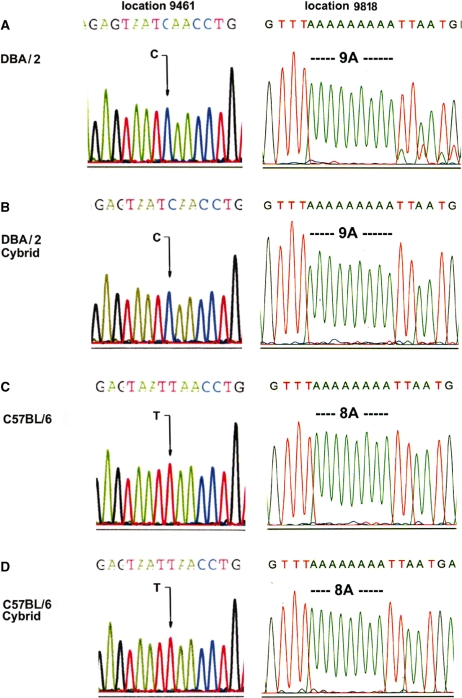
To confirm that the cybrids we generated did carry the mtDNA from the specific muscle tissues, we did a pilot experiment utilizing two species of mice, C57BL/6 and DBA/2, with distinctive mtDNA sequences. Six clones each were selected from the fusion of C57BL/6 and DBA/2 skeletal muscle to LL/2-m21 ρ0 cells. As two sequencing variances between C57BL/6 and DBA/2 have been identified in the ND3 and tRNAArg genes (25,26), we determined the sequence of two DNA fragments covering these genes in the putative transmitochondrial clones containing the mtDNA from the muscle (Figure 2B and D). The mtDNA from the original muscle samples was also sequenced as a control (Figure 2A and C). Representative sequencing results are shown in Figure 2. As indicated by the signature sequences at positions 9461 and 9818 (25,26), the clones derived from the fusion products of muscle from DBA/2 carried a C at position 9461, and were with a stretch of 9A from position 9821 to 9829; while clones from C57BL/6 muscle exhibited a T at position 9461 and were with 8A from position 9821 to 9828; both matched the mtDNA from the original muscle samples, respectively. It is interesting to note that the DBA/2 mice in this study were heteroplasmic at 9821 with about 10% carrying 10A (Figure 2A), and the heteroplasmy was lost during mitochondria-mediated transformation as observed previously (21). Thus, our fusion approach has been validated.
Figure 2.
Validation of transfer of mtDNA from muscle to cybrids. Representative chromatograms of signature sequences of mtDNA at positions 9461 and 9818 and surrounding areas of DBA/2-muscle sample (A), cybrids derived from fusion of mitochondria from DBA/2 muscle and ρ0 cells (B), C57BL/6-muscle (C) and cybrids derived from fusion of mitochondria from C57BL/6 muscle and ρ0 cells (D). An arrow shows the base difference at 9461, and the different lengths of the A-stretch are also indicated.
Production of cybrids with muscle mtDNA from mice of different ages
C57BL/6 mice at the age of 26 months (old) and 6 months (young), six per group, were selected to investigate potential age-associated mtDNA alterations. Using the novel method described herein, we have thus obtained 24 and 25 cybrid clones carrying mtDNA derived from the skeletal muscles from young and old mice, respectively.
Respiration properties of trans-mitochondrial clones
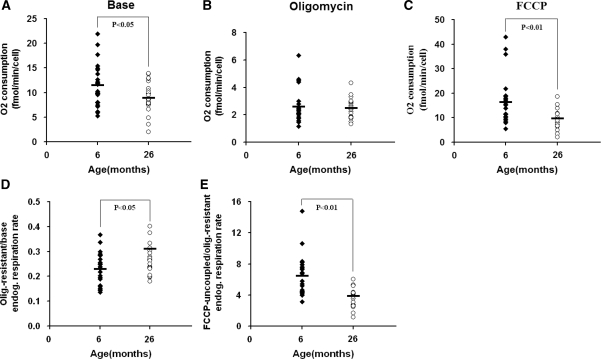
To assess the mitochondrial function of the cybrids carrying mtDNA from muscles of young and old mice, the respiration properties were investigated. First, we measured the endogenous respiration activity of intact cells. As shown in Figure 3A, there was a decrease in average base endogenous respiration in cybrids of old groups compared with that of young group. We then quantified oxygen consumption in the presence of oligomycin A, a chemical inhibitor that can block adenosine triphosphate (ATP)-synthase, or in the presence of the chemical uncoupler FCCP, which can completely uncouple oxidation from phosphorylation in mitochondria and raise the respiratory capacity to a maximal level. The difference between two age groups was more pronounced (P < 0.01) in the maximal respiratory capacity as revealed by oxygen consumption with FCCP (Figure 3C). However, no significant differences in uncoupled respiration were detected when ATP synthase was inhibited (Figure 3B).
Figure 3.
Oxygen consumption measurement. Six and 26 months represent 24 and 22 cybrids per age group, respectively. Endogenous respiration was measured in intact cells (A), after treatment with the ATP synthase inhibitor oligomycin at 2.5 µg/ml (B), and with the uncoupler FCCP at 0.5 µM (C). The activities were determined with about 5 × 106 cells. Three determinations were made for each cybrid and the average values are shown. Ratios of oligomycin-resistant to native endogenous respiration rates (D) and FCCP-treated fully uncoupled to oligomycin-resistant respiration rates (E) of each age group were also calculated.
To more carefully examine how respiration is coupled with ATP synthesis in these cybrids, we then calculated the ratio of oligomycin resistant to base endogenous respiration. Compared with the young group, the average of the old group exhibited a higher value (Figure 3D), indicating more relaxed coupling of oxidative phosphorylation associated with mtDNA from old mice. The capacity of regulation over oxidative phosphorylation was also monitored by calculating the ratio of FCCP-uncoupled to oligomycin-resistant respiration. As shown in Figure 3E, cybrids containing mtDNA from the old group exhibited a lower average value, suggesting compromised control of mitochondrial membrane potential over respiration associated with mtDNA from muscle cells of old mice.
Growth and viability analysis
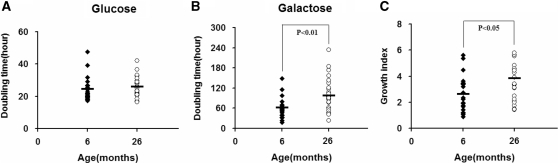
Mammalian cells rely on both mitochondrial oxidative phosphorylation and glycolysis to generate ATP to support various cellular pathways. However, they cannot utilize galactose efficiently in the glycolytic pathway (27). Figure 4 shows the study of growth properties of the cybrids in glucose- and galactose-containing media. As observed with respiratory activities, measurements of population doubling times revealed substantial variances among the same age group. Nevertheless, overall, a significant difference between old and young groups was observed. In particular, in regular glucose medium there was no significant difference in the average doubling time between cells derived from the young and old groups (Figure 4A). However, in the galactose medium, where ATP production predominantly relies on mitochondrial oxidative phosphorylation, the average doubling time of old group cells was markedly longer than that of young group cells (Figure 4B). In order to reduce the background variability caused by other factors, the ratio of doubling time in galactose medium to that in glucose medium was calculated as indicated by the growth index. As shown in Figure 4C, the difference between the two age groups persisted, although with somewhat reduced statistical significance (P < 0.05 versus P < 0.01).
Figure 4.
Growth capacity measurement. Six and 26 months represent 24 and 25 cybrids per age group, respectively. Cells were grown in glucose-containing DMEM for 24 h, then in glucose medium (A) or in galactose medium (B) for another 72 h. Doubling times were calculated as: 72 h × Log (2)/[Log (total cell number at the end)-Log (total cell number when media were changed)]. Three determinations were made for each cybrid. Growth index was calculated as the ratio of doubling time in galactose-containing medium to that in glucose medium (C).
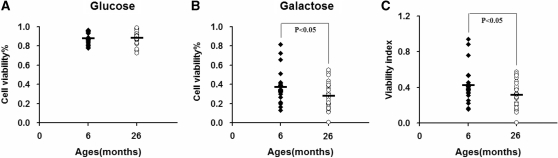
As mitochondrial dysfunction has been associated with apoptosis, we further investigated cell viability in glucose- and galactose-containing media following 3 days of culturing in the respective media. As shown in Figure 5A, in the regular glucose medium, there was no significant difference in cell viability between the young group and the old group. However, in the galactose medium, the cell viability of the old group was significantly lower than that of the young group (P < 0.05) (Figure 5B). The ratio of viability in galactose to that in glucose medium, indicated as viability index, was significantly lower for the old group relative to the young group cells (P < 0.05) (Figure 5C).
Figure 5.
Cell viability analysis. Six and 26 months represent 24 and 25 cybrids per age group, respectively. Cells were grown in glucose-containing DMEM for 24 h, then in glucose medium (A) or in galactose medium (B) for another 72 h. Cell viabilities were analyzed with a Vi-cellTM XR cell viability analyzer (Beckman Coulter). Three determinations were made for each cybrid. Viability index was calculated as the ratio of percentage of cell viability in galactose-containing medium to that in glucose medium (C).
mtDNA content alteration and its implication
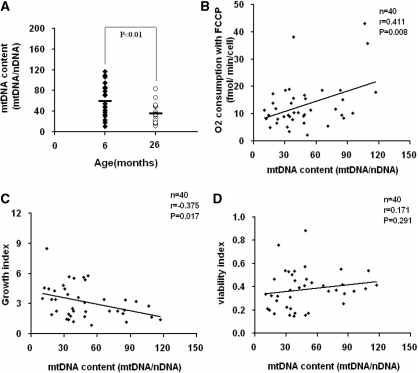
To investigate the alteration of mtDNA content as a result of changes in mtDNA sequences associated with replication or turnover, the mtDNA copy number of the transformants was measured by quantitative real- time PCR method. While a considerable variability among the clones derived from the same age group was revealed, there is a clear tendency of decrease of mtDNA content in the cybrids from old mice (Figure 6A). To further determine if the decrease in mtDNA copy number has any effects on the compromised mitochondrial function we observed, we plotted the maximal respiration rate measured by oxygen consumption with FCCP treatment (Figure 6B), growth index as defined by the ratio of doubling time of cybrids in galactose medium to that in glucose medium (Figure 6C) and cell viability index as the ratio of cell viability assayed in galactose medium to that in glucose medium (Figure 6D), to mtDNA copy number, respectively. We found that the maximal respiration rate declined as the mtDNA copy number reduced (Figure 6B). Similarly, the normalized growth capacity in galactose medium also reduced with the decline of the mtDNA copy number (Figure 6C). However, no significant effect was observed in the range of the mtDNA copy number changes in this study on the cell viability in galactose medium (Figure 6D), while we did observe a decease in cell viability with the same assay in cybrids carrying mtDNA from muscle of old mice (Figure 5C),
Figure 6.
mtDNA copy number analysis. (A) Six and 26 months represent 24 and 20 cybrids per age group. Relative mtDNA copy number (mtDNA/nDNA) was the mtDNA content normalized by nuclear DNA encoded β-actin gene. (B) Relationship between the relative mtDNA copy number and the maximal respiration as measured by oxygen consumption with uncoupler FCCP in 40 individual transformants of both age groups. (C) Relationship between the relative mtDNA copy number and the ratio of doubling time (in galactose/in glucose media) in 40 individual transformants of both age groups. (D) Relationship between the relative mtDNA copy number and the ratio of viability (in galactose/in glucose media). In (B–D), n includes 22 cybrids of 6-month-young mice, and 18 cybrids of 26-month-old mice.
These results indicate that the declined mtDNA copy number might contribute to the compromised respiration rate and oxidative phosphorylation capacity associated with the mtDNA from the skeletal muscle of the old mice. In addition, other age-related mtDNA mutations in mouse skeletal muscle could also play a role in age-related pathogenesis.
DISCUSSION
The successful generation of muscle cybrids provided a unique opportunity to recover and analyze somatic mtDNA mutations directly from fresh muscle samples. This new technology also provided an opportunity to further our ability to investigate diseases or age-related mtDNA mutations. In addition, since low-occurrence mutations within the heteroplasmic mtDNA populations could be enriched in culture cells (24), one great advantage of this new approach is the potential to study the low level pathogenic mutations which could otherwise be very difficult to carry out sophisticated studies on at the molecular level in tissues.
Studies of the role of mitochondrial dysfunction and mtDNA mutations in pathogenesis associated with diseases and aging in skeletal muscle is of particular interest since: (i) muscle is one of the most targeted tissues by mitochondrial deficiency. Actually, one of the earliest identified hallmarks of mitochondrial disease is the characterization of ragged red fibers (RRG) in biopsied skeletal muscle, and interestingly most have been associated with abnormal mtDNA (28). (ii) Skeletal muscle predominantly relies on oxidative phosphorylation and at the same time, suffers rather marked age-related degeneration (29). (iii) Skeletal muscle is arguably the best system where the causative relationship among age-related degeneration, mitochondrial dysfunction, and mtDNA alteration, mtDNA deletions in particular, has been established, owing largely to a collection of excellent work from Judd Aiken and colleagues (30–34).
Our results revealed that respiration measured with FCCP was more sensitive in detecting in mitochondrial function decline associated with aging. The other significant result in the present study was the demonstration that the loss of coupling and regulation of oxidative phosphorylation associated with mtDNA alteration during aging. Such an age-related compromise in coupling efficiency of respiration was reported previously in a large scale investigation with fibroblasts, mostly skin-derived, from human subjects with a large range of age, by measurement of P:O ratios (35).
As previously reported with human skeletal muscle (36) and mitochondrial transformants with mtDNA derived from human fibroblasts (37), an age-related decline of mtDNA contents was revealed with mouse skeletal muscle in this study. However, while we observed the declines in respiration and growth capacity associated with the decrease in mtDNA copy number, we did not see a significant correlation between mtDNA content and cell viability in galactose medium. As shown with the mutator mice, a premature aging mouse model with an increase in mtDNA mutations due to the expression of a proof-reading-deficient version of mtDNA polymerase (38,39), increased mtDNA mutations have been associated with enhanced apoptosis. Our results also suggested that there was an age-related increase in mtDNA mutations which might also play a role in the decreased viability in galactose medium associated with the cybrids of old mice.
As a significant difference between young and old age groups was only detected when cybrids were grown in galactose medium, but not in the regular glucose medium where cells can obtain ATP through both oxidative phosphorylation and glycolysis, these findings suggested that the decrease in mitochondrial ATP production in old mitochondria could be compensated for by an upregulation in the glycolytic pathway. It is interesting to note that such a bioenergetic switch is also a signature of cancer cells (40), as aging is the biggest risk factor for cancer occurrence.
FUNDING
The National Institute of Aging/National Institutions of Health (grants R01 AG025223 and R03AG024640 to Y.B.). Funding for open access charges: National Institutions of Health (R01 AG025223).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This article is dedicated to the late Dr Giuseppe Attardi, a pioneer in mitochondrial genetics research. This work was initiated in the laboratory of Dr Giuseppe Attardi at Caltech where the corresponding author was a postdoctoral fellow. Dr Xiufeng Song, Jin Chen and Qi Zhao participated in early stage of this project. We thank Dr Brian Herman for sharing the aged animals utilized in this study.
REFERENCES
- 1.Attardi G, Schatz G. Biogenesis of mitochondria. Annu. Rev. Cell. Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 2.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Lenaz G. The mitochondrial production of reactive oxygen species: mechanisms and implications in human pathology. IUBMB Life. 2001;52:159–164. doi: 10.1080/15216540152845957. [DOI] [PubMed] [Google Scholar]
- 4.Miquel J, de Juan E, Sevila I. Oxygen-induced mitochondrial damage and aging. EXS. 1992;62:47–57. doi: 10.1007/978-3-0348-7460-1_5. [DOI] [PubMed] [Google Scholar]
- 5.Miquel J, Economos AC, Fleming J, Johnson JE., Jr Mitochondrial role in cell aging. Exp. Gerontol. 1980;15:575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- 6.Navarro A, Lopez-Cepero JM, Sanchez del Pino MJ. Skeletal muscle and aging. Front. Biosci. 2001;6:D26D44. doi: 10.2741/navarro. [DOI] [PubMed] [Google Scholar]
- 7.DiMauro S, Tanji K, Bonilla E, Pallotti F, Schon EA. Mitochondrial abnormalities in muscle and other aging cells: classification, causes, and effects. Muscle Nerve. 2002;26:597–607. doi: 10.1002/mus.10194. [DOI] [PubMed] [Google Scholar]
- 8.Aiken J, Bua E, Cao Z, Lopez M, Wanagat J, McKenzie D, McKiernan S. Mitochondrial DNA deletion mutations and sarcopenia. Ann. NY Acad. Sci. 2002;959:412–423. doi: 10.1111/j.1749-6632.2002.tb02111.x. [DOI] [PubMed] [Google Scholar]
- 9.Cao Z, Wanagat J, McKiernan SH, Aiken JM. Mitochondrial DNA deletion mutations are concomitant with ragged red regions of individual, aged muscle fibers: analysis by laser-capture microdissection. Nucleic Acids Res. 2001;29:4502–4508. doi: 10.1093/nar/29.21.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wanagat J, Wolff MR, Aiken JM. Age-associated changes in function, structure and mitochondrial genetic and enzymatic abnormalities in the Fischer 344 x Brown Norway F(1) hybrid rat heart. J. Mol. Cell. Cardiol. 2002;34:17–28. doi: 10.1006/jmcc.2001.1483. [DOI] [PubMed] [Google Scholar]
- 11.Masoro EJ. Subfield history: caloric restriction, slowing aging, and extending life. Sci. SAGE KE. 2003;2003:RE2. doi: 10.1126/sageke.2003.8.re2. [DOI] [PubMed] [Google Scholar]
- 12.Masoro EJ. Dietary restriction and aging. J. Am. Geriatr. Soc. 1993;41:994–999. doi: 10.1111/j.1532-5415.1993.tb06767.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee CM, Aspnes LE, Chung SS, Weindruch R, Aiken JM. Influences of caloric restriction on age-associated skeletal muscle fiber characteristics and mitochondrial changes in rats and mice. Ann. NY Acad. Sci. 1998;854:182–191. doi: 10.1111/j.1749-6632.1998.tb09901.x. [DOI] [PubMed] [Google Scholar]
- 14.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 15.Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int. Rev. Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 16.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am. J. Med. Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 17.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 18.Chomyn A, Lai ST, Shakeley R, Bresolin N, Scarlato G, Attardi G. Platelet-mediated transformation of mtDNA-less human cells: analysis of phenotypic variability among clones from normal individuals—and complementation behavior of the tRNALys mutation causing myoclonic epilepsy and ragged red fibers. Am. J. Hum. Genet. 1994;54:966–974. [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S, Ohta S, Nishimaki K, Kagawa Y, Soma R, Kuno SY, Komatsuzaki Y, Mizusawa H, Hayashi J. Functional integrity of mitochondrial genomes in human platelets and autopsied brain tissues from elderly patients with Alzheimer’s disease. Proc. Natl Acad. Sci. USA. 1999;96:2099–2103. doi: 10.1073/pnas.96.5.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue K, Ito S, Takai D, Soejima A, Shisa H, LePecq JB, Segal-Bendirdjian E, Kagawa Y, Hayashi JI. Isolation of mitochondrial DNA-less mouse cell lines and their application for trapping mouse synaptosomal mitochondrial DNA with deletion mutations. J. Biol. Chem. 1997;272:15510–15515. doi: 10.1074/jbc.272.24.15510. [DOI] [PubMed] [Google Scholar]
- 21.Bai Y, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum. Mol. Genet. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Y, Hu P, Park JS, Deng JH, Song X, Chomyn A, Yagi T, Attardi G. Genetic and functional analysis of mitochondrial DNA-encoded complex I genes. Ann. NY Acad. Sci. 2004;1011:272–283. doi: 10.1007/978-3-662-41088-2_26. [DOI] [PubMed] [Google Scholar]
- 24.Bai Y, Shakeley RM, Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell. Biol. 2000;20:805–815. doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goios A, Pereira L, Bogue M, Macaulay V, Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai Y, Park JS, Deng JH, Li Y, Hu P. Restoration of mitochondrial function in cells with complex I deficiency. Ann. NY Acad. Sci. 2005;1042:25–35. doi: 10.1196/annals.1338.003. [DOI] [PubMed] [Google Scholar]
- 28.Morgan-Hughes JA. Mitochondrial diseases of muscle. Curr. Opin. Neurol. 1994;7:457–462. doi: 10.1097/00019052-199410000-00014. [DOI] [PubMed] [Google Scholar]
- 29.Hiona A, Leeuwenburgh C. The role of mitochondrial DNA mutations in aging and sarcopenia: implications for the mitochondrial vicious cycle theory of aging. Exp. Gerontol. 2008;43:24–33. doi: 10.1016/j.exger.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pak JW, Vang F, Johnson C, McKenzie D, Aiken JM. MtDNA point mutations are associated with deletion mutations in aged rat. Exp. Gerontol. 2005;40:209–218. doi: 10.1016/j.exger.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbst A, Pak JW, McKenzie D, Bua E, Bassiouni M, Aiken JM. Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:235–245. doi: 10.1093/gerona/62.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKiernan SH, Tuen VC, Baldwin K, Wanagat J, Djamali A, Aiken JM. Adult-onset calorie restriction delays the accumulation of mitochondrial enzyme abnormalities in aging rat kidney tubular epithelial cells. Am. J. Physiol. Renal. Physiol. 2007;292:F1751–F1760. doi: 10.1152/ajprenal.00307.2006. [DOI] [PubMed] [Google Scholar]
- 34.McKiernan SH, Colman R, Lopez M, Beasley TM, Weindruch R, Aiken JM. Longitudinal analysis of early stage sarcopenia in aging rhesus monkeys. Exp. Gerontol. 2009;44:170–176. doi: 10.1016/j.exger.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greco M, Villani G, Mazzucchelli F, Bresolin N, Papa S, Attardi G. Marked aging-related decline in efficiency of oxidative phosphorylation in human skin fibroblasts. FASEB J. 2003;17:1706–1708. doi: 10.1096/fj.02-1009fje. [DOI] [PubMed] [Google Scholar]
- 36.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl Acad. Sci. USA. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laderman KA, Penny JR, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent functional alterations of mitochondrial DNA (mtDNA) from human fibroblasts transferred into mtDNA-less cells. J. Biol. Chem. 1996;271:15891–15897. doi: 10.1074/jbc.271.27.15891. [DOI] [PubMed] [Google Scholar]
- 38.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 39.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 40.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]