Abstract
Duplicated sequences are substrates for the emergence of new genes and are an important source of genetic instability associated with rare and common diseases. Analyses of primate genomes have shown an increase in the proportion of interspersed segmental duplications (SDs) within the genomes of humans and great apes. This contrasts with other mammalian genomes that seem to have their recently duplicated sequences organized in a tandem configuration. In this review, we focus on the mechanistic origin and impact of this difference with respect to evolution, genetic diversity and primate phenotype. Although many genomes will be sequenced in the future, resolution of this aspect of genomic architecture still requires high quality sequences and detailed analyses.
SDs and dynamism in the genomes
Over 40 years ago, Ohno and colleagues postulated the importance of duplications in the evolution of new gene functions [1]. Since then, our knowledge and understanding of the evolution of genes and genomes has increased enormously. Both computational and experimental approaches indicate that gene loss and gain have been common within the primate lineage [2,3,5–10] and that much of this occurs within or is mediated by duplicated sequences. The dynamism and complexity of these changes has complicated molecular comparative genomic studies. Nevertheless, the available data clearly indicate that this variation is critical for understanding the evolution and phenotypic variation of our species. The study of SDs, defined as highly identical duplicated DNA fragments greater than 1 Kb, is relevant for two basic reasons. First, SDs are hotbeds for genome structural change between and within species. Regions of SDs are preferred sites of copy number polymorphism [13–15], disease-causing rearrangement [16,17] and evolutionary breakpoints during primate genome evolution [18,19]. Duplicated sequences – by nature of the sequence homology that promotes unequal crossover during meiosis – are mutagenic with both beneficial (genome flexibility) and damaging (human disease) consequences. Second, because primate SDs are particularly enriched for transcripts, there is the potential for gene innovation and rapid adaptation as a result of the accelerated tempo of mutation within these regions [20–22].
To date, the sequences of three primate genomes have been published: human [23], chimpanzee [24] and macaque [25]. Sequencing of the orangutan and marmoset genomes is near completion, with additional genomes from other branch points of the primate phylogeny being targeted. More than 20 other mammalian genomes have been earmarked for whole-genome sequencing at various levels of coverage. Although many draft mammalian genomes have been published, only one (mouse) [26] is comparable in quality to the human genome because it was completed using a hierarchical bacterial artificial chromosome (BAC)-based approach [27]. The resources [28] developed as part of these projects have begun to provide a new framework for researchers to understand the evolution of genomes. However, most available genome assemblies have been resolved from a whole-genome shotgun (WGS) approach, rendering the mapping and interrogation of SDs a more challenging task. Computational methods have been developed to identify duplicated sequences independently from the genome assembly, and experimental methods (FISH and array comparative genomic hybridization (array-CGH)) have been used to validate and explore the distribution and organization of these sequences [21]. In this review, we will address the various methods of detection, organization and impact of SDs toward primate evolution and human disease.
Patterns of mammalian SDs
Characterization of the human genome revealed that SDs are large, highly identical and interspersed [2,8,29], usually separated by >1Mb of unique sequences. Their distribution is largely nonrandom, with peculiar clustering observed near the subtelomeric and pericentromeric regions in addition to enrichments within the euchromatic portions of specific human chromosomes. The majority of human SDs map to ~400 distinct regions of the genome (termed “duplication blocks”) [10]. Within these duplication blocks, SDs are organized into complex mosaics where individual ancestral duplicated segments (termed “duplicons”) are juxtaposed adjacent to other duplicons of diverse origin. Evolutionary studies of these duplication blocks suggest additional rounds of duplication among the duplication blocks creating a complex pattern of duplications-within-duplications and a bedazzling complexity of interspersed duplications – a hallmark feature of hominoid duplication organization [30].
Building on this shared evolutionary history among the duplication blocks, Jiang and colleagues applied a de Bruijn graph approach (a model applied to resolve a complicated set of sequence alignments using a data structure from combinatorial mathematics) to construct an evolutionary framework for the origin and relationship of duplication blocks [12]. There were three important conclusions: (i) most human duplication blocks can be grouped into 24 distinct clades based on the sharing of at least one duplicon; (ii) pericentromeric and subtelomeric duplications evolved independently from intrachromosomal duplications; and (iii) the analysis pinpointed 14 specific gene-rich sequences called “core duplicons” ranging in length from 5–30 Kb that were found to be associated with most of the intrachromosomal duplications within specific human chromosomes. It was proposed that these core duplicons were the focal point for duplication expansion, driving the duplication of other segments in a stepwise process during hominid evolution (“core duplicon hypothesis”). A detailed study of one of these “core” duplicons LCR16a in humans and African apes supports the notion that these sequences were the catalyst for independent and recent expansions of duplications within different ape lineages [30].
Despite the draft nature of non-human primate genomes, it has been possible to estimate and compare SD content using both experimental and computational methods. Using an assembly-independent method (Box 1), it was found that both human and chimpanzee show an overall similar content of SDs (~5%). Although the majority are shared between the two species (~66%), differences in duplication content and copy number account for more genetic differences between the two species (~2.5%) than single basepair substitutions (~1%) [8]. Analysis of orangutan duplications performed by mapping orangutan WGS reads to the human assembly found ~40% less duplication in the Asian ape than either human or chimpanzee. The draft sequence of the macaque genome shows a substantial reduction of SDs (~2.4% of the genome) using three different methods: two based on the genome assembly and one independent of the genome assembly [25]. A small proportion of SDs is shared between human and macaque. These data support a model where SD activity increased after the divergence of African great apes (chimpanzee, gorilla and human) from the Asian great ape (orangutan) [21] (Figure 1). Numerous studies tracking the history of specific duplicons by comparative FISH analyses among primates provide additional support for this burst of activity (e.g. SLC6A18, NF1, CHEK2 [31,32]). Interestingly, this two- to fourfold acceleration seems to have occurred at a time when other mutational processes were slowing down (such as point mutations or retrotransposon activity). This finding is also consistent with the statistical mode of sequence divergence among highly identical human intrachromosomal SDs (97–99%) [29].
Box 1. Methods to detect duplications.
Despite extensive progress in genome sequencing, only the human genome can be considered reliable in terms of the assembly of high identity duplications among the primates. SDs are complicated to detect for several reasons. First, by definition, they are highly identical (>90% similarity), making them challenging to distinguish. Thus, SDs are either under- or over-represented owing to the inherent nature of whole-genome assembly-based methods. Second, these analyses are frequently interfered by common repeats dispersed throughout the genome. Third, the mosaic architecture of duplications derived from various chromosomal locations by incomplete or partially duplicated transposition complicates SD detection.
Two main methods are broadly used to detect SDs: an assembly-based method (WGAC or whole-genome assembly comparison [2]) and an assembly-independent method (WSSD or whole-genome shotgun sequence detection [3]). WGAC was first used to describe global duplication content in the preliminary version of the human genome assembly [2]. Briefly, the genome assembly is partitioned into shorter segments (400 Kb). Because common repeats complicate self-alignment, repeats are repeat-masked and removed, leaving “unique” genomic segments that are compared to identify large regions of high identity. Once seed duplication alignments are identified, local pairwise alignments are computed with their common repeats reinserted and the endpoints defined using a reiterative heuristic method that maps within common repeats. We initially reported alignments longer than 1000 bases aligned with >90% identity (hence, the formal definition of SD was created). Assuming neutrality and a molecular clock, this level of sequence identity within the human genome should detect all duplications since the split of New and Old World monkeys (35 million years ago (mya)). This method has the advantage of providing an absolute estimate of copy number, the structural details of duplicated regions and the location of all duplications. Obviously, this method depends on the quality of the assembly. If sequences are missing, collapsed or not correctly assembled, duplication content can be over-or underestimated.
A more versatile method, one not directly dependent on a finished genome assembly, is WSSD [3]. This method aligns whole-genome shotgun (WGS) reads against a reference assembly (with a defined identity threshold, usually 94%). The idea is straightforward. Because duplicated reads will map to both the paralogous and the orthologous location in the assembly, duplications will be detected as an excess of read-depth even if that duplication has not been resolved. The number of reads and average sequence identity are calculated across window intervals, and the boundaries of SDs are then determined by defining transitions in read-depth across smaller window intervals. This method requires that the WGS reads be randomly distributed and that all sequences are represented at least once within the reference assembly. The method does not provide information on the location of paralogous copies, details about their structure or the sequence identity between paralogs. However, it is a method with high sensitivity for high identity duplications larger than 20 Kb and can be used to detect duplications in other species that are genetically similar to the assembly reference without that genome being fully resolved [8,20–22]. Of course, the greater the evolutionary distance, the more difficult the mapping, but it has performed well in all the great ape WGS sequences mapped to the human assembly. As expected, duplication copy number correlates well with depth of read coverage, allowing copy number differences between primate genomes to be predicted (Figure I).
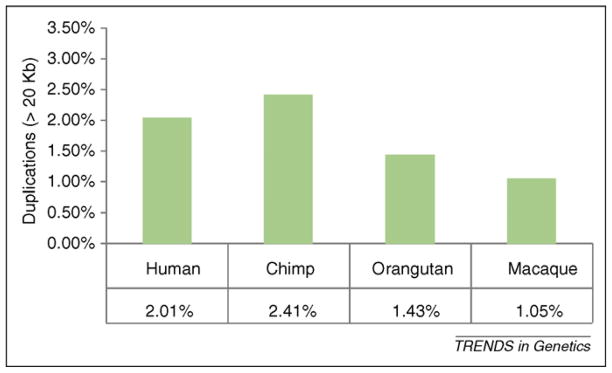
Figure 1.
SDs in different primate species. The proportion of large (>20Kb) and high identity duplications are given for four primate genomes. Estimates were based on identifying regions of excess read-depth (Figure I in Box 1) after copy number correction to avoid the bias of non-human-specific SDs [21]. The genomes of human and chimpanzee show twice the number of duplicated basepairs. This observation was also supported by experimental analysis [9]. FISH analysis from 384 randomly selected BACs in chimpanzee, baboon and marmoset estimated 7.73%, 4.39% and 2.00% of duplications, respectively.
The duplication content in non-primate mammalian genomes is much less clear owing to the draft nature of most current genome assemblies and the greater evolutionary distance from humans, which complicates the mapping of WGS sequence reads to the human reference. Estimates of duplication content based on the extant assemblies range widely among the different mammalian genomes (Table 1). The observation that an appreciable fraction of the assembly-detected duplications are not supported by assembly-independent methods and that 30–40% of validated duplications cannot be assigned to a chromosome clearly indicate that too much stock should not be put into current assembly-based estimates [25,26,33–36]. Only the mouse genome (C57BL/6J) is comparable in quality to that of the human genome [27]. As the mouse genome assembly progressed from a WGS assembly to a clone-ordered assembly, the duplication content more than doubled. Both human and mouse C57BL/6J genomes show similar levels of recent duplication (~5%); however, the two genomes differ radically in the organization of these sequences. In mouse, 88% of the larger duplications (>20Kb) are organized in tandem (as opposed to just 33% in the human genome) [37]. Experimental analyses of duplications in dog [38] and cow suggest that an abundance of tandem duplications represent the mammalian archetype. In total, these data imply a fundamental shift in the organization and evolution of primate SDs in which the mosaic architecture and expansion of high identity interspersed intrachromosomal duplications seem to be most pronounced in human and great ape genomes [29].
Table 1.
Mammalian genomes and SDs
| Human (hg18) | Chimpanzee (panTro2) | Orangutan (panAbe2) | Macaque (rheMac2) | Mouse (mm8) | Rat (rn4) | Dog (canFam2) | Cow (bosTau4) | Opossum (monDom4) | |
|---|---|---|---|---|---|---|---|---|---|
| Total content of assembly-detected SDs | 5.32% | 3.78% | 1.18% | 1.55% | 6.13% | 1.60% | 0.82% | 5.63% | 2.70% |
| Assembly-independent SDs | 4.60% | 5.14% | 6.23% | 1.02% | 5.48% | N/A | 3.96% | 2.66% | N/A |
| Overlap of assembly-dependent and -independent methods >10Kb | 106,042,326 (86.21%) | 75,175,287 (82.7%) | 48,552,031 (80.48%) | 4,320,656 (56.59%) | 83,135,830 (85.39%) | N/A | 35,692,222 (79.66%) | 42,369,544 (20.88%) | N/A |
| Size of the non-gapped genome (Mb) | 2881 | 2909 | 3124 | 2646 | 2567 | 2566 | 2384 | 2850 | 3405 |
The link between SD, evolution and disease
Unequal crossovers between directly orientated duplicated sequences can predispose to disease in two distinct ways. First, they can directly increase or decrease the copy number of a particular gene or parts of a gene embedded within SDs [90]. This local expansion or contraction leads to dosage changes or the altered functional properties of a gene (Table 2). Most gene copy number polymorphisms associated with human diseases belong to this category. Second, duplicated sequences can sense particular unique regions of the genome to duplicate or delete because they are bracketed by interspersed duplications [16]. Dosage imbalance or gene disruption of one or more genes leads to a highly penetrant rare allele. Most of the recurrent, large copy number variants associated with neurocognitive disease belong to this second category. The characterization of human and great ape SDs (http://humanparalogy.gs.washington.edu) allows researchers to reconstruct the evolutionary framework for any duplicated region of interest. For instance, a comparison of the duplication maps of the spinal muscular atrophy (SMA) region (5q13.2) among primates shows that the SMN2 (survival of motor neuron 2) gene is duplicated in both human and chimpanzee [39]. Humans have had the most dramatic expansion of SMN2. Interestingly, when SMN1 is deleted the severity of SMA is determined by the number of remaining copies of SMN2 [40]. Similarly, the lipoprotein Lp (a) gene (6q25) partially overlaps with a SD that is shared by human, chimpanzee and macaque and at less extent in orangutan (in which there seems to be a reduction of tandem repeats). This particular duplication is a tandem expansion of two exons within a 4.5 Kb segment and has increased in copy among humans compared with other primates (Table 2). Expansion of this cassette reduces serum levels of lipoprotein A, which is protective for coronary heart disease.
Table 2.
Pathogenic copy number polymorphisms and SDs
| Genes/loci | Description | Human disease or predisposition | Gene ID | SD size (Kb) | Copy number change | Classification |
|---|---|---|---|---|---|---|
| GSTM1 | Glutathione S-transferase M1 isoform | Squamous cell carcinoma | NM_000561 | 18 | Decrease | Great ape/OWM-duplication |
| CYP2D6 | Cytochrome P450, subfamily II D, polypeptide 6 | Drug metabolism | NM_000106 | 5 | Increase | Human-specific |
| CYP21A2 | Cytochrome P450, family 21 subfamily A | Congenital adrenal hyperplasia | NM_000500 | 35 | Increase | Great ape-specific |
| GSTT1 | Glutathione S-transferase theta 1 (GSTT1) | Halothane/epoxide toxicity | NM_000853 | 54.3 | Decrease | Non-duplicated |
| OPN1MW | Opsin 1 (cone pigments) medium wave sensitive | X-linked color blindness | NM_000513 | 38 | Decrease | Great ape/OWM-duplication |
| FCGR3B* | Fc fragment of IgG, low affinity 3a receptor | SLE and glomerulonephritis | NM_000570 | No SD | Decrease | African ape ancestor |
| IRGM* | Immune-related GTPase family M | Crohn’s disease | NM_001145805 | No SD | Decrease | Non-duplicated |
| SNCA | Alpha-synuclein | Parkinson’s disease | NM_00345.2 | No SD | Increase | Non-duplicated |
| NEGR1* | Neuronal growth regulator 1 | Obesity | NM_173808 | No SD | Decrease | Non-duplicated |
| LPA | Lipoprotein Lp (a) | Coronary heart disease | NM_005577 | 5.5 | Decrease | Great ape/OWM-duplication |
| RHD | Rhesus blood group, D antigen | Rhesus blood group | NM_016124 | 60 | Decrease | African ape ancestor |
| CFH | Complement factor H | Age-related macular degeneration | NM_000186 | 28 | Increase | Great ape/OWM-duplication |
| C4A/B | Complement component 4B preprotein | Systemic lupus erythematosis | NM_007293 | 32.8 | Decrease | Great ape/OWM-duplication |
| DEFB4 | Defensin, beta 4 | Psoriasis/Crohn’s disease | NM_004942 | 310 | Increase/decrease | Great ape/OWM-duplication |
| CCL3L1 | Chemokine ligand 3-like1 precursor | HIV/AIDS | NM_021006 | 64 | Decrease | Great ape/OWM-duplication |
These three Copy Number Polymorphisms (CNPs) are associated with more ancient SDs; IRGM was a duplicated gene family that subsequently contracted within the anthropoid lineage [85], whereas FCGR3B is associated with a more divergent tandemly duplicated gene family.
From the perspective of the evolution of genomic disorders, we can now determine the most probable age of appearance of disease-predisposing duplications (Figure 2). For example, lineage-specific amplification of the 24 Kb SDs flanking the Charcot–Marie–Tooth disease region (17p12) CMT1A-REP occurred more recently in the hominoid common ancestor after the divergence of chimpanzees and humans [41]. The LCR22 duplications flanking the DiGeorge syndrome region (22q11.2) expanded after the divergence of hominoids from Old World monkeys [42]. SDs flanking the Angelman/Prader–Willi region (15q11–q13) began to expand before the divergence of the Old World monkeys, whereas the Smith-Magenis syndrome-specific SD blocks (17p11.2) SMS-REPS date back to after the separation of the New World monkeys [43]. Interestingly, these comparative analyses suggest that the predisposing genomic architecture for most genomic disorders emerged during the past 25 million years. In the few cases where detailed large-scale clone-based sequencing has been performed, humans and chimpanzees show the greatest complexity of structure [36]. A corollary of this research is that these specific molecular causes of complex diseases such as schizophrenia, epilepsy, intellectual disability and developmental delay are, in part, the result of relatively hominid-specific duplication architectures that emerged during the evolution of our species.
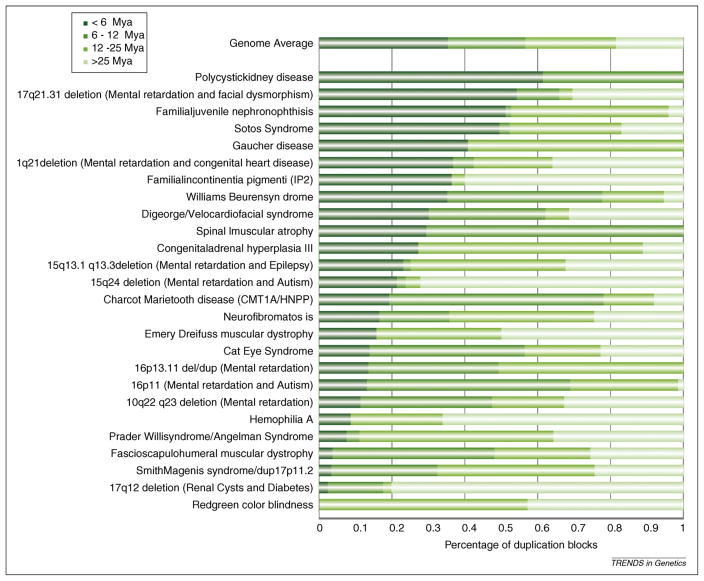
Figure 2.
Comparative analysis of disease-associated SDs. The breakpoint regions of genomic loci associated with SDs and human disease were comparatively analyzed among the primates [21]. The evolutionary age of the duplicated basepairs was inferred based on whether human SDs mapping to each region were shared or lineage-specific (i.e. <6 mya for human-specific SDs, 6–12 mya for duplications shared with chimpanzee, 12–25 mya for those shared with orangutan and >25 mya for those shared with macaque). With a few exceptions, the analysis shows that most of the complex duplication architecture that promotes rearrangement has evolved relatively recently (i.e. <12 mya).
How do SDs influence gene evolution?
Genome duplication is a classically accepted mechanism for the birth of new genes and the functional diversification and expansion of gene families. The outcome of a gene duplication event is contingent on the nature of the duplication and lineage-specific selection. Natural selection operates independently on the new copy of the duplicated genes such that the new duplicate can acquire a novel (neofunctionalization) or modified (subfunctionalization) function [44,45]. The latter frequently results in tissue-specificity or partitioning of the function from the ancestral single-copy genes. Because the process of duplication is no respecter of gene structure, partial or incomplete gene duplications are more common and these segments are, by definition, born “dead” and decay naturally within the genome as unprocessed pseudogenes. The duplication blocks of the human genome, thus, can be regarded as graveyards of exon-rich DNA from which evolutionary innovations occasionally arise.
Certain SDs undergo lineage-specific expansions in copy numbers before fixation by positive selection; although, in some lineages fixation never occurs and the gene continues to vary in copy. Interspecific variation in copy numbers is potentially essential for the evolution of species-specific adaptive traits. Trichromatic color vision, a trait essential for distinguishing red, blue and green, arose by X-linked gene duplication after the divergence of New World monkeys [46,47]. Variation in copy numbers between populations within species has resulted in dosage-sensitive effects for certain diseases (Table 2). For example, an increased CCL3L1 (chemokine (CC motif) ligand 3-like1) copy is associated with a significant reduction in susceptibility to HIV infection and the progression of AIDS in humans [48]. and other gene duplications are associated to basic biologic phenomena such as lactation in mammals [49] or the presence of venom in monotremes [50]. Copy number differences within macaque populations also affect the rate of progression of simian AIDS; Indian-origin macaques with fewer CCL3L1 copies showed shorter post-infection survival rates than Chinese-origin macaques containing higher amounts of CCL3L1 copies [51]. Copy number variation of human FCGR3 (Fc receptor, IgG, low affinity III) also seems to determine susceptibility to immune-system-mediated glomerulonephritis [52]. Notably, an increased copy number of beta-defensins is associated with a significant risk of psoriasis [53], whereas a decreased copy number predisposes to Crohn’s disease of the colon [54]. It is, therefore, evident that genes within SDs contribute to human morbidity, in addition to providing the raw material for evolutionary novelties.
In principle, the interspersed architecture of the human and great ape genomes offers tremendous evolutionary potential. The mosaic architecture of the duplication blocks in these genomes means that disparate segments can be juxtaposed, essentially shuffling different functional segments of the genome in combinations that are not found in ancestral species. Although most of these juxtapositions are non-functional, occasionally an evolutionary novel “fusion gene” can arise with functional importance (TRE2/USP6) (Box 2). Numerous, exon–intron structures with unknown functions have also been identified within SDs [4,32,55–61]. It is noteworthy that many of the core duplicons (see above) that seem to be central in the great ape expansion of SDs also harbor rapidly evolving genes and gene families. Several show evidence of positive selection, changes in gene structure or radical differences in gene expression compared with their ancestor genes [4,56,57,59,61], including NPIP (nuclear pore complex interacting protein)/morpheus (Box 2), RANBP2 (RAN binding protein 2) and the DUF1220 domain containing NBPF11 (neuroblastoma breakpoint family 11) gene. These “genes” have expanded in the human–great ape lineage to show variation in copy numbers and content within and between primate populations, and are the source of recurrent rearrangements associated with disease.
Box 2. SD-mediated genic evolution by inter/intrachromosomal remodeling.
More than a dozen genes within SDs have been identified to have undergone rapid diversification and apparent neofunctionalization. Intrachromosomal remodeling by SDs usually involves euchromatic regions of chromosomes. The transposed blocks of SDs are 96–98% identical and deciphering the ancestral “duplication core” is nontrivial. Interestingly, many of these genes map to paralogous regions that are responsible for recurrent disease-associated rearrangement events (16p11.2 autism deletion, 16p13.11 deletion/duplication, Williams, Prader–Willi, velocardiofacial/DiGeorge, neurofibromatosis, spinal muscular atrophy and Smith-Magenis syndromes). A classic example of intrachromosomal remodeling is the formation of the morpheus gene. A 20 Kb segment of chromosome 16 (LCR16a) proliferated from 1–2 copies in Old World monkeys to 15–20 copies in humans and chimpanzees [4]. The morpheus gene family was identified within these LCR16a duplication blocks, Furthermore, evolutionary analysis of the protein coding segments of morpheus showed an enhanced rate of adaptive evolution, with an excess of non-synonymous substitutions compared with synonymous substitutions for certain exons (Ka/Ks = 35) after the separation of human and great ape lineages from the Old World monkey. Notably, this exquisite pattern of evolutionary dynamics has given rise to the diversification of the expression profile for this gene family, from testis-specific mRNA expression in baboons to ubiquitous expression in humans and closely related primates (Johnson and Eichler, unpublished results).
Interchromosomal remodeling involves the coalescing of duplicated segments from disparate chromosomal regions, occasionally leading to distinct functional roles. For example, the trypsinogen IV gene on chromosome 9p13 is formed by a fusion of PRSS3 (encoding mesotrypsinogen) from 7q35 and LOC120224 from 11q24 [11,12]. The first exon of trypsinogen IV is derived from the non-coding first exon of LOC120224, whereas exons 2–5 are derived from PRSS3. This interchromosomal juxtaposition of SDs from chromosomes 7 and 11 occurred after the divergence of hominids from Old World monkeys. Furthermore, the two variants of PRSS3 mesotrypsinogen and trypsinogen IV exhibit tissue-specific expression differences, suggesting different selective constraints on functionality.
How are these SDs fixed in the genomes? One possibility is that the negative effect of these core duplicon expansions is offset by the advantage of newly minted genes located within these regions (“core duplicon hypothesis”) [12,30]. An alternative explanation that would help fix SDs even if they are slightly deleterious is a reduction in the effective population size of primate hominid populations. This hypothesis has already been proposed to account for the burst of nuclear mitochondrial insertion sequences at the prosimian–anthropoid divergence [62]. If we assume that most large SDs are weakly deleterious, such variants might be disproportionately fixed because of the whims of genetic drift as opposed to being eliminated by purifying selection in a large effective population size. Such an excess of deleterious mutations has been seen in certain cases, such as gene control regions in comparisons between humans and chimpanzees [63] or, at a smaller scale, in human populations experiencing a bottleneck [64].
The role of SDs in evolutionary rearrangements
The role of SDs in evolutionary rearrangements has supported a nonrandom “fragile–breakage” model for chromosomal rearrangements in mammals [19,65,66]. The association between clusters of SDs and evolutionary chromosomal breakpoints is strong and has been observed in most mammalian genomes [67,68]. Overall, about half (51%) of human–mouse breakpoints of conserved synteny are associated with SDs, significantly more than by random chance (2%) [18]. An important outcome of this non-random model is the propensity for evolutionary “re-use” of chromosomal breakpoints; supporting this, approximately 20% of evolutionary breakpoints from eight mammalian genomes showed evolutionary re-use [68]. In primates, lineage-specific hyperexpansion of SDs might be the consequence of the intrinsic fragility of certain chromosomal sites for rearrangements or, alternatively, this instability could lead to SD hyperexpansion (see below). Unsurprisingly, six of the nine breakpoints of the large cytological pericentric inversions that distinguish the karyotype of humans and chimpanzees map within SD duplication blocks. Furthermore, some of the species-specific SD-mediated inversions (chromosomes 4, 5, 9, 12, 15, 16 and 17 for chimpanzee and chromosomes 1 and 18 for human) also map within species-specific SDs (chr12 in chimpanzee and chr1 and chr18 in human). The breakpoints of the inversions that do not map to SDs are enriched for common repeats (SINE, LINEs), among which non-allelic homologous recombination (NAHR) events might also have occurred (for a review on the genomic comparison between humans and chimpanzees see [69]).
Notably, the great apes and the lesser apes (gibbons) show apparent contrasting trends in terms of chromosomal evolution, with a slow rate of rearrangement in the African great apes and a rapid karyotypic evolution in the gibbon lineage giving rise to four species and 12 sub-species. In contrast to humans and great apes, a smaller fraction (~46%) of Nomascus leucogenys (NLE) gibbon rearrangement breakpoints map to SDs in the human lineage [70]. If SDs are more common in humans and great apes and they associate with rearrangement, one might expect the African great ape lineage to show more rearrangements as opposed to the fourfold excess of rearrangements in the gibbon lineage [70]. One possible explanation for this paradox is that the paucity of SDs in ancestral gibbon genomes diverted rearrangement pathways away from homology-mediated events, favoring alternative replication-based mechanisms (e.g. MMIR, FoSTeS, break-induced replication) for a review on specific non-homology-mediated replication based mechanisms see [91]. If we assume that the rate of rearrangement is uniform among all ape genomes, but that fewer SDs drive fewer homology-mediated events, we would expect non-homology-based mechanisms to contribute more significantly, manifesting as larger chromosomal rearrangements in gibbons. The abundance of duplication blocks dispersed through great ape/human chromosomes might have promoted many more regional and smaller structural rearrangement events (<1Mb) that have a transparent cytogenetic resolution [71]. Moreover, given that NAHR events are often associated with breakpoint re-use [18,36,68] at a constant rearrangement rate, the great apes would show apparently fewer structural changes because of the recurrent rearrangements involving “local” chromosomal segments. Therefore, with the same effective number of events, gibbons with fewer SDs would tend to have more distinct, cytogenetically visible “global” structural changes. In support of this model, no excess of smaller regional structural rearrangements has been reported in gibbons despite a genome-wide survey for such events using BAC-end sequence pairs [72].
Origin of SDs
The origin and mechanism of the dispersion of SDs is still unclear. Different models of SD formation have been suggested for pericentromeric, subtelomeric or general interstitial SDs [73]. Within subtelomeric regions, a translocation-based model was proposed wherein recurrent unequal non-homologous end-joining or non-homologous end joining (NHEJ)-mediated translocations followed by the serial transfer of sequences generated the complex blocks of subtelomeric duplication [74]. A common observation is that SD breakpoints are enriched for SINE repeats (especially Alus) [75–77]. This has opened the possibility that the expansion of Alu elements within the primate lineage might have shaped the ancestral human genome, making it particularly susceptible to Alu–Alu-mediated rearrangement events, which, in turn, promoted the expansion of SDs and their subsequent role in NAHR [76]. Notably, the timing of the burst of Alu repeats (~35 million years ago (mya)) is dated earlier than the expansion of SDs in the human and great ape ancestry (10–20 mya). High resolution sequencing of primate genomes for some of these complex regions has suggested the possibility that specific sequences might be apt to duplicate themselves and flanking sequences to new locations. For example, the LCR16a core duplicon has moved independently in both orangutan and human lineages to new locations, acquiring its own suite of lineage-specific duplications on its flanks [30]. The independent expansion of the gorilla and chimpanzee chr10 duplicon (Figure 3) [21] might represent another manifestation of this core duplicon-flanking transposition model. Interestingly, many core duplicons, such as LCR16a, are particularly Alu-repeat-rich and also the source of primate gene innovations (see above).
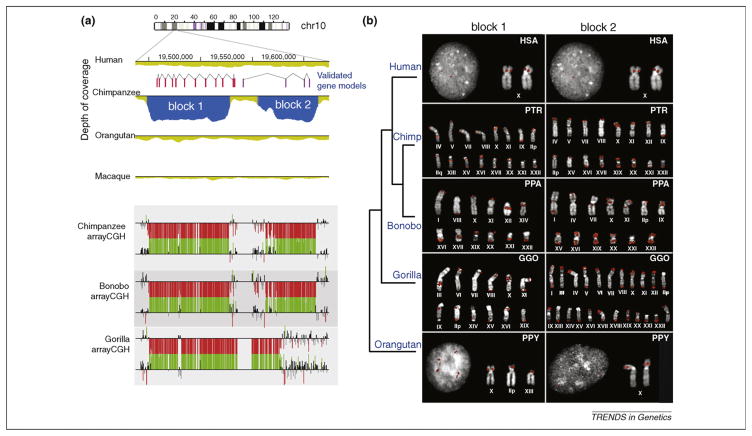
Figure 3.
A recurrent segmental duplication specific to African great apes. (a) Initial WSSD analysis of the chimpanzee genome predicted two chimpanzee-specific duplications (depicted as block 1 and block 2 in blue). The duplication was confirmed by comparative array-CGH (using the human genome as a reference). Note that probes with log2 ratios above (increased copies) or below (decreased copies) 1.5 standard deviations from the normalized log2 ratio are colored green or red, respectively. Array- CGH analysis revealed that both bonobo and gorilla also carried the duplication. Two genes were predicted to map to the duplicated segments. (b) Fluorescence in situ hybridization showed that the duplications (i.e. blocks 1 and 2) had expanded in copy among all African great apes but not in humans. Interestingly, experimental and computational data suggest that all derivative locations between chimpanzee and gorilla are non-orthologous.
Two studies on replication-based mechanisms in yeast and high quality sequencing of the human–NLE gibbon breakpoints of synteny have provided additional insights into the nature of the formation of SDs [72,78]. An experiment designed to study single gene amplification and gene dosage in Saccharomyces cerevisiae led to the serendipitous observation of spontaneous duplication of multiple large inter- and intrachromosomal DNA segments encompassing several dozens of genes [79]. Furthermore, even when all potential DNA repair pathways (homologous recombination and NHEJ/MMEJ (Micro homology Mediated End Joining) pathways) were suppressed, SD formation was observed, suggesting alternative replication-mediated events [78]. These duplicated blocks are essentially formed by replication accidents as other recombination-based repair mechanisms were suppressed [78]. The proposed model suggested that following a double-strand break (DSB) originating from a collapsed replication fork, the free end of the DNA spontaneously invades a suitable template strand with low complexity (polyA/T) sequences or micro-homology, followed by reassembly of a new replication fork. The template switching mechanism can be favored by the presence of microhomology or microsatellite (MMIR).
Sequencing analysis of human–NLE gibbon rearrangements (regions specifically selected because they did not carry SDs) identified mosaic new insertions in ~40% of NLE precisely at the breakpoint of synteny [72] (Figure 4). Similar to the duplication blocks, these mosaic segments originated as small duplications from disparate locations that were both intra- and interchromosomal. The presence of sequence microhomology, topoisomerase binding sites and mosaic architecture at the larger breakpoint intervals suggested a replication-based mechanism for these rearrangements. A subset of these mosaic insertions were, in fact, SDs that had amplified specifically within the gibbon lineages (Figure 4). A notable example is the presence of a 4.2 Kb gibbon-specific SD mapping precisely at the translocation fusion point between chromosomes 3 and 12. Sequence analysis revealed that this SD actually consisted of duplicatively transposed sequences mapping 72 Kb and 64.5 Kb further upstream of the point of fusion on chromosome 3 [72] (Figure 4). Thus, regions of rearrangement are indeed a source of new duplications [80,81]. These data support an alternative model that associates SDs and rearrangements and reinforces that DSBs can generate SDs [79,82,83]. Consequently, regions of genome rearrangement might, in effect, promote the formation of SDs in other regions of the genome as opposed to SDs being the cause of evolutionary rearrangements.
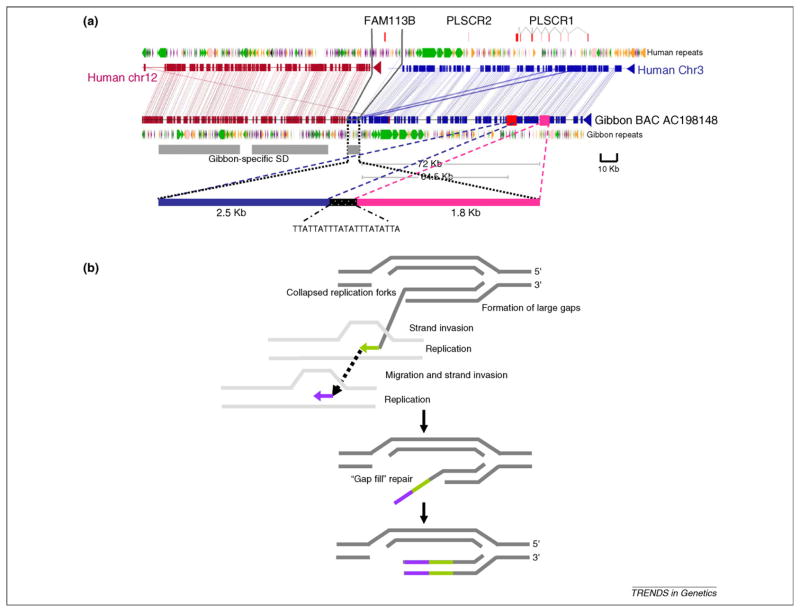
Figure 4.
Sequence analysis of human–gibbon breakpoints of synteny reveals potential mechanisms for SD formation. (a) Insertion of a 4.3 Kb sequence at the human–NLE gibbon breakpoint is shown. Note that the 4.3 Kb sequence block at the breakpoint is derived from ~2.5 Kb and 1.8 Kb blocks that originated 72 Kb and 64.5 Kb upstream, respectively. The grey bar denotes gibbon-specific SD, as assessed by WSSD and validated by FISH using fosmid probes. (b) A replication-based model for formation of SDs is shown [78,83,88]. Large gaps are generated by DSBs (because of a possible collapsed or stalled replication forks) at rearrangement sites. Replication is initiated by recurrent strand invasion and replication to repair the gap. Consequent to a series of strand invasion, replication and uncoupling of the replication machinery, the gap is filled by a mosaic of sequence segments.
Regions of SDs are doomed to endless cycles of rearrangement. If duplication events are not eliminated by selection, they can promote additional rounds of inversions, duplications and deletions with an increased probability of further rearrangements as a direct function of the complexity and homology of the flanking duplications. Not surprisingly, unique genes mapping adjacent to ancestral duplications have a 10-fold higher likelihood of being duplicated – a phenomenon described as “duplication shadowing” [8,21]. Given the high dynamism of these regions, it is common to find recurrent events at nearly identical locations within the genome. A 150 Kb human polymorphic inversion on chromosome Xq28, for example, has been shown to be recurrently inverted in eutherian evolution at least a dozen times [84] owing, in part, to the presence of a diverse array of duplicated sequences located at the inversion breakpoint in almost every mammalian species. Similarly, a 970 Kb inversion polymorphism on human chromosome 17q21.31 is predicted to have inverted at least three times independently in the orangutan, human and chimpanzee lineages [36,85]. In humans, the inverted haplotype (referred to as H2) enriched in European populations, is associated with increased fecundity and is a predisposing factor to recurrent deletions found in handicapped children with the 17q21.31 deletion syndrome [17,86,87]. Both the evolutionarily recurrent inversion and predisposition to recurrent microdeletions in European populations are consequences of the recent duplication architecture that evolved within the human–great ape lineage (Figure 5). This example highlights the complexity of these regions and the importance of high quality final sequences for understanding the role of SDs in human disease, evolution and diversity.
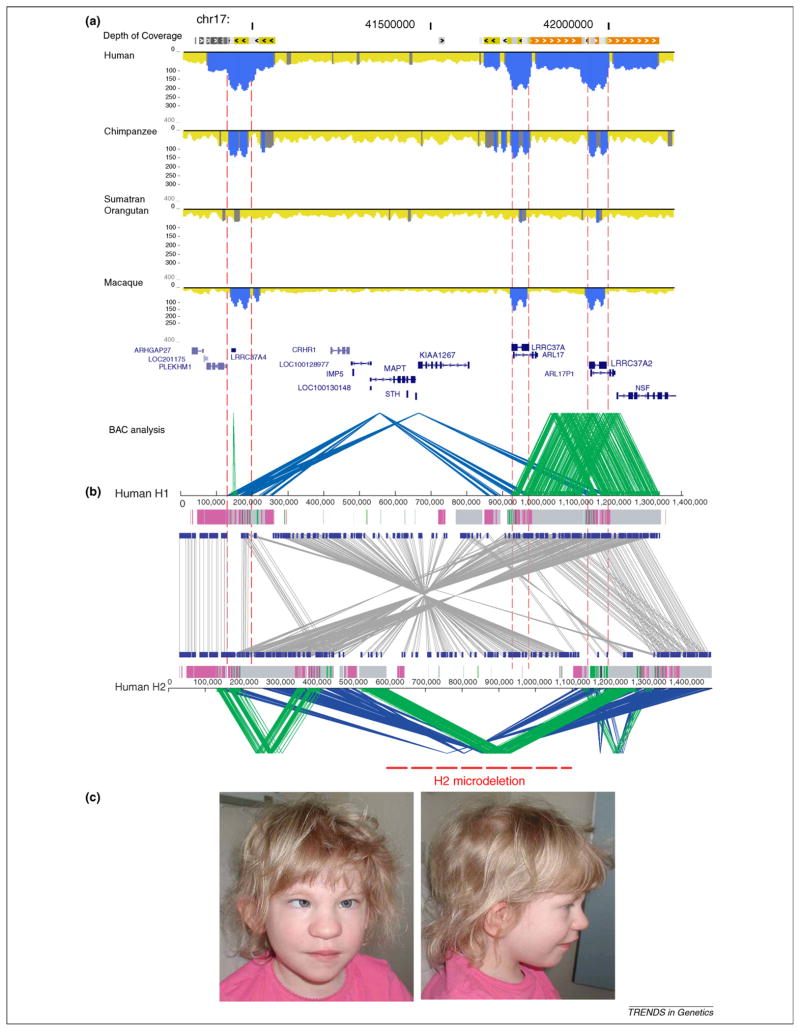
Figure 5.
Comparative duplication architecture of 17q21.31. (a) The schematic shows the extent of duplication for a 1.5 Mb genomic region among human, chimpanzee, orangutan and macaque as determined by WSSD (blue excess read-depth). Dashed lines show the position of the “core” duplicon region corresponding to the LRRC37A gene family. The complexity of the region was not revealed until a complete high quality sequence contig was generated in BAC clones [36]. (b) The inverted sequence organization (grey lines) between two human haplotypes H1 (non-inverted) and H2 (inverted) is shown. Direct (green) and inverted (blue) SDs are depicted for both haplotypes. The H2 haplotype has larger, more identical and directly orientated duplications flanking a suite of neurological genes. It has increased in frequency in the European population presumably as a result of positive selection [85]. The different pattern of duplications in H2 leads to pathogenic microdeletions associated with the 17q21.31 deletion syndrome [17,87,89]. This region clearly highlights the complexity of the duplicated regions and the importance for high quality sequences to understand disease and human evolution. (c) A photograph of a child with cognitive disabilities and developmental delay carrying a 17q21.31 microdeletion is shown. Note the characteristic features including a bulbous nose, and silvery depigmentation of the hair and eyes.
Concluding remarks
Gene duplication is considered the primary means by which new genes and gene families evolve. Until recently, considerations of the birth–death process of gene duplications uncoupled these events from the underlying genomic duplication events. Recent published data suggest that dynamic structural changes mediated by duplication are intricately intertwined with the emergence of functional novelty. Primates provide a unique opportunity to study this aspect of biology. First, there has been an excess of interspersed SD relatively recently in evolution, which provides ample substrate for novel juxtapositions and selection. These studies have also suggested a nonuniform rate of duplication throughout primate evolution with an excess of duplication rate at the time of the hominoid common ancestor. Second, the human genome sequence is arguably the best functionally annotated and assembled reference sequence. Finally, genomic resources (BAC libraries, cDNAs, etc.) and sequences are available to characterize these complex regions of dynamism with precision.
Primate genomes, therefore, provide an opportunity to understand the evolutionary history and mechanism of SDs and how these events precipitated the emergence of novel genes. Such analyses, we believe, are beginning to have far-reaching implications. Recent research is revealing more genetic dissimilarity between humans and the great apes than previously anticipated, leading to the identification of novel human genes, many of which lack antecedents in other mammalian species, and suggesting mechanisms of evolutionary plasticity. Finally, it is apparent that SDs mediate genomic instability associated with disease. Understanding the dynamics of this process is, therefore, critical in assessing its impact on human health.
In this era of massive parallel sequencing, there is the promise that the genomes of most extant primate taxa will ultimately be sequenced. Simply sequencing greater diversity without a focus on the complex duplicated regions of our genome is shortsighted because it will limit our understanding of disease and the origin of our species. Without high quality sequences, it will be difficult to provide a comprehensive and functional understanding of lineage-specific duplicated genes that have been important, if not critical, in our adaptation. Not only the sequence but the diversity of these regions must be systematically understood to accurately genotype and determine their phenotypic consequences within our species, which requires accurately predicting copy, content and structure of these duplicated regions. Comparative high quality sequences of these regions among primates will provide insight into the mechanisms of their dispersion in different lineages (primates vs. other mammalian species) and the mode of selection acting on these regions. Focused efforts on these complex duplicated regions will enhance our understanding of the structure of primate genomes and their dynamic integration within the full spectrum of evolutionary change. Such studies bring to light their potential impact in evolution, variation and disease.
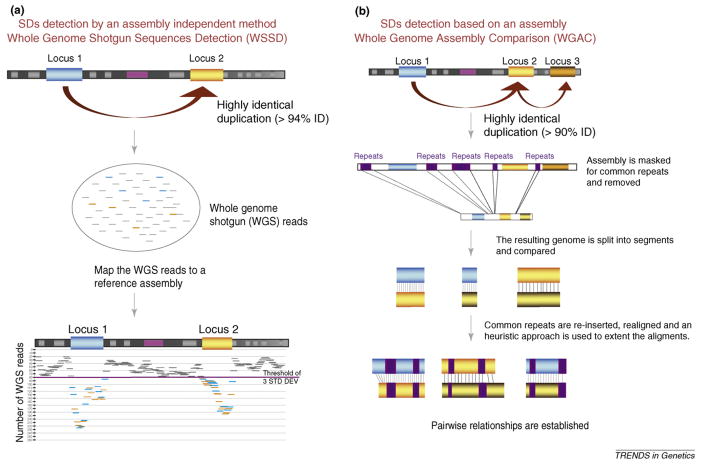
Figure I.
Strategies for duplication detection (WSSD and WGAC). (a) A schematic representation of the whole-genome shotgun sequence detection (WSSD) method to detect recent duplications. In short, whole-genome shotgun reads are mapped against a reference assembly and duplication is detected by the excess of read-depth. Thresholds for duplication detection are estimated from known single-copy BACs. (b) A whole-genome assembly comparison (WGAC) strategy where the genome is segmented, repeats are extracted and the remaining genome segments are compared to identify high identity pairwise alignments.
Acknowledgments
We thank Jeff Kidd, Lin Chen, Ze Cheng, Heather Mefford, Leslie Emery, and Tonia Brown for valuable comments and help in the preparation of this manuscript. This work was supported, in part, by NIH grants GM058815 and HG002385 to E.E.E. T.M.-B. is supported by a Marie Curie fellowship. E.E.E. is an investigator of the Howard Hughes Medical Institute. The authors declare no conflicts of interest.
References
- 1.Ohno S, et al. Evolution from fish to mammals by gene duplication. Hereditas. 1968;59:169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x. [DOI] [PubMed] [Google Scholar]
- 2.Bailey JA, et al. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 4.Johnson ME, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 5.Fortna A, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn MW, et al. Accelerated rate of gene gain and loss in primates. Genetics. 2007;177:1941–1949. doi: 10.1534/genetics.107.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumas L, et al. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Z, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 9.She X, et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She XW, et al. The structure and evolution of centromeric transition regions within the human genome. Nature. 2004;430:857–864. doi: 10.1038/nature02806. [DOI] [PubMed] [Google Scholar]
- 11.Rowen L, et al. Interchromosomal segmental duplications explain the unusual structure of PRSS3, the gene for an inhibitor-resistant trypsinogen. Mol Biol Evol. 2005;22:1712–1720. doi: 10.1093/molbev/msi166. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 13.Sharp AJ, et al. Segmental duplications and copy number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 15.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 16.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 17.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 18.Bailey JA, et al. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armengol L, et al. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum Mol Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- 20.Han MV, et al. Adaptive evolution of young gene duplicates in mammals. Genome Research. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang JZ, et al. Positive Darwinian selection after gene duplication in primate ribonuclease genes. P Natl Acad Sci U S A. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium HGS. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 24.Consortium, C.S.a.A. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 25.Consortium MGS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 26.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 27.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osoegawa K, et al. Construction of bacterial artificial chromosome (BAC/PAC) libraries. Curr Protoc Hum Genet. 2001;Chapter 5(Unit 5–15) doi: 10.1002/0471142905.hg0515s21. [DOI] [PubMed] [Google Scholar]
- 29.She XW, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 30.Johnson ME, et al. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc Natl Acad Sci U S A. 2006;103:17626–17631. doi: 10.1073/pnas.0605426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regnier V, et al. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Human Molecular Genetics. 1997;6:9–16. doi: 10.1093/hmg/6.1.9. [DOI] [PubMed] [Google Scholar]
- 32.Munch C, et al. Evolutionary analysis of the highly dynamic CHEK2 duplicon in anthropoids. BMC Evol Biol. 2008;8:269. doi: 10.1186/1471-2148-8-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 34.Mikkelsen TS, et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 35.Gibbs R, et al. Bovine genomic sequencing initiative Cattleizing the human genome. White paper 2004 [Google Scholar]
- 36.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.She X, et al. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas TJ, et al. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Research. 2009;19:491–499. doi: 10.1101/gr.084715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rochette CF, et al. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 40.Morrison KE. Advances in SMA research: review of gene deletions. Neuromuscul Disord. 1996;6:397–408. doi: 10.1016/s0960-8966(96)00368-9. [DOI] [PubMed] [Google Scholar]
- 41.Reiter LT, et al. The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet. 1997;6:1595–1603. doi: 10.1093/hmg/6.9.1595. [DOI] [PubMed] [Google Scholar]
- 42.Babcock M, et al. Hominoid lineage specific amplification of low-copy repeats on 22q11.2 (LCR22s) associated with velocardiofacial/DiGeorge syndrome. Human Molecular Genetics. 2007;16:2560–2571. doi: 10.1093/hmg/ddm197. [DOI] [PubMed] [Google Scholar]
- 43.Park SS, et al. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 45.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 46.Dominy NJ, Lucas PW. Ecological importance of trichromatic vision to primates. Nature. 2001;410:363–366. doi: 10.1038/35066567. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs GH, et al. Trichromatic color vision in New World monkeys. Nature. 1996;382:156–158. doi: 10.1038/382156a0. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 49.Lemay DG, et al. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biology. 2009;10:R43. doi: 10.1186/gb-2009-10-4-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whittington CM, et al. Defensins and the convergent evolution of platypus and reptile venom genes. Genome Research. 2008;18:986–994. doi: 10.1101/gr.7149808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Degenhardt JD, et al. Copy number variation of CCL3-like genes affects rate of progression to simian AIDS in Rhesus Macaques (Macaca mulatta) PLoS Genet. 2009;5:e1000346. doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aitman TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 53.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fellermann K, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn’s disease of the colon. American Journal of Human Genetics. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bosch N, et al. Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum Mol Genet. 2007;16:2572–2582. doi: 10.1093/hmg/ddm209. [DOI] [PubMed] [Google Scholar]
- 56.Paulding CA, et al. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc Natl Acad Sci U S A. 2003;100:2507–2511. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popesco MC, et al. Human lineage-specific amplification, selection and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 58.Symmons O, et al. How segmental duplications shape our genome: recent evolution of ABCC6 and PKD1 Mendelian disease genes. Mol Biol Evol. 2008;25:2601–2613. doi: 10.1093/molbev/msn202. [DOI] [PubMed] [Google Scholar]
- 59.Vandepoele K, et al. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol Biol Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- 60.Yu YH, et al. Evolution of the DAZ gene and the AZFc region on primate Y chromosomes. BMC Evol Biol. 2008;8:96. doi: 10.1186/1471-2148-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciccarelli FD, et al. Complex genomic rearrangements lead to novel primate gene function. Genome Research. 2005;15:343–351. doi: 10.1101/gr.3266405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gherman A, et al. Population bottlenecks as a potential major shaping force of human genome architecture. PLoS Genet. 2007;3:e119. doi: 10.1371/journal.pgen.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keightley PD, et al. Evidence for widespread degradation of gene control regions in hominid genomes. Plos Biol. 2005;3:282–288. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–995. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey JA, et al. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 2004;14:789–801. doi: 10.1101/gr.2238404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elsik CG, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy WJ, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 69.Kehrer-Sawatzki H, Cooper DN. Structural divergence between the human and chimpanzee genomes. Hum Genet. 2007;120:759–778. doi: 10.1007/s00439-006-0270-6. [DOI] [PubMed] [Google Scholar]
- 70.Carbone L, et al. A high resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2006;2:e223. doi: 10.1371/journal.pgen.0020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newman TL, et al. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Girirajan S, et al. Sequencing human–gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Research. 2009;19:178–190. doi: 10.1101/gr.086041.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 74.Linardopoulou EV, et al. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Babcock M, et al. Shuffling of genes within low-copy repeats on 22qll (LCR22) by Alu-mediated recombination events during evolution. Genome Research. 2003;13:2519–2532. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey JA, et al. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Payen C, et al. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koszul R, et al. Eukaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kehrer-Sawatzki H, et al. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am J Hum Genet. 2002;71:375–388. doi: 10.1086/341963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ranz JM, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim PM, et al. Analysis of copy number variants and segmental duplications in the human genome: evidence for a change in the process of formation in recent evolutionary history. Genome Res. 2008 doi: 10.1101/gr.081422.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith CE, et al. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 84.Caceres M, et al. A recurrent inversion on the eutherian X chromosome. P Natl Acad Sci U S A. 2007;104:18571–18576. doi: 10.1073/pnas.0706604104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bekpen C, et al. Death and Resurrection of the Human IRGM Gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nature Genetics. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 87.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nature Genetics. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 88.Lee JA, et al. A DNA replication mechanism for generating non-recurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 89.de Vries BB, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lupski JR, Stankiewicz P. Molecular mechanisms for rearrangements and their conveyed phenotypes in genomic disorders. PLoS Genet. 2005;1:627–633. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hastings PJ, et al. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]