SUMMARY
Enhancers integrate spatiotemporal information to generate precise patterns of gene expression. How complex is the regulatory logic of a typical developmental enhancer, and how important is its internal organization? Here, we examine in detail the structure and function of sparkling, a Notch- and EGFR/MAPK-regulated, cone cell-specific enhancer of the Drosophila Pax2 gene, in vivo. In addition to twelve previously identified protein binding sites, sparkling is densely populated with previously unmapped regulatory sequences, which interact in complex ways to control gene expression. One segment is essential for activation at a distance, yet dispensable for other activation functions and for cell-type patterning. Unexpectedly, rearranging sparkling’s regulatory sites converts it into a robust photoreceptor-specific enhancer. Our results show that a single combination of regulatory inputs can encode multiple outputs, and suggest that the enhancer’s organization determines the correct expression pattern by facilitating certain short-range regulatory interactions at the expense of others.
INTRODUCTION
Enhancers, or cis-regulatory elements, are the primary determinants of spatiotemporal patterns of gene expression. In order to properly regulate their target genes, enhancers must perform a number of functions, such as identifying and communicating with the promoter, sometimes over great distances, and triggering transcription in certain cells, but not in others. Many enhancers are capable of driving a heterologous promoter in the proper pattern when removed from their normal genomic context. This autonomy implies that enhancers can assemble a complete set of biochemical activities that together are sufficient for robust, patterned transcriptional activation at a remote promoter. Do different DNA-binding factors recruit distinct types of activation activities, or must the enhancer merely accumulate enough of a single, limiting activity to exceed a threshold for activation?
Different types of studies reach widely divergent conclusions about enhancer complexity. For example, Eric Davidson and colleagues, combining reporter assays with affinity purification in an extensive study of cis-regulatory logic in the sea urchin Endo16 gene, identified 55 binding sites for 16 regulatory proteins, which form an intricate regulatory computer spanning 2300 bp of DNA (Davidson, 1999). On the other hand, most developmental genetics-based enhancer studies culminate in models requiring no more than three to five different regulators (often only one or two), binding within ~300–1000 bp of DNA, to explain the activity and specificity of a seemingly typical enhancer. In the very rare cases where the question of sufficiency is addressed in vivo, the defined regulatory sites are generally insufficient to properly reconstitute enhancer function, and an unknown activator “X” is added to the model (reviewed by Barolo and Posakony, 2002). How many cis-regulatory sites are sufficient, when combined, to recapitulate normal enhancer function, in the context of a chromosome in a normal cell?
We have pursued a bottom-up approach to these questions by taking a previously well-characterized developmental enhancer and exhaustively dissecting it in vivo, both to discover the extent of its regulatory complexity and to determine whether different enhancer sub-elements perform distinct functions. We chose to study the sparkling (spa) enhancer of the dPax2 gene, which is necessary and sufficient to specify the cone cell fate in certain multipotent cells in the developing Drosophila eye (Fu and Noll, 1997; Fu et al., 1998; Flores et al., 2000; Shi and Noll, 2009). spa drives cone cell-specific dPax2 expression in response to four direct regulators, acting through twelve transcription factor binding sites (TFBSs): Suppressor of Hairless [Su(H)], under the control of Notch signaling; two Ets factors, the activator PointedP2 (Pnt) and the repressor Yan, both controlled by EGFR/Ras/MAPK signaling; and the Runx-family protein Lozenge (Lz) (Fu et al., 1998; Flores et al., 2000; Tsuda et al., 2002) (Figure 1A). In their report describing the direct regulation of the spa enhancer by Su(H), Lz, and Ets factors, Flores et al. (2000) proposed a model in which a combinatorial code, Lz + EGFR/Pnt/Yan + Notch/Su(H), determines the cell type specificity of spa activity. The authors were careful to state that “the model…reflects requirements rather than sufficiency for cell fate specification.” Despite this caveat, the Lz+Ets+Su(H) code is now considered to “define the combinatorial input required for cone cell specification” (Voas and Rebay, 2004; see also Pickup et al., 2009; Shi and Noll, 2009).
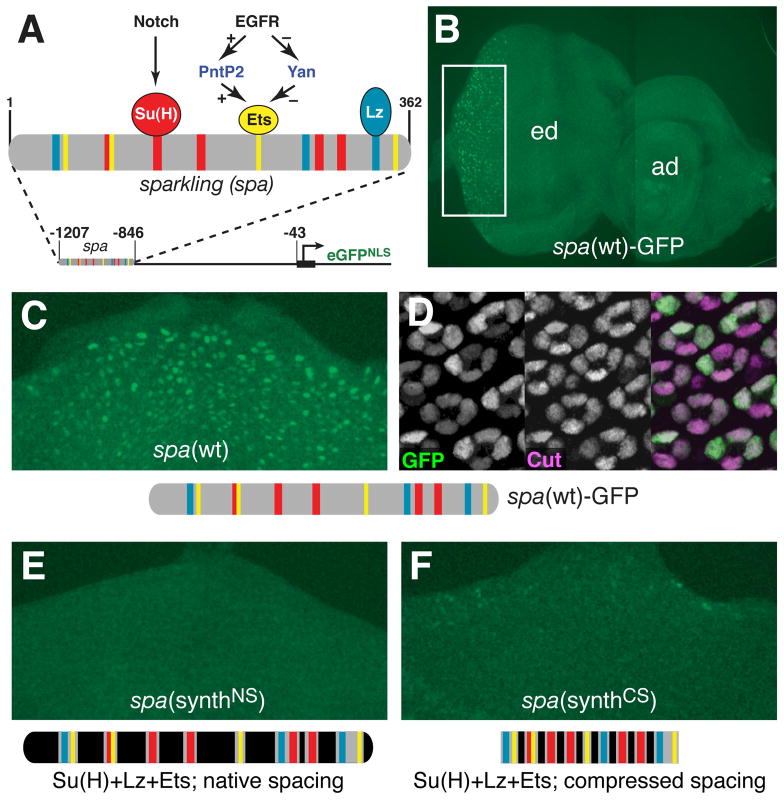
Figure 1. The Known Regulators of spa Are Insufficient for Transcription in Cone Cells.
(A) Summary of the known regulatory inputs of the sparkling (spa) cone cell enhancer of dPax2. Defined TF binding sites (TFBSs) are shown as colored bars; uncharacterized sequences are grey. The enhancer is placed 846 bp upstream of the transcription start site in all transgenic constructs, except those in Figure 4.
(B–D) Expression of a GFP transgene under the control of spa. (B) Eye-antennal imaginal disc from a spa-GFP transgenic larva. (C) The posterior of an eye disc, corresponding approximately to the boxed area in (B). Posterior is to the top. (D) Eye of a 24-hour pupa carrying spa(wt)-GFP, stained with antibodies against GFP (green) and the cone cell nuclear marker Cut (magenta).
(E) spa(synthNS), in which the previously uncharacterized sequences have been altered (black), but the 12 defined TFBSs are present in their native arrangement and spacing. (F) spa(synthCS), containing the 12 TFBSs in compressed spacing.
Because the spa enhancer is small (362 bp), and because the known regulatory inputs could, in theory, explain its cell-type specificity (Flores et al., 2000), we considered it an ideal test case for a comprehensive structure-function analysis. Here, we report the results of our initial tests, which reveal several surprising aspects of spa enhancer function in vivo.
RESULTS
For our in vivo analysis of the spa enhancer, we used a specially built Gateway reporter transgene vector, Ganesh-G1, in which enhancers are placed upstream of a minimal, TATA-containing promoter taken from the Drosophila Hsp70 gene, driving an EGFP-NLS reporter (Swanson et al., 2008). An important feature of this vector is that the enhancer is placed 846 bp upstream from the transcription start site (Figure 1A), so that in all experiments presented here (except those in Figure 4), the enhancer is forced to act at a moderate distance from the promoter. We do not consider this an unfair test of enhancer activity, given that, in its native genomic context, spa is located >7 kb from the dPax2 promoter (Fu et al., 1998). We generated at least 4 independent transgenic lines for each reporter construct. Because line-to-line variability was generally low, we found that examination of 3–5 independently derived lines was sufficient for most constructs. For constructs with more variable expression (usually those with low activity), we examined additional lines (10–14) to ensure that our conclusions were not based on rare insertion effects. Supplemental Table S1 lists all transgenic lines and their expression levels.
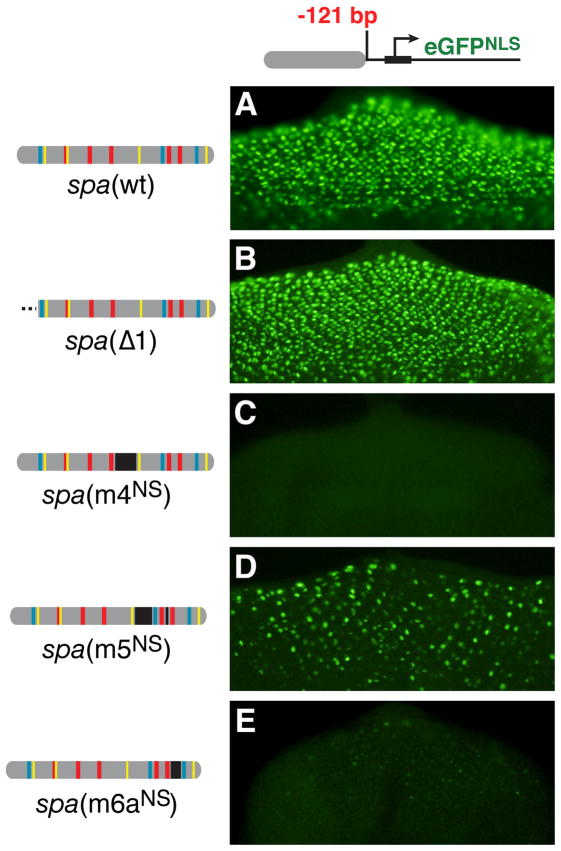
Figure 4. Region 1 Is Required for Activation at a Distance, But Not for Patterning.
(A–E) Transgenic larval eye discs. In this figure, all enhancers are proximal to the minimal Hsp70 promoter, at position −121 from the transcription start site, compared to −846 in all other figures. Because spa drives stronger expression from a promoter-proximal position, these images were collected at a lower exposure setting than those in other figures.
When placed in Ganesh-G1, spa drives cone cell-specific GFP expression in developing retinas of transgenic larvae and pupae (Figures 1B-1D). This and previous work by Flores et al. (2000) demonstrate that the 362-bp spa enhancer contains all sequences necessary to (1) activate gene expression in vivo and (2) restrict this activation to developing cone cells.
The [Lz + Pnt + Su(H)] Code Is Insufficient to Specify Cone Cell Expression
All three of the known positive regulators of the spa enhancer are required for its activity and cone cell specificity. This suggested a “combinatorial code” model for dPax2 regulation, in which the combined activities of Lz, Pnt, and Su(H), acting through binding sites in spa, cooperatively activate dPax2 expression specifically in cone cells (Flores at al., 2000; Tsuda et al., 2002; Nagaraj and Banerjee, 2007). We began our analysis by testing the simplest form of such a model, which predicts that the binding sites within spa that mediate those three regulatory inputs should suffice, in combination, to drive gene expression in cone cells.
First, we built a synthetic spa enhancer construct in which all twelve of the defined binding sites for Lz, Su(H), and Pnt/Yan within spa are intact (along with 3–4 flanking base pairs to either side), and are placed in their native arrangement and spacing—but in which all other enhancer sequences are mutated by altering every second base pair. This construct, called spa(synthNS) because of the native spacing of its TFBSs, fails to activate gene expression in vivo (Figure 1E). A second version of spa(synthNS), in which the opposite set of base pairs was mutated, produced the same result (not shown). We also created spa(synthCS), a compressed-spacing construct containing the same twelve sites, in which inter-site sequences of >12 bp have been reduced to 12 bp. spa(synthCS) also fails to act as a cone cell enhancer, although weak GFP expression can be detected in a few non-cone cells (Figure 1F). Based on these findings, we hypothesized that additional sequences, besides the twelve defined regulatory sites, are necessary for proper transcriptional regulation mediated by spa.
Numerous Regulatory Sites Within spa, in Addition to the Known Binding Sites, Are Required for Cone Cell Activation
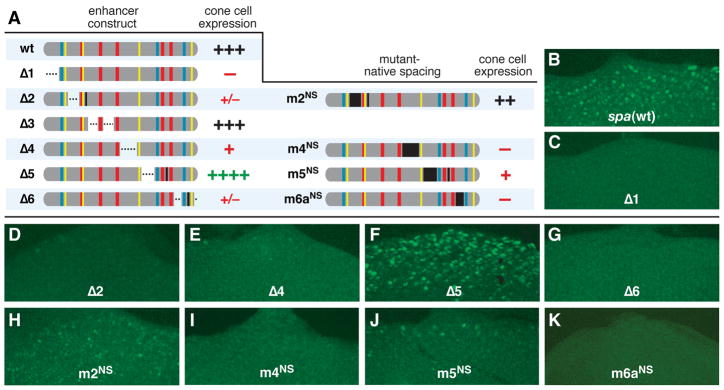
In order to pinpoint the regulatory sites within spa that make essential contributions to enhancer activity in vivo, we conducted a systematic mutational analysis of all previously uncharacterized sequences within spa. These sequences were divided into regions 1 through 6, and each region was deleted in turn, leaving the known TFBSs intact in all cases (Figure 2A). Of all segments mutated in this manner, only region 3 makes no significant contribution to cone cell expression. Deleting regions 1, 2, 4, or 6 causes total or near-total loss of gene expression in vivo; conversely, deleting region 5 enhances expression in cone cells (Figures 2A–2G).
Figure 2. Sequence and/or Spacing Constraints Apply to Multiple Segments of spa.
(A) Diagrams of spa enhancer constructs and summary of their cone cell activity in larval eye discs. Dotted lines indicate deletions; black bars indicate mutations that preserve native spacing (NS). In each case, the 12 known TFBSs are preserved. +++, wild-type levels and pattern of expression in cone cells; ++, moderately reduced; +, severely reduced; +/−, detectable in very few cells;−, no detectable expression; ++++, augmented levels of expression.
(B–K) GFP expression in eye imaginal discs driven by the wild-type spa enhancer (B) and mutant enhancers (C–K) carrying deletions or NS mutations in previously uncharacterized sequences, numbered 1 through 6.
Spacing vs. Sequence
Internal deletions of enhancer DNA cause two simultaneous changes: loss of the deleted sequence, and altered relative spacing of the sites to either side. To distinguish between these two types of effects, we made native-spacing (NS) mutations in which a specific sequence was altered, but its length was preserved. In regions 4 and 6, native-spacing alterations and deletions have similar effects, indicating that the sequence content of these regions is functionally significant (Figures 2D, 2G, 2H, and 2K). However, a native-spacing mutation in region 2 has a less severe effect than a deletion (Figure 2H; cf. Figure 2D), from which we infer that much of the regulatory contribution of region 2 can be attributed to its length, rather than its sequence.
Within region 5, deleting the DNA and altering its sequence have opposing effects. Deleting region 5 augments cone cell expression, while a native-spacing mutation causes a severe loss of activity (Figures 2F and 2J). The simplest interpretation of these results is that region 5 harbors positive regulatory sequences that are normally required, but that the deletion brings together sites on either side of region 5, increasing synergy between TFs and thus compensating for the loss of regulators normally binding to region 5. Consistent with this interpretation is the fact that Pnt and Lz, which bind to either side of region 5, physically interact and synergistically activate transcription, as can mammalian orthologs of these factors (Flores et al., 2000; Behan et al., 2005 and references therein). The fact that multiple smaller-scale NS mutations within region 5 impair spa function, while none augment expression (see Figure 3), further supports this conclusion.
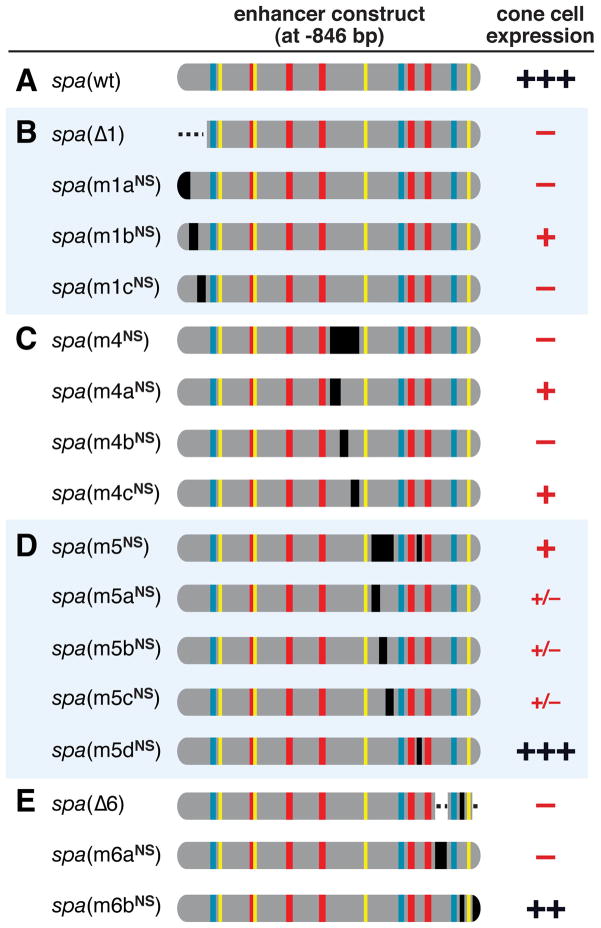
Figure 3. Most of spa Is Composed of Critical Regulatory Sequences.
(A–E) Diagrams of mutated spa enhancer constructs. Blue, yellow and red bars indicate defined binding sites for Lz, Pnt/Yan, and Su(H), respectively. Dotted lines indicate deletions; black bars indicate mutations that preserve native spacing (NS). GFP expression in larval cone cells is summarized as in Figure 2.
spa Is Densely Packed With Regulatory Sites
The above analysis demonstrates that, in addition to the defined TFBSs, regions 1, 4, 5, and 6 of spa (and to a lesser extent region 2) are essential for its proper function. Each of these segments is large enough to contain several protein binding sites of typical size. To determine what proportion of these sequences has a regulatory role, we made native-spacing mutations to smaller segments (10 bp, on average) within regions 1, 4, 5, and 6. Of these 12 finer-scale mutations, ten cause severe or total loss of gene expression in cone cells (Figure 3). In addition, results described below indicate the presence of repressive regulatory site(s) within spa, but outside of regions 1/4/5/6. Given that the consensus binding sites for the known regulators of spa are <9 bp in length, there is room for many regulatory sites within these regions. Together, the regulatory sites described here and the previously described TFBSs densely populate spa, with apparent “junk” or “spacer” sequences constituting a small proportion of the enhancer.
To investigate the possibility that the regulatory sites in regions 1, 4, 5, and 6 act by facilitating binding of the known activators to nearby binding sites, and the related possibility that these regions contain cryptic or non-canonical binding sites for the known activators, we tested the ability of Lz and Su(H) to bind to sites within spa in vitro. In all cases, mutating the newly characterized essential regulatory sequences did not significantly reduce the affinity of Lz or Su(H) for nearby binding sites, as determined by EMSA competition experiments (Table S2). Pnt does not bind in vitro to any sites flanking regions 1, 4, 5, or 6 (Flores et al., 2000). Therefore, in subsequent experiments we pursued the possibility that the newly characterized regions of spa have functions that differ from those of the Lz/Ets/Su(H) binding sites.
Evidence for a Special Type of Regulatory Site, Specifically Mediating Action at a Distance
The mutational analysis described above defined many regulatory sites of equal importance to the known Lz/Ets/Su(H) sites. We next attempted to isolate and study an important but poorly understood function of the enhancer: activation at a distance. As mentioned above, all of the enhancer constructs described thus far were placed 846 bp upstream of the promoter, thus forcing them to act over a moderate distance. If we could rescue the activity of a mutant enhancer by moving it close to the promoter, we reasoned, the mutated region is likely to specifically mediate remote enhancer-promoter interactions. Conversely, if a mutation cannot be rescued by promoter-proximal placement, it is likely to mediate a different step in gene activation.
The wild-type spa enhancer drives the same pattern from −121 bp as from −846 bp (Figure 4A), although activation is noticeably more robust from the more proximal position. A mutant spa enhancer lacking region 1 [spa(Δ 1)], which is transcriptionally dead at −846 bp (Figure 2C), is completely rescued by placement at position −121, driving robust gene expression in the normal pattern (Figure 4B). By contrast, enhancers with mutations in regions 4, 5, or 6a remain unable to drive wild-type levels or patterns of gene expression at −121 (Figures 4C–4E). Interestingly, each of these constructs partially recovers cone-cell activity by mid-pupal stages (not shown), suggesting that these regions may be more critical for initiation than for maintenance of gene expression. Similarly, Lz, Pnt, and Su(H) binding sites are required even when spa is promoter-proximal (Flores et al., 2000). Of all regulatory sites within spa, only region 1 is both dispensable for enhancer activity and patterning in a promoter-proximal position, and essential for activation at a distance.
To our knowledge, this is the first case of a regulatory element found within an enhancer that specifically mediates action from a remote position, with no apparent role in patterning of gene expression or other basic activation functions (see Discussion). We therefore refer to region 1 as a “remote control” element to functionally distinguish it from patterning elements within spa, which include the defined TFBSs as well as newly mapped patterning sites to be discussed below. Future experiments will test the range, potential promoter preferences, and functional properties of this intriguing regulatory element.
Unlike the Known TFs, Region 1 Acts Independently of Its Position Within spa
Having mapped all essential regulatory sites within spa, we could then ask whether their linear organization influences gene expression in vivo. First, we tested the structural flexibility of region 1, the remote control element (RCE), by moving it from the 5′ end to the 3′ end of the enhancer. This rearranged enhancer performs normally at −846 bp (Figure 5G), which indicates that the precise position of the RCE, relative to the other regulatory sites within spa, is not a critical factor in its remote activation function. Future experiments will determine the distance, relative to the enhancer and to the promoter, over which the RCE can act.
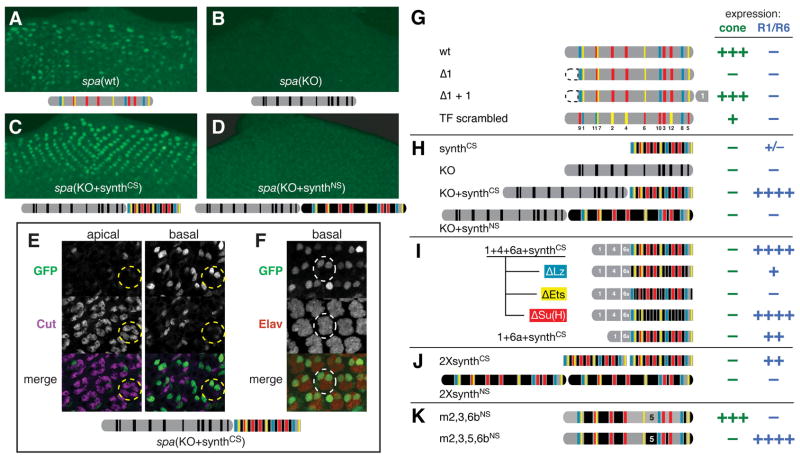
Figure 5. Cell-Type Specificity of spa Is Controlled by the Arrangement of Its Regulatory Sites.
(A–D) GFP expression driven by spa enhancer constructs in larval eye discs. All constructs shown here are placed at −846 bp. (A) spa(wt). (B) spa(KO), in which all 12 Lz/Ets/Su(H) sites are mutated. (C) A rearranged version of spa, in which spa(KO) is placed next to the 12 TFBSs to create spa(KO+synthCS). (D) spa(KO+synthNS), in which the TFBSs are placed in their native spacing next to spa(KO).
(E and F) spa(KO+synthCS) is expressed specifically in photoreceptors (PRs), but not in cone cells, in 24-hour pupae. (E) Confocal images at two different planes, in retinas stained with antibodies against GFP (green) and the cone cell nuclear marker Cut (magenta), show GFP in two nuclei per ommatidium, located basally to cone cells. Posterior is to the top. (F) GFP driven by spa(KO+synthCS) co-localizes with the PR marker Elav (red).
(G–J) Organization of regulatory elements within spa is critical for both transcriptional activity and cell-type specificity. (G) Effects of relocating region 1 (the remote control element or RCE), or of scrambling the locations of the known TFBSs, on enhancer function. (H) Rearranging the regulatory sites of spa converts its cell-type specificity. (I) Creation of a minimal synthetic R1/R6-specific element. (J) 2XsynthCS and 2XsynthNS, both of which contain two copies of all known TFBSs.
(K) Region 5 of spa mediates repression in PRs, as well as activation in cone cells.
By contrast, the Lz/Ets/Su(H) binding sites show strong position dependence. We rearranged these sites within spa by moving each TFBS (along with flanking sequences) to the position of another, randomly chosen, TFBS. The resultant construct, spa(TF scrambled), is only weakly active in cone cells (Figure 5G). Thus, unsurprisingly but in contrast to the RCE, the configuration of the known TFBSs within spa plays an important role in enhancer function in cone cells.
Cell Type Specificity Is Controlled by the Structural Organization of spa
The diminished activity of spa(TF scrambled), along with the altered gene expression resulting from deletions in regions 2 and 5, suggest that the spatial organization of spa impacts its transcriptional activity. We next took a different approach to investigate the relationship between structure and function within spa.
As we have demonstrated, the 12 defined TFBSs within spa are insufficient for cone cell enhancer activity, even when combined. Likewise, when these TFBSs are mutated, the remaining sequences are incapable of driving transcription [spa(KO), Figure 5B]. Since these two constructs, taken together, include all sequences from spa, we tested whether combining them would reconstitute enhancer activity. The resultant rearranged spa construct, KO+synthCS, drives strong gene expression in the eye (Figure 5C).
Three aspects of this finding are worth noting. First, the activity driven by KO+synthCS is robust, exceeding spa(wt) in intensity (Figure 5C; cf. panel A). The defined TFBSs, therefore, are capable of acting synergistically with newly mapped activator sites in spa, even when the enhancer is reconfigured. This, combined with the in vitro binding data mentioned above, strongly suggests that the regulatory sites we have identified are not merely extended binding sequences for Lz/Pnt/Su(H).
Second, when the TFBSs adjacent to spa(KO) are spread out to mimic their native spacing, gene expression is lost (KO+synthNS, Figure 5D). The activity of spa is apparently highly dependent on close proximity, among the known TFs and/or between those TFs and previously uncharacterized regulatory sites. Since KO+synthCS and KO+synthNS differ by only 29% in total length, and because KO+synthNS, at 730 bp, is not large compared to many enhancers, this extreme dependence on short-range interactions was surprising.
Third, and most importantly, the pattern of gene expression driven by the rearranged enhancer spa(KO+synthCS) differs from that of spa(wt)—in fact, the two elements drive completely non-overlapping expression patterns. Unlike spa(wt), whose activity co-localizes with the cone cell marker protein Cut (Figure 1D), KO+synthCS-GFP is expressed only in nuclei located basally to Cut+ cells (Figure 5E). KO+synthCS is active in a subset of basal cells expressing Elav, a marker of photoreceptor cell fate (Robinow and White, 1988). Based on the position of the two GFP+ cells within the Elav+ photoreceptor cluster, spa(KO+synthCS)’s activity is restricted to photoreceptors 1 and 6 (R1/R6) (Figure 5F). Thus, merely re-arranging the regulatory sites within spa is sufficient to cleanly switch its cell-type specificity in vivo.
Ectopic Photoreceptor-Specific Transcription Depends on Lz + Ets Sites, Multiple Newly Mapped Regulatory Sequences, and Tight Clustering of Regulatory Sites
We next attempted to identify the regulatory sites responsible for ectopic activity of spa in photoreceptors (PRs). Combining regions 1, 4, and 6a with the known TFBSs (1+4+6a+synthCS) results in strong R1/R6 expression; removing region 4 from this construct weakens its activity (Figure 5I). By selectively mutating TFBSs, we found that R1/R6 expression requires Lz and Ets sites, but not Su(H) sites (Figure 5I). This is consistent with the fact that R1/R6 receive MAPK signaling and express Lz at high levels, but do not respond to Notch signaling (reviewed by Voas and Rebay, 2004).
Based on our remote vs. proximal enhancer analysis (Figure 4), we hypothesized that different regulatory sequences within spa contribute distinct activities to gene activation. If this is so, one type of activity may not be able to functionally substitute for another. We tested this idea by creating tandem repeats of the synthCS and synthNS constructs, which contain two copies of each known TFBS, in compressed or native spacing, respectively. 2XsynthCS is inactive in cone cells and relatively weakly active in PRs, while 2XsynthNS is inactive in all cell types (Figure 5J). We therefore conclude that the Lz+Ets+Su(H) combination is insufficient for cone cell activation. Further, the fact that additional Lz/Ets/Su(H) sites fail to compensate for the missing sequences adds support to the idea that some parts of the enhancer perform functions in transcriptional activation that are qualitatively distinct from those of the known regulators.
Interestingly, when spa(synthNS) is placed at −121 bp, we observe occasional position-effect-dependent activity in cone cells (1 out of 7 lines) or PRs (1 of 7 lines) (Figure S1). The pattern of gene expression in these two lines depends on the site of transgene insertion, which is consistent with the conclusion that Lz+Ets+Su(H) can contribute to gene expression in multiple cell types, but only in combination with additional regulatory inputs.
A Short-Range, Cell Type-Specific Repressor Activity Prevents spa Activation in Photoreceptors
In both spa constructs driving strong ectopic R1/R6 activity, spa(KO+synthCS) and spa (1+4+6a+synthCS), the configuration of defined TFBSs differs from wild-type in two respects: their spacing relative to one another is reduced, and their linear order and position relative to the newly mapped regulatory sequences is altered. Ectopic photoreceptor expression, then, could have three possible (non-exclusive) causes: (1) tight TF clustering may increase synergy by Lz and Pnt in R1/R6, or altered spacing between TFs and newly mapped sites may cause (2) inappropriate synergistic activation and/or (3) weakened repressive interactions in PRs. In order to test these models, and to further explore the role of enhancer structure, we generated compound mutations in multiple regions of spa, while keeping the spacing/arrangement of the remaining sequences intact.
First, we simultaneously mutated regions 2, 3, and 6b of spa, none of which are essential for cone cell expression. This construct, spa(m2,3,6bNS), is comparable to spa(wt) in its pattern and levels of expression (Figure 5K). Next, we additionally mutated region 5 in this construct to create spa(m2,3,5,6bNS). Remember that when region 5 alone is mutated, cone cell expression is severely reduced, and no ectopic expression is seen (Figures 2J and 3D). However, when region 5 is mutated simultaneously with 2/3/6b, a discrete switch from cone cell- to R1/R6-specific expression occurs (Figure 5K). Therefore, region 5 mediates repression in PRs, in addition to activation in cone cells. This repressive activity must be redundant with additional repressor site(s) in regions 2/3/6b. It must also have a very limited range of action, since moving Lz and Ets sites to the 3′ end of the enhancer, without altering the repressor sites (KO+synthCS), de-represses spa in R1/R6.
spa Enhancer Evolution: Function Is Conserved Despite Rapid Turnover of Regulatory Sequences
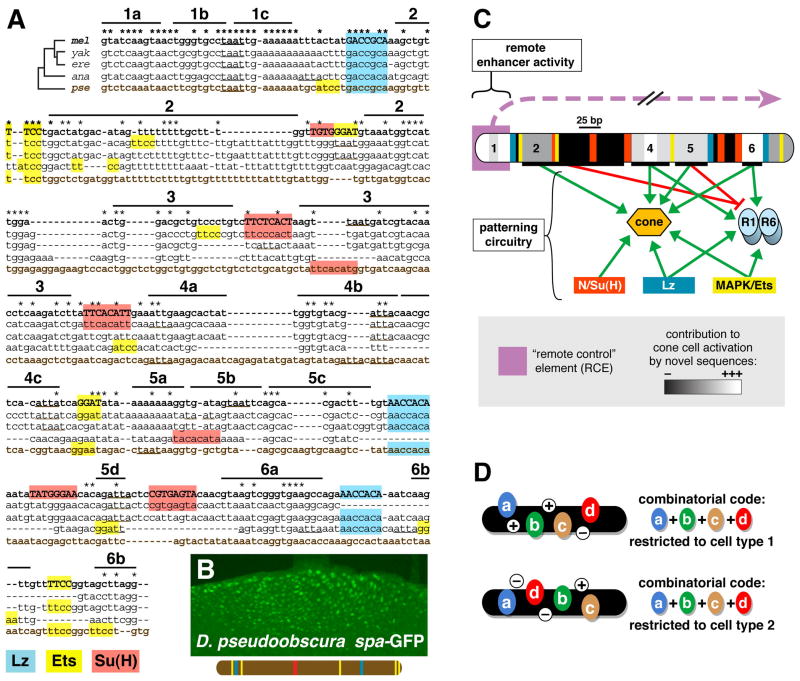
Taking this study and previous work into account, spa is among the most finely mapped enhancers with respect to regulatory sites essential for function in vivo. We made use of the recent sequencing of multiple Drosophila species genomes (Drosophila 12 Genomes Consortium, 2007) to investigate the evolutionary history of spa. We will focus on the D. melanogaster-D. pseudoobscura (mel-pse) comparison, which is commonly used to study cis-regulatory sequence evolution; the two populations diverged ~25 million years ago (e.g., Ludwig et al., 2005 and references therein). As we will discuss below, blocks of sequence conservation between melanogaster and pseudoobscura spa are relatively few and short, and most TFBSs and newly mapped regulatory sites were not alignable (Figure 6A). We were therefore surprised to find that a 409-bp pseudoobscura sequence we identified as the putative ortholog of spa was able to drive cone cell-specific reporter gene expression in transgenic D. melanogaster, indistinguishably in pattern and intensity from melanogaster spa, even from −846 bp (Figure 6B).
Figure 6. spa Enhancer Function Is Evolutionarily Conserved, Despite Rapid Sequence Divergence.
(A) Alignment of the spa enhancer of D. melanogaster (mel) and orthologous sequences from D. yakuba (yak), D. erecta (ere), D. ananassae (ana), and D. pseudoobscura (pse). Binding sites for Lz, Pnt/Yan, and Su(H), and predicted orthologous sites, are highlighted in color. Regions 1 through 6 are labeled with black bars. TAAT motifs are underlined. Conserved bases are indicated with asterisks.
(B) The 409-bp D. pseudoobscura sequence shown in panel A drives robust cone cell-specific gene expression in eye discs of transgenic D. melanogaster from −846 bp.
(C) Summary of spa regulation: at least two functionally distinct classes of regulatory sites govern the enhancer activity of spa in vivo. spa requires the presence and proper arrangement of many regulatory sub-elements for its transcriptional activity and cell-type specificity. Region 1 appears to be required for remote enhancer activity, but dispensable for patterning. In addition, proper cell-type patterning of spa in the developing eye is considerably more complex than previously thought, and depends on short-range interactions among many regulatory sites. Green arrows indicate activation mediated by sites within spa; red bars indicate cell-type-specific repression activities.
(D) A simple “combinatorial code” model is insufficient to explain the cell type specificity of spa, as the same regulatory elements can be rearranged to generate transcription in either cone cells or photoreceptors. Thus any model describing cone cell-specific transcriptional activation by spa must also incorporate rules of spatial organization.
We wish to point out several notable aspects of spa sequence evolution. First, its distribution of sequence conservation appears to be unusual among developmental enhancers. When total mel-pse sequence identity is considered, spa falls only slightly below the range of six well-studied Drosophila enhancers we analyzed for comparison (Table S3). However, spa is relatively poor in extended blocks of conserved sequence; it contains only one block of 100% conservation of ≥10 bp in length (located in region 1, the RCE), constituting 3.9% of total enhancer sequence. By contrast, in the six reference enhancers, an average of 52% of sequence lies in perfectly conserved blocks of ≥10 bp (range is 37% to 75%). Even more strikingly, in the six reference enhancers, an average of one-third of the sequence is in perfectly conserved blocks of ≥20 bp, while spa has no conserved blocks of this length (Table S3). Lack of sequence conservation does not appear to result from a reshuffling of regulatory sequences, as melanogaster vs. pseudoobscura dot-plot analysis does not detect any rearrangements within spa (data not shown).
Second, of the 12 identified binding sites for Lz, Pnt/Yan, and Su(H), only three can be unambiguously aligned with orthologous predicted binding sites in pseudoobscura. Four other predicted binding sites for these TFs were found in the pseudoobscura enhancer, but had no definitive orthologs in melanogaster spa, due to significant differences in sequence and/or position (Figure 6A). Overall, pseudoobscura spa contains fewer predicted TFBSs than melanogaster spa: 1 vs. 5 Su(H) sites, 2 vs. 3 Lz sites, and 5 GGAW consensus Ets sites vs. 6 in melanogaster.
Third, with respect to the previously uncharacterized sequences within spa, we do not observe a strong correlation between functional significance and sequence conservation. Of the essential, previously unmapped sequences identified in this report (regions 1/4/5abc/6a), the total mel-pse sequence identity is not greatly higher than that of sequences making little or no contribution to activation (regions 2/3/5d/6b) (65% vs. 58% identity). Thus, in the context of the spa enhancer, we find evolutionary sequence conservation to be a poor indicator of functional importance in transcriptional regulation.
DISCUSSION
The goal of this study was to use a well-characterized, signal-regulated developmental enhancer to examine, in fine detail, the regulatory interactions and structural rules governing transcriptional activation in vivo. Taking the elegant work of Flores and colleagues (2000) as a starting point, we have used functional in vivo assays to test the power of the proposed combinatorial code of “Notch/Su(H) + Lz + MAPK/Ets” to explain the activity and cell-type specificity of the spa cone cell enhancer of dPax2. In the course of this work, we have discovered several surprising properties of spa that are not accounted for in current models of enhancer function.
The spa Patterning Code Is Massively Combinatorial
We chose the spa enhancer for our fine-scale analysis because (1) the known direct regulators and their binding sites are well defined, (2) they could in theory constitute the sum total of the patterning information received by the enhancer, and (3) the enhancer, at 362 bp, is relatively small, simplifying mutational analyses. To our surprise, a large proportion of the previously uncharacterized sequence within spa is vital for normal enhancer activity in vivo, and of that subset, a large proportion directly influences cell-type specificity. These findings are summarized in Figure 6C.
Activation in cone cells
In addition to necessary inputs from Lz, Pnt, and Su(H), we have identified three segments of spa, regions 4, 5, and 6, that make essential contributions to gene expression in cone cells. In addition, region 2 makes a relatively minor contribution. (Region 1, another essential domain, will be discussed separately.) Fine-scale mutagenesis reveals that within regions 4/5/6, very little DNA is dispensable for cone cell activation. The previously uncharacterized regulatory sites in spa are very likely bound by factors other than Lz/Pnt/Su(H), for the following reasons: no sequences resembling Pnt/Lz/Su(H) binding sites reside in these regions; mutations in the newly mapped sites have different effects than removing the defined TFBSs or the proteins that bind them; doubling the known TFBSs fails to compensate for the loss of the newly mapped sequences; and most importantly, mutating the newly mapped regulatory regions does not significantly affect binding of the known activators to nearby binding sites in vitro (Table S2). We cannot tell whether the proposed novel regulators are cone cell-specific, eye-specific, or ubiquitous in their expression—we only know that the newly mapped sites are necessary both for normal cone cell expression and ectopic PR expression. Besides Lz, Pnt, and Su(H), we know of no transcriptional activators present in cone cells; Cut, Prospero, and Tramtrack are expressed in cone cells, but are thought to act as transcriptional repressors (e.g., Lai and Li, 1999; Cook et al., 2003; Sato et al., 2006). The transcription factor Hindsight is required for dPax2 expression and cone cell induction, but acts indirectly, activating Delta in R1/R6 to induce Notch signaling in cone cells (Pickup et al., 2009).
Unsurprisingly, placing the enhancer closer to the promoter boosts expression of spa(wt), as well as some of the impaired mutants (Figure 4). Remember that spa is located at +7 kb in its native locus, and that nearly all mutational studies place the enhancer immediately upstream of the promoter. If our entire analysis had been performed at −121 bp, we would have underrated the functional significance of several critical regulatory sequences, and would have dismissed region 1 entirely as non-regulatory DNA. Other well-characterized enhancers, which have been analyzed in a promoter-proximal position only, may therefore contain more critical regulatory sites than is currently realized.
Like many transcriptional activators, all three known direct activators of spa (or their orthologs) recruit p300/CBP histone acetyltransferase coactivator complexes (e.g., Kitabayashi et al., 1998; Barolo and Posakony, 2002). Doubling the number of binding sites for these TFs (to 6 Lz, 8 Ets, and 10 Su[H] sites) does not suffice to drive cone cell expression in the absence of the newly mapped regulatory regions (Figure 5). It may be, then, that factors recruited to the newly mapped regulatory sites within spa employ mechanisms that are distinct from those of the known activators. The remote activity of spa, mediated by region 1, appears to be an example of such a mechanism.
Cell-type specificity
We were able to convert spa into a photoreceptor R1/R6-specific enhancer in three ways: (1) by moving the defined TFBSs to one side of the enhancer in a tight cluster; (2) by placing Lz and Ets sites next to regions 1/4/6a; and (3) by mutating regions 2/3/5/6b within spa while maintaining the native spacing of all other sites. From these experiments, we conclude that spa contains short-range repressor sites that prevent ectopic activation in PRs by Lz + Pnt + regions 4 + 6a. spa contains at least two redundant repressor sites, since both region 5 and regions 2/3/6b must be mutated to attain ectopic R1/R6 expression.
klumpfuss, which encodes a putative transcriptional repressor, is directly activated by Lz in R1/R6/R7, but is also present in cone cells (Wildonger et al., 2005, and references therein), making it an unlikely repressor of spa. seven-up, another known transcriptional repressor, is expressed in R3/R4/R1/R6 and could therefore act to repress spa in photoreceptors (Mlodzik et al., 1990; Cooney et al., 1993). However, we did not identify putative seven-up binding sites within spa. Phyllopod, an E3 ubiquitin ligase component, represses dPax2 and the cone cell fate in R1/R6/R7, but the transcription factor mediating this effect is not yet known (Shi and Noll, 2009). Perhaps the best candidate for a photoreceptor-specific direct repressor of spa is Bar, which encodes the closely related and redundant homeodomain TFs BarH1 and BarH2. Bar expression is activated by Lz in R1/R6 and is required for R1/R6 cell fates (Higashijima et al., 1992; Crew et al., 1997). Furthermore, misexpression of BarH1 in presumptive cone cells can transform them into photoreceptors (Hayashi et al., 1998). It is unclear whether Bar-family proteins act as repressors, activators, or both. BarH1/2 can bind sequences containing the homeodomain-binding core consensus TAAT (Noyes et al., 2008), and region 5 of spa contains two TAAT motifs (underlined in Figure 6A). Future studies will explore the possibility that Bar directly represses spa in photoreceptors.
The combinatorial code of spa, then, requires multiple inputs in addition to Lz, MAPK/Ets, and Notch/Su(H). Indeed, our data suggest that the known regulators can contribute to expression in multiple cell types, depending on context. The newly mapped control elements we have identified within spa are necessary not only to facilitate transcriptional activation, but also to steer the Lz/Ets/Su(H) code toward cone cell-specific gene expression.
Functional Evidence for a Special Enhancer Regulatory Element, Mediating Remote Interactions But Not Patterning
Enhancers are often located many kilobases from the promoters they regulate. Enhancer-promoter interactions over such distances are very likely to require active facilitation (Rippe, 2001). Even so, few studies have focused specifically on transcriptional activation at a distance, and the majority of this work involves locus control regions (LCRs) and/or complex multigenic loci, which are not part of the regulatory environment of most genes and enhancers (e.g., Yoshida et al., 1999; Carter et al., 2002; Song et al., 2007). Like spa, many developmental enhancers act at a distance in their normal genomic context, yet can autonomously drive a heterologous promoter in the proper expression pattern, without requiring an LCR or other large-scale genomic regulatory apparatus. However, in nearly all assays of enhancer function, the element to be studied is placed immediately upstream of the promoter. In such cases, regulatory sites specifically mediating remote interactions cannot be identified. Because our initial mutational analysis of spa was performed on enhancers placed at a moderate distance from the promoter (−846 bp), we were able to screen for sequences required only at a distance, by moving crippled enhancers to a promoter-proximal position. Only one segment of spa, region 1, was absolutely essential at a distance but completely dispensable near the promoter. This region, which contains the only block of extended sequence conservation within spa, plays no apparent role in patterning, or in basic activation at close range. We therefore call this segment of spa a “remote control” element (RCE).
The remote enhancer regulatory activity described here differs from previously reported long-range regulatory mechanisms in two important ways. First, the remote function of spa does not require any sequences in or near the dPax2 promoter. This functionally distinguishes spa from enhancers in the Drosophila Hox complexes that require promoter-proximal “tethering elements” and/or function by overcoming insulators (e.g., Calhoun et al., 2002; Chen et al., 2005, Akbari et al., 2008). This distal activation mechanism also likely differs from enhancer-promoter interactions mediated by proteins that bind at both the enhancer and the promoter, as occurs in looping mediated by ER, AR, and Sp1 (Wang et al., 2005; Williams et al., 2007; Pan et al., 2008). Second, studies of distant enhancers of the cut and Ultrabithorax genes have revealed a role for the cohesin-associated factor Nipped-B, especially with respect to bypassing insulators (Misulovin et al., 2008, and references therein), but it has not been demonstrated that Nipped-B, or any other enhancer-binding regulator, is required only when the enhancer is remote.
To our knowledge, the spa RCE is the first enhancer sub-element demonstrated to be essential for enhancer-promoter interactions at a distance, but unnecessary for proximal enhancer function and cell-type specificity. However, the present work contains only a limited examination of this activity, as part of a broader study of enhancer function. We are currently extending these functional studies, testing for potential promoter preferences and distance limitations, and pursuing the identities of factors binding to the RCE.
Enhancer Structure: Shaped and Constrained by Short-Range Patterning Interactions
As discussed above, it is fairly easy to switch spa from cone cell expression to R1/R6 expression (though, curiously, we have yet to generate a construct that is active in both cell types). Our results show that multiple regions of spa mediate a repression activity in R1/R6 but not in cone cells. We further conclude that these spa-binding repressor(s) act in a short-range manner; that is, they must be located very near to relevant activator binding sites, since moving Lz and Pnt sites to one side of spa, without removing the repressor sites (KO+synthCS), abolishes repression. Despite this failure of repression, synergistic interactions among Lz + Ets sites and the newly mapped sites still occur in this re-organized enhancer—at least in R1/R6 cells. Cone cell-specific expression is lost, however, revealing (along with other experiments) that transcriptional activation in cone cells is highly sensitive to the organization of regulatory sites within spa. Slightly wider spacing of regulatory sites (KO+synthNS) kills the enhancer altogether, suggesting that synergistic positive interactions within spa, though apparently longer in range than repressive interactions, are severely limited in their range. The structural organization of spa, then, appears to be constrained by a complex network of short-range positive and negative interactions (Figure 6D). Activator sites must be spaced closely enough to trigger synergistic activation in cone cells; at the same time, repressor sites must be positioned to disrupt this synergy in non-cone cells, preventing ectopic activation.
Recent work by Crocker et al. (2008) has shown that changes to enhancer organization can “fine-tune” the output of a combinatorial code, subtly changing the sensitivity of the enhancer to a morphogen. Given the importance of the structure of the spa enhancer for its proper function, we propose that any combinatorial code model, no matter how complex, is insufficient to describe the regulation of spa, since the same components can be rearranged to produce drastically different patterns.
Conservation of spa Function Despite Lack of Sequence Conservation: Insights Into Enhancer Structure
One might expect that the regulatory and organizational complexity of the spa enhancer, and its extreme sensitivity to mutation, would be reflected in strict evolutionary constraints upon enhancer sequence and structure. Yet we observe very poor conservation of spa sequence, both in the known TFBSs and in most of the newly mapped essential regulatory elements. The reduced presence of Lz/Ets/Su(H) sites in D. pse could potentially be attributed to redundancy of those sites in D. mel, or to compensatory gain of binding sites for alternate factors in the D. pse enhancer. Perhaps more difficult to understand is the apparent loss of critical regulatory sequences in regions 4, 5, and 6a in D. pse; our experiments in D. mel suggest that the absence of those inputs would result in loss of cone cell expression and/or ectopic activation. It remains possible that many of these inputs are in fact conserved, but that conservation is not obvious due to binding site degeneracy and/or rearrangement of elements within the enhancer. Fine-scale comparative studies are ongoing.
spa is by no means the first example of an enhancer that is functionally maintained despite a lack of sequence conservation (for a review of this topic, see Wittkopp, 2006). The most thoroughly characterized example of this phenomenon is the eve stripe 2 enhancer; its function is conserved despite changes in binding site composition and organization (Ludwig et al., 2000; Ludwig et al., 2005; Hare et al., 2008). Note, however, that spa has undergone much more rapid sequence divergence than eve stripe 2 (Figure 6; Table S3), with no apparent change in function. In general, the ability of an enhancer to maintain its function in the face of rapid sequence evolution suggests that enhancer structure must be quite flexible. These observations support the “billboard” model of enhancer structure, which proposes that as long as individual regulatory units within an enhancer remain intact, the organization of those units within the enhancer is flexible (Arnosti and Kulkarni, 2005). Yet our findings concerning the importance of local interactions among densely clustered, precisely positioned transcription factors are more consistent with the tightly structured “enhanceosome” model (Thanos and Maniatis, 1995). Further structure-function analysis will be necessary to fully understand the players and rules governing this regulatory element.
EXPERIMENTAL PROCEDURES
Generation of Enhancer Constructs
The 362-bp sparkling enhancer was amplified from w1118 genomic DNA with the following primers: 5′-CACCGGATCCgtatcaagtaactgggtgcctaattg-3′; 5′-GGGTCTAGAcctaagctaccggaaaacaacttg-3′. The 409-bp D. pseudoobscura spa enhancer was PCR-amplified from genomic DNA with the following primers: 5′-CACCGGATCCgtctcaaataacttcgtgtc-3′; 5′-GGGTCTAGAcacaggaagccggaaactg-3′. Lower-case sequence is homologous to genomic DNA.
Most mutant spa constructs were generated by one of three PCR techniques: (1) amplification of spa(wt) with tagged primers to create mutations at the 5′ or 3′ end; (2) overlap extension (sewing) PCR to generate internal mutations; or (3) assembly PCR to synthesize enhancers with multiple mutations. See Supplemental Experimental Procedures for complete sequences of all enhancer constructs.
Mutagenesis by Overlap Extension PCR (Sewing PCR)
When targeting mutations in the interior of spa, such as in constructs m4A, m4-rs, etc., we separately amplified 5′ and 3′ fragments, using overlapping tagged primers to integrate mutated sequence, and then joined the fragments using overlap extension (Swanson et al., 2008 and references therein). In our sewing PCR protocol, the 5′ and 3′ fragments (which overlap by 20 bp) were separately PCR amplified and gel purified. We combined 3 μl of each gel purified fragment with 33.5 μl water, 1.5 μl of 10 μm dNTPs, and 5 μl 10X PCR buffer (Roche Expand High Fidelity PCR System). This mix was incubated at 90°C 10 min, then cooled one degree per min to 72°C. 1 μl of polymerase mix (Roche Expand High Fidelity PCR System) was then added, followed by incubation for 10′ at 72°C. Finally, 1.5 μl of each the flanking 5′ and 3′ primers (15 pmol each) was added and the full-length construct was amplified in our standard PCR program (94°C for 2′; 10 cycles of (94°C for 15″, 55°C for 30″, 72°C for 45″); 20 cycles of (94°C for 15″, 55°C for 30″, 72°C for 45″+5″/cycle); 72°C for 7′).
Assembly PCR
In constructs with extensive mutated sequence (such as spa[mut] and spa[synth]), constructs were built by annealing overlapping 40 bp oligonucleotides to create the full-length construct by assembly PCR (Swanson et al., 2008 and references therein). We combined 2.5 μl of each flanking primer (10 μM), 1 μl internal primer mix (each primer at 0.25 μM), 1 μl of 10 μM dNTPs, and 18 μl sterile water in the template mix. The enzyme mix contained 19.25 μl sterile water, 5 μl 10X PCR buffer, and 0.75 μl DNA polymerase (Roche Expand High Fidelity PCR System). The template mix and enzyme mix were combined immediately before amplification in our standard PCR program (see above).
In mutating previously uncharacterized enhancer sequences, we made non-complementary transversions to every other base pair. We left 2–4 bp of non-mutated sequence to either side of every TFBS (as defined by consensus sequences), to avoid interfering with TF binding. In mutating TFBSs, we converted Lz sites from RACCRCA to RAAARCA; Ets sites from GGAW to TTAW; and Su(H) sites from YGTGDGAA (or related sequence) to YGTGDCAA; these changes eliminate TF binding in vitro (Barolo et al., 2000; Flores et al., 2000; references therein).
Enhancer Cloning, Vectors, and Transgenesis
PCR-amplified enhancer constructs were TOPO-cloned into the pENTR/D-TOPO vector (Invitrogen). spa(synthCS) was created by annealing two complementary oligonucleotides and ligation into the Gateway donor vector pBS-ENTR-TOPO (Swanson et al., 2008). Subcloned constructs were then Gateway-cloned into the Ganesh-G1 GFP reporter vector (Swanson et al., 2008) via LR recombination (Invitrogen), with the following exception: constructs placed at −121 bp from the promoter (Figure 4) were Gateway-cloned into Ganesh-G2, which lacks the 0.7-kb spacer sequence between the recombination cloning site and the promoter (Swanson et al., 2008). P element transformation was performed essentially as described by Rubin and Spradling (1982). w1118 flies were used for transgenesis.
Tissue Preparation, Staining, and Microscopy
Eye tissues were dissected from transgenic third-instar larvae or 24-hour pupae and fixed in 4% formaldehyde in PBS for 30 minutes at room temperature. For larval imaginal discs, GFP fluorescence was imaged with an Olympus BX51 microscope and an Olympus DP70 digital camera. Pupal eyes were stained with antibodies to GFP (see below) and imaged with an Olympus IX71 inverted microscope and an Olympus FV500 confocal system. Primary antibodies used: rabbit anti-EGFP (a gift from B. Novitch), diluted 1:100; mouse anti-Cut 2B10 (a gift from K. Cadigan), diluted 1:100; mouse anti-Elav 9F8A9 (Developmental Studies Hybridoma Bank), diluted 1:100.
DNA Sequence Alignment
The sparkling multi-species alignment is based on BLASTZ alignments and was taken from the UCSC genome browser (http://genome.ucsc.edu). Pairwise mel-pse enhancer alignments were performed using zPicture (Ovcharenko et al., 2004; http://zpicture.dcode.org).
Supplementary Material
Acknowledgments
This research was supported in part by a Center for Organogenesis Training Grant (5T32HD007505) to C.I.S. and by NIH grant GM076509 to S.B. We thank Ying Zhao, Lisa Johnson, Andy Vo, Zeeshaan Bhatti, Iehsus Flores-Pérez, and Trish Hinrichs for research support, and Ben Novitch and Ken Cadigan for sharing reagents. We are grateful to the following people for helpful discussions: Ken Cadigan, Tim Blauwkamp, Deneen Wellik, Ben Novitch, Doug Engel, Tom Glaser, Trisha Wittkopp, Jim Posakony, Billy Tsai, Albert Erives, Robert Drewell, and many members of the lab.
Footnotes
Supplemental Data include Experimental Procedures (including annotated sequences of all enhancer constructs), one figure, and three tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbari OS, Bae E, Johnsen H, Villaluz A, Wong D, Drewel RA. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development. 2008;135:123–131. doi: 10.1242/dev.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky A, Freschi G, Keil T, Posakony JW. A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Lin Q, Zhou J. Multiple Promoter Targeting Sequences exist in Abdominal-B to regulate long-range gene activation. Dev Biol. 2005;286:629–636. doi: 10.1016/j.ydbio.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- Cooney AJ, Leng X, Tsai SY, O’Malley BW, Tsai M. Multiple mechanisms of Chicken Ovalbumin Upstream Promoter Transcription Factor-dependent repression of transactivation by the vitamin D, thyroid hormone, and retinoic acid receptors. J Biol Chem. 1993;268:4152–4160. [PubMed] [Google Scholar]
- Crew JR, Batterham P, Pollock JA. Developing compound eye in lozenge mutants of Drosophila: lozenge expression in the R7 equivalence group. Dev Genes Evol. 1997;206:481–493. doi: 10.1007/s004270050079. [DOI] [PubMed] [Google Scholar]
- Crocker J, Tamori Y, Erives A. Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol. 2008;6:e263. doi: 10.1371/journal.pbio.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. A view from the genome: spatial control of transcription in sea urchin development. Curr Opin Genet Dev. 1999;9:530–541. doi: 10.1016/s0959-437x(99)00013-1. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Duan H, Frei E, Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943–2950. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PloS Genetics. 2008;4(6):e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Kojima T, Saigo K. Specification of primary pigment cell and outer photoreceptor fates by BarH1 homeobox gene in the developing Drosophila eye. Dev Biol. 1998;200:131–145. doi: 10.1006/dbio.1998.8959. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Kojima T, Michiue T, Ishimaru S, Emori Y, Saigo K. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992;6:50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Li Y. Tramtrack69 is positively and autonomously required for Drosophila photoreceptor development. Genetics. 1999;152:299–305. doi: 10.1093/genetics/152.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, Macarthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the Steroid Receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. Picture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 2004;14(3):472–477. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YF, Wansa KDSA, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E. Regulation of Estrogen Receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283:32977–32988. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- Pickup AT, Ming L, Lipshitz HD. Hindsight modulates Delta expression during Drosophila cone cell induction. Development. 2009;136:975–82. doi: 10.1242/dev.027318. [DOI] [PubMed] [Google Scholar]
- Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem Sci. 2001;26:733–740. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Seto H, Hayashi Y, Kwon E, Taguchi O, Yamaguchi M. Antagonistic regulation of the Drosophila PCNA gene promoter by DREF and Cut. Genes Cells. 2006;11:499–512. doi: 10.1111/j.1365-2443.2006.00956.x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Noll M. Determination of cell fates in the R7 equivalence group of the Drosophila eye by the concerted regulation of D-Pax2 and TTK88. Dev Biol. 2009;331:68–77. doi: 10.1016/j.ydbio.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Hinrichs T, Johnson LA, Zhao Y, Barolo S. A directional recombination cloning system for restriction- and ligation-free construction of GFP, DsRed, and lacZ transgenic Drosophila reporters. Gene. 2008;408:180–186. doi: 10.1016/j.gene.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wildonger J, Sosinsky A, Honig B, Mann RS. Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 2005;19:1034–1039. doi: 10.1101/gad.1298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AO, Isaacs RJ, Stowell KM. Down-regulation of human topoisomerase IIα expression correlates with relative amounts of specificity factors Sp1 and Sp3 bound at proximal and distal promoter regions. BMC Mol Biol. 2007;8:36. doi: 10.1186/1471-2199-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ. Evolution of cis-regulatory sequence and function in Diptera. Heredity. 2006;97:139–147. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- Yoshida C, Tokumasu F, Hohmura KI, Bungert J, Hayashi N, Nagasawa T, Engel JD, Yamamoto M, Takeyasu K, Igarashi K. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells. 1999;4:643–655. doi: 10.1046/j.1365-2443.1999.00291.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.