Abstract
Replication Protein A (RPA) is the primary eukaryotic ssDNA binding protein utilized in diverse DNA transactions in the cell. RPA is a heterotrimeric protein with seven globular domains connected by flexible linkers, which enable substantial inter-domain motion that is essential to its function. Small angle X-ray scattering (SAXS) experiments on two multi-domain constructs from the N-terminus of the large subunit (RPA70) were used to examine the structural dynamics of these domains and their response to the binding of ssDNA. The SAXS data combined with molecular dynamics simulations reveal substantial interdomain flexibility for both RPA70AB (the tandem high affinity ssDNA binding domains A and B connected by a 10-residue linker) and RPA70NAB (RPA70AB extended by a 70-residue linker to the RPA70N protein interaction domain). Binding of ssDNA to RPA70NAB reduces the interdomain flexibility between the A and B domains, but has no effect on RPA70N. These studies provide the first direct measurements of changes in orientation of these three RPA domains upon binding ssDNA. The results support a model in which RPA70N remains structurally independent of RPA70AB in the DNA bound state and therefore freely available to serve as a protein recruitment module.
RPA is the primary eukaryotic ssDNA binding protein utilized for diverse DNA transactions in the replication and maintenance of the genome (reviewed by Fanning and coworkers [1]). RPA functions by binding and protecting ssDNA from degradation by endonucleases, inhibiting formation of ssDNA secondary structure, and providing a scaffold for DNA processing machinery by interacting with numerous DNA processing proteins. RPA biochemical functions and biological activities have been intensively investigated and the structures of its domains determined [2–9]. Despite this detailed information, the mechanisms for RPA function remain poorly understood, largely due to the inherent difficulties of characterizing proteins with modular organization and the fact that RPA function is integrated within complex multi-protein machinery.
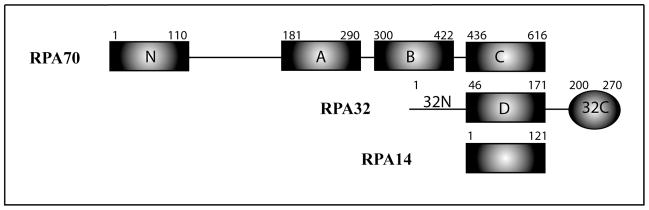
RPA is a modular 116 kDa heterotrimer composed of seven structured globular domains and one disordered domain (Figure 1). The trimer subunits are named on the basis of their approximate molecular weights: RPA70, RPA32 and RPA14. The RPA70 subunit contains four oligonucleotide-oligosaccharide binding (OB-fold) domains: RPA70N, RPA70A, RPA70B and RPA70C. RPA70N is linked to RPA70A by a 70-residue linker, which in turn is connected to RPA70B by a 10-residue linker. RPA70B is connected to RPA70C by a 15-residue linker. The RPA32 subunit contains a 45-residue unstructured N-terminal domain (RPA32N), along with a central OB-fold domain (RPA32D) and a C-terminal winged helix domain (RPA32C), which are separated by a 23-residue linker. The RPA14 subunit consists of a single OB-fold domain. High-resolution structures have been determined by X-ray crystallography or NMR spectroscopy for all of the globular domains [10]. However, knowledge of the spatial organization of the domains in the intact protein is lacking. Such information is important because remodeling of RPA ‘architecture’ constitutes an essential element of its function in DNA processing machinery.
Figure 1.
Domain organization of RPA. Rectangles are OB-fold domains, and the elipsoid is a winged helix domain. RPA32 has one unstructured N-terminal domain (RPA32N). Trimerization occurs via non-covalent interactions between RPA70C, RPA32D and RPA14 domains. ssDNA binds to RPA70A, RPA70B, RPA70C and RPA32D.
RPA ssDNA binding activity is associated with the A-D domains of RPA70 and RPA32. The binding of ssDNA by RPA occurs with a 5′ → 3′ molecular polarity in the order A, B, C, and D [11, 12]. The directional binding to ssDNA is the result of differences in the binding affinity of the four ssDNA binding domains. The RPA70A and RPA70B domains have the highest ssDNA affinity and serve as the anchor for all ssDNA binding activities. Two X-ray crystal structures of RPA70AB have been determined, one without ssDNA [8] and another in the presence of d-CCCCCCCC [3]. In the complex with ssDNA, RPA70A and RPA70B are aligned with the two binding loops wrapped around and nearly encircling the DNA. In contrast, two very different orientations of RPA70A with respect to RPA70B were observed in the structure of the free protein. Moreover, NMR analysis indicated the two domains of RPA70AB are structurally independent and implied that the two domains are attached by a flexible tether [13]. However, no direct information has been obtained about interdomain orientations in solution and the effect of ssDNA on RPA70AB and the relationship to the rest of the protein.
RPA70N is suggested to have weak ssDNA binding activity that is important for the DNA unwinding activity of RPA [14, 15]. However, the ssDNA binding affinity of this domain is more than 1000-fold weaker than that of RPA70AB, and RPA70N is generally accepted to be a protein interaction module targeting transcription factors and checkpoint proteins such as p53 and ATRIP [16–18]. The only RPA70N structure is of the isolated domain [5, 16], so there is no information available on its disposition with respect to the rest of RPA and hence its availability to influence the DNA binding properties of the protein. NMR spectroscopy on a construct containing the RPA70N, RPA70A and a portion of RPA70B suggested that RPA70N does not interact with the RPA70A domain [19]. An NMR study of full-length RPA and larger multi-domain constructs suggest the motion of RPA70N is independent of the remaining DNA-binding domains, both in the absence and presence of ssDNA [13, 20].
To expand these initial observations and obtain direct information on interdomain orientation, we have turned to small angle X-ray scattering (SAXS), which is a powerful approach to study proteins under native solution conditions and extract low-resolution spatial information for dynamic systems such as RPA. To experimentally address the structural dynamics of the RPA70A, RPA70B and RPA70N domains, the impact of ssDNA binding on inter-domain flexibility, and the effect of RPA70N on the ssDNA binding activity of the tandem high affinity RPA70AB domains, we purified RPA70AB and RPA70NAB and examined them with and without ssDNA in solution by small angle X-ray scattering (SAXS). Analysis of SAXS data for these dynamic systems was facilitated by the generation of large ensembles of structures with different inter-domain orientations using rigid body molecular dynamics simulations. The results show RPA70N is flexibly linked to RPA70AB and has no influence on the binding of ssDNA, and have general implications for RPA dynamic architecture and functions.
MATERIALS AND METHODS
Materials
Fragments of human RPA were expressed from a pSV281 RPA70AB plasmid containing a TEV cleavable 6X-His tag at the N-terminus and a pBG100 RPA70NAB plasmid containing an H3C 6X-His tag also at the N-terminus. TEV and H3C proteases are produced in-house. ssDNA oligomers d(CCACCCCC) and d(AAAAAACCACCCCC) purchased from Integrated DNA Technologies were desalted, lyophilized and re-suspended into autoclaved distilled water.
Expression of RPA proteins
Recombinant RPA70AB (RPA70181–422) and RPA70NAB (RPA701–422) constructs were prepared as described [13, 20]. Proteins were expressed in the Escherichia coli host Rosetta (DE3) cells (Novagen, Madison, WI). Cells were grown in LB medium containing kanamycin at 37 ºC, induced with 0.1 M IPTG when the O.D. reached 0.6, and harvested after 3 hours using a JLA 8.1 Beckman rotor at 7,500 r.p.m. at 4 ºC. Pellets were stored at −20 ºC.
Protein purification
RPA70AB samples were purified using Nickel affinity chromatography (NiNTA) in 10 mM Hepes at pH 7.5, 500 mM NaCl, 5 mM BME and 10% glycerol using an elution gradient of 20–300 mM imidazole. Cleavage of the His-tag with TEV protease was performed through overnight dialysis in a buffer containing 10 mM Hepes at pH 7.5, 200 mM NaCl, 5 mM BME, 200 mM L-arginine and 10% glycerol. A second NiNTA purification step was used to remove the His-tag. Size exclusion chromatography (SEC) using a Superdex S75 column equilibrated with the dialysis buffer was used as a last step of purification. Protein was concentrated and stock solutions were frozen in a dry ice/ethanol bath and kept at −80 °C. The same procedure was used for RPA70NAB samples using NiNTA buffer of 30 mM MES at pH 6.5, 500 mM NaCl, 10 mM BME, and 5 mM MgCl2, and cleaving with H3C protease. The SEC step was performed using a Superdex S200 column and a buffer containing 30 mM MES at pH 6.5, 200 mM NaCl, 10 mM BME, 10% glycerol and 5 mM MgCl2.
Preparation of protein-DNA complexes
RPA70AB or RPA70NAB was incubated in the presence of 1.2–1.5 molar excess of d(CCACCCCC) or d(AAAAAACCACCCCC) for 20 minutes on ice. 500 μL of sample was purified by SEC using S75 (RPA70AB) or S200 (RPA70NAB) resin. The samples eluted as one peak for the complex followed by a DNA-only peak.
Size exclusion chromatography - multi-angle light scattering
The monodispersity of each sample was verified by multi-angle light scattering connected in line with SEC (SEC-MALS). All experiments were performed using a Wyatt Technology instrument and data were analyzed using ASTRA v16.25. Samples were analyzed using a 2.4 mL Superdex75 column. Only samples that exhibited monodispersity were selected for data collection.
Small angle x-ray scattering
SAXS data of the various RPA constructs were collected at the SIBYLS 12.3.1 beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory. Scattering measurements were performed on 20 μl samples at 15 °C using a Hamilton robot for loading samples from a 96 well plate into a helium-purged sample chamber. Protein-DNA samples were further purified on a 24 ml SEC column just prior to data collection to eliminate any free protein or DNA. Data were collected on both the original gel filtration fractions and samples concentrated ~2x-8x from individual fractions. Fractions prior to the void volume and concentrator eluates were used for buffer subtraction.
The experiments on RPA70AB used an X-ray beam from a single crystal monochomator of 11 keV, covering a momentum transfer range 0.007 Å−1 < q < 0.35 Å−1 (q = 4πsinΘ/λ, where 2Θ is the scattering angle). Sequential exposures (6 s, 6 s, 60 s, 6 s, 200 s, 6 s) were taken, and data were monitored for radiation-dependent aggregation. SAXS experiments for all RPA70NAB samples and for RPA70AB with DNA were acquired using an X-ray beam from a multilayer monochromator of 12 keV covering a momentum transfer range of 0.012 Å−1 < q < 0.317 Å−1. The multilayer provides increased X-ray flux allowing higher signals for lower protein concentrations. Sequential exposures (0.5 s, 0.5 s, 5 s, 0.5 s) were taken, and data were monitored for radiation-dependent aggregation. All SAXS data were collected using the MarCCD 165 detector in fast frame transfer mode, and reduced by normalizing to the incident beam intensity. Buffer scattering was subtracted from the protein scattering. This was followed by azimuthal averaging to obtain the intensity I(q) versus q scattering plot visualized by xmgrace. The data were analyzed using PRIMUS (Primary Analysis & Manipulations with Small-Angle Scattering Data) from version 3.0 ATSAS 2.0 [21], from which Guinier, Kratky, P(r) and CRYSOL plots were generated.
For each sample, multiple experiments were acquired over multiple runs. Experiments providing the highest signal to noise and that remained consistent with the relative concentrations from original gel filtration fractions were selected for further analysis. For RPA70AB alone, the final data used for analysis were merged between a 60 s exposure at 147 μM and a 200 s exposure at 331 μM. The concentration of the sample used for analysis for RPA70AB with 8mer DNA, RPA70AB with 14mer DNA, RPA70NAB alone, RPA70NAB with 8mer DNA, RPA70NAB with 14mer DNA were 71 μμM, 81 M, 163 μM, 80 μM, and 98 μM, respectively, each with a 5 seconds exposure time.
Computational modeling
Ab initio shape envelopes were calculated with the program GASBOR [22]. Ten GASBOR runs were merged using the DAMAVER suite. Overlay with PDB coordinates were done using SUPCOMB. PDB coordinates from the RPA70N NMR structure (1EWI) and RPA70AB ssDNA bound X-ray crystal structure (1JMC) were used to construct a model for RPA70NAB. From these coordinates, multiple conformers were generated by rigid body molecular dynamics simulations with the program BILBO-MD [23]. For the DNA complexes, ssDNA coordinates were removed from the RPA70AB model. For RPA70NAB, the connecting linker between domains 70N and 70A was built using the Biopolymer module of Insight II [Accelrys, Inc.: San Diego, CA, USA, 2005], followed by refinement with Rosetta [24]. To generate RPA70NAB-ssDNA complexes for back calculation of scattering profiles, the DNA coordinates were added back using Molecular Operating Environment, MOE 2010.09, (Chemical Computing Group, Montreal, Canada) and Chimera [25]. Molecular graphics imagines were generated using PYMOL (DeLano Scientific, Palo Alto, CA, USA).
RESULTS
To characterize effects on the structural dynamics of RPA70A and RPA70B as they bind ssDNA and investigate the influence of RPA70N, small angle X-ray scattering (SAXS) experiments were performed on RPA70AB and RPA70NAB in the absence and presence of ssDNA. SAXS measures the electron pair distribution and is well suited for characterizing the architecture of molecules in solution [26]. Since scattering data is distorted by scattering from small amounts of aggregation, the monodispersity of the samples was carefully monitored by multi-angle light scattering (MALS) of the peaks eluted by size exclusion chromatography (SEC) (Supplementary Figures 1 and 2) and by verification of the data in the Guinier analysis (Supplementary Figure 3). To ensure mono-dispersity and remove any free DNA for protein-DNA complexes, each sample was treated by SEC just prior to data collection.
Structural dynamics of RPA70AB from analysis of SAXS
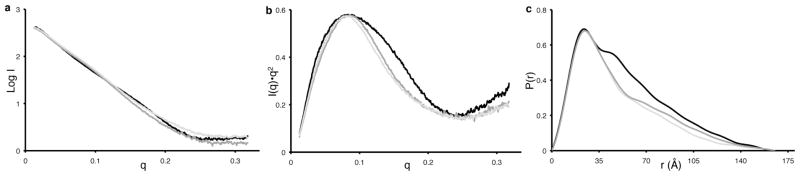
The RPA70AB scattering profile (Figure 2A) reveals the high quality of the SAXS data obtained after optimization of the sample and acquisition parameters. A Kratky analysis of the data is consistent with the two globular well-folded domains connected by a 10-residue linker (Figure 2B). The use of the Kratky plot of SAXS data to detect flexibility in proteins is well established (see Figure 24 in Ref. 31). A Guinier analysis provided a radius of gyration (Rg) of RPA70AB in solution of 25.6 Å. The data were also analyzed with GNOM to derive the probability distribution function P(r), which reflects the distribution of inter-atomic distances in the molecule (Figure 2C). The main peak in the P(r) function corresponds to scattering between atoms within the globular RPA70A and RPA70B domains, which have similar shape and size. The shoulder at longer distances corresponds to scattering between atoms in one domain and atoms in the other. The maximum distance (Dmax) in the P(r) function is 100 Å and the Rg value derived from P(r) is 25.7 Å, consistent with the reciprocal space Rg derived directly from the scattering data (Table 1).
Figure 2.
Scattering curves (a), kratky analysis (b) and P(r) functions (c) for RPA 70AB in absence (black) and presence (gray) of d-(CCACCCCC).
Table 1.
SAXS measurements.
| Proteins | AB | AB-8mer | AB-14mer | NAB | NAB-8mer | NAB-14mer |
|---|---|---|---|---|---|---|
| Rg (Å), Guinier Analysis | 25.6 | 23.4 | 24.4 | 39.5 | 37.4 | 37.8 |
| Rg (Å), P(r) analysis | 25.7 | 23.2 | 24.7 | 41.6 | 40.6 | 41.0 |
| Dmax (Å), P(r) analysis | 100 | 87 | 100 | 165 | 165 | 165 |
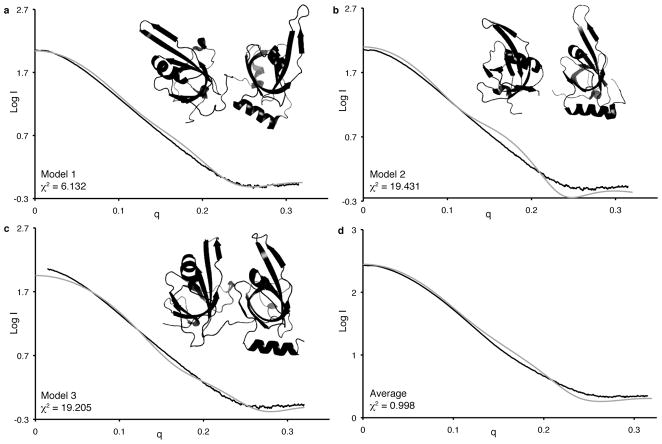
To examine the implications of the SAXS data for the solution structure, scattering curves were back calculated from three crystallographic models with the program CRYSOL and superimposed on the experimental data (Figures 3A–C). Models 1 and 2 correspond to the two different molecules in the asymmetric unit of the RPA70AB X-ray crystal structure, which have very different orientations of the two domains. Model 3 was obtained from the X-ray crystal structure of RPA70AB bound to d-CCCCCCCC, from which the ssDNA coordinates were removed. The χ2 parameter reflecting the fit of the calculated curve to the experimental data is also included for each model. Although χ2 is dependent in part on the experimental noise, a lower χ2 value generally corresponds to a better fit of the model(s) to the experimental data. This analysis shows that although Model 1 provides a better fit than the other two models, none of the models fit especially well to the data. Similarly, each of the crystallographic models poorly fit the ab initio envelope predicted from the scattering data, with model 1 being most similar. These results suggest two possibilities: (i) the structure is not accurately represented by any of these three specific models or (ii) the two domains occupy multiple inter-domain orientations.
Figure 3.
CRYSOL fit to experimental data for each of the crystal structure models and their average. Comparison of RPA70AB experimental SAXS scattering curves (black) and back-calculated scattering curves (gray) for models 1 (a), 2 (b), 3 (c), and the average (d). Models 1 and 2 correspond to the two molecules in the unit cell of RPA70AB (1FGU.pdb). Model 3 is the protein molecule after ssDNA atoms were extracted from RPA70AB/d-C8 (1JMC.pdb).
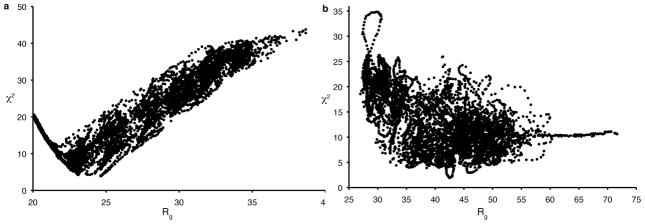
To distinguish these two possibilities, we employed rigid body molecular dynamics simulations with the program BILBO-MD [23] to generate 6400 conformers with a wide range of inter-domain orientations. For these calculations, the RPA70A and RPA70B domains were treated as rigid bodies and the linker between them was allowed to fully sample conformational space (Figure 4A). The radius of gyration was allowed to vary between 20 and 40 Å. The shape of the distribution of data in this plot is a reflection of the RPA70AB structure. Despite there being small gaps in the plot due to incomplete sampling of the conformational space accessible to RPA70AB, the sampling is sufficient to draw conclusions about the fit to the experimental data. The observation of a wide χ2 minimum around 4 implies the data cannot be properly represented by a single structure.
Figure 4.
Plot of 2 fit parameter versus radius of gyration for RPA70AB (a) and RPA70NAB (b) conformers generated by bilbomd. The calculated scattering curve for each conformer was generated by CRYSOL.
To analyze the ensemble of structures, we employed a genetic algorithm to determine if select groups of conformers can fit the data better than single conformers [23]. The best fit obtained taking two or three conformers provides χ2 values of 2.1 and 1.6, respectively, indicating that multiple conformers represent the data far better than any single conformer. Figure 3D shows the improved fit to the experimental RPA70AB scattering obtained for the combination of three RPA70AB conformers comprised of the models 1, 2 and 3 described above. Overall, analyses of the SAXS data show directly that the two domains in RPA70AB are not fixed in space but rather occupy a range of inter-domain orientations.
ssDNA binding to RPA70AB
To determine the effect of binding ssDNA on the structural dynamics of RPA70AB, the scattering measurements were repeated in the presence of d-CCACCCCC. Previous studies have established that RPA70AB binds oligomers of 8–10 nucleotides with an affinity in the high nM range [13]. The tight binding affinity for ssDNA implies that a mono-disperse protein-DNA complex can be prepared and characterized, and the monodispersity was confirmed by SEC-MALS (Supplementary Figures 1 and 2).
When the scattering data analyzed in the same manner as for free RPA70AB are compared to the free protein, it is evident that binding of ssDNA significantly reduces the protein’s structural dynamics (Figure 2A). The Rg value derived directly from the Guinier analysis was 23.4 Å, 2.2 Å less than the value determined for the free protein (Table 1). The range of inter-atomic distances reflected in the P(r) function is significantly decreased relative to the free protein (Figure 2C). In addition, the Dmax value is 87 Å, a reduction of ~ 13 Å relative to that determined for the free protein. Notably, the Dmax is significantly larger than the 67.4 Å Dmax measured directly from the crystal structure (1jmc.pdb), even when taking into account the missing residues at the N and C terminus. The lower value of Rg, and the narrowing of the curve and lower Dmax in the P(r) function all indicate there is an overall compaction of RPA70AB upon binding ssDNA. The reduction in the Rg in particular directly reflects that the two domains are on average closer to each other when ssDNA is bound. This interpretation is consistent with the ssDNA serving to further tether the two domains together [3, 27]. The SAXS data show directly that inter-domain dynamics is quenched relative to free RPA70AB.
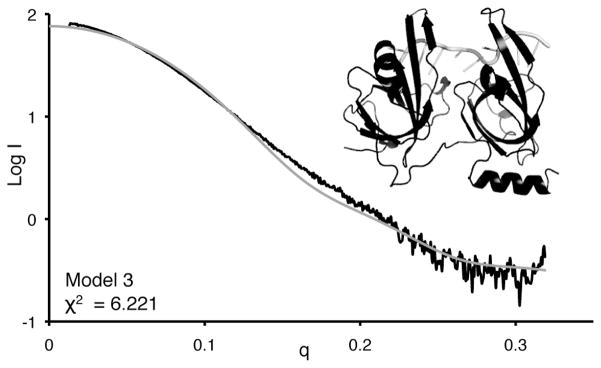
We next asked if the dynamic architecture revealed by the solution scattering data was accurately represented by the X-ray crystal structure of RPA70AB in complex with d-CCCCCCCC. Gasbor calculations were first performed to determine the conformational envelope generated from the data. While the fits to the crystal structure was reasonable in this case, back-calculation proves a more direct and quantifiable assessment of the fits to atomic resolution models. We therefore turned to back-calculating the scattering curve from the coordinates of the crystal structure using CRYSOL, and the results were plotted and compared to the experimental data (Figure 5). Overall, there is agreement between the experimental scatter and the crystal structure, which is consistent with the value of 20.0 Å for Rg calculated from the crystal structure. However, the χ2 fitting parameter is 6.22, which indicates inconsistencies between the X-ray crystal structure and the SAXS data. This result suggests either the complex has a different structure in solution or, as for free RPA70AB, that the complex cannot be adequately represented by a single structure. The latter explanation is supported by the observation in the Kratky analysis that the curve does not completely return to baseline at higher scattering angles, which indicates that some disorder or flexibility is still present even when DNA is bound (Figure 2B). The inter-domain flexibility probably arises from the flexible linker between domains and from torsional inter-domain motions around the bound ssDNA and suggests RPA70AB complex retains conformational flexibility that is not evident in the crystal structure. However, due to the low resolution of SAXS data, we cannot completely rule out the possibility that the complex can be represented by a single structure that is different from the crystal structure.
Figure 5.
CRYSOL fit to experimental RPA70AB-8mer data (black) for the crystal structure model of RPA70AB bound to dC8 (gray). Comparison of experimental scattering curve for the RPA70AB-8mer complex (black), versus the back-calculated scattering curve for the crystal structure (1JMC).
Small angle X-ray scattering of RPA70NAB
To examine RPA70AB structural dynamics and investigate the influence of RPA70N, SAXS data were acquired for the RPA70NAB construct (Figure 6). An Rg value of 39.5 Å was derived directly from the data based on the Guinier analysis, which is over 14 Å greater than the Rg for RPA70AB. The P(r) function shows the same primary peak centered at ~24 Å as observed for RPA70AB, reflecting scattering within the globular OB-fold domains (Figure 6C). Where the scattering curves differ substantially is at longer distances. For example, Dmax extends out to 165 Å. Calculation of ab initio molecular envelopes directly from the scattering data using GASBOR, did not converge. This observation suggests that envelope representations are problematic for highly flexible systems in which domain orientations may vary considerably between conformers. Together the results from the SAXS analysis indicate that the conformational space sampled by RPA70NAB is substantially larger than that observed for RPA70AB.
Figure 6.
Comparison of Scattering curves (a), kratky analysis (b), and P(r) funditon (c) for RPA70NAB in absence (black) and presence (dark and light gray, respectively) of d-(CCACCCCC) and d-(AAAAAACCACCCCC).
To test the implications of the SAXS data with respect to the structural dynamics of RPA70NAB, BILBO-MD was used to generate 6000 conformers of RPA70NAB. For these calculations each globular domain was treated as an independent rigid body, but the N-A and A-B linkers were allowed to sample conformational space freely (Figure 4B). Notably, χ2 does not reach a specific minimum but rather plateaus through a wide range of Rg values, even more so than was observed for RPA70AB. This observation of a broad plateau for the χ2 minimum indicates the data is not properly represented by a single structure and suggests that RPA70NAB has substantial inter-domain flexibility.
To obtain further insight into the solution structure of RPA70NAB, we examined representative conformers from the ensemble generated by BILBO-MD, including fully extended and closely packed arrangements of the three domains. CRYSOL was used to back-calculate scattering curves for each conformer selected (Figure 7). The calculated Rg values are ~45 Å for extended and ~29 Å for the closely packed conformations. The Rg value for both of these conformers is far from the experimentally observed value and the scattering curves do not match the experimental data. Notably, poor fits to the scattering curve were also obtained even for specific conformers that closely match the experimentally observed Rg value. Thus, the SAXS data indicate that RPA70NAB has extensive inter-domain flexibility, which is substantially larger than RPA70AB as a result of the long flexible linker between the N and A domains.
Figure 7.
Comparison of experimental scattering curves for RPA70NAB (black) against back-calculated scattering curves of selected BILBOMD generated models (gray). Cartoon representations are shown for each of the models, along with the corresponding χ2 fit parameter and Rg values.
Effect of ssDNA binding on the structural dynamics of RPA70NAB
To investigate the effects of binding ssDNA on the structural dynamics of RPA70NAB, experiments were performed with an 8-mer and a 14-mer ssDNA oligonucleotide. The 8-mer corresponds to the excluded site size of RPA70AB and is designed as a control to characterize the effect of ssDNA binding to these high affinity domains. The 14-mer was designed to determine if RPA70N is able to modulate the ssDNA binding activity by providing 6 extra nucleotides 5′ to the high affinity RPA70AB domains. The SAXS experiment is ideally suited to detect ssDNA binding by RPA70N in the context of RPA70NAB because any appreciable interaction would produce a pronounced compaction of the molecule as a result of the alignment of RPA70N with RPA70AB. In particular, since RPA70AB is already strongly bound to 8 nucleotides of ssDNA, the binding of RPA70N to the remaining overhang would result in a substantial reduction in the Rg of RPA70NAB to a value in the vicinity of 30 Å (see below).
Two important considerations guided the design of the 14-mer oligonucleotide used for these experiments: (i) the number of nucleotides had to allow binding of an additional OB-fold domain without enabling binding of a second molecule, and (ii) the position of RPA70AB on the ssDNA needed to be biased to the 3′ end of the oligo to maximize the availability of free ssDNA for binding to RPA70N. The X-ray crystal structure of RPA70AB bound to d-CCCCCCCC shows each OB-fold domain makes contact with three nucleotides, and two nucleotides bridge between the RPA70A and RPA70B domains [3]. Thus, the length of the oligonucleotide needed to be less than 16 to preclude binding of two protein molecules on the DNA. A ssDNA 14-mer was therefore selected because it is too short for binding two molecules yet it provides six extra nucleotides for RPA70N to bind. The sequence d-CCACCCCC was used at the 3′ end of the 14-mer oligonucleotide based on previous analysis of the sequence preferences of RPA70AB (E. Bochkareva, A.I. Arunkumar, W.J. Chazin and A. Bochkarev, unpublished results). These studies showed that RPA70AB binds more strongly to cytosine rich sequences than adenine rich sequences and that placement of a single adenine at position 3 in d-CCCCCCCC further enhances binding affinity. To bias RPA70AB to the 3′ end of the oligonucleotide, the 14-mer was constructed by adding 6 adenines 5′ of the high affinity sequence, resulting in d-AAAAAACCACCCCC. Although RPA binds poly-adenine more weakly than poly-pyrimidines, the affinity is nonetheless in the nM range [28]. Moreover, six free adenines is adequate for binding because the tethering to RPA70AB means RPA70N is present in a higher local concentration relative to free diffusion. All studies with 8-mer oligonucleotides used the sequence d-CCACCCCC to ensure accuracy of the comparisons between binding of RPA70NAB to 8-mer and 14-mer ssDNA.
Scattering measurements for RPA70NAB bound to d-CCACCCCC (8-mer) or d-AAAAAACCACCCCC (14-mer) were acquired and analyzed following the strategy for free protein and the RPA70AB-ssDNA complex. Comparison of the data with free RPA70NAB reveals that binding of ssDNA alters the structural dynamics of the protein, although in a relative sense, the effect on the scattering curve is not as great as for RPA70AB (Figure 6A). A Kratky analysis indicated little change in the relative amount of unordered polypeptide in RPA70NAB when either ssDNA oligomer was bound (Figure 6B). The Rg values from the Guinier analysis for the 8-mer and 14-mer are 37.4 Å and 37.8 Å, respectively, reflecting a reduction of 2.1 Å to 1.7 Å relative to the free protein. As was observed for RPA70AB, the peak range of inter-atomic distances reflected in the P(r) function decreases significantly (Figure 6C). These observations indicate the reductions in Rg and Dmax upon binding of ssDNA closely parallel what is observed for RPA70AB. Thus, the data indicate a compaction of the RPA70AB domains as they bind the ssDNA within the complex, which is correlated with quenching of the inter-domain dynamics of these domains. However, there is no indication of further compaction of the protein, which would result from the RPA70N domain also engaging the ssDNA. On the contrary, the SAXS data shows that RPA70N remains as flexible in the DNA-bound state as in the free protein, confirming the hypothesis proposed based on indirect NMR evidence in our study of intact RPA [11, 12, 29].
To further analyze the data, large ensembles of conformations for both RPA70NAB-ssDNA complexes were generated using BILBO-MD, then the scattering was back calculated using CRYSOL and compared to the experimental data. The initial model was built using the coordinates from the X-ray crystal structures of RPA70N and RPA70AB bound to d-CCCCCCCC, along with a purely modeled N-A linker. For these calculations the entire RPA70AB module was treated as a single rigid body, RPA70N was treated as a second rigid body, and the N-A linker was allowed to sample conformational space freely. For a select number of conformers, 14-mer ssDNA was added back and CRYSOL was used to back calculate scattering profiles. An examination of a range of different conformers including highly extended and closely packed arrangements of the three domains shows that in fact, scattering is dominated by the relative position of the three globular domains (Supplementary Figure 4).
The key to our interpretation is the fact that all of the observed Rg values for RPA70NAB experiments (Table 1) are consistent only with RPA70N populating interdomain orientations where it is distant from RPA70AB. In particular, in experiments with the 14-mer, if RPA70N were interacting with the ssDNA it would be closely packed to RPA70AB and the overall shape of the RPA70NAB molecule would be substantially more compact and globular (Figure 7). This would result in Rg values significantly lower than those observed in the control experiments with the free protein and the 8-mer ssDNA. The models of the complex of RPA70NAB with the 14-mer in which RPA70N is packed in near the DNA have Rg values of the order of 29 Å, which is far off from the experimentally determined value of 41 Å. Thus, our analysis shows that RPA70N does not become ordered even when ssDNA is bound to RPA70NAB.
DISCUSSION
RPA ssDNA binding occurs with 5′ to 3′ directionality, initiated by the high affinity ssDNA binding domain RPA70A [11, 12, 29]. The existence of the short 10-residue A-B linker increases the effective concentration of RPA70B, which promotes its binding to ssDNA [13]. Binding of DNA to RPA70AB is followed in turn by rearrangement of RPA70C and RPA32D, respectively. The trajectory of binding is therefore in opposition to the orientation of RPA70N toward the 5′ end of ssDNA sequences.
It has been proposed that RPA70N plays a direct role in binding ssDNA [11, 12, 29]. The ssDNA binding affinity to the isolated RPA70N domain is extremely weak, which based on the evidence in the literature [16–18, 30] has a lower limit for the dissociation constant (Kd) in the millimolar range. Our study was designed to determine the effect of RPA70N in the context of ssDNA binding to the adjacent RPA70 A and B domains, which more directly addresses the hypothesis put forth in the literature. The effective local concentration of ssDNA in the vicinity of RPA70N is maximized in the experiment with the ssDNA 14-mer, therefore providing every opportunity for the RPA70N domain to engage the ssDNA. If RPA70N had any role to play at all in binding ssDNA, this would have been reflected in a change in the distribution of conformational states occupied by RPA70NAB relative to the distributions observed in the control experiments on free RPA70NAB and the complex with the ssDNA 8-mer. The fact that there is no indication of RPA70N interaction with ssDNA in the context of the RPA70NAB is convincing evidence against the proposal that RPA70N plays a direct role in the ssDNA binding activity of RPA.
The 70-residue linker between the N and A domains suggests there would be little correlated movement of the two domains, as suggested by NMR relaxation analysis of a construct containing RPA70N, RPA70A, and a portion of RPA70B [19]. However, it is difficult to draw firm conclusions from that study because the RPA70A domain in the absence of the RPA70B domain has only very weak affinity for ssDNA. Our studies show directly, and in the physiologically relevant context of high affinity binding of ssDNA, that the dynamic and flexible N-A linker enables a wide range of RPA70N orientations relative to RPA70AB. Thus, the long linker provides the large degree of freedom to RPA70N that is critical to this domain’s participation in the recruitment of partner proteins. RPA70N binds multiple proteins involved in DNA replication, damage response and repair mechanisms including p53, ATRIP, MRE11, and NBS1 [16–18, 30]. Hence, our results support models of RPA function in which RPA70N acts as a general protein recruitment module.
SAXS is an emerging technique in structural biology that measures electron scattering intensities to obtain interatomic distances for molecules in solution [23, 31]. For globular proteins, which have fixed rather than variable conformations, it is possible to convert this distance distribution into molecular envelopes that reflect the average shape of a protein or protein complex in solution [31]. Such coarse-grained structural envelopes are invaluable complements to atomic-resolution information. For systems with high degrees of inter-domain flexibility, such as RPA70NAB, a single, ‘averaged’ conformation fails to provide an adequate description of an intrinsically time-varying architecture. The lack of statistically significant correlation reflected by high χ2 values between experimental scattering profiles from RPA70NAB and those calculated for individual models readily illustrates this point, and defines this as a flexible region. Proper interpretation of scattering data from these flexible systems requires an ensemble approach – both to describe the population of feasible conformations and to characterize their relative frequency within the ensemble at a given moment in time. BILBO-MD [23] allowed us to search a broad range of accessible RPA70NAB conformational space, and examine in detail a subset of models (including some that represent the extreme conformations) and their capacity to recapitulate the scattering data. Notably, while averaging theoretical scattering profiles from multiple conformations improves the goodness-of-fit to the experimental data, the challenge of distinguishing the relative merit of one conformational subset over another remains.
Methods/programs are available to select combinations of conformers that provide better fits to the data, including the minimum ensemble (MES) approach that is part of BILBO-MD [23]. However, while it is possible to define combinations of structures that give improved fits to the data, these combinations are not necessarily unique but represent a minimal identified ensemble that bits the data. In the case of highly flexible proteins there are many combinations of conformers that fit the data equally well. Consequently, the main conclusion that can be drawn from this type of analysis of a complex highly flexible system such as RPA70NAB is that the protein contains substantial degrees of conformational heterogeneity, a point that is best made directly from data. Nevertheless, ensemble fitting does provide valuable insight into the dynamics of the native, solution architectures of proteins and macromolecular complexes.
The key role of dynamics in facilitating the organization and progression of large multi-protein machines is increasingly recognized particularly for DNA replication and repair machinery that requires precise coordination in order to efficiently preserve genome integrity. Our results suggest SAXS offers a robust approach to characterizing protein structural dynamics in solution without the complications of isotopic enrichment or spin labeling required by spectroscopic methods. RPA70N is found to be structurally independent of RPA70AB in the DNA bound state and therefore able to act flexibly as a protein recruitment module. Notably, this flexible attachment of the RPA DNA and protein binding domains, elucidated by SAXS, enables the interactions of RPA with diverse DNA substrates and protein partners required for effective orchestration of DNA replication and repair. Similar flexible attachments joining protein and DNA binding domains were recently discovered for DNA-PK and polynucleotide kinase [32, 33]. In fact, such RPA domain structural flexibility as experimentally defined here is essential to enable efficient protein handoffs and interface exchanges, as proposed for FEN1-PCNA [34] and BRCA2-Rad51 [35]. RPA binding protein partners, such as the Mre11-Rad50-Nbs1 complex have similar ordered and flexible domains, as shown by Nbs1 SAXS and crystal structures [36]. Such dynamic character may be a hallmark for scaffold proteins such as RPA.
The role of dynamics in facilitating recruitment, organization, and exchange of DNA processing factors has been characterized in several model systems, most notably in a recent study of homotetrameric E. coli SSB diffusion dynamics along ssDNA [37]. In that study, SSB diffusion was shown to be critical for resolving DNA secondary structures to enable RecA filament formation. Unlike the modular, multi-domain RPA, the homotetrameric single domain SSB does not utilize pre-existing structural dynamics to facilitate organizing strands of ssDNA for DNA processing. Instead, the compact, globular SSB homotetramer is encircled by the ssDNA and is conjectured to ‘roll’ along the template via a consecutive unwrapping/wrapping of ssDNA. Thus, while the structural organization of these two SSB systems remains fundamentally different, dynamic motion would appear to be integral aspects of both. Specifically, the nature of the structural dynamics of linked, ordered and flexible RPA domains as identified here appears critical to accommodating the large-scale complex conformational changes proposed to regulate RPA related functions, while preserving the integrity of DNA and protein partner interactions to maintain genetic fidelity.
Supplementary Material
Acknowledgments
We thank Kristian Kaufmann for guidance in implementing the Rosetta refinement of the RPA70NAB model, Seung Joong Kim, Dina Schneidman and Andrej Sali for ideas about generating models, and Michal Hammel for guidance in running BILBO-MD and other aspects of data analysis. We also thank Aravinda Raghavan and Kevin Weiss for initial SAXS studies of RPA70AB.
This research has been funded by the National Institutes of Health (RO1 GM65484). Additional NIH support was provided by the Structural Biology of DNA Repair Machines Program (PO1 CA92584), Vanderbilt Molecular Biophysics Training Program (T32 GM08320), the Vanderbilt Center in Molecular Toxicology (P50 ES00267), and the Vanderbilt-Ingram Cancer Center (P30 CA68485). The X-ray scattering technology and applications to the determination of macromolecular shapes and conformations at the SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory, are supported in part by the U.S. Department of Energy program Integrated Diffraction Analysis Technologies (IDAT) and the DOE program Molecular Assemblies Genes and Genomics Integrated Efficiently (MAGGIE) under Contract Number DE-AC02-05CH11231.
Abbreviations
- BME
beta-mercapto ethanol
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- MALS
Multi-Angle Light Scattering
- NMR
nuclear magnetic resonance
- RPA
Replication Protein A
- SAXS
Small Angle X-ray Scattering
- SEC
Size Exclusion Chromatography
- SEC-MALS
SEC using a Multi-Angle Light Scattering detection system
- ssDNA
single stranded DNA
Footnotes
Experimental and theoretical scattering profiles, P(r) functions, SAXS envelopes and atomic models will be deposited in the BIOISIS database (www.bioisis.net) under accession code 61.
SUPPORTING INFORMATION AVAILABLE. SEC profiles and Guinier analysis for RPA70AB, RPA70NAB and their ssDNA complexes. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34(15):4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochkareva E, et al. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21(7):1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochkarev A, et al. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385(6612):176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 4.Bochkarev A, et al. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18(16):4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs DM, et al. Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J Biomol NMR. 1999;14(4):321–331. doi: 10.1023/a:1008373009786. [DOI] [PubMed] [Google Scholar]
- 6.Pfuetzner RA, et al. Replication protein A. Characterization and crystallization of the DNA binding domain. J Biol Chem. 1997;272(1):430–434. doi: 10.1074/jbc.272.1.430. [DOI] [PubMed] [Google Scholar]
- 7.Deng X, et al. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J Mol Biol. 2007;374(4):865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- 8.Bochkareva E, et al. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20(3):612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mer G, et al. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103(3):449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 10.Mer G, et al. Three-dimensional structure and function of replication protein A. Cold Spring Harb Symp Quant Biol. 2000;65:193–200. doi: 10.1101/sqb.2000.65.193. [DOI] [PubMed] [Google Scholar]
- 11.Iftode C, Borowiec JA. 5′--> 3′ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry. 2000;39(39):11970–11981. doi: 10.1021/bi0005761. [DOI] [PubMed] [Google Scholar]
- 12.de Laat WL, et al. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12(16):2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arunkumar AI, et al. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J Biol Chem. 2003;278(42):41077–41082. doi: 10.1074/jbc.M305871200. [DOI] [PubMed] [Google Scholar]
- 14.Binz SK, et al. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J Biol Chem. 2003;278(37):35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- 15.Binz SK, Wold MS. Regulatory functions of the N-terminal domain of the 70-kDa subunit of replication protein A (RPA) J Biol Chem. 2008;283(31):21559–21570. doi: 10.1074/jbc.M802450200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bochkareva E, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005;102(43):15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005;16(5):2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, et al. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol. 2008;28(24):7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daughdrill GW, et al. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29(15):3270–3276. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosey CA, et al. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J Am Chem Soc. 2009;131(18):6346–6347. doi: 10.1021/ja9013634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konarev PV, VVV, Sokolova A, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36(Part 5):1277–1282. [Google Scholar]
- 22.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28(2):174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohl CA, et al. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55(3):656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya S, et al. Characterization of binding-induced changes in dynamics suggests a model for sequence-nonspecific binding of ssDNA by replication protein A. Protein Sci. 2002;11(10):2316–2325. doi: 10.1110/ps.0209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C, Snyder RO, Wold MS. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12(7):3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolpashchikov DM, et al. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 2001;29(2):373–379. doi: 10.1093/nar/29.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oakley GG, et al. Physical interaction between replication protein A (RPA) and MRN: involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry. 2009;48(31):7473–7481. doi: 10.1021/bi900694p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putnam CD, et al. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein NK, et al. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, Polynucleotide Kinase. Nucleic Acids Res. 2009;37(18):6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammel M, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 285(2):1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapados BR, et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116(1):39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 35.Shin DS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22(17):4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139(1):87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy R, et al. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461(7267):1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.