The lack of ERAD determinants and a powerful ER exit signal mediate Wsc1p's evasion of ER folding surveillance systems. As a consequence, Wsc1p traffics to Golgi apparatus whether folded or not, making it an obligate substrate of Golgi quality control.
Abstract
In the endoplasmic reticulum (ER), most newly synthesized proteins are retained by quality control mechanisms until folded. Misfolded molecules are sorted to ER-associated degradation (ERAD) pathways for disposal. Reports of mutant proteins degraded in the vacuole/lysosome suggested an independent Golgi-based mechanism also at work. Although little is understood of the post-ER pathway, the growing number of variants using it suggests a major role in quality control. Why seemingly redundant mechanisms in sequential compartments are needed is unclear. To understand their physiological relationship, the identification of endogenous pathway-specific substrates is a prerequisite. With ERAD substrates already well characterized, the discovery of Wsc1p as an obligate substrate of Golgi quality control enabled detailed cross-pathway analyses for the first time. By analyzing a panel of engineered substrates, the data show that the surveillance mode is determined by each polypeptide's intrinsic design. Although most secretory pathway proteins can display ERAD determinants when misfolded, the lack thereof shields Wsc1p from inspection by ER surveillance. Additionally, a powerful ER export signal mediates transport whether the luminal domain is folded or not. By evading ERAD through these passive and active mechanisms, Wsc1p is fully dependent on the post-ER system for its quality control.
INTRODUCTION
Newly synthesized proteins fold into their correct three-dimensional (3D) structures to be functional. The fidelity of the process is so fundamentally important that multiple cellular strategies have evolved to monitor protein folding. These mechanisms are collectively termed protein quality control (PQC). A key feature of PQC is the integration of turnover mechanisms that eliminate misfolded and unassembled proteins. The stringent surveillance is needed because of the potential toxicity of aberrant proteins.
The best-studied PQC pathways are found in the endoplasmic reticulum (collectively termed ER quality control or ERQC). Most proteins synthesized in the ER function in other parts of the cell or outside in the case of secreted proteins. For this reason, a major role of ERQC is to recognize and retain conformational intermediates until they fold. Misfolded proteins are targeted for destruction by ER-associated degradation pathways (ERAD). Multiple ER-localized E3 ubiquitin ligases organize cofactors to recognize, extract, and ubiquitinate substrates. Degradation takes place in the cytosol by the 26S proteasome (for reviews, see Sifers, 2004; Romisch, 2005; Anelli and Sitia, 2008; Vembar and Brodsky, 2008).
A number of mutant proteins escape detection by ERQC. In budding yeast, some variants of the plasma membrane Pma1p and Ste2p are degraded by ERAD, whereas others (Pma1-7p and Ste2-3p) are diverted to the vacuole (the yeast lysosome) and degraded (Chang and Fink, 1995; Jenness et al., 1997). Although the structural basis for this difference is unknown, restoring plasma membrane localization of Pma1-7p and Ste2-3p through vacuolar sorting mutants fully reversed their loss-of-function phenotypes (Luo and Chang, 1997; Li et al., 1999). These results demonstrate that the variants are not grossly misfolded and suggest that Golgi surveillance is more sensitive to subtle structural aberrations than ERAD.
Post-ER surveillance is not restricted to yeast. In mammals, gap junction proteins that fail to assemble are degraded in lysosomes and specific disease variants of the human prion protein (PrP) also use a lysosomal route (VanSlyke et al., 2000; Ashok and Hegde, 2009). Of the two systems, PrP was more extensively studied. Certain PrP disease variants and ∼10% of wild-type PrP are degraded by ERAD, indicating that PrP folding is initially monitored by ER quality control (Ma and Lindquist, 2001; Yedidia et al., 2001). However, a recent study raised questions over which mechanism predominates over PrP quality control (Ashok and Hegde, 2009). A number of mutations in the PrP C-terminal globular domain caused the formation of alternate, aggregate-like forms that evade ER surveillance. It was hypothesized that these aberrant PrP structures are “invisible” to ERAD. These findings are reminiscent of studies in yeast and mammals demonstrating that ERAD cannot process protein aggregates, causing them to use autophagy as an alternative route (Kamimoto et al., 2006; Kruse et al., 2006).
Foreign proteins can subvert ERAD in budding yeast. A fusion protein consisting of the N-terminal fragment of the phage λ repressor and native yeast invertase is secreted. Point mutations that thermodynamically destabilize λ repressor in bacteria, if introduced into the hybrid molecule, divert it to the vacuole (Hong et al., 1996). Its dependence on the Golgi-localized sorting factor Vps10p, led investigators to propose that the mutations caused unfolded structures that mimic sorting signals of normal vacuolar proteins (Marcusson et al., 1994; Cooper and Stevens, 1996). A similar example comes from experiments using a Y35L variant of bovine pancreatic trypsin inhibitor (BPTI). It is a folded protein but displays more flexibility in its carbon backbone than wild type (Zhang and Goldenberg, 1993; Hanson et al., 2003). Remarkably, instead of being secreted, it is transported to the vacuole for degradation (Coughlan et al., 2004). As foreign proteins, do they pass ERQC because of absence of ERAD determinants or are their structural aberrations too subtle for ERAD detection?
An example of a grossly misfolded protein trafficking through the Golgi is the forced exit of the classical ERAD substrate CPY* (a point mutant of vacuolar carboxypeptidase Y; Finger et al., 1993). At moderate expression levels, nearly 100% of CPY* is retained in the ER and degraded by ERAD. Its overexpression or attachment to a cytoplasmic ER exit signal causes its transport to the Golgi, where it is sorted to the vacuole instead of being retrieved to the ER (Spear and Ng, 2003; Kincaid and Cooper, 2007). These studies show that a Golgi-based mechanism can detect grossly misfolded proteins that contain determinants normally recognized by ERAD.
Typically, PQC pathways are delineated by first defining their substrates (Sifers et al., 1988; Finger et al., 1993; Hampton and Rine, 1994; McCracken and Brodsky, 1996a; Loayza et al., 1998; Hill and Cooper, 2000; Hosokawa et al., 2001; Rabinovich et al., 2002; Molinari et al., 2003). Along these lines, we sought to develop Wsc1p as a model. Wsc1p is a nonessential single-span type I transmembrane protein that functions as a sensor for plasma membrane/cell wall integrity signaling (Verna et al., 1997; Philip and Levin, 2001). The extracellular/luminal domain lacks N-linked glycans but is heavily O-mannosylated. The extension of these glycans in the Golgi apparatus provides a convenient marker for transport (Lommel et al., 2004). Its lack of N-linked glycans makes Wsc1p particularly interesting because most luminal model substrates in budding yeast require them for recognition (Knop et al., 1996; Jakob et al., 1998; Nakatsukasa et al., 2001). The one exception is the nonglycosylated variant of the pro-α-factor, a soluble protein. This unusual substrate uses a mechanism that requires the proteasome but is independent of other known ERAD factors (Brodsky and McCracken, 1999; Wahlman et al., 2007). Because Wsc1p differs from existing ERAD model substrates, we reasoned its analysis could reveal a different mode of PQC.
In this study, a panel of Wsc1p variants was created that severely disrupt folding of the luminal domain. The mutants did not localize to the plasma membrane and were rapidly degraded, indicative of an effective quality control mechanism. The variants were not subject to ERAD but transported to the vacuole by way of the Golgi apparatus. Detailed analyses show that Wsc1p is an obligate substrate of Golgi quality control, demonstrating that the pathway can function as a primary inspection site for some proteins. Combining Wsc1p and the classical ERAD substrate CPY* to perform a cross-pathway analysis showed that pathway utilization is rooted in the intrinsic design of client proteins. Taken together, the study reveals the interplay between the pathways in substrate management and emphasizes the importance of post-ER mechanisms in regulating protein biosynthesis in the secretory pathway.
MATERIALS AND METHODS
Strains and Antibodies
Yeast strains used in this study are listed in Table S1. Anti-hemagglutinin (HA) mAb (HA.11) and anti-HA–conjugated agarose (sc-7392) were purchased from Covance Research Products (Madison, WI) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Kar2p polyclonal antiserum was provided by Peter Walter (University of California, San Francisco, San Francisco, CA). Anti-CPY antiserum was a kind gift of Dr. Reid Gilmore (University of Massachusetts, Worcester, MA). The 13D11 anti-V-ATPase (60-kDa subunit) was purchased from Molecular Probes (Eugene, OR).
Plasmids Used in This Study
Plasmids used in this study and were constructed using standard protocols (Sambrook et al., 1989). All plasmid inserts were confirmed by DNA sequence analysis. Substrate constructs were engineered with an HA epitope tag at their C-termini. Unless otherwise noted, all recombinant genes are driven by the PRC1 promoter. pDN436 is the CPY*-HA expression vector described previously (Ng et al., 2000). The construction of pWX75 containing the ERAD determinant of CPY (the CPY C-terminal domain [CTD]) fused with the Kar2 signal sequence was described previously (Xie et al., 2009).
pSW3.
The WSC1 open reading frame lacking its terminator codon was amplified from genomic DNA using SWN1 and SWN2 and digested with BamHI and NcoI. The fragment was used to replace DN-α-F in pDN333 (Ng et al., 1996) to generate an HA-epitope tagged Wsc1p driven by the TDH3 promoter (Wsc1p-HA).
pSW5.
pSW3 was digested with BamHI and XbaI to release the Wsc1p-HA open reading frame. The fragment was ligated into pSM36, a YCp50-based plasmid containing the PRC1 promoter and the ACT1 terminator.
pSW100.
pSW5 was digested with AatII, and the ends were filled using T4 DNA polymerase (New England Biolabs, Ipswich, MA). The DNA was digested again with SalI to release the gene encoding Wsc1p-HA. This fragment was ligated into pRS315 digested with SmaI and SalI to generate pSW100.
pSW144 and pSW145.
pSW144: In the first step, a 1.4-kb fragment was amplified using SWN84 and SWN85 primers and pWX75 as the template (Xie et al., 2009). This contains the PRC1 promoter upstream of sequences encoding the Kar2p signal sequence fused to the CPY CTD (R370 to L532). In the second step, a 1.6-kb fragment was amplified from pSW104 with primers SWN86 and SWN50. This fragment contains Wsc1p coding sequences after its signal sequence (Bendtsen et al., 2004), an HA epitope tag, and the ACT1 terminator. The two fragments were digested with NotI and SalI, respectively, and inserted into pRS315 digested with the same enzymes to generate pSW144 (ED-Wsc1-L63R). pSW145 (ED-Wsc1-Δ68-80) was constructed like pSW144 except that the second fragment was amplified using pSW113 as a template.
pSW147, pSW148, and pSW149 encode Wsc1p, Wsc1-L63R, and Wsc1-Δ68-80 driven by the GAS1 promoter, respectively. A 600-base pair fragment containing the GAS1 promoter was amplified from genomic DNA using SWN87/SWN88 primers and digested by NotI and BamHI. Second, 1.7-kb BamHI/SalI fragments were released from pSW100, pSW104, and pSW113. They contain the Wsc1p-HA, Wsc1-L63R-HA, or Wsc1-Δ68-80-HA gene followed by the ACT1 terminator sequence. The two fragments were ligated into pRS315 digested with NotI and SalI.
Site-directed Mutagenesis
Plasmids were modified by site-directed mutagenesis as described previously (Sawano and Miyawaki, 2000) and are listed in Table S2.
Cell Labeling and Immunoprecipitation Analysis
Metabolic Pulse-Chase Analysis.
Metabolic pulse-chase experiments were carried out as described previously (Ng et al., 2000). Typically, 3.0 OD600 U of log phase cells were labeled with 82.5 μCi using [35S]methionine/cysteine (EasyTag EXPRESS 35S Protein Labeling Mix, Perkin Elmer, Wellesley, MA). Immunoprecipitated proteins were analyzed by SDS-PAGE and exposure to phosphor screens. Visualization and quantification of resolved proteins were performed using a Typhoon 8600 scanner and ImageQuant TL software (GE Healthcare Biosciences, Piscataway, NJ). All quantification data reflect three independent experiments with mean ± SD indicated.
PEGylation-based Protein-folding Assay.
Cells were labeled for 5 min using [35S]methionine/cysteine and chased for the times indicated in figures. Proteins were precipitated in 10% trichloroacetic acid (TCA), pelleted, washed in cold acetone, and solubilized by boiling 5 min in TCA resuspension buffer (100 mM Tris, pH 11.0, 3% SDS, 1 mM PMSF) or Mal-PEG reaction buffer (100 mM Tris, pH 7.4, 2% SDS, 1 mM PMSF) containing 5 mM methoxypolyethylene glycol 5000 maleimide (Mal-PEG, Fluka, Buchs, Switzerland). The samples were incubated on ice for 1 h. Control and reaction detergent lysates were used for immunoprecipitation as described previously (Ng et al., 2000). Immunoprecipitated proteins were analyzed as described above. To reduce proteins in vivo, dithiothreitol (DTT) was added to media 30 min before and during cell labeling.
Indirect Immunofluorescence.
Indirect immunofluorescence experiments were performed as described previously (Spear and Ng, 2003) with the following modifications. After fixation, cells were incubated in spheroplasting buffer (1 mg/ml zymolase 20T [US Biological], 0.1 M potassium phosphate, pH 7.5, 1.2 M sorbitol) for 20–40 min. Next, spheroplasts were adhered to poly-l-lysine–coated 10-well slides for 10 min and washed once with TBS (50 mM Tris-HCl, pH 7.4, 150 mM NaCl). The slides were immersed in cold methanol (−20°C) for 6 min and then in cold acetone (−20°C) for 30 s. The blocking buffer (TBS, 0.05% Tween-20, 5% nonfat dry milk) was applied onto each well for 30 min and was used for subsequent antibody dilutions. Primary antibodies were incubated overnight at 4°C, and secondary antibodies were incubated for 90 min. For differential interference contrast (DIC) images, samples were blocked in TBS block (3% BSA, TBS). Slides were washed with TBS buffer twice after each application. Primary antibody dilutions were HA.11 mAb (1:200), anti-V-ATPase (1:100), and rabbit anti-Kar2p (1:1000). Secondary antibody dilutions were Alexa Fluor 488 goat anti-mouse (1:500), Alexa Fluor 546 goat anti-mouse (1:500), and Alexa Fluor 594 goat anti-rabbit (1:1000; Molecular Probes). Samples were visualized using a Zeiss Axiovert 200M microscope with a 100× 1.25 NA oil Achroplan objective (Carl Zeiss MicroImaging). For DIC images, an Axio Imager.M1 microscope with a 100× 1.4 NA oil DIC Plan-Apochromat objective was used (Carl Zeiss MicroImaging). Images were archived using Adobe Photoshop CS3 (Adobe Systems, San Jose, CA).
Coimmunoprecipitation and Western Blotting
Coimmunoprecipitation experiments were performed as described previously (Xie et al., 2009). The clarified supernatant was incubated with anti-HA–conjugated agarose or anti-Kar2p and protein A-Sepharose beads (Sigma-Aldrich, St. Louis, MO) under native conditions. The immunoprecipitates were subjected to 8% SDS-PAGE and immunoblot analysis.
RESULTS
Generation of Wsc1p Misfolded Variants
The folding of secretory pathway proteins is highly sensitive to mutation (Zhang et al., 1997). Although many model ERQC substrates were discovered as loss-of-function mutations (Sifers et al., 1988; Finger et al., 1993; Sommer and Jentsch, 1993; Loayza et al., 1998), some were easily created by introducing structurally disruptive mutations (McCracken and Brodsky, 1996b; Fujita et al., 2006). Without existing folding-defective alleles of WSC1, we constructed a series of mutants targeting its luminal domain. Six were generated: three containing nonconservative point mutations (L31R, L63R, and C98R/G99R) and three bearing short deletions (Δ34-44, Δ68-80, Δ96-110; Figure 1A and Figure S1A).
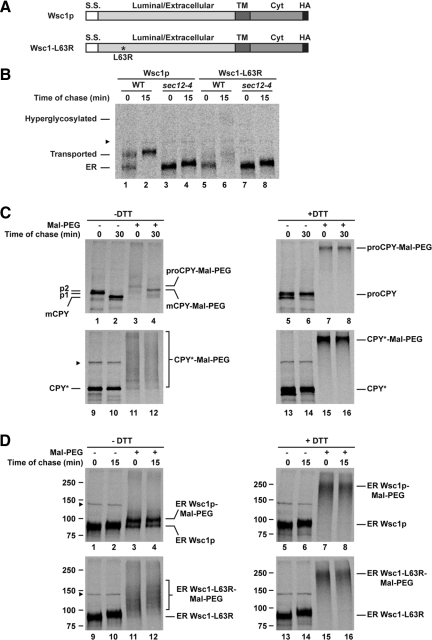
Figure 1.
A misfolded variant of Wsc1p. (A) Schematic representation of HA-tagged Wsc1p and Wsc1-L63R. The asterisk marks the position of the L63R mutation. S.S., signal sequence; TM, transmembrane domain; Cyt, cytoplasmic domain. (B) Wild-type (WT) and sec12-4 cells expressing Wsc1p or Wsc1-L63R were grown to log phase at 23°C and shifted to 37°C for 20 min before a 5-min pulse-label with [35S]methionine/cysteine and a 15-min chase. Wsc1p and Wsc1-L63R were immunoprecipitated from detergent lysates using anti-HA antibody and resolved by SDS-PAGE. (C) Wild-type cells incubated in the presence or absence of 10 mM DTT were pulse-labeled for 5 min and chased for 0 and 30 min at 30°C. Proteins were incubated with or without Mal-PEG after TCA precipitation. CPY and CPY* were immunoprecipitated from detergent lysates using anti-CPY and anti-HA antisera, respectively. The positions of ER (p1), Golgi (p2), and mature vacuolar form (mCPY) of CPY are indicated. (D) The sec12-4 strain expressing Wsc1p or Wsc1-L63R was labeled for 5 min and chased for 15 min as described in B. PEGylation-based folding assay was performed as described in C. Immunoprecipitated Wsc1p and Wsc1-L63R were resolved by SDS-PAGE. The arrowhead denotes the position of a nonspecific protein that cross-reacts with the anti-HA antibody.
To directly analyze the effects of the mutations on Wsc1p folding, a chemical-based assay was developed. It is based on a 5-kDa modifier, Mal-PEG, that covalently attaches to free sulfhydryls but not to disulfides (Roberts et al., 2002). Disulfide bonds often stabilize cysteine-containing membrane proteins like Wsc1p (Wsc1p contains nine cysteines, eight in the luminal domain, and one in the transmembrane). Thus, the extent of unpaired cysteines, a measure of misfolding, can be monitored by gel mobility shift after chemical modification. To determine efficacy, the assay was applied to wild-type CPY and to its misfolded variant CPY*, a well-characterized ERAD model substrate (Finger et al., 1993). Folded CPY has five disulfide bonds and one free cysteine (Endrizzi et al., 1994).
Wild-type CPY is initially synthesized as glycosylated proCPY (p1) in the ER (Figure 1C, lane 1). Extension of core glycans in the Golgi causes a slight shift in mobility to the p2 form (Figure 1C, the p2 form is not visible after the short pulse). Proteolytic cleavage of the prodomain in the vacuole yields mature CPY that is visible after a short chase (Figure 1C, lane 2; Stevens et al., 1982). To monitor folding, immunoprecipitated CPY was reacted with Mal-PEG after a metabolic pulse chase. After a 5-min pulse, PEGylated proteins migrated mostly as a high-molecular-weight smear indicative of folding intermediates (Figure 1C, cf. lane 1 with lane 3). A minor 5-kDa mobility-shifted species represents the folded fraction (Figure 1C, lane 3). After a 30-min chase, PEGylating mCPY resulted in the disappearance of folding intermediates and the appearance of a singly modified species (Figure 1C, cf. lanes 3 and 4). As a control, the assay was performed on proCPY reduced in vivo by DTT treatment. PEGylated reduced proCPY migrated as a single high-molecular-weight band (Figure 1C, lanes 7 and 8). We next applied the assay to the folding mutant CPY*. After the pulse-label, PEGylated CPY* migrated as a broad smear that persisted after the chase (Figure 1C, lanes 11 and 12). This experiment shows that CPY, except for a single free cysteine, becomes Mal-PEG resistant after folding. Its misfolded counterpart CPY*, however, exhibits disulfide bond heterogeneity that does not resolve over time.
Application of the assay to Wsc1p variants required an adjustment. In pulse-chase experiments, Wsc1p variants convert to higher molecular weight forms, reminiscent of Golgi O-mannose modifications of the wild type (Figure 1B, cf. lanes 2 and 6). If these are Golgi modifications, blocking ER-to-Golgi transport using a COPII vesicle budding mutant should prevent the shift (Nakano et al., 1988; Barlowe and Schekman, 1993). As shown in Figure 1B and Figure S1B, the sec12-4 allele blocked the conversion indicating that the variants are transported from the ER via COP II vesicles. Because protein folding occurs in the ER, the folding assay was performed in this strain to make PEGylated forms more discernable. Applying the assay to Wsc1p, the 90-kDa ER form shifts to a doublet comprising the major singly modified species and minor unmodified form was already observed during the pulse, indicating rapid folding (Figure 1D, lanes 3 and 4). This is consistent with the rapid rate of conversion to the mature Golgi form (Figure 1B, lane 1). When the assay was applied to Wsc1p mutants, only higher molecular weight heterogeneously modified forms were observed at both times points (Figure 1D, lanes 11 and 12, and Figure S1C). Note that these forms migrate faster that the DTT-treated control, where a single broad ∼250-kDa PEGylated species results from the modification of all nine cysteine residues (Figure 1D, lanes 15 and 16). This mirrors the pattern of CPY* and provides direct evidence that Wsc1p variants are severely misfolded.
Degradation of Wsc1p Variants Is ERAD Independent
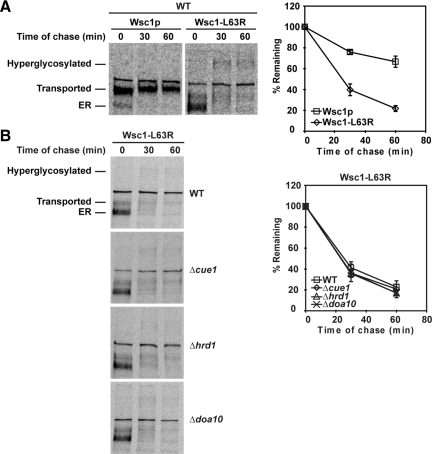
The finding that misfolded Wsc1p traffics from the ER seems surprising but some ERAD substrates can be retrieved from the Golgi for degradation (Caldwell et al., 2001; Vashist et al., 2001). To determine if degradation is via the ER quality control system, Wsc1p variants were expressed in a panel of ERAD-specific mutants. By quantitative pulse-chase analysis, we determined that wild-type Wsc1p is a moderately stable protein (Figure 2A). By contrast, Wsc1p variants degraded rapidly with half-lives of ∼20 min (Figure 2A and Figure S2). The exception is the Δ68-80 variant, which was degraded more slowly, with a half-life of ∼40 min (Figure S2D). We tested the turnover of each variant in a panel of mutants that disrupt the Hrd1 pathway (Δhrd1), the Doa10 pathway (Δdoa10), or both pathways (Δcue1; Carvalho et al., 2006; Denic et al., 2006). Degradation of all six variants was unaffected in these mutants, demonstrating that their turnover does not require ERAD (Figure 2B and Figure S2). In addition, degradation of misfolded Wsc1p is unaffected in cells treated with the proteasome inhibitor MG132 (data not shown).
Figure 2.
Wsc1-L63R is degraded rapidly using an ERAD-independent pathway. (A) Wild-type cells expressing Wsc1p or Wsc1-L63R were pulse-labeled at 30°C with [35S]methionine/cysteine for 10 min followed by a cold chase for times indicated. Proteins were resolved by SDS-PAGE and quantified by phosphorimager analysis. The data plotted reflect three independent experiments with mean ± SD indicated. (B) Wsc1-L63R turnover was measured in wild-type, Δcue1, Δhrd1, and Δdoa10 strains by pulse-chase analysis as described in A.
Misfolded Wsc1p Traffics through the Golgi for Turnover in the Vacuole
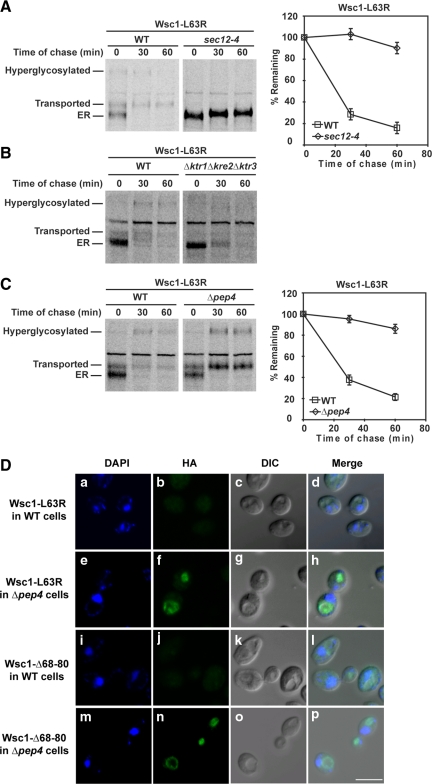
Like wild-type Wsc1p, the variants trafficked out of the ER shortly after synthesis (Figure 2 and Figure S2). The Wsc1p variants also exhibited strong stabilization if transport was inhibited using the sec12-4 block (L63R and Δ68-80 shown in Figure 3A and Figure S3). We noted that the transport block eliminated conversion to high-molecular-weight forms indicative of Golgi modification of O-linked glycans. This was confirmed by pulse-chase analysis in a strain lacking KTR1, KRE2, and KTR3, genes encoding the Golgi-localized 1,2-mannosyltransferases (Lussier et al., 1997). As shown in Figure 3B, conversion to the two major higher molecular weight species was nearly eliminated in this strain. Interestingly, unlike the sec12-4 block, disrupting Golgi glycosylation had no effect on mutant Wsc1p degradation.
Figure 3.
Wsc1-L63R is transported to the vacuole via the Golgi for degradation. (A) Stability of Wsc1-L63R was examined in wild-type and sec12-4 cells using pulse-chase analysis as described in Figure 2A, except both strains were grown to log phase at 23°C and shifted to 37°C for 20 min before labeling. (B and C) Pulse-chase analysis was performed in wild-type, Δktr1Δkre2Δktr3, and Δpep4 cells expressing Wsc1-L63R as described in Figure 2A. (D) Wild-type and Δpep4 cells expressing Wsc1-L63R or Wsc1-Δ68-80 were fixed and permeabilized. Staining was performed using anti-HA primary antibodies followed by Alexa Fluor 488 goat anti-mouse secondary antibodies. DAPI staining marks the position of nuclei. Cells were visualized by confocal and DIC microscopy. Scale bar, 5 μm.
Because of the vacuole's well-known role in catabolism, we measured turnover in Δpep4 mutants specifically defective in vacuolar proteolysis (Ammerer et al., 1986). As shown in Figure 3C and Figure S4, all six variants were strongly stabilized in the strain. Rapid and complete conversion to Golgi forms of mutant Wsc1p indicated that transport was not impeded. This shows that ER quality control mechanisms neither retain nor degrade Wsc1p variants.
To determine the steady-state localization of Wsc1p variants, indirect immunofluorescence microscopy was performed with wild-type and Δpep4 cells. In wild-type cells, signals were barely above background levels reflecting the efficiency of turnover (Figure 3D, b and j). By contrast, stabilization resulted in strong vacuolar staining in the Δpep4 cells (Figure 3D, f and n, and Figure S5). Taken together, these data show that misfolded Wsc1p is not subject to ERQC. Instead, it exits the ER via COPII vesicles to the Golgi, where it is diverted to the vacuole for degradation.
Autophagy is sometimes used to eliminate protein aggregates in the cytosol and the ER (Ravikumar et al., 2002; Kruse et al., 2006; Sarkar et al., 2007). Because misfolded Wsc1p is degraded in the vacuole, we wondered if autophagy contributes to the process. For this, turnover was measured in the cells lacking APG8, a gene required for autophagasome formation (Ravikumar et al., 2002). Wsc1p mutant turnover was unaffected in Δapg8 cells, indicating that an autophagy-independent mechanism is used for their sorting (Figure S6A).
Misfolded Wsc1p Is Sorted for Degradation in the Golgi Apparatus
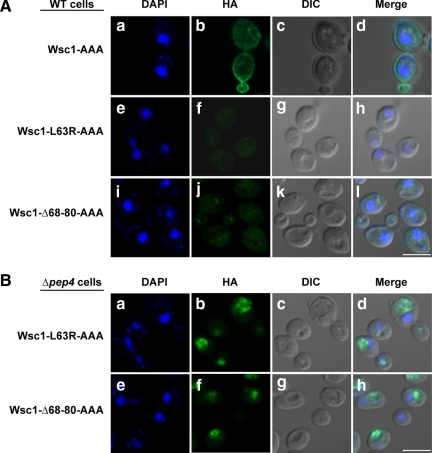
The absence of Wsc1p variants on the plasma membrane suggests a direct route from the Golgi to the vacuole (Figure 3D). However, because the wild type protein undergoes receptor recycling at the plasma membrane there is the possibility of an alternative route. A recent study identified an NPFX(1,2)D endocytic signal in the cytoplasmic domain of Wsc1p (Piao et al., 2007). Specific point mutations altering its NPFDD signal abolish endocytosis. To test whether a portion of Wsc1p variants can use its endocytic signal to reach the vacuole, the signal was destroyed in Wsc1p (Wsc1-AAA), L63R (Wsc1-L63R-AAA), and Δ68-80 (Wsc1-Δ68-80-AAA). In agreement with Piao et al. (2007), Wsc1-AAA is localized exclusively to the plasma membrane in wild-type cells (Figure 4Ab). By contrast, the staining patterns of Wsc1-L63R-AAA and Wsc1-Δ68-80-AAA did not change from the original unmodified forms (cf. Figures 4 and 3D). We conclude from these data that Wsc1p misfolded luminal domain variants traffic directly from the Golgi to the vacuole.
Figure 4.
Misfolded Wsc1p is degraded by Golgi QC. (A and B) Wild-type and Δpep4 cells expressing Wsc1-AAA, Wsc1-L63R-AAA, or Wsc1-Δ68-80-AAA proteins driven by the GAS1 promoter were processed as described in Figure 3D. Staining was performed using anti-HA primary antibodies and Alexa Fluor 488 goat anti-mouse secondary antibodies. Images were captured by confocal and DIC microscopy. Scale bars, 5 μm.
Devoid of ERAD Determinants, Wsc1p Is Undetected by ER Quality Control
The results from our analyses validate Wsc1p as a bona fide endogenous, obligate substrate of Golgi quality control. As such, the most immediate question is why ERQC fails to detect Wsc1p misfolding. One possibility is that the cytoplasmic domain carries a dominant ER exit signal recognized by the COPII transport machinery. Cooper and colleagues showed that anchoring CPY* to the membrane with a powerful cytoplasmic ER exit signal can drive its export (Kincaid and Cooper, 2007). If the Wsc1p cytoplasmic domain directs its exit in a way that bypasses ERAD, uncoupling the molecule from the signal might restore ERAD of the misfolded luminal domain.
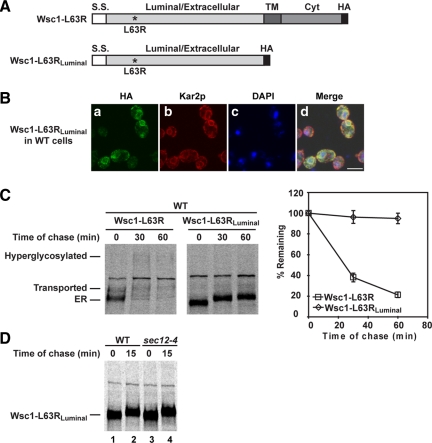
To test this idea, the L63R and Δ68-80 variants were converted to soluble luminal proteins like CPY* by deleting transmembrane and cytosolic segments (Figure 5A and Figure S7A). By indirect immunofluorescence microscopy, both luminal variants were detected strongly in the ER (Figure 5B and Figure S7B). This experiment suggests that the deleted sequences removed a signal required for the transport of misfolded variants. This conclusion was confirmed by pulse-chase analysis of wild-type cells. Wsc1-L63RLuminal and Wsc1-Δ68-80Luminal proteins failed to convert from their ER forms throughout the chase (Figure 5C and Figure S7C). The slight mobility shift is due to ER modification because the same was observed for variants in the sec12-4 mutant (Figure 5D and Figure S7D). Surprisingly, both molecules are completely stable despite their accumulation in the ER (Figure 5C and Figure S7C). Two possible reasons can account for their avoidance of ERAD. The Wsc1p luminal domain could simply lack ERAD determinants or part of its structure/composition is inhibitory for ERAD. Integrating a bona fide ERAD determinant into Wsc1 variants would address these possibilities.
Figure 5.
Wsc1-L63R lacks an ERAD determinant in its luminal domain. (A) A schematic diagram of Wsc1-L63R and Wsc1-L63RLuminal. Asterisk indicates the L63R mutation. S.S., signal sequence; TM, transmembrane domain; Cyt, cytoplasmic domain. (B) Wild-type cells expressing Wsc1-L63RLuminal were processed as in Figure 3D. The cells were stained with anti-HA and anti-Kar2p primary antibodies followed by Alexa Fluor 488 goat anti-mouse and Alexa Fluor 594 goat anti-rabbit secondary antibodies. Cells were visualized by confocal microscopy. Scale bar, 5 μm. (C) The turnover of Wsc1-L63R and Wsc1-L63RLuminal in the wild-type strain was measured by pulse-chase analysis as described in Figure 2A. (D) Pulse-chase analysis was performed in wild-type and sec12-4 cells expressing Wsc1-L63RLuminal as described in Figure 1B.
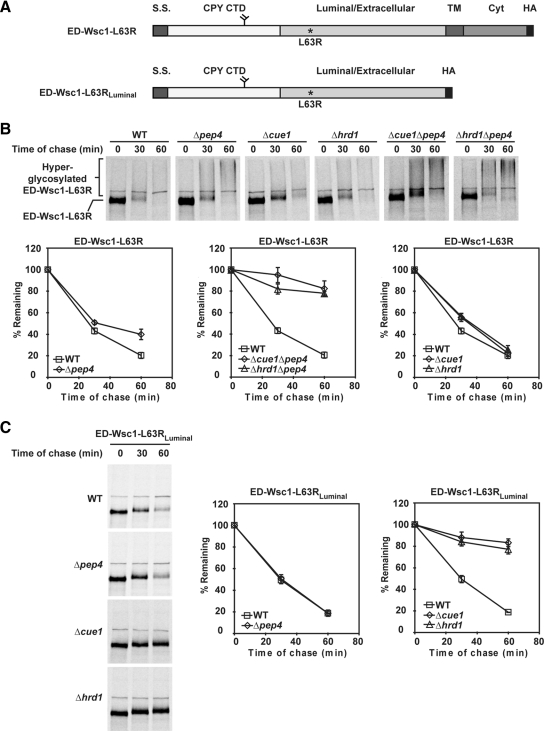
The CPY CTD was appended to the full-length (L63R and Δ68-80) and soluble (L63RLuminal and Δ68-80Luminal) Wsc1 variants in their luminal domains (Figure 6A and Figure S8A). This domain forms a glycan-dependent ERAD signal if it cannot integrate into the tertiary structure of CPY (Kostova and Wolf, 2005; Spear and Ng, 2005; Xie et al., 2009). The degradation profile of each test molecule was determined in wild-type, Δpep4 (defective in vacuolar degradation), Δhrd1 (defective in ERAD-L and ERAD-M pathways), Δcue1 (all known ubiquitin-dependent ERAD pathways), and Δpep4Δhrd1 and Δpep4Δcue1 strains. As shown in Figure 6B and Figure S8B, the full-length ED-Wsc1-L63R and ED-Wsc1-Δ68-80 (ED denotes ERAD Determinant) degraded rapidly in wild-type cells and partly stabilized in the Δpep4 strain. This result demonstrates that the fused signal directed degradation to another pathway but a small, but significant fraction still uses the vacuolar pathway. Their strong stabilization in the Δpep4Δhrd1 and Δpep4Δcue1 double mutants shows that degradation is primarily via ERAD (Xie et al., 2009). Notably, the stabilized substrates converted to a high-molecular-weight smear indicative of heterogeneous outer-chain glycosylation of the appended N-glycan (Figure 6B and Figure S8B; Spear and Ng, 2003). This indicates that blocking ERAD does not prevent their export. The rapid turnover of ED-Wsc1-L63R and ED-Wsc1-Δ68-80 in the Δhrd1 and Δcue1 strains confirms that they are able to use the vacuolar pathway efficiently when ERAD is unavailable (Figure 6B and Figure S8B). This was surprising because the CPY CTD binds the ER chaperone BiP (immunoglobulin heavy chain binding protein)/Kar2p strongly as part of its role in ERAD (Xie et al., 2009). The data show that Wsc1p's intrinsic export signal can overcome the ER retention activity of the appended ERAD determinant (see next section).
Figure 6.
ED-Wsc1-L63R and ED-Wsc1-L63RLuminal are ERAD substrates. (A) Schematic representations of ED-Wsc1-L63R and ED-Wsc1-L63RLuminal. S.S., signal sequence from Kar2p; CPY CTD, CPY C-terminal domain; TM, transmembrane domain; Cyt, Wsc1p cytoplasmic domain; HA, hemagglutinin tag. (B) The turnover of ED-Wsc1-L63R was examined in the wild-type and mutant strains using pulse-chase analysis as described in Figure 2A. (C) Degradation rates of ED-Wsc1-L63RLuminal were determined in wild-type and mutant cells by pulse-chase analysis as in Figure 2A.
When the same analysis was performed with its luminal counterparts ED-Wsc1-L63RLuminal and ED-Wsc1-Δ68-80Luminal, the dependency on ERAD was absolute. The molecules degraded rapidly and independently of PEP4 (Figure 6C and Figure S8C). In Δhrd1 and Δcue1 ERAD mutants, the soluble substrates are completely stable, demonstrating that adding the determinant produces a bona fide ERAD substrate when the competing export signal is eliminated. These experiments demonstrate that Wsc1p is an obligate substrate of Golgi quality control because 1) it lacks ERAD determinants and 2) a strong ER exit signal promotes its export whether it is folded or not.
Misfolded Wsc1p Does Not Bind the ERAD Chaperone Protein Kar2p
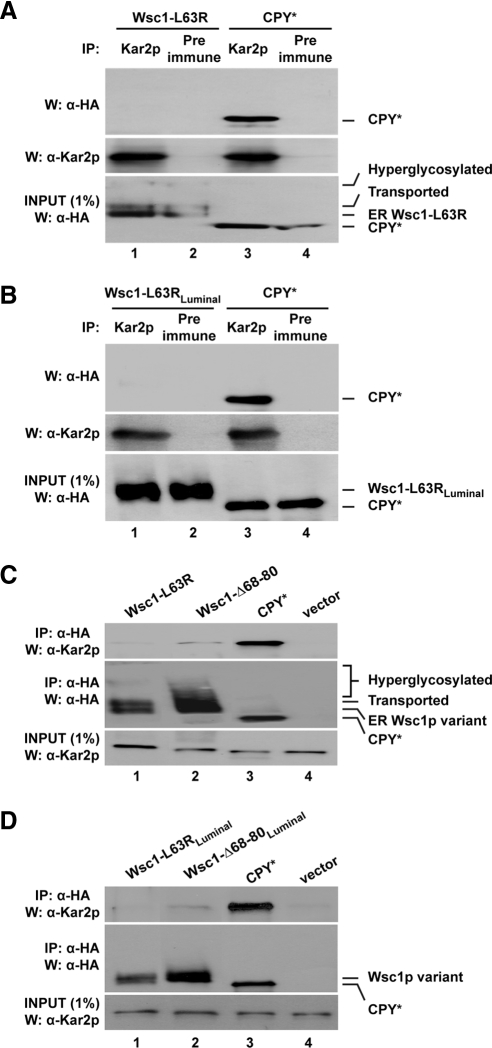
An oft-reported characteristic of folding intermediates and misfolded proteins in ER is their physical interaction with the chaperone BiP (also known as Kar2p in yeast; Haas and Wabl, 1983; Gething et al., 1986; Hurtley et al., 1989). The function of the chaperone is required for quality control of ERAD-L substrates like CPY* (Knittler et al., 1995; Nishikawa et al., 2001; Kabani et al., 2003). To extend our understanding of Wsc1p's quality control mechanism, we analyzed how this central ERQC factor interacts with misfolded variants. Coimmunoprecipitation experiments were performed from wild-type cells expressing the L63R, L63RLuminal, Δ68-80, and Δ68-80Luminal variants, and their “ED” appended versions. CPY* serves as a positive control for Kar2p/substrate binding.
Immunoprecipitation using Kar2p antisera identified a strong interaction with CPY* as previously reported (Figure 7, A and B; Xie et al., 2009). By contrast, no association was detected for Wsc1-L63R under the same assay conditions (Figure 7A). The identical result was obtained with Wsc1-L63RLuminal, which is stable and accumulates in the ER (Figure 7B). Coimmunoprecipitation experiments were also performed in reverse using anti-HA antibody-conjugated agarose to pull down substrates. Again, an interaction between Kar2p and CPY* was easily detectable (Figure 7, C and D). Little Kar2p was detected for Wsc1-L63R and Wsc1-L63RLuminal. A signal, albeit weak, was detected for the Δ68-80 and Δ68-80Luminal variants (Figure 7, C and D). These data show that, unlike the bona fide ERAD substrate CPY*, Kar2p does not interact appreciably with misfolded Wsc1p.
Figure 7.
Misfolded Wsc1p is not recognized by the major ER chaperone BiP/Kar2p. (A and B) Kar2p was immunoprecipitated from wild-type cells expressing Wsc1-L63R, Wsc1-L63RLuminal, or CPY* under native conditions. Protein complexes were resolved by SDS-PAGE and analyzed on immunoblots. Blots were probed using anti-HA antibody for substrates (top), stripped, and reprobed with anti-Kar2p antiserum as a control (middle). One percent of each lysate was loaded and analyzed in parallel to measure the relative amounts of input substrate used for each experiment (bottom). (C and D) Lysates from wild-type cells expressing the indicated substrates were subjected to native immunoprecipitation with anti-HA resin and Western blot analysis. Coimmunoprecipitated Kar2p was detected by anti-Kar2p immunoblots (top). The same blots were reprobed with anti-HA antibody as a control (middle). 1% of the lysates was analyzed in parallel and probed with anti-Kar2p antibody as an input control (bottom).
We repeated the analysis with the ED-appended Wsc1p variants. Here, both ED-Wsc1-L63R and ED-Wsc1-L63RLuminal were recovered in a complex with Kar2p as strongly as CPY* (Figure 8, A and B). The reverse immunoprecipitations of ED-Wsc1-L63R, ED-Wsc1-Δ68-80, ED-Wsc1-L63RLuminal, and ED-Wsc1-Δ68-80Luminal each recovered as much Kar2p as CPY* (Figure 8, C and D). This is no surprise because these variants are ERAD substrates and the CPY CTD itself binds Kar2p (Xie et al., 2009). These data show that the Wsc1p luminal domain is a poor substrate for Kar2p binding when misfolded. Because Kar2p plays an important role in ERQC and ERAD, this explains in part why Wsc1p is an obligate substrate of Golgi quality control.
Figure 8.
Misfolded Wsc1p fused with an ERAD determinant binds Kar2p efficiently. Lysates from wild-type cells expressing the indicated substrates were subjected to immunoprecipitation with anti-Kar2p antibody (A and B) or anti-HA resin (C and D) followed by Western blotting as described in Figure 7.
DISCUSSION
The role of the ER in protein quality control is well established with conserved mechanisms in all eukaryotes (for reviews see Ellgaard and Helenius, 2003; Romisch, 2005; Vembar and Brodsky, 2008). As the site of secretory and membrane protein folding, its deployment is needed to prevent the premature exit of proteins, which could be detrimental to cell. With protein quality control well situated in the ER, the logic of placing an outpost in the Golgi apparatus is not obvious. It also requires that the most basic tenet of ER quality control—the retention of unfolded proteins—be broken. Nevertheless, reports from several laboratories have provided evidence to support such a mechanism (see Introduction). What remained unclear was how the two independent pathways function as a system in maintaining secretory protein quality control.
Wsc1p Is an Obligate Substrate of Golgi Quality Control
In this communication, we report the first endogenous protein that is an obligate client of Golgi quality control. Its discovery establishes that the pathway can serve as a primary quality control mechanism for some proteins. More importantly, detailed analyses provided the mechanistic basis for why ERAD fails to recognize Wsc1p. At the most basic level, Wsc1p simply lacks determinants recognizable by ERAD. This finding supports the provocative view that ER quality control deploys a less stringent sieve than the post-ER mechanism. This likely reflects fundamental differences in how the two compartments detect misfolding. Recent reports demonstrate that ERAD uses signal-receptor based mechanisms for recognition. For glycan-dependent ERAD, the Htm1/Mnl1p mannosidase trims N-linked glycans of substrates to produce a terminal α1,6-linked mannose. This residue is the ligand of the Yos9p receptor in the Hrd1p complex (Quan et al., 2008; Clerc et al., 2009). This signal is so critical that deleting the CPY* C-terminal glycan blocks its degradation even as the protein remains severely misfolded (Kostova and Wolf, 2005; Spear and Ng, 2005). In mammalian cells, a cysteine residue within a 20-amino acid degron is critical for ERAD of secretory IgM μs chain (Shapira et al., 2007). Although misfolded Wsc1p contain free cysteines, the data show that they are not in a proper context to signal ERAD. The requirement for specific signals might explain why some foreign proteins are not detected by ERAD, even if aberrant (Hong et al., 1996; Coughlan et al., 2004). From this perspective, the existence of endogenous proteins lacking ERAD determinants due to functional or structural constraints should not be surprising. By necessity, such molecules require an alternative to ERAD for their quality control.
To rely on a post-ER quality control mechanism, substrates must retain the capacity to exit the ER when misfolded. Luminal cargo proteins concentrate into COPII vesicles by binding transport receptors (Kuehn et al., 1998; Belden and Barlowe, 2001; Malkus et al., 2002). The misfolded luminal domain of Wsc1p lacks functional export signals so it must use a different mechanism. To transport some membrane proteins, COPII components bind their cytoplasmic export signals (for reviews see Barlowe, 2003; Lee et al., 2004). Interestingly, the Wsc1p cytosolic domain contains di-acidic (DXE; E295XD297, E300XE301, E333XE335) and di-hydrophobic motifs (V368L369, V371V372) that conform to COPII signals. Consistent with this, the domain is required for misfolded Wsc1p transport. Our data show the Wsc1p exit signal to be remarkably robust. The ED-Wsc1-L63R and ED-Wsc1-Δ68-80 chimeras exit the ER efficiently after ERAD is knocked out (Figure 6 and Figure S8). This occurs despite their interaction with the ER resident Kar2p, which retains unfolded proteins as part of its function (Figure 8; Simons et al., 1995; Nishikawa et al., 2001). In this way, the strength of the Wsc1p export signal may have precluded ERAD for its quality control even if it could incorporate ERAD signals. On the other hand, the lack of ERAD determinants makes Wsc1p completely reliant on its powerful export signal for quality control.
Candidate Golgi Sorting Factors for Misfolded Proteins
How the Golgi recognizes and sorts misfolded proteins is unclear. Indeed, additional endosomal sorting steps for substrates en route to the vacuole have not been ruled out. At present, there are three candidates. Vps10p remains a strong contender for detecting unfolded proteins in the Golgi lumen. Although best known as a sorting factor for the folded proteins CPY and proteinase A, it was proposed that their sorting signals are displayed to Vps10p unfolded (Marcusson et al., 1994; Jorgensen et al., 1999). Thus, it seems possible that Vps10p also has the capacity to recognize destabilized proteins trafficking through the Golgi (Hong et al., 1996; Jorgensen et al., 1999). Interestingly, the degradation of misfolded Wsc1 proteins is partly compromised in cells lacking Vps10p (Figure S6B). Could it be that Vps10p evolved for protein quality control and some vacuolar proteins evolving signals mimicking unfolded proteins?
For integral membrane proteins, Pelham and colleagues have proposed that a Golgi-localized E3 ubiquitin ligase Tul1p might serve a role to detect unassembled membrane proteins (Reggiori and Pelham, 2002). Tul1p detects transmembrane domains (TMDs) with polar residues, which are commonly found in proteins bearing multiple TMDs or as part of multisubunit complexes. Thus, it seems to have the right activity to seek out proteins bearing polar or charged TMDs incorrectly configured or disassembled post-ER. Although an intriguing hypothesis, a bona fide substrate remains to be identified. Pma1-7p (Pizzirusso and Chang, 2004) and misfolded Wsc1p variants (our unpublished results) are unaffected in Δtul1 cells.
Pma1-7p vacuolar targeting requires the Rsp5p/Bul1p/Bul2p E3 ubiquitin ligase complex (Pizzirusso and Chang, 2004). This prompted authors to propose that the complex might be involved in a post-ER mechanism in quality control. Interestingly, the phenotype mirrors the regulated trafficking of the folded general amino acid permease (Gap1p) depending on the nitrogen availability (Roberg et al., 1997). Like Gap1p, Pma1-7p is stable at the plasma membrane in the absence of the complex. As a functional protein, it will be interesting to determine whether the pma1-7 mutation results in a Gap1p-like trafficking signal or if it causes a minor structural change detected by Golgi quality control and not ERAD. Because both mechanisms are important and unsolved, there is no doubt that Pma1-7p will continue to be a valuable tool in efforts to decipher them.
A post-ER quality control mechanism requires a reassessment of the long-held belief that misfolded proteins in the ER be retained to prevent them from doing harm elsewhere (Sitia and Braakman, 2003; Nakatsukasa and Brodsky, 2008). Indeed, the data support an alternative view. In the normal course of ERAD, a fraction of the substrates recycles between the ER and Golgi before they are degraded (Hsu et al., 1991; Hammond and Helenius, 1994; Caldwell et al., 2001; Vashist et al., 2001; Yamamoto et al., 2001). If ERAD is saturated, excess CPY* traffics efficiently to the vacuole for degradation (Spear and Ng, 2003). Surprisingly, increasing the flow of misfolded proteins into this pathway is well tolerated, whereas increasing the concentration of misfolded proteins in the ER is toxic (Spear and Ng, 2003; Haynes et al., 2004). Wsc1p's quality control mechanism demonstrates that the endomembrane system can accommodate the passage of severely misfolded proteins under normal conditions. These data do not contradict the view that the best way to abrogate proteotoxicity is to eliminate them at the source. Instead, they demonstrate that the cell's arsenal against misfolded secretory pathway proteins is more extensive than just ERAD.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Ng lab for discussion and comments. We thank Chia Sing Tan, Yu Jun Tan, Jeremy Brodhead, and the TLL/NUS core facilities for providing excellent technical support. The parental strain of SWY653 was constructed by Shilpa Vashist when she was a member of the Ng laboratory. This work was supported by funds from the Temasek Trust and by a grant from the Singapore Millennium Foundation to S.W. (Predoctoral fellowship).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0910) on February 3, 2010.
REFERENCES
- Ammerer G., Hunter C. P., Rothman J. H., Saari G. C., Valls L. A., Stevens T. H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T., Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A., Hegde R. S. Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog. 2009;5:e1000479. doi: 10.1371/journal.ppat.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 2003;13:295–300. doi: 10.1016/s0962-8924(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Barlowe C., Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Belden W. J., Barlowe C. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 2001;294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., McCracken A. A. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Caldwell S. R., Hill K. J., Cooper A. A. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chang A., Fink G. R. Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. A., Stevens T. H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan C. M., Walker J. L., Cochran J. C., Wittrup K. D., Brodsky J. L. Degradation of mutated bovine pancreatic trypsin inhibitor (BPTI) in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 2004;279:15289–15297. doi: 10.1074/jbc.M309673200. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Endrizzi J. A., Breddam K., Remington S. J. 2.8-Angstrom structure of yeast serine carboxypeptidase. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- Finger A., Knop M., Wolf D. H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko O. T., Jigami Y. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 2006;17:834–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., McCammon K., Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986;46:939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Haas I. G., Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Hammond C., Helenius A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J. Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R. Y., Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J. Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson W. M., Beeser S. A., Oas T. G., Goldenberg D. P. Identification of a residue critical for maintaining the functional conformation of BPTI. J. Mol. Biol. 2003;333:425–441. doi: 10.1016/j.jmb.2003.08.023. [DOI] [PubMed] [Google Scholar]
- Haynes C. M., Titus E. A., Cooper A. A. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Hill K., Cooper A. A. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E., Davidson A. R., Kaiser C. A. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V. W., Yuan L. C., Nuchtern J. G., Lippincott-Schwartz J., Hammerling G. J., Klausner R. D. A recycling pathway between the endoplasmic reticulum and the Golgi apparatus for retention of unassembled MHC class I molecules. Nature. 1991;352:441–444. doi: 10.1038/352441a0. [DOI] [PubMed] [Google Scholar]
- Hurtley S. M., Bole D. G., Hoover-Litty H., Helenius A., Copeland C. S. Interactions of misfolded influenza virus hemagglutinin with binding protein (BiP) J. Cell Biol. 1989;108:2117–2126. doi: 10.1083/jcb.108.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C. A., Burda P., Roth J., Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness D. D., Li Y., Tipper C., Spatrick P. Elimination of defective alpha-factor pheromone receptors. Mol. Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen M. U., Emr S. D., Winther J. R. Ligand recognition and domain structure of Vps10p, a vacuolar protein sorting receptor in Saccharomyces cerevisiae. Eur. J. Biochem. 1999;260:461–469. doi: 10.1046/j.1432-1327.1999.00176.x. [DOI] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimoto T., Shoji S., Hidvegi T., Mizushima N., Umebayashi K., Perlmutter D. H., Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J. Biol. Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- Kincaid M. M., Cooper A. A. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol. Biol. Cell. 2007;18:455–463. doi: 10.1091/mbc.E06-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler M. R., Dirks S., Haas I. G. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Hauser N., Wolf D. H. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast. 1996;12:1229–1238. doi: 10.1002/(sici)1097-0061(19960930)12:12<1229::aid-yea15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J. Cell Sci. 2005;118:1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- Kruse K. B., Brodsky J. L., McCracken A. A. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol. Biol. Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M. J., Herrmann J. M., Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Li Y., Kane T., Tipper C., Spatrick P., Jenness D. D. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D., Tam A., Schmidt W. K., Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel M., Bagnat M., Strahl S. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 2004;24:46–57. doi: 10.1128/MCB.24.1.46-57.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Chang A. Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J. Cell Biol. 1997;138:731–746. doi: 10.1083/jcb.138.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M., Sdicu A. M., Bussereau F., Jacquet M., Bussey H. The Ktr1p, Ktr3p, and Kre2p/Mnt1p mannosyltransferases participate in the elaboration of yeast O- and N-linked carbohydrate chains. J. Biol. Chem. 1997;272:15527–15531. doi: 10.1074/jbc.272.24.15527. [DOI] [PubMed] [Google Scholar]
- Ma J., Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc. Natl. Acad. Sci. USA. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkus P., Jiang F., Schekman R. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J. Cell Biol. 2002;159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson E. G., Horazdovsky B. F., Cereghino J. L., Gharakhanian E., Emr S. D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 1996a;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 1996b;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Nakano A., Brada D., Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J. L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Nishikawa S., Hosokawa N., Nagata K., Endo T. Mnl1p, an alpha -mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem. 2001;276:8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- Ng D. T., Brown J. D., Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. T., Spear E. D., Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip B., Levin D. E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 2001;21:271–280. doi: 10.1128/MCB.21.1.271-280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H. L., Machado I. M., Payne G. S. NPFXD-mediated endocytosis is required for polarity and function of a yeast cell wall stress sensor. Mol. Biol. Cell. 2007;18:57–65. doi: 10.1091/mbc.E06-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzirusso M., Chang A. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol. Biol. Cell. 2004;15:2401–2409. doi: 10.1091/mbc.E03-10-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Frohlich K. U., Diamant N., Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Duden R., Rubinsztein D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Pelham H. R. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- Roberg K. J., Rowley N., Kaiser C. A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. J., Bentley M. D., Harris J. M. Chemistry for peptide and protein PEGylation. Adv. Drug Deliv. Rev. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. M., Maniatis T. NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual, Plainview. [Google Scholar]
- Sarkar S., et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira I., Charuvi D., Elkabetz Y., Hirschberg K., Bar-Nun S. Distinguishing between retention signals and degrons acting in ERAD. J. Cell Sci. 2007;120:4377–4387. doi: 10.1242/jcs.011247. [DOI] [PubMed] [Google Scholar]
- Sifers R. N. Insights into checkpoint capacity. Nat. Struct. Mol. Biol. 2004;11:108–109. doi: 10.1038/nsmb0204-108. [DOI] [PubMed] [Google Scholar]
- Sifers R. N., Brashears-Macatee S., Kidd V. J., Muensch H., Woo S. L. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J. Biol. Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Simons J. F., Ferro-Novick S., Rose M. D., Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R., Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- Sommer T., Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Spear E. D., Ng D. T. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell. 2003;14:2756–2767. doi: 10.1091/mbc.E02-11-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear E. D., Ng D. T. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J. Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- VanSlyke J. K., Deschenes S. M., Musil L. S. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verna J., Lodder A., Lee K., Vagts A., Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J., DeMartino G. N., Skach W. R., Bulleid N. J., Brodsky J. L., Johnson A. E. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Kanehara K., Sayeed A., Ng D. T. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol. Biol. Cell. 2009;20:3317–3329. doi: 10.1091/mbc.E09-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Fujii R., Toyofuku Y., Saito T., Koseki H., Hsu V. W., Aoe T. The KDEL receptor mediates a retrieval mechanism that contributes to quality control at the endoplasmic reticulum. EMBO J. 2001;20:3082–3091. doi: 10.1093/emboj/20.12.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia Y., Horonchik L., Tzaban S., Yanai A., Taraboulos A. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 2001;20:5383–5391. doi: 10.1093/emboj/20.19.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. X., Braakman I., Matlack K. E., Helenius A. Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol. Biol. Cell. 1997;8:1943–1954. doi: 10.1091/mbc.8.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. X., Goldenberg D. P. Amino acid replacement that eliminates kinetic traps in the folding pathway of pancreatic trypsin inhibitor. Biochemistry. 1993;32:14075–14081. doi: 10.1021/bi00214a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.