Abstract
Homologous recombination plays pivotal roles in DNA repair and in the generation of genetic diversity. To locate homologous target sequences at which strand exchange can occur within a timescale that a cell’s biology demands, a single-stranded DNA-recombinase complex must search among a large number of sequences on a genome by forming synapses with chromosomal segments of DNA. A key element in the search is the time it takes for the two sequences of DNA to be compared, i.e. the synapse lifetime. Here, we visualize for the first time fluorescently tagged individual synapses formed by RecA, a prokaryotic recombinase, and measure their lifetime as a function of synapse length and differences in sequence between the participating DNAs. Surprisingly, lifetimes can be ∼10 s long when the DNAs are fully heterologous, and much longer for partial homology, consistently with ensemble FRET measurements. Synapse lifetime increases rapidly as the length of a region of full homology at either the 3′- or 5′-ends of the invading single-stranded DNA increases above 30 bases. A few mismatches can reduce dramatically the lifetime of synapses formed with nearly homologous DNAs. These results suggest the need for facilitated homology search mechanisms to locate homology successfully within the timescales observed in vivo.
INTRODUCTION
Homologous recombination plays a fundamental role in the generation of genetic diversity, and as a pathway for DNA repair in both prokaryotes and eukaryotes. During horizontal gene transfer processes in bacteria, exogenous pieces of DNA thousands of bases long are imported into a cell and incorporated into the chromosome at specific loci of sufficiently high homology (1). Both the search for homology along a chromosome and the strand exchange process that ensues after homology is found are catalyzed by the RecA protein (2). Since chromosomal DNAs are very long molecules, the number of sequences that must be probed until homology is found can be exceedingly large. Yet, the search for homology is completed within a cell cycle, leading to the consideration of facilitated search processes (3,4), similarly to site-specific proteins searching for their targets (5,6).
The elementary step in the homology search is the formation of a complex (synapse) between a segment of the genomic double-stranded DNA (dsDNA) and a RecA-covered single-stranded tract of DNA (ssDNA). It is within a synapse that the two sequences are compared. Little is known about the natural variability of synapse lengths and the time needed to carry out a sequence comparison as a function of both length and the degree of sequence similarity (7–9). The large excess of heterologous sequences that must be scanned during a homology search suggests that for search processes to be efficient the lifetime of synapses that do not lead to strand exchange must be very short (10). According to the prevailing view, homology is found by a series of random collisions (11,12), implying search events as short as 2.5 × 10−4 s for eukaryotes such as yeast (13). A similar estimation for E. coli yields times of the same order of magnitude. However, ensemble kinetic experiments indicate that synapses may last much longer, ∼15 s (14). The combination of long synapse lifetimes with a pure three-dimensional diffusion-limited search may be prohibitively long. The knowledge of how homology is found in a cell remains scant (4,13).
In this study, we report the observation of individual synapses as they formed and dissociated in real time, allowing for a direct measurement of their lifetime. Short, anchored dsDNA molecules were epi-illuminated by total internal reflection (Figure 1A). RecA–ssDNA complexes formed with fluorescently tagged ssDNA molecules were then introduced and monitored as they formed synaptic intermediates with the anchored dsDNA. Our goal was to test for the existence of long events and measure their lifetime as a function of synapse length and differences in sequence between the participating ssDNA and dsDNA molecules. We supplement these results with measurements of the average lifetime of heterologous synapses in large ensembles of molecules in solution using fluorescence resonance energy transfer (FRET) methods (14), that probe the lifetime distribution over a range from milliseconds and above.
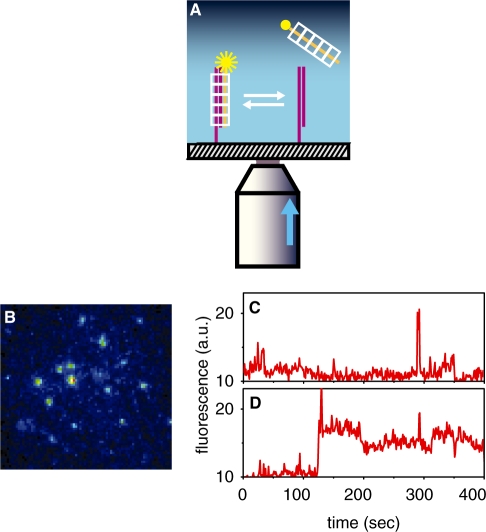
Figure 1.
Experimental scheme for observing individual synapses and sample results. (A) Fluorescently-tagged ssDNA–RecA complexes are observed only when forming synaptic intermediates with dsDNA anchored on a glass surface. The exponentially decaying evanescent light produced by a narrow laser beam (entering a high numerical aperture objective off-axis as illustrated by the arrow), illuminates only a ∼200 nm thick layer above the glass. (B) Snapshot of synaptic intermediate complexes, ∼25 µM across. (C) Fluorescence versus time of a single synapse formed with a fully heterologous, 50-bp-long ssDNA–RecA complex, or (D), with a fully homologous ssDNA–RecA complex. Traces and snapshot represent raw, unfiltered data.
MATERIALS AND METHODS
Experimental setups
An objective-type total internal reflection microscope (TIRF) was constructed using published methods (15). Evanescent illumination was achieved using a high numerical aperture, immersion oil objective (Alpha-Plan-Fluar 1.45/100x, Carl Zeiss, Oberkochen, Germany). The 514-nm line of an Argon laser (Melles Griot, Irvine, CA) was used for excitation of the TAMRA fluorophores. All experiments were carried out at 37°C. The fluorescent signal was imaged with an intensified CCD camera (PentaMAX, Princeton Instruments, Trenton, NJ), and frames were acquired continuously, with an exposure time of 2 s per frame to achieve a good signal-to-noise ratio.
DNA sequences
HPLC purified, 5′ TAMRA tagged, ssDNA oligonucleotides, as well as 5′ biotin modified 100-nt oligonucleotides, were purchased from IDTDNA (Coralville, IA) and from Metabion (Martinsried, Germany). Unmodified oligonucleotides were synthesized and purified by the Biological Services Department of the Weizmann Institute of Science. All ssDNA sequences were checked against secondary structure formation using the Mfold v3.2 software (16).
Sample preparation for single molecule assays
All chemicals were purchased from Sigma-Aldrich unless specified. All dsDNA constructs were attached to coverslips via their biotinylated tails, using previously described methods (17), but injecting dsDNA at 25 nM in EB3 buffer (50-mM Tris–HCl, 20-mM MgCl2) into the chamber. To form the duplexes, biotin-modified 100-mers (sequence a1, see Supplementary Data), with either of the non-tagged oligomers 21a2, 50a2 and 70a, were mixed, at a molar ratio of 2:3, respectively, in 10-mM Tris–HCl, 100-mM NaCl, 10-mM MgCl2 at pH 7.5, and were hybridized by cooling from 90°C to 20°C over 5 h. The suffix a2 was added to denote strands fully complementary to a portion of a1 beginning at the 3′-end of a1, so that the a1 and a2 hybridization results in a duplex having a biotin-modified single-stranded overhang through which the duplex is attached to the glass surface (Figure 1A). In the changing synapse length experiment, constructs were prepared by hybridization of a1 with 21a2, 50a2 or 70a2. All the rest of the experiments were carried out with a duplex formed by a1 and 50a2. For experiments testing synapse lifetime as a function of the length of homologous tract at the 3′-end of the incoming strand, the incoming strands HOM0, HOM15, HOM26, HOM35 and HOM50 were used. For experiments with a varying length of homologous tract at the 5′-end, the incoming strands HOM0, 5HOM26, 5HOM36 and HOM50 were used. For experiments testing the effects of point mutations on synapse lifetime, the incoming strands MM1 and MM3 were used. The sequences that were used are in the Supplementary Data.
Sample preparation for FRET assays
dsDNA was prepared by hybridization of the tagged oligomers Phi50 and Phi60 (see Supplementary Data). The competitor dsDNA was prepared by hybridization of the oligomers COMS and COMAS. The complementary strands were mixed in a 1:1 molar ratio in 10-mM HEPES–KOH (pH 7.5), 100-mM NaCl, 10-mM MgCl2, and were hybridized by cooling from 90°C to 20°C over 5 h. The concentrations of the invading ssDNAs (PhiH50) were 2-fold those of tagged duplexes. FRET measurements were carried out in a reaction buffer containing 20 mM HEPES–KOH (pH 7.5), 3.3 mM ATP and 20 mM MgCl2. In these assays, 200 nM ssDNA and 3.5 µM E. coli RecA (New England Biolabs, Ipswich, MA, USA) were pre-incubated for 5 min at 37°C in the reaction buffer. Following pre-incubation, 100 nM of dsDNA tagged at both strands and the indicated ratio of competitor dsDNA were added and then injected into the measuring chamber as described previously (14). Experiments were carried out at 28°C. The sequences that were used are in the Supplementary Data.
Reaction conditions for single molecule assays
ssDNA (1–5 nM) having the same length as the short strand in the duplex, tagged with TAMRA at its 5′-end and bearing the sequence relevant to each of the experiments, was mixed with 1.5 µM RecA and 2 mM ATP in EB3 buffer containing an oxygen scavenging system [2.5-mM protocatechuic acid, 30 nM protocatechuate 3,4-dioxygenase (18), 1-mM Trolox (19)]. The mixture was then pre-incubated for 10 min at 37°C, in order to allow the pre-synaptic complexes to form. The mixture was then injected into the reaction cell. Using the oxygen scavenging system, the characteristic bleaching time of the fluorophores was ∼15 min.
Data acquisition and analysis of single-molecule data
Frames acquired by the ICCD camera were captured by WinView32 software (Princeton Instruments, Trenton, NJ). The intensity of the molecules was tracked by a version of the ‘IDL Particle Tracking’ software (http://www.physics.emory.edu/∼weeks/idl; 20) adapted to MatLab (Mathworks, Natick, MA, USA) by D. Blair and E. Dufresne (http://physics.georgetown.edu/matlab). Briefly, the images were first spatially filtered with a bandpass filter to eliminate pixel noise and spatial variations in the illumination over the field of view. Then, the tracking procedure was used to generate intensity versus time information of each identified DNA site. The events were then counted and put into a histogram of lifetimes. The process was tested by marking specific molecules manually and acquiring their time traces. In compiling histograms of lifetimes, events lasting for only one frame were not included in order to avoid including camera shot noise (our frame integration time was 2 s, chosen to give a high signal-to-noise ratio).
RESULTS
In this study, we have measured the lifetime of synaptic complexes by visualizing them directly, at the single-molecule level. This technique offers a number of significant advantages over FRET (14) in measuring synapse lifetimes. The estimate based on FRET is indirect, as it is inferred from the retardation effected by a heterologous DNA competitor on a strand-exchange reaction. Moreover, the FRET measurements do not separate between the on rate of the reaction (synapse formation by molecular encounters in solution) and the off rate (reflecting the synapse lifetime). While the FRET estimate was made under the assumption that the rate of molecular encounters is much faster than the inverse lifetime (14), the single molecule study proves directly that the synapse lifetime is a rate limiting step. In order to monitor synaptic complexes for as long as they last, surface anchoring and TIRF, as well as conditions enabling a long lifespan for the fluorophores were chosen.
In order to measure synapse lifetime at the single-molecule level, we used the scheme depicted in Figure 1A. The capture of tagged ssDNA–RecA complexes from solution by anchored duplexes resulted in the appearance of bright spots (Figure 1B). The spots were monitored over time until their disappearance (Supplementary videos 1 and 2 online show typical movies in the heterologous and homologous cases, respectively), and the duration of such events, which we refer to as the synapse lifetime was recorded. Typical intensity versus time traces of events corresponding to ssDNAs heterologous and homologous to the target duplexes are shown in Figure 1C and D, respectively.
The lifetime of heterologous synapses and its dependence on synapse length
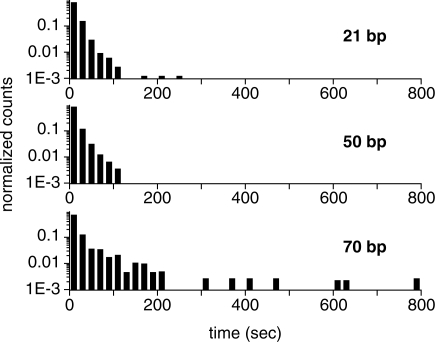
We measured the lifetime of synapses formed between highly heterologous sequences for three synapse lengths: 21, 50 and 70 bp. The corresponding histograms, shown in Figure 2, have been normalized to unit area after neglecting events lasting only one frame (‘Materials and Methods’ section). In these experiments, the incoming and the short, non-anchored strand on the duplex had the same length (Figure 1A), and therefore the lifetime of synaptic complexes of smaller length for which these two strands were not in full registry–i.e. overlapping only along part of their full length—was also probed. It is noteworthy that the lifetimes of events of long duration are of the order of ∼10 s irrespective of synapse length, in agreement with the estimates of the off rate of the synapse formation reaction (14) and FRET assay results presented below, and contradicting assertions that scanning non-specific sites must be extremely fast (10).
Figure 2.
Histograms of lifetimes of individual, fully heterologous synapses versus synapse length. Three different synapse lengths were tested: 21, 50 and 70 bp (sequences 21SR, 50SR and 70SR, respectively; Supplementary Data). Each histogram (total area normalized to 1) comprises a few thousand events, obtained in at least two independent experimental runs.
We note that to accumulate significant statistics in the 21 bp synapse experiment a concentration of ssDNA–RecA complexes up to 30 times higher than in the 50- and 70 bp experiments had to be used (few events were observed when the concentration was similar to that used in experiments with longer synapses). The integration time of our camera (∼2 s) limits us to observing only long events. For a given concentration of incoming ssDNA–RecA complexes, viewing fewer events for the 21-bp case may imply a decreased binding rate, as well as a shift of the lifetime distribution towards shorter times. In conclusion, synapse lifetime increases with length, although few events with lifetimes beyond ∼200 s are seen.
In control experiments, the absence of RecA yielded approximately two events, in comparison to thousands in its presence. To assess the degree of non-specific binding of ssDNA–RecA complexes to the surface, experiments in the absence of anchored duplexes were carried out. We observed events of a wide range of durations, but their number was ∼6% compared to experiments in the presence of duplexes. Taken together, these measurements demonstrate that even in the case of heterology, in which synapse lifetimes should be the shortest, lifetimes can still be seconds long.
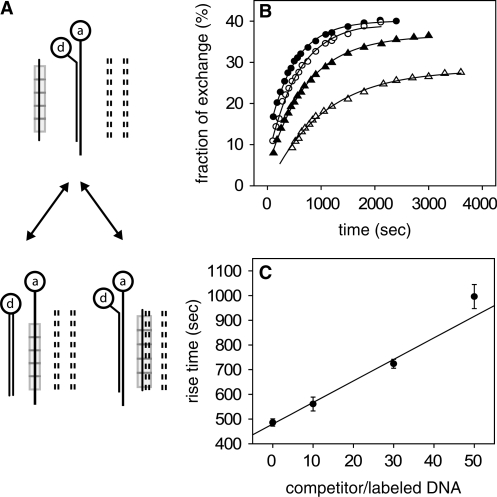
Measurement of average lifetime of heterologous synapses by ensemble FRET
In order to ascertain that the second-long events observed in the single-molecule experiments are representative of the average synapse lifetime, we have extended the FRET measurements in Sagi et al. (14), probing events down to the millisecond timescale. Briefly, 50 bp dsDNA targets tagged at the same end with donor and acceptor moieties were combined with non-tagged fully homologous 50 nt ssDNA–RecA complexes in solution, in the presence of various concentrations of competing, fully heterologous non-tagged dsDNA (Figure 3A). The strand exchange reaction between the targets and the ssDNA–RecA complexes was monitored over time by following the decrease in the measured FRET efficiency, and the time by which this reaction was slowed down by the presence of the heterologous competitor dsDNA was measured. Under the assumption that the rate of the strand exchange reaction is limited by synapse lifetime and not by the rate of molecular encounters, it was proposed in (14) that the increase in the fraction of targets having undergone strand exchange, deduced from the FRET efficiency measurements, should be characterized by a rise time given approximately by
| (1) |
where τ is the rise time in the presence of competitor and τ0 in its absence, nc/n0 is the molar ratio between competitor and target dsDNA concentrations and τs is the average synapse lifetime. Note that the linearity of the rise time with competitor concentration implied by Equation (1) was not previously checked experimentally (14). Figure 3B shows plots of the fraction of strand exchange versus time for the reaction, in the presence of different heterologous competitor concentrations. The decrease in the saturation value with increasing competitor concentration (Figure 3B) is due to the reduction in the effective steady-state concentration of ssDNA by the competitor dsDNA. The balance is between the strand exchange and its opposite reaction, in which tagged ssDNA strands replace non-tagged ones in homologous duplexes. Solid lines represent exponential fits from which the rise time, τ, was extracted. A plot of τ as a function of nc/n0 is shown in Figure 3C. The data are consistent with a linear dependence on nc/n0 as predicted by Equation (1), and the slope of the linear fit to the data yields τs = 10 ± 1 s and is in agreement with results obtained in the single-molecule measurements. We conclude that the long events observed in the single-molecule experiments yield a good estimate of the average synapse lifetime.
Figure 3.
Effects of double-stranded heterologous competitor on the kinetics of strand exchange between fully homologous ssDNA and dsDNA. (A) Experimental scheme to study RecA-induced recombination in the presence of non-tagged competing DNA duplexes (dashed lines) by FRET: an ssDNA substrate covered with RecA (rectangles) is combined with dsDNA tagged at both strands on the same side of the duplex with a donor-acceptor pair, d and a, respectively. The products include both duplexes having undergone strand exchange as well as synaptic complexes with competitor dsDNA. After strand exchange, no energy transfer takes place. RecA monomers dissociate and are then able to reploymerize on ssDNA. (B) Fraction of tagged duplexes having undergone strand exchange as a function of time for different ratios of competitor to tagged dsDNA concentrations, as obtained from FRET experiments: no competitor (full circles); 10-fold (empty circles); 30-fold (full triangles) and 50-fold (empty triangles). The solid lines are exponential fits to the data. (C) Rise time of the curves in B as function of competitor concentration. The straight line is a linear fit to the data. Error bars represent the uncertainties derived from the fits in B.
The effects of sequence homology at the ends of an invading strand on synapse lifetime
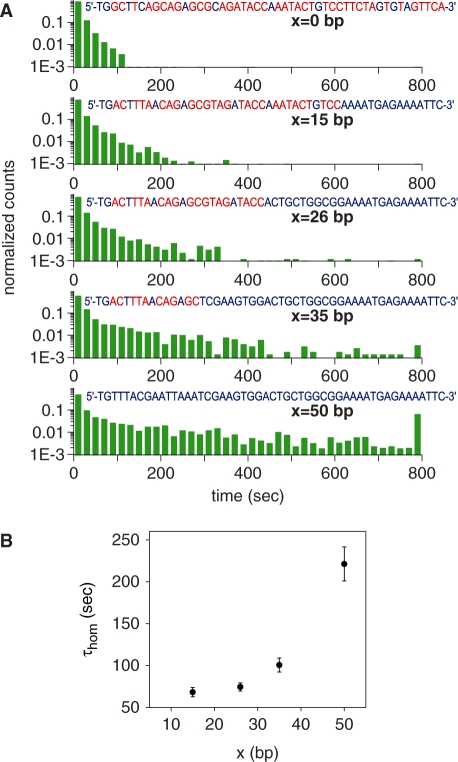
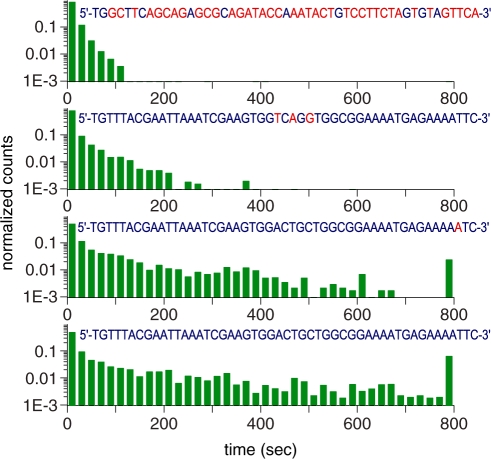
The point of initial pairing between the pre-synaptic complex and the duplex may occur randomly along the length of DNAs involved (21). If local homology is found, a strand exchange reaction proceeds outwards from that point (see ‘Discussion’ section). In addition, it has been shown that there is a strong dependence of the end product of the recombination reaction on the different ends of an invading strand (14). We therefore investigated how synapse lifetimes change when a segment of full homology is included at either the 3′- or 5′-end of the incoming strand. Lifetime measurements were carried out on 50-bp-long synapses formed with invading ssDNAs whose sequences were chosen so that the first x bases at the 3′ end were fully homologous with the target dsDNA, while the rest (50- x bases) shared no homology with the target. As Figure 4A clearly shows, events of long duration become more frequent as x increases (note that the last column in all the histograms includes also all events with lifetimes longer than 800 s). Similar results were observed when 26- or 36-base-long segments of homology were present at the 5′-end of the invading ssDNA strand (see Supplementary Figure S1). A control experiment in which the fluorophore was located at the opposite end of the invading strand yielded similar results. We conjecture that the enhanced stability of synapses formed with a region of homology at the incoming strand is associated with the stability of a nascent duplex.
Figure 4.
Dependence of the lifetime of individual synapses on differences in sequence between the anchored and incoming DNAs. (A) Normalized histograms of lifetimes for five values of x, the length of homologous tract (blue) at the 3′-end of the incoming strand whose sequence is shown (sequences HOM0, HOM15, HOM26, HOM35 and HOM50, see Supplementary Data). Heterologies are shown in red. The last column in the histogram includes also all events with lifetimes longer than 800 s. (B) Dependence of τhom on the length of homology x at the 3′-end of the incoming strands [see Equation (2) and accompanying text]. Error bars represent the uncertainties derived from the fits of the data in A with Equation (2).
Both in vivo and in vitro studies have revealed the existence of a minimum length of homology (∼30 bp), known as the minimum efficient processing segment or MEPS, below which homologous recombination is inefficient (14,22). In order to determine whether our data bear any signature of the MEPS, we assume a minimal model in which observed events are divided into those for which the incoming strand is in full registry with the displaced strand and those for which it is not. There are about 200 configurations out of registry compared to one in registry for 50-base-long strands, considering both parallel and anti-parallel alignments. Within the model, we assume that out-of-registry configurations, all of which are highly heterologous, are characterized by a typical synapse lifetime τhet, which we determine by a single exponential fit to the x = 0 histogram, yielding τhet = 10.45 ± 0.13 s. Assuming a typical synapse lifetime τhom for in-registry events, we make fits of the normalized histograms for 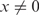 with a sum of two exponentials:
with a sum of two exponentials:
| (2) |
with Ahet, Ahom and τhom as free parameters and τhet as fixed. As shown in Figure 4B, τhom increases rapidly with x from ∼30 bp, in full agreement with results in vivo and in vitro FRET measurements (14,22), and at x = 50 the histogram has becomes massively populated at long times. In this particular case, the fluorescence signal represents mostly events in which strand exchange has been completed and not synapses involving the tagged ssDNA–RecA and the anchored duplex. The duration of events in our measurements is limited from above both by photobleaching and by the time of observation, which cut long events short. We note that τhom does not represent the average synapse lifetime due to these effects, as well as due to the insensitivity of our measurements to events whose duration is shorter than the integration time. Two interesting features of the fits of the data with Equation (2) are noteworthy: the ratio of weights of the two exponentials, 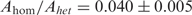 , is nearly constant for all
, is nearly constant for all 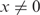 , and its value agrees within an order of magnitude with the ratio of multiplicities of in-registry to out-of-registry configurations (note that of all possible out-of-registry configurations, only those with high enough overlap contribute significantly, as the experiments with 21-bp-long heterologous synapses demonstrate). Allowing τhet to vary as a free parameter in the double-exponential fits yields values that differ from 10.45 s by not more than ∼10%, demonstrating that the separation of timescales between heterologous and partially homologous events is justified.
, and its value agrees within an order of magnitude with the ratio of multiplicities of in-registry to out-of-registry configurations (note that of all possible out-of-registry configurations, only those with high enough overlap contribute significantly, as the experiments with 21-bp-long heterologous synapses demonstrate). Allowing τhet to vary as a free parameter in the double-exponential fits yields values that differ from 10.45 s by not more than ∼10%, demonstrating that the separation of timescales between heterologous and partially homologous events is justified.
Full homology is crucial for a stable nascent duplex
The efficiency of strand exchange can be exquisitely sensitive to the presence of even a single mismatch, and few clustered mismatches reduce significantly the efficiency of strand exchange (14). Here, we tested the hypothesis that these effects should also manifest themselves as an inhibition of the formation of long-lived synapses. The histograms of synapse lifetimes displayed in Figure 5 illustrate the effects of one and three clustered mismatches together with data from fully homologous and highly heterologous sequences, also shown for comparison. Fitting the data with Equation (2) indeed shows that one mismatch reduces τhom from ∼221 s in the fully homologous case to ∼145 s, while a cluster of three mismatches separated from each other by less than three bases, starting at position 25, behaves nearly as if no homology were present after the cluster (compare with x = 15 and 26, Figure 4A). Hence, full homology during the initial stages of the process is a crucial determinant in promoting efficient strand exchange.
Figure 5.
Comparison between normalized lifetime histograms for sequences with either three clustered mismatches at positions 25, 27 and 29 (MM3; Supplementary Data), or one mismatch at position 48 (MM1, Supplementary Data), and histograms for highly heterologous and fully homologous sequences, as observed in single-molecule experiments. Mismatches or heterologies in the invading sequences are shown in red, whereas homologies are in blue. The last column in the histogram includes also all events with lifetimes longer than 800 s.
DISCUSSION
The present study demonstrates that the lifetime of fully heterologous synapses tens of base pairs long can be ∼10 s or longer, posing a severe challenge in a cellular context. This result has been obtained independently by interrogating molecular ensembles as well as single synapse. Our direct single molecule assay allows us to monitor synapses while they last, and to isolate the measurement of their lifetimes from any kinetics of molecular encounters, demonstrating that homology readout can be an intrinsically long process, independent of side reactions such as free RecA oligomerization (3).
The inclusion of a region of homology at either the 3′ or 5′ end of the incoming strand leads in general to the appearance of events with longer lifetimes, hinting at an enhancement of synaptic stability. The enhancement is, however, mild, in spite of the fact that the region of full homology can extend up to ∼30 bases, suggesting the synaptic complex is still unstable and dissociates within few tens of seconds. We conjecture that the enhanced stability of synapses formed with a region of homology at the ends of the incoming strand is associated with the stability of a nascent duplex, and these considerations determine the MEPS lengthscale (14,22).
RecA-mediated strand exchange is a process that consists of several steps. Our experiments show that the early step of synapse formation is not sensitive to whether a homologous tract is located at the 3′- or 5′-end of the invading strand. However, the final result of the process, namely the yield of a heteroduplex and a fully displaced strand, has been found to be much more sensitive to mismatches near the 3′-end of the invading strand rather than near the 5′-end (14). Consistent with this, it has also been reported that a homologous 3′-end is more invasive than a homologous 5′ one (23–25). This has been attributed to the higher likelihood of the 3′-end to be covered by RecA polymerization, which proceeds in the 5′- to 3′-direction (26,27). Other experiments have reported a higher invasiveness of the 5′-end (28). Our single molecule experiments, conducted under conditions in which the high concentration of RecA most likely ensured the complete coverage of the ssDNA, did not reveal a difference in invasiveness between the 3′- and 5′-ends.
The directionality of RecA-mediated homologous recombination has been extensively discussed in the literature. Initial pairing can occur at random points along homologous regions of the participating DNAs and extends in both directions from the point of initial pairing (21). The initial pairing involves local strand exchange (29). While our results do not rule out nascent duplex formation occurring randomly along the homologous tract on the ssDNA, we believe it is more likely that invasion occurs preferably from the incoming strand’s ends, due to the short length of the DNA used [for an extensive discussion see (29)].
Consistent with the nascent duplex interpretation, the presence of a single base mismatch in an otherwise fully homologous invading strand has a noticeable effect suppressing events of long duration. In addition, an incoming strand with three closely clustered mismatches so that adjacent ones were separated by no more than three bases (the site covered by each RecA monomer) reduced significantly synapse lifetimes. Thus, synapse lifetime is highly sensitive to differences in sequence and is an important element in the comparison of sequences during the search for homology.
It has previously been reported that RecA-mediated strand exchange can traverse substitutional heterologies when using non-hydrolyzable ATP analog, ATPγS, instead of ATP (30). We found in our single-molecule experiments that ATPγS induced an irreversible aggregation of labeled oligomers on the non-labeled dsDNA anchored on the surface (data not shown) and individual synapses could not be resolved. Similar effects were previously observed in ensemble experiments in solution (14).
Synapse lifetimes are observed to increase when the synapse length is increased between 21 and 70 bp in the case of highly heterologous synapses. It is noteworthy that in the 21-long synapse case events of long enough duration to be detected were rare unless the concentration of pre-synaptic complexes was increased significantly. This suggests that heterologous synapses of this length are highly unstable.
Taken together, our data led us to suggest that facilitated/parallel search mechanisms may be essential to locate homology along chromosomes: the combination of long synapse lifetimes together with homology search by random collisions as commonly held (11,12) may take prohibitively long times. While sliding appears not to occur (3), the simultaneous formation of synapses between a RecA-covered ssDNA and a number of chromosomal segments, intersegmental transfer and inchworming may be crucial mechanisms to overcome slow homology readout (4). It remains a challenge to study if these or other mechanisms are operative in the cell. One cannot, however, dismiss the possibility that in vivo, not all the genome is available for scanning during the search for homology, due to packing considerations and the association of genomic DNA with other proteins (31), and that homology may only be found in a subpopulation of the cells.
The present experiments pave the way for carrying out similar investigations into the behavior of eukaryotic recombinases. In contrast to RecA, homology search processes in eukaryotic organisms may be assisted not only by recombinases, but by additional factors (12,32,33), although evidence exist that a Rad51–ssDNA nucleofilament is sufficient to capture nucleosomal homology during double-strand break repair by recombination (34). Further studies will elucidate whether eukaryotic and prokaryotic organisms employ the same search strategies. Be that as it may, synapse lifetimes are a fundamental ingredient that must be taken into account in the construction of models of homology search. Long synapse lifetimes will provide strong constraints on such models.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Minerva Foundation with funding from the Federal German Ministry for Education and Research.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Amnon Amir and Asaf Tal for very helpful comments and suggestions.
REFERENCES
- 1.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia-coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adzuma K. No sliding during homology search by RecA protein. J. Biol. Chem. 1998;273:31565–31573. doi: 10.1074/jbc.273.47.31565. [DOI] [PubMed] [Google Scholar]
- 4.Dutreix M, Fulconis R, Viovy J.-L. The search for homology: a paradigm for molecular interactions? ComPlexUs. 2003;1:89–99. [Google Scholar]
- 5.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 6.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee AM, Xiao J, Singleton SF. Origins of sequence selectivity in homologous genetic recombination: insights from rapid kinetic probing of RecA-mediated DNA strand exchange. J. Mol. Biol. 2006;360:343–359. doi: 10.1016/j.jmb.2006.04.065. [DOI] [PubMed] [Google Scholar]
- 8.Folta-Stogniew E, O'M;alley S, Gupta R, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E-coli RecA protein. Mol. Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Rould E, Muniyappa K, Radding CM. Unwinding of heterologous DNA by RecA protein during the search for homologous sequences. J. Mol. Biol. 1992;226:127–139. doi: 10.1016/0022-2836(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 10.Yancey-Wrona JE, Camerini-Otero RD. The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr. Biol. 1995;5:1149–1158. doi: 10.1016/s0960-9822(95)00231-4. [DOI] [PubMed] [Google Scholar]
- 11.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 12.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 13.Barzel A, Kupiec M. Finding a match: how do homologous sequences get together for recombination? Nat. Rev. Genet. 2008;9:27–37. doi: 10.1038/nrg2224. [DOI] [PubMed] [Google Scholar]
- 14.Sagi D, Tlusty T, Stavans J. High fidelity of RecA-catalyzed recombination: a watchdog of genetic diversity. Nucleic Acids Res. 2006;34:5021–5031. doi: 10.1093/nar/gkl586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokunaga M, Kitamura K, Saito K, Iwane AH, Yanagida T. Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem. Biophys. Res. Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 18.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 20.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J. Coll. Inter. Sci. 1996;179:298–310. [Google Scholar]
- 21.Morel P, Stasiak A, Ehrlich SD, Cassuto E. Effect of length and location of heterologous sequences on RecA-mediated strand exchange. J. Biol. Chem. 1994;269:19830–19835. [PubMed] [Google Scholar]
- 22.Shen P, Huang HV. Homologous recombination in Escherichia-coli - dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konforti BB, Davis RW. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc. Natl Acad. Sci. USA. 1987;84:690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konforti BB, Davis RW. The preference for a 3′ homologous end is intrinsic to RecA-promoted strand exchange. J. Biol. Chem. 1990;265:6916–6920. [PubMed] [Google Scholar]
- 25.Friedman-Ohana R, Cohen A. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc. Natl Acad. Sci. USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Register JC, 3rd, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 27.Stasiak A, Egelman EH, Howard-Flanders P. Structure of helical RecA-DNA complexes. III. The structural polarity of RecA filaments and functional polarity in the RecA-mediated strand exchange reaction. J. Mol. Biol. 1988;202:659–662. doi: 10.1016/0022-2836(88)90293-8. [DOI] [PubMed] [Google Scholar]
- 28.McIlwraith MJ, West SC. The efficiency of strand invasion by Escherichia coli RecA is dependent upon the length and polarity of ssDNA tails. J. Mol. Biol. 2001;305:23–31. doi: 10.1006/jmbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 29.Adzuma K. Stable Synapsis of Homologous DNA-Molecules Mediated by the Escherichia-Coli Reca Protein Involves Local Exchange of DNA Strands. Genes Dev. 1992;6:1679–1694. doi: 10.1101/gad.6.9.1679. [DOI] [PubMed] [Google Scholar]
- 30.Bucka A, Stasiak A. RecA-mediated strand exchange traverses substitutional heterologies more easily than deletions or insertions. Nucleic Acids Res. 2001;29:2464–2470. doi: 10.1093/nar/29.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stavans J, Oppenheim A. DNA-protein interactions and bacterial chromosome architecture. Phys. Biol. 2006;3:R1–R10. doi: 10.1088/1478-3975/3/4/R01. [DOI] [PubMed] [Google Scholar]
- 32.Aylon Y, Kupiec M. DSB repair: the yeast paradigm. DNA Repair (Amst) 2004;3:797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 34.Sinha M, Peterson CL. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol. Cell. 2008;30:803–810. doi: 10.1016/j.molcel.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.