Abstract
Particle-based simulators represent molecules of interest with point-like particles that diffuse and react in continuous space. These simulators are often used to investigate spatial or stochastic aspects of biochemical systems. This article presents new particle-based simulation algorithms for modeling interactions between molecules and surfaces; they address irreversible and reversible molecular adsorption to, desorption from, and transmission through membranes. Their central elements are: (i) relationships between adsorption, desorption, and transmission coefficients on the one hand, and simulator interaction probabilities on the other, and (ii) probability densities for initial placements of desorbed molecules. These algorithms, which were implemented and tested in the Smoldyn simulator, are accurate, easy to implement, and computationally efficient. They allow longer time steps and better address reversible processes than an algorithm that Erban and Chapman recently presented (Physical Biology 4:16-28, 2007). This article also presents a method for simulating unbounded diffusion in a limited spatial domain using a partially absorbing boundary, as well as new solutions to the diffusion differential equation with reversible Robin boundary conditions.
1. Introduction
Cell modeling studies typically represent molecular interactions with mass action methods. These methods are simple, well supported by software (e.g. Copasi [21] and Virtual Cell [32]), and fully adequate for a wide variety of studies. However, they cannot represent the natural stochasticity that arises within and between cells [38], nor the intricate intracellular spatial organization that most cell systems exhibit. To investigate these topics, many scientists use more detailed modeling methods (reviewed in [4]).
Particle-based simulation is one of these more detailed methods. It represents individual molecules with point-like particles that behave in accordance with elementary processes, including diffusion and chemical reactions. For typical cell biology systems, particle-based simulation can support spatial resolution to scales of several nanometers [1, 14], can simulate up to tens of minutes of real time [23], and can represent hundreds of thousands of simulated molecules [2]. Freely available particle-based simulators include Cell++ [39], ChemCell [34], MCell [22], and Smoldyn [2]. Using these tools, researchers have shown, for example, that neural signaling may depend upon neurotransmitter release from sites that are away from the synapse [14] and that stable protein concentration gradients may arise in bacterial cells [31].
Recently, Erban and Chapman developed particle-based algorithms for simulating molecular adsorption to surfaces [18] (Figure 1A). They addressed the following problem: given a typical particle-based simulator design [3, 22, 35] (Figure 2), in which molecules diffuse in discrete steps and either adsorb to (with adsorption probability Pa) or reflect from any surface they encounter, how should the simulator compute this adsorption probability from the surface's physical adsorption coefficient? Their result (Eq. 10 of [18]) is
| (1) |
where κ is the adsorption coefficient (defined below and in the glossary), Δt is the simulation time step, and D is the molecular diffusion coefficient. Bartol and coworkers [7] derived essentially the same equation 16 years earlier (Eq. 6a of [7]) for the closely related problem of simulating reactions between solution-phase and surface-bound molecules. Also, Singer and coworkers recently generalized Eq. 1 to multi-dimensional systems with molecular drift and anisotropic diffusion matrices [41].
Figure 1.
(A) Model system for adsorption and desorption. Molecules, shown with dots, adsorb to the grey surface with adsorption coefficient κ and desorb with rate constant k. (B) Model system for partial transmission. Molecules transmit from the front side of the surface (x > 0) to the back side (x < 0) with permeability coefficient κF and transmit from back to front with permeability coefficient κB.
Figure 2.
Particle-based simulator design assumed throughout this work. The probability, P, refers to the adsorption, desorption, or partial transmission probability, as appropriate, and “rand()” is a uniformly distributed random number between 0 and 1. This article presents molecule-surface interaction probabilities (P) with which simulators following this design will adsorb, desorb, and transmit molecules at the correct rates.
All of these derivations of Eq. 1 assumed well-mixed systems, in which molecular concentrations are spatially uniform throughout the solution. An assumption about the average molecular positions is required for the adsorption probability to be independent of the precise position that each molecule diffused from. However, the well-mixed assumption is inconsistent with the problem definition because irreversible molecular adsorption to surfaces necessarily creates a local concentration gradient. Regardless of the starting state, this gradient develops rapidly in the immediate vicinity of the surface and approaches the time-invariant steady state (see section 3.3.1 of [16] and section 5, below). As a result, molecular concentrations near adsorbing surfaces, whether physical or simulated, are likely to spend the vast majority of their time much closer to the steady state than to the well-mixed state.
In this article, we re-investigate the problem that Erban and Chapman posed, but for steady-state systems rather than well-mixed systems. The relationship we find between the adsorption probability and the adsorption coefficient enables substantially more accurate simulations than Eq. 1 does. We also develop related algorithms for simulating irreversible desorption, reversible adsorption, and either irreversible or reversible transmission through permeable surfaces. Together, these algorithms enable quantitative simulation of most elementary molecule-surface interactions. Examples include peripheral membrane protein binding and release, membrane permeability, and heterogeneous catalysis. Where coarse-grained approximations are appropriate, our algorithms also support simulation of ligand binding to receptors or molecular transport through channels or pores. In addition, our adsorption algorithm enables a method for efficiently modeling unbounded diffusion in a limited spatial domain, which we discuss near the end of this article.
A specific example that illustrates the need for accurate adsorption algorithms arises in the yeast pheromone response system (reviewed in [6]), a model eukaryotic signaling system. Upon pheromone stimulation, pheromone-sensitive cells execute a series of actions, of which a central one is that the Ste5 protein translocates from the cytoplasm to the plasma membrane [37]. Colman-Lerner and coworkers recently recorded microscopy image sequences that show this translocation [15]. Because their data are two-dimensional images of three-dimensional cells, including in-focus and out-of-focus portions, they require further analysis to determine the adsorption coefficient between Ste5 and the membrane, which could lend insights into the interaction mechanisms. This analysis could be performed relatively easily by adjusting a simple computer model until it agreed with the microscope images; the simulator that ran this model would require accurate adsorption algorithms. Afterwards, the adsorption coefficient between Ste5 and the plasma membrane could be used in new spatial simulations of the pheromone response system. The resulting simulations, which would again require accurate adsorption algorithms, might lend further insights into the yeast signaling system dynamics or signal processing (see e.g. [24, 28]).
2. Model systems
We explore one model system for adsorption and desorption and another for partial transmission (Figure 1). The adsorption/desorption system extends over all x > 0 while the partial transmission system extends over all space. Both active surfaces are at x = 0. While these models are three-dimensional for convenient visualization, they can be described equally well with fewer dimensions because their average molecular concentrations are translationally invariant parallel to the surfaces; the mathematics throughout this article uses one dimension.
We assume that solution-phase molecules move solely by diffusion. This means we treat water and other un-modeled species only through their implicit contributions to the Brownian dynamics of the modeled molecules. It also means we ignore intermolecular interactions between modeled molecules, which is typically valid when their concentrations are low. These approximations allow us to describe diffusion with Fick's laws [11, 16]:
| (2) |
| (3) |
where J is the average molecular flux towards positive x and C(x,t) is the average concentration at position x and time t. We call C(x,t), which is technically a distribution function [26], the concentration profile.
Adsorption/desorption model system definition
The net adsorption rate in the adsorption/desorption system is
| (4) |
where Ca(t) is the average concentration of molecules that are adsorbed to the surface at time t, and k is the desorption rate constant. The first equality expresses the conservation of molecules between the solution and surface phases. More precisely, it states that the rate of adsorption onto the surface equals the flux of solution-phase molecules towards the surface, measured adjacent to the surface (see Eq. 2). The second equality expresses the adsorption rate in terms of the solution and surface concentrations. The first term on the right side of the equation is the Robin or radiation boundary condition [12, 16, 19], in which the adsorption rate is proportional to the concentration of solution-phase molecules at the surface. The proportionality constant, κ, is the adsorption coefficient. In this model system, κ can range from zero, for a completely inert surface, to infinity, for a surface that adsorbs molecules immediately upon contact. Physically, κ is limited to about the thermal velocity of potential adsorbents [13], which is (kBT/m)½, where kB is Boltzmann's constant, T is the temperature, and m is the molecule's mass, because this velocity determines how frequently molecules collide with the surface (for a 50 kDa protein at 37°C, κ < 7×106 μm/s). Note that the Robin boundary condition does not depend on the surface-bound concentration, so it does not account for surface saturation (in contrast to the Langmuir equation [27]). Also, note that authors frequently use the term “adsorption coefficient” to mean other things, such as the adsorption equilibrium constant [9, 10, 40] (see below) or an exponent for a variant of the Langmuir equation [20]. The second term on the right side of Eq. 4 is the desorption rate. Its proportionality constant, k, can also range from zero to infinity, now representing no desorption and instantaneous desorption, respectively. According to Chou and D'Orsogna [13], desorption rates are typically between 10−4 s−1 and 104 s−1.
Partial transmission model system
The net flux of molecules through the permeable surface, from front to back, is
| (5) |
In analogy to prior definitions, κF and κB are the permeability coefficients from the front of the surface (which faces the positive x-axis) to the back, and from the back to the front, respectively. Eq. 5 specifies the surface sides with +0 for the front and –0 for the back because the concentration profile is typically discontinuous at x = 0. As before, the first equality expresses the conservation of molecules and the second equality represents the Robin boundary condition. Again, this equation ignores saturation; for example, it does not account for ion channels operating at capacity. In general, permeability coefficients are large for molecules that tend to partition into the membrane as well as for molecules with high diffusion coefficients within the membrane (see [8]). Paula et al. [33] found that permeability coefficients across a 27 Å phosphatidylcholine bilayer are about 3.5×10−8 μm/s for potassium ions, 0.014 μm/s for urea, 0.027 μm/s for glycerol, and 150 μm/s for water.
3. Simulator design
Figure 2 presents the simulator design assumed throughout this work. Smoldyn [2] follows this design, and MCell [22] and ChemCell [35] use similar approaches.
In Step 1, the simulator diffuses molecules for a fixed amount of time with Gaussian-distributed random displacements, while ignoring any surfaces or bimolecular interactions. The standard deviation of these Gaussian distributions, called the root mean square (rms) step length, is
| (6) |
This is a characteristic length for the simulation, against which we previously compared intermolecular interaction distances [3] and surface curvature radii [2]. Molecules move about an rms step length at each time step, so the planar surfaces in our model systems accurately represent membranes that have curvature radii which are much larger than the rms step length, and are less accurate for more tightly curved membranes. For either model system, a diffusive step transforms a starting concentration profile, C(x,t), to
| (7) |
The star represents convolution and the term on its right is a Gaussian with unit area and standard deviation s.
In Step 2, the simulator addresses any solution-phase molecules that diffused across the surface in Step 1, as well as the possible desorption of any surface-bound molecules. The former molecules adsorb to the surface with probability Pa, transmit through it with probability Pt, and otherwise reflect off of it using ballistic reflection [3]. The latter molecules desorb with probability Pd; the simulator displaces desorbed molecules away from the surface according to the probability density p(x). We solve for Pa, Pt, Pd, and p(x) below. The simulator does not perform multiple surface interactions, such as both adsorption and desorption, on the same molecule so as to improve computational efficiency and to make their order of execution irrelevant (however, the algorithms do account for the multiple surface interactions that physical molecules may undergo). Step 2 transforms the adsorption/desorption system concentration profile to
| (8) |
It also transforms the adsorbed concentration to
| (9) |
The analogs of these equations for the partial transmission system are not shown because they are more complicated and less useful.
This design for Step 2 incorporates two simplifications for good computational efficiency. First, Pa and Pt do not depend on how far molecules are from surfaces, but instead depend only on static properties, such as diffusion and adsorption coefficients. This allows a simulator to compute these probabilities just once for a simulation, rather than once for each molecule-surface interaction. Secondly, the simulator ignores all molecules that diffused across the surface during Step 1, but that returned back to the side where they started before the end of the time step (see section 3.2 of [3] and section 2.2 of [18]). In preliminary work, accounting for these additional surface contacts proved computationally intensive, complicated to implement, and minimally more accurate.
In Step 3, the simulator performs chemical reactions [3]. We assume that any reactions are slow enough that their effects on concentrations near surfaces are negligible, where the rms step length provides the necessary length scale. In Step 4, the simulator increments the simulation time. This updates the concentration profile and surface concentration to
| (10) |
| (11) |
Eq. 10 applies to both model systems, while Eq. 11 only applies to the adsorption/desorption system. By definition, C(x,t+Δt) equals the starting concentration profile, C(x,t), if and only if they represent a steady state. Ordinarily, the adsorbed concentration, Ca(t), would also have to be constant at steady-state. In fact, we require this time independence for reversible adsorption but not for irreversible adsorption. We do not require it for irreversible adsorption because (i) we wish to investigate a constant adsorption flux, and (ii) the adsorbed concentration does not affect other system aspects. Finally, the simulator performs any observations of the system in Step 5. It then returns to Step 1 to start the next time step.
4. Interaction probabilities
In this section, we relate the molecule-surface interaction probabilities to their respective coefficients for accurate steady-state simulation. We also derive the necessary probability densities for desorbed molecule displacements. These results are presented as five algorithms, along with several variants. Figure 5 presents the main results graphically, the Supplementary Information presents them in tabular form, and several equations below approximate them with interpolating functions. Also, the Supplementary Information includes the C language library files that Smoldyn uses to perform the necessary table look-up and interpolation. Except as noted, most results in this section were computed with Mathematica software [44].
Figure 5.
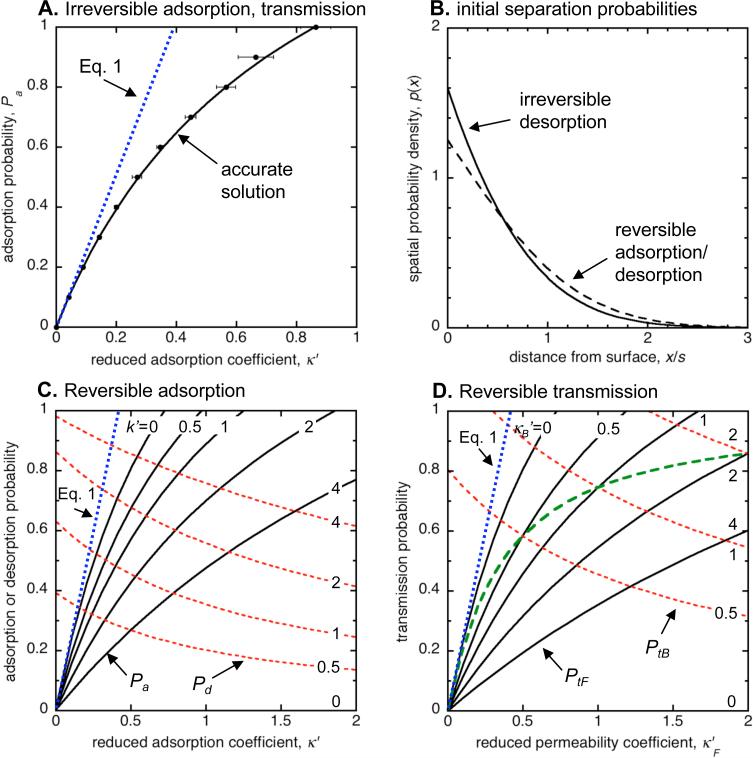
Relationships between transition coefficients and transition probabilities, and initial separations between surfaces and desorbed molecules. (A) Relationship between κ′ and Pa for irreversible adsorption. The solid black line, calculated by the simulation emulator and approximated by Eq. 21, enables accurate adsorption simulation. Points with one standard deviation error bars represent results from stochastic simulations of a steady-state system (see Supplementary Information for details). (B) Probability densities for the initial separation between a desorbed molecule and the surface that it desorbed from. The solid line represents the density that should be used for irreversible desorption (Eq. 25) and the dashed line represents the density for reversible adsorption (Eq. 33). (C) Relationships between Pa (solid black lines), Pd (dashed red lines), κ′ (abscissa), and k′ (marked on figure) for reversible adsorption; these are also given in Eqs. 32 and 37. The k′ = 0 desorption probability line is coincident with the abscissa. (D) Relationships between PtF (solid black lines), PtB (dashed red lines), κF′ (abscissa), and κB′ (marked on figure) for reversible transmission; these are also given in Eqs. 47 and 48. The κB′ = 0 transmission probability line is coincident with the abscissa. The green bold long-dashed line presents the transmission probabilities for the condition that κB′ = κF′. Where present, the dotted blue line represents the relationship given by Eq. 1.
Unitless reduced variables, shown with a prime symbol, simplify several results. We use the simulation time step as the standard time unit and the rms step length as the standard length unit. From these, the reduced adsorption coefficient, desorption rate constant, and diffusion coefficient are:
| (12) |
| (13) |
| (14) |
Reduced permeability coefficients, κF′ and κB′, are analogous to the reduced adsorption coefficient (Eq. 12).
4.1 Algorithm I: Irreversible adsorption
Consider the adsorption/desorption system and assume irreversible adsorption (κ > 0 and k = 0). Solving Eqs. 3 and 4 shows that the model steady-state concentration profile, C(x), increases linearly with distance away from the surface and the surface concentration increases at a constant rate (Figure 3A),
| (15) |
| (16) |
where C0 is the solution-phase concentration at x = 0. (Eq. 3 and the outer portions of Eq. 4 are the equations of motion for C(x,t) and Ca(t), respectively, while the latter two portions of Eq. 4 give the necessary boundary condition.) Eq. 15 becomes physically unreasonable as the system increases in size because C(x) is unbounded; however, this model is likely to be physically correct near the surface, so it is adequate for our work. As before, note that the concentration profile and adsorption flux are time-invariant at steady state.
Figure 3.
Steady-state concentration profiles for model systems that (A) reversibly or irreversibly adsorb molecules to a surface or (B) reversibly transmit molecules through a surface. The grey line at x = 0 represents the surface.
In an exact algorithm, meaning one in which the simulated system is theoretically indistinguishable from the underlying model system, the average simulated steady-state concentration profile would equal Eq. 15. We determine whether this is possible by seeing if our simulator design can preserve the model steady-state profile over one time step. Step 1 (Eq. 7) transforms Eq. 15 to
| (17) |
Steps 2 to 4 (Eqs. 8 and 10) then transform this profile to
| (18) |
for x > 0. Eq. 18 is the concentration profile at the end of the time step. Comparing Eqs. 15 and 18 shows that they cannot equal each other for arbitrary κ′ and x values. Thus, Eq. 15 is a steady state for the model system but not for our simulator design, which means that our simulator design cannot treat irreversible adsorption exactly. (The simplifications in Step 2, which we included for computational efficiency, cause this inexactness.)
Knowing that simulated concentration profiles would necessarily be inexact, I computed the adsorption probabilities that yield the correct adsorption rates with a “simulation emulator” (see Figure 4A and the Supplementary Information). Each emulator run started with a fixed adsorption probability and a well-mixed numerical concentration profile (which extended from −6 to +10 rms step lengths and was tabulated at 401 equally spaced points, plus additional points immediately above and below 0 for higher accuracy). The emulator iterated Steps 1 and 2, using Eqs. 7-9, until the system was essentially at steady state (as determined by the adsorption rate changing by less than 0.01% between sequential iterations). It addressed the region with x < −6s by extrapolating the concentration profile with an error function, and it addressed the region with x > 10s by fixing these concentrations to 1. Once at steady state, it fit a straight line to the final concentration profile (x ranged from 3s to 7s to avoid edge effects), from which it calculated the effective reduced adsorption coefficient using
| (19) |
Eq. 19 derives from Eq. 15, with m as the concentration profile slope (C0κ/D) and C0 as the intercept. Because the emulator could not actually reach the steady state, it performed the above routine twice, starting from different states; it started with C(x,0) equal to either 0 or 1 so that it would approach the steady-state profile from both above and below. Finally, the emulator averaged the two results to yield a final κ′ value. These κ′ values are almost certainly accurate to within ±10–6 because (i) the two intermediate solutions never differed by more than 10–7, and (ii) more conservative choices for emulator parameters (number of tabulated points, size of x-domain, and number of iterations) affected results by even smaller amounts.
Figure 4.
Steady-state concentration profiles at different steps of the simulation process. These apply to (A) irreversible adsorption and (B) reversible adsorption. Solid lines represent the profiles at both the beginning and end of time steps, long-dashed lines represent the profiles after the simulator's Step 1, and the short-dashed line in Panel B represents the profile after the reflection and adsorption portions of Step 2, but before desorption (this situation doesn't apply to Panel A). Areas on the figure represent molecular populations, as labeled. The grey line at x = 0 represents the surface. Panel (A) results were computed with the simulation emulator (see the main text and Supplementary Information) and Panel (B) with Mathematica software [44].
Figure 5A shows the emulator results, oriented so that Pa depends on κ′. Figure 5A also shows stochastic simulation results, generated by Smoldyn, for systems that were similar to those that the emulator quantified (Supplementary Information). The close agreement between the two sets of results lends confidence in the emulator software, the Smoldyn software, and the mathematical analysis. When reduced adsorption coefficients are small, the emulator results agree with Eq. 1. This makes sense because small κ′ values imply either: (i) small concentration gradients (because κ is small or D is large), so the steady state is similar to the well-mixed state, or (ii) a small simulation time step, so molecules diffuse over short distances and the concentration gradient affects the simulated adsorption rate minimally. On the other hand, the emulator results are up to a factor of two smaller than Eq. 1 for larger κ′ values. This implies that simulators basing their adsorption probabilities on Eq. 1 would simulate adsorption too quickly in many situations. Finally, Figure 5A shows that κ′ values larger than about 0.86 cannot be modeled with our simulator design because the adsorption probability cannot exceed 1. This means that large adsorption coefficients can only be simulated quantitatively by using small time steps. (This limitation arises from simplifications that we made for computational efficiency in the simulator's Step 2.)
Polynomial fits to the emulator's relationship between κ′ and Pa, which may be more convenient than the raw data, are:
| (20) |
| (21) |
The initial terms were constrained with Eq. 1 to make these fits exact as κ′ or Pa tend toward 0. These equations agree with the tabulated data to within 1% over their entire domains.
4.2 Algorithm II: Irreversible desorption
Continuing with the adsorption/desorption system, we now consider irreversible desorption (κ = 0 and k > 0). Desorption is a simple first order process, like a first order chemical reaction, so a molecule should desorb during a time step with probability [3, 42]
| (22) |
This is exact for all possible desorption rate constants and simulation time steps.
The more difficult question concerns where the simulator should place desorbed molecules. In one algorithm variant, the simulator places them adjacent to the surface. This ignores diffusion occurring between when a molecule desorbs and the end of the time step, which does not affect the accuracy of the total surface- and solution-phase concentrations, but means that this algorithm variant yields inexact concentration profiles.
A better variant displaces desorbed molecules away from the surface. To calculate the probability density for this displacement, consider a molecule that desorbs at time t, where t is between the start of the time step at time 0 and the end at time Δt. The probability density for this molecule's position at time Δt is [16]
| (23) |
We denote the probability density that the molecule desorbs at time t, given that it desorbs between 0 and Δt, as pt(t). Treating pt(t) as a weighting factor for Eq. 23, the spatial probability density for a molecule that desorbs at an unknown time between 0 and Δt is
| (24) |
To be consistent with Eq. 22, pt(t) should equal k/Pd exp(–kt). However, this makes p(x,Δt) depend on k, which is inconvenient. So, we make the approximation that the molecule is equally likely to desorb at any time during the time step, which simplifies pt(t) to 1/Δt. This approximation is valid when kΔt << 1 because the exact exponential function approaches 1/Δt in this limit. Because Eq. 22 already ensures accurate surface and solution concentrations and we are just refining the solution concentration profile here, this approximation is also likely to be acceptable for substantially larger values of kΔt as well. Using the approximation, Eq. 24 integrates to
| (25) |
Figure 5B shows this displacement probability density with a solid line.
A final task makes Eq. 25 useful. We need a relationship with which a simulator can transform random numbers that are uniformly distributed between 0 and 1 into random numbers that obey the probability density in Eq. 25. We do this by integrating and inverting Eq. 25 [36]. The integral, from 0 to x, is
| (26) |
This cannot be solved for x analytically, so I inverted it numerically. A least-squares rational function fit to the result is
| (27) |
Eq. 27 converts a uniform deviate, P, into the desired deviate, x. Compared to the numerical solution, it is in error by less than 0.02s for nearly all P values.
4.3 Algorithm III: Reversible adsorption/desorption
If adsorption is reversible (κ > 0 and k > 0), the adsorption and desorption rates equal each other for the system to be at steady state. Adding this condition to Eqs. 3 and 4 yields the steady-state concentrations (Figure 3A),
| (28) |
| (29) |
where C∞ is the solution concentration. In contrast to the preceding investigations of irreversible processes, this steady state is a well-mixed state. This steady state is also an equilibrium state, because there are no net molecular fluxes. The ratio κ/k, which clearly equals Ca/C∞, is the adsorption equilibrium constant; it is analogous to the familiar equilibrium constant for reversible reactions [5].
As in section 4.1, we determine whether our simulator design can treat reversible adsorption exactly by seeing if it can preserve the model concentration profile over one time step (see Figure 4B). Transforming Eq. 28 with Eq. 7 yields
| (30) |
The adsorption and reflection portions of Step 2 (see Eq. 8) then transform the concentration profile to
| (31) |
Finally, the desorption portion of Step 2 transforms this result back to the starting condition, if and only if:
| (32) |
| (33) |
Thus, our simulation design can simulate reversible adsorption exactly under equilibrium conditions. Eq. 32, which relates the adsorption and desorption probabilities, arises from the constraint that equal numbers of molecules must adsorb and desorb during each time step, on average. Eq. 33, which is the probability density for desorbed molecule displacements and is shown in Figure 5B with a dashed line, causes desorbed molecules to exactly replace those that adsorbed. This density is the ratio of the function C(x,t + Δt) – C(2a)(x,t) and its integral (the “desorbed molecules” region of Figure 4B), so that the probability density has unit area.
We integrate and invert Eq. 33 to make it useful. Integration yields
| (34) |
A rational function fit to the numerically calculated inverse function is
| (35) |
As before, P is a uniform deviate between 0 and 1 and x is the initial displacement for a desorbed molecule. This fit is in error by less than 0.02s for nearly all P values.
The remaining question is how Pa or Pd should be chosen. Their solutions for irreversible adsorption (Eq. 21) and irreversible desorption (Eq. 22) do not simultaneously satisfy the equilibrium condition of Eq. 32 because the prior results did not consider molecules that both adsorb and desorb during a single time step. We address them with the following conceptual scheme: we label all molecules as being solution-phase or surface-bound at time 0, the system evolves over one time step, and we then use the labels to see how many molecules had a net adsorption transition by time Δt. We solve this problem in Appendix A. The answer, Eq. A12, represents the model adsorbed molecules at the end of a time step. Meanwhile, the expectation simulated adsorbed molecules for one time step (see Figure 4B and Eq. 9) is
| (36) |
Equating these amounts and solving for Pa, yields the adsorption probability,
| (37) |
| (38) |
| (39) |
Figure 5C shows this adsorption probability and the corresponding desorption probability (from Eq. 32) for different κ′ and k′ values. As before, (i) the adsorption probability approaches Eq. 1 as κ′ is reduced to zero, (ii) Eq. 1 simulates adsorption too quickly with larger κ′ values, and (iii) κ′ values that exceed some cut-off, which now depends on k′, cannot be simulated quantitatively with our simulator design because adsorption probabilities cannot exceed 1. In the limit that κ′ is reduced to zero, Pd approaches its value for irreversible desorption (Eq. 22), as one would expect. In contrast though, Pa does not approach the irreversible adsorption probability (Eq. 21) as k′ is reduced to zero. This is because the concentration profile remains well-mixed for the reversible adsorption case, even as k′ is reduced towards zero, which is a consequence of the fact that the reversible adsorption steady state is an equilibrium state (as k′ is reduced towards zero, the adsorption equilibrium constant and the adsorbed concentration increase towards infinity); in contrast, the concentration profile is sloped for the irreversible adsorption case.
4.4 Algorithm IV: Irreversible transmission
The partially transmitting system (Figure 1B), with irreversible transmission from the front face to the back face (κF > 0 and κB = 0), is essentially identical to the irreversible adsorption system. Its steady-state concentration profile is
| (40) |
where C+0 represents the concentration at the front of the surface and is used if x > 0, and C–0 represents the concentration at the back of the surface and is used if x < 0. As we found before: our simulation design cannot simulate concentration profiles exactly for steady-state irreversible transmission, but it can achieve accurate concentrations on each side of the surface; Figure 5A shows the transmission probability as a function of the transmission coefficient; and Eqs. 20 and 21 are polynomial fits to the relationship between Pt and κF.
The question remains where the simulator should place transmitted molecules. It can, of course, place them at the back side of the surface, although this ignores diffusion occurring between transmission and the end of the time step. A better algorithm variant accounts for this diffusion using the probability density given in Eq. 25. Whereas this density was inexact before, it is exact now because a steady-state irreversibly transmitting surface acts as a constant molecular source to the region with x < 0. In yet a third algorithm variant, the simulator does not move transmitted molecules, but simply leaves them where they ended up after Step 1 of the simulation process. The resulting probability density (which can be approximated with the x < 0 portion of Eq. 17) is very similar to the exact result. This is the most computationally efficient variant.
4.5 Algorithm V: Reversible transmission
Finally, consider the reversible partially transmitting system (κF > 0 and κB > 0). From Eqs. 3 and 5, the model steady state is well-mixed on each side of the surface but is likely discontinuous at the surface,
| (41) |
| (42) |
This steady state is an equilibrium state, for which each side of Eq. 42 represents the chemical partition coefficient [5]. Physically, the concentration discontinuity can be maintained actively, such as by trans-membrane molecular pumps, or passively, such as by chemical partitioning between different solvents.
Our simulator design can simulate reversible transmission exactly under equilibrium conditions. The starting concentration profile (Eq. 41) diffuses to
| (43) |
where the first term represents molecules that diffuse from x < 0 and the second term represents molecules that diffuse from x > 0. In Step 2, the simulator transmits molecules that diffused across the surface with probabilities PtF and PtB for the front and back faces, respectively. The number of molecules that the simulator transmits from the front to the back is the integral of the second term of Eq. 43 from –∞ to 0 (see Eq. 36), times PtF. This result, and the analogous one for transmission the other way, is
| (44) |
| (45) |
These are equal at steady state. Combining them with Eq. 42 yields
| (46) |
To return the concentration profile to the steady state by the end of the time step, the simulator can displace transmitted molecules away from the surface according to Eq. 25 or just leave them where they ended up after Step 1. These produce the same steady-state concentration profiles and so are equally accurate at steady state, although the latter variant is more computationally efficient.
We find PtF much as we found Pa for the reversible adsorption problem. Now, we solve: given an initially well-mixed concentration profile for x > 0, no concentration for x < 0, and a reversible partially transmitting boundary, what is the total number of molecules in the region x < 0 at time Δt? Appendix B presents the result. The solution, Eq. B12, represents the number of molecules that the simulator should transmit over one time step. We equate it to Eq. 44, which represents the number that the simulator does transmit, and solve for the transmission probabilities. They are:
| (47) |
| (48) |
Figure 5D presents these probabilities.
The results are familiar: transmission probabilities approach Eq. 1 as κF′ and κB′ approach zero; Eq. 1 simulates transmission too quickly with larger κF′ and κB′ values; and, in most cases, a simulator can only quantitatively simulate transmission up to finite κF′ and κB′ values. The exception to the last result occurs if κF′ = κB′ (the bold dashed line in Figure 5B), which is typical for passive membranes.
5. Algorithm verification and comparison
All of the algorithms presented above rest on firm theoretical foundations that ensure high accuracy for steady-state systems. Here, we investigate their accuracy away from steady state. Figure 6 shows results from several algorithm tests, run with the Smoldyn simulator. Each test started with 20,000 molecules that were well-mixed within box-shaped regions and that had 5 μm2/s diffusion coefficients (see Figure 1; x extended from 0 to 2 μm and y and z were each 1 μm wide). These are reasonably typical parameters for proteins in bacteria [17, 43]. Initially, no molecules were adsorbed to a surface at x = 0 and there were no molecules in the region x < 0. Simulation time steps were 1 ms, which is larger than the 0.25 μs to 0.1 ms time steps that modelers typically use [14, 25, 29-31], both because this tested the algorithms where they are likely to be weakest and because a goal of this work is to enable accurate simulations with longer time steps. Depending on the specific test, molecules interacted with the surface with irreversible adsorption, reversible adsorption, or reversible transmission. These tests used either “fast” rates, with the maximum possible adsorption or transmission coefficients that the time step allowed, or “slow” rates, with coefficients that led to adsorption or transmission probabilities to be about 0.1 (the Figure 6 caption presents the coefficients). Figure 6A compares these stochastic simulation test results with exact calculations, where the latter are:
| (49) |
(Eq. 3.37 of [16]) for irreversible adsorption, Eq. A12 for reversible adsorption, and Eq. B12 for reversible transmission. In all cases, the simulation results agreed essentially perfectly with the model ones. This is true at all time points, despite the fact that the system was far from steady state for the initial ones. Additional tests (not shown) performed equally well for smaller adsorption and transmission coefficients.
Figure 6.
Algorithm tests for systems that started with well-mixed states. Dots represent stochastic simulation results and lines represent predictions for the corresponding model systems. (A) Number of molecules adsorbed to a surface or transmitted through the surface, as appropriate, in tests of irreversible adsorption (IA, circles), reversible adsorption (RA, triangles), or reversible transmission (RT, diamonds). Upper black points are for “fast” rates: κ = 85.9 μm/s for irreversible and reversible adsorption (Pa = 1), k = 276 s−1 for reversible adsorption (Pd = 0.128), and κF = κB = 5045 μm/s for reversible transmission (PtF = PtB ≈ 1). Lower red points are for “slow” rates: κ = 4.23 μm/s for irreversible and reversible adsorption (Pa = 0.1), k = 28 s−1 for reversible adsorption (Pd = 0.026), and κF = κB = 4.36 μm/s for reversible transmission (PtF = PtB = 0.1). Every third simulation data point is shown for clarity. (B) Concentration profiles at several time points for the irreversible adsorption test with the “fast” rate. Black circles represent time 0, red triangles represent time 1 ms (1 time step), green diamonds represent time 10 ms, and blue squares represent time 100 ms. See Supplementary Information for details.
Figure 6B shows several snapshots of molecular spatial distributions from the irreversible adsorption test that used the “fast” rate. Comparison with the model profiles,
| (50) |
(Eq. 3.35 of [16]), again showed excellent agreement at all time points. As before, additional tests that used smaller adsorption coefficients performed equally well. Note that even though the system shown in Figure 6B was well-mixed initially, its concentration profile evolved rapidly in the immediate vicinity of the surface (the rms step length for this simulation was 0.1 μm) toward a steady state profile; this is the reason why we developed our algorithms for steady-state systems.
These tests indicate that our irreversible adsorption, reversible adsorption, and reversible transmission algorithms are accurate not only for steady-state systems, but also for those away from steady state. Furthermore, they are accurate over the entire range of adsorption or transmission coefficients accessible for a given simulation time step. Considering the other two algorithms, the irreversible desorption probability presented above (Eq. 22) is accurate for all systems because it does not rely on the steady-state assumption, and the irreversible transmission algorithm is essentially identical to the irreversible adsorption algorithm, which we just verified.
In contrast, simulations based on Eq. 1 performed less well. First, they could not be tested with the “fast” set of adsorption or transmission coefficients because Eq. 1 does support such large coefficients (Eq. 1 only allows κ′ values up to 0.399, whereas they can reach 0.859 for our irreversible adsorption algorithm; see Figure 5A). For the “slow” set of coefficients, adsorption was about 6% too fast and partial transmission about 10% too fast, at all time points. With larger adsorption or transmission coefficients, use of Eq. 1 led simulations to irreversibly adsorb molecules up to 2.1 times too fast, or to reversibly adsorb or transmit molecules with even larger errors (the maximum errors depended on the rates of the reverse processes).
6. Simulating unbounded diffusion with partial absorption
Our final algorithm, which applies to all spatial simulation methods, builds on the irreversible adsorption algorithm. It addresses the following situation: suppose a modeler wants to investigate biochemical interactions in unbounded space, such as between extracellular pheromones and cell-surface receptors, but wants to limit the simulated spatial domain for computational efficiency [2]. The modeler needs to know how much space to simulate explicitly and how to treat its boundary. Clearly, the boundary should be neither completely reflective, which would keep molecules from diffusing away permanently, nor completely absorbing, which would inaccurately represent those molecules that diffuse away and then return. We propose the intermediate solution that the boundary should absorb molecules according to whatever absorption coefficient causes the simulated concentrations to equal those for the unbounded model system. We solve for this absorption coefficient here.
We derive the absorption coefficient for three-dimensional systems with stationary sources that emit molecules at constant rates, without significant obstacles to diffusion, and without significant molecular turnover through reactions. We number the sources with index j: source j is located at rj and emits molecules with rate qj. For our unbounded model, the steady-state molecular concentration at location r is [16]
| (51) |
The flux at r is computed from the concentration gradient using Fick's law, as in Eq. 2,
| (52) |
The Robin boundary (Eq. 4) relates the flux into the surface with the concentration at the surface, which we generalize to more than one dimension to yield
| (53) |
where ñ(r) is the unit outward normal for the surface at r; also, note that κ, the absorption coefficient, is now made a function of r. We equate the fluxes in Eqs. 52 and 53 and solve for the adsorption coefficient,
| (54) |
Thus, if a surface absorbs molecules with this absorption coefficient, the concentration profile will be the same as if the system were unbounded.
Figure 7 illustrates this algorithm. Using the Smoldyn program, I defined two molecular sources and bounded the system with a cube that was 5 μm on each side. Each cube face was tiled with 25 panels, each 1 μm square. Each of these panels absorbed molecules according to the coefficient given in Eq. 54, calculated with r set to the panel center. The upper portion of Figure 7 shows a slice through the middle of the system and the lower portion compares average steady-state molecular concentrations in a band through the system with model concentrations from Eq. 51. The simulated and model systems agree well.
Figure 7.
Simulation of effective unbounded diffusion with a partially absorbing boundary. (A) a 0.5 μm thick slice, taken parallel to the x,y-plane, through the middle of a cubical simulated system. Large dots represent molecular emitters, small dots represent molecules, and the grey band across the system (5 μm long by 0.5 μm wide and thick) represents the volume in which molecules were counted. Each face of the system is assembled from a grid of 25 panels, each with an absorption coefficient from Eq. 54; panels with larger absorption coefficients are depicted with thicker boundary lines. (B) Dots represent simulated molecule concentrations, measured along the grey band from panel A, at steady-state and averaged over 1000 time steps. Dashed lines represent theoretical concentrations from each individual source, along the grey band, for the unbounded model system. The solid line represents the sum of the dashed lines, which is the total theoretical concentration for the unbounded model system. See Supplementary Information for details.
7. Discussion and conclusions
Erban and Chapman's adsorption algorithm for particle-based simulators [18], based on Eq. 1, is simple to understand, easy to implement, and fast to simulate. These qualities make it ideal for “quick-and-dirty” simulations. However, it is only quantitatively accurate if (i) the modeled system is kept well-mixed, even in the immediate vicinity of adsorbing surfaces, or (ii) simulation time steps or adsorption coefficients are very small (more precisely, κ′ << (2π)−1/2). We built on their result here with algorithms that yield more accurate adsorbed concentrations and solution-phase concentration distributions, and also with new algorithms for desorption and partial transmission. We derived these algorithms for steady-state systems, but also showed that they are very accurate for systems that start well-mixed. They require essentially the same computational effort as Erban and Chapman's method at each simulation time step but may enable faster simulations overall because they maintain their accuracy as time steps are made longer.
This work shares two important themes with our prior work on simulating bimolecular reactions [3]. First, we designed both sets of algorithms to yield correct rates when the systems are at steady state rather than when they are well-mixed. We did this because the adsorption and reaction processes we wished to model cause systems to evolve rapidly away from well-mixed states (or other initial states) and towards steady states. As a result, portions of these systems close to adsorbing surfaces or reactive molecules, which are the regions that determine adsorption and reaction rates, spend nearly all of their time close to steady state. As we saw above and in prior work [3], steady-state investigations do not lead to the simple analytical equations that other researchers found using well-mixed assumptions [18, 22, 41], but are nevertheless necessary for accurate simulations. Secondly, we found that reversible interactions need to be considered as special cases, rather than as independent forward and reverse processes. We saw that here for reversible adsorption and reversible transmission, and previously for reversible bimolecular chemical reactions [1, 3]. Treating the processes as independent forward and reverse processes leads to incorrect transition rates and, more importantly, incorrect equilibrium constants.
The algorithms presented here will, hopefully, make it easier for researchers to quantify biological adsorption, desorption, and transmission rates by using computational modeling. This will help address the current paucity of quantitative data for these interactions. Combining these algorithms with the better data should also enable particle-based biological simulations that are faster and more accurate than current ones. These investigations may help scientists better understand dynamic protein localization, intracellular molecular gradients, and related topics. To promote these research directions, all algorithms presented here are implemented in a code library that is supplied with the Supplementary Information, and in the general-purpose Smoldyn biochemical simulator (available at http://www.smoldyn.org).
Supplementary Material
Acknowledgements
This publication was made possible by Grant Number P50 HG002370 from the NHGRI, and also by MITRE Corp. contract number 77459; R. Brent was the principle investigator for both grants. Its contents are solely the responsibility of the author and do not necessarily represent the official views of the NIH. I thank Attila Szabo for help with the Appendix results; Radek Erban and Jon Chapman for helpful discussions; Nathan Addy, Esther Mecking, and Orna Resnekov for editing assistance; and anonymous reviewers for exceptionally helpful reviews.
Nomenclature
Units are given in parentheses, assuming a 3-dimensional system; L = length, T = time, and 1 = unitless.
Roman symbols
- C(x,t)
concentration in solution; time is omitted for steady-state (L−3)
- Ĉ(x, z)
Laplace transform of solution concentration (L−3T)
- C0, C±∞
concentration adjacent to and infinitely far from the surface (L−3)
- Ca(t)
concentration adsorbed to the surface; time is omitted for steady-state (L−2)
- D, D′
normal and reduced diffusion coefficient (L2T−1, 1)
- J, J(r)
flux parallel to x-axis and flux vector (L−2T−1)
- k, k′
normal and reduced desorption rate constant (T−1, 1)
- ñ(r)
unit outward normal for bounding surface (1)
- Pa, Pd
adsorption and desorption probability (1)
- PtF, PtB
transmission probability for front or back (1)
- p(x)
spatial probability density (L−1)
- qj
emission rate of source j (T−1)
- r, rj
position on bounding surface and of source j (L)
- s
root mean square step length (L)
- t, Δt
time and simulation time step (T)
- x
position on x-axis (L)
- z
Laplace transform conjugate of time (T−1)
Greek symbols
- κ
adsorption coefficient (L T−1)
- κF
permeability coefficient from front side (L T−1)
- κB
permeability coefficient from back side (L T−1)
- κ′, κF′, κB′
reduced adsorption or permeability coefficient (1)
Glossary
- Adsorption coefficient
The proportionality constant in the Robin boundary condition that expresses the rate at which solution-phase molecules that are adjacent to a surface adsorb to the surface. It is defined here by κ in Eq. 4 and has units of length/time. Other authors frequently define the term differently, depending on their adsorption model.
- Adsorption probability
The probability with which molecules that diffused across a surface during the prior simulation time step adsorb to that surface.
- Concentration profile
The expectation molecular concentration as a function of the distance away from the surface. It is a distribution function.
- Desorption rate constant
The probability with which a molecule desorbs from a surface during a simulation time step.
- Exact algorithm
An algorithm for which all of its results are theoretically indistinguishable from those of the underlying model system.
- Particle-based simulation
A simulation method in which individual molecules of interest are represented with point-like particles in continuous space. This method accounts for stochastic and spatial detail.
- Permeability coefficient
The proportionality constant in the Robin boundary condition that expresses the rate at which solution-phase molecules that are adjacent to a surface diffuse through the surface. It is defined here by κF and κB in Eq. 5 and has units of length/time.
- Robin boundary condition
A weighted combination of Dirichlet and Neumann boundary conditions in which the gradient of a function at a surface is directly proportional to its value at the surface. This is also called the radiation boundary condition.
- Root mean square (rms) step length
The average length of a step for a molecule in a Brownian dynamics simulation. This is often a characteristic length for the simulation algorithms.
- Steady-state system
A system in which expectation molecular concentrations are constant with respect to time, although there may be constant molecular fluxes.
- Well-mixed system
A system in which the expectation solution-phase molecular concentration is uniform throughout space.
Appendix A
This section solves the following problem. In the adsorption/desorption model system, defined by Eqs. 3 and 4, suppose the solution is well-mixed with concentration C∞ and there are no adsorbed molecules at time 0. What is the adsorbed concentration at time t?
We Laplace transform Eqs. 3 and 4 to yield
| (A1) |
| (A2) |
A “hat” symbol denotes a Laplace transformed function and z is the Laplace conjugate of the time (we use z instead of s, which is conventional, because we use s for the rms step length). In addition, three boundary conditions are
| (A3) |
| (A4) |
| (A5) |
Substitution of Eq. A3 into Eq. A1, followed by solution of the differential equation, yields the general solution
| (A6) |
where a and b are constants of integration. Using Eq. A5, a equals 0. Meanwhile, the first and last portions of Eq. A2, with substitution from Eq. A4, yield
| (A7) |
Combination with Eq. A6 simplifies the result to
| (A8) |
Using the middle and last portions of Eq. A2, along with substitutions from Eqs. A6 and A8, yields
| (A9) |
We solve this for b to find
| (A10) |
This result is substituted into Eqs. A6 and A8 to yield the Laplace transformed solution for the surface concentration,
| (A11) |
Using the method of partial fractions, Eq. A11 inverse Laplace transforms to yield the surface concentration as a function of time
| (A12) |
| (A13) |
| (A14) |
where c1 and c2 are variables that the partial fraction expansion introduced, but which do not have a clear physical interpretation. This solution applies for all physically reasonable (i.e. non-negative) values of the parameters, κ, k, D, and t. However, it may not be particularly convenient for many parameter choices because the constants c1 and c2 are complex for many reasonable values of κ and k. Note that the solution-phase concentration profile can be found from results presented in section 12.4.III.iii of [12].
Appendix B
This section is very similar to Appendix A. In the partial transmission model system, defined by Eqs. 3 and 5, suppose the concentration is 0 for all x < 0 and is C∞ for all x > 0 at time 0. How much substance is in the x < 0 portion of the system at time t?
We Laplace transform Eqs. 3 and 5 to yield
| (B1) |
| (B2) |
| (B3) |
CB(x,t) and CF(x,t), along with their Laplace transformed versions, are the concentration profiles on the back and front sides of the surface, respectively. Some additional boundary conditions are
| (B4) |
| (B5) |
| (B6) |
| (B7) |
The first two of these boundary conditions simplify the differential equations, Eqs. B1 and B2. They are then solved (see Eq. A6) and simplified further with the latter two boundary conditions to yield
| (B8) |
| (B9) |
In these equations, a and b are constants of integration. These general solutions are combined with the Robin boundary condition in Eq. B3 and simplified to yield solutions for a and b,
| (B10) |
Substitution of Eq. B10 into Eqs. B8 and B9 yields the Laplace transforms of the concentration profiles. The inverse Laplace transform, which would yield the time-dependent concentration profiles appears to be intractable. Instead, we work towards the desired solution by integrating the concentration profile on the back side of the surface to find the total number of transmitted molecules. Its Laplace transform is
| (B11) |
This inverse Laplace transforms to yield the desired solution
| (B12) |
This solution applies to all physically reasonable values of the parameters (κF > 0, κB > 0, t > 0, and D > 0).
Footnotes
Supplementary Information
The Supplementary Information for this article includes (i) details about the simulation emulator, (ii) tables that relate adsorption coefficients to adsorption probabilities for irreversible and reversible adsorption, (iii) details about stochastic simulations used in Figures 5A, 6 and 7, and (iv) C source code that converts between molecule-surface interaction coefficients and their simulation probabilities, and also includes the simulation emulator, and (v) documentation for the source code.
References
- 1.Andrews SS. Serial rebinding of ligands to clustered receptors as exemplified by bacterial chemotaxis. Phys. Biol. 2005;2:111–122. doi: 10.1088/1478-3975/2/2/004. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SS, Addy NJ, Brent R, Arkin AP. Detailed simulation of cell biology with Smoldyn 2.1. PLoS Comput. Biol. 2010 doi: 10.1371/journal.pcbi.1000705. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews SS, Bray D. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys. Biol. 2004;1:137–151. doi: 10.1088/1478-3967/1/3/001. [DOI] [PubMed] [Google Scholar]
- 4.Andrews SS, Dinh T, Arkin AP. Stochastic modeling of biochemical reaction networks. In: Meyers RA, editor. Encyclopedia of Complexity and System Science. Vol. 9. Springer; Heidelberg: 2009. pp. 8730–8749. [Google Scholar]
- 5.Atkins PW. Physical Chemistry. third ed. W.H. Freeman and Co.; New York: 1986. [Google Scholar]
- 6.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartol TMJ, Land BR, Salpeter EE, Salpeter MM. Monte Carlo simulation of miniature endplate current generation in the vertebrate neuromuscular junction. Biophys. J. 1991;59:1290–1307. doi: 10.1016/S0006-3495(91)82344-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bemporad D, Luttmann C, Essex JW. Computer simulation of small molecule permeation across a lipid bilayer: dependence on bilayer properties and solute volume, size, and cross-sectional area. Biophys. J. 2004;87:1–13. doi: 10.1529/biophysj.103.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Tal N, Honig B, Bagdassarian CK, Ben-Shaul A. Association entropy in adsorption processes. Biophys. J. 2000;79:1180–1187. doi: 10.1016/S0006-3495(00)76372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benz R, McLaughlin S. The molecular mechanism of action of the proton ionophore FCCP (carbonylcyanide p-trifluoromethoxyphenylhydrazone) Biophys. J. 1983;41:381–398. doi: 10.1016/S0006-3495(83)84449-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg HC. Random Walks in Biology. 2nd ed. Princeton Univ. Press; Princeton, NJ: 1993. [Google Scholar]
- 12.Carslaw HS, Jaeger JC. Conduction of Heat in Solids. 2nd ed. Clarendon Press; Oxford: 1959. [Google Scholar]
- 13.Chou T, D'Orsogna MR. Multistage adsorption of diffusing macromolecules and viruses. J. Chem. Phys. 2007;127:105101. doi: 10.1063/1.2764053. [DOI] [PubMed] [Google Scholar]
- 14.Coggan JS, et al. Evidence for ectopic neurotransmission at a neuronal synapse. Science. 2005;309:446–451. doi: 10.1126/science.1108239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colman-Lerner A. 2008. Personal communication.
- 16.Crank J. The Mathematics of Diffusion. 2nd ed. Oxford Univ. Press; Oxford: 1975. [Google Scholar]
- 17.Elowitz MB, Surette MG, Wolf P-E, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erban R, Chapman SJ. Reactive boundary conditions for stochastic simulations of reaction-diffusion processes. Phys. Biol. 2007;4:16–28. doi: 10.1088/1478-3975/4/1/003. [DOI] [PubMed] [Google Scholar]
- 19.Gustafson K, Abe T. The third boundary condition – was it Robin's. The Mathematical Intelligencer. 1998;20:63–71. [Google Scholar]
- 20.Hlady V, Buijs J, Jennissen HP. Methods for studying protein adsorption. Methods in Enzymology. 1999;309:402–429. doi: 10.1016/s0076-6879(99)09028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoops S, et al. COPASI – a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 22.Kerr RA, et al. Fast Monte Carlo simulation methods for biological reaction-diffusion systems in solution and on surfaces SIAM. J. Sci. Comput. 2008;30:3126–3149. doi: 10.1137/070692017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr RA, Levine H, Sejnowski TJ, Rappel W-J. Division accuracy in a stochastic model of Min oscillations in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2006;103:347–352. doi: 10.1073/pnas.0505825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kofahl B, Klipp E. Modelling the dynamics of the yeast pheromone pathway. Yeast. 2004;21:831–850. doi: 10.1002/yea.1122. [DOI] [PubMed] [Google Scholar]
- 25.Koh X, Srinivasan B, Ching HS, Levchenko A. A 3D Monte Carlo analysis of the role of dyadic space geometry in spark generation. Biophys. J. 2006;90:1999–2014. doi: 10.1529/biophysj.105.065466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo R. Statistical Mechanics. An Advanced Course with Problems and Solutions. North-Holland; Amsterdam: 1965. [Google Scholar]
- 27.Langmuir I. The constitution and fundamental properties of solids and liquids. I. Solids. J. Am. Chem. Soc. 1916;38:2221–2295. [Google Scholar]
- 28.Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipkow K. Changing cellular location of CheZ predicted by molecular simulations. PLoS Comp. Biol. 2006;2:e39. doi: 10.1371/journal.pcbi.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipkow K, Andrews SS, Bray D. Simulated diffusion of CheYp through the cytoplasm of E. coli. J. Bact. 2005;187:45–53. doi: 10.1128/JB.187.1.45-53.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipkow K, Odde DJ. Model for protein concentration gradients in the cytoplasm. Cellular and Molecular Bioengineering. 2008;1:84–92. doi: 10.1007/s12195-008-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. TRENDS Biotech. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- 33.Paula S, Volkov AG, van Hoek AN, Haines TH, Deamer DW. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 1996;70:339–348. doi: 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plimpton S, Slepoy A. ChemCell: A Particle-based model of protein chemistry and diffusion in microbial cells. Sandia National Laboratory; 2003. [Google Scholar]
- 35.Plimpton SJ, Slepoy A. Microbial cell modeling via reacting diffusive particles. J. Phys.: Conf. Ser. 2005;16:305–309. [Google Scholar]
- 36.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipies in C. The Art of Scientific Computing. Cambridge University Press; Cambridge, UK: 1988. [Google Scholar]
- 37.Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the G-beta-gamma complex underlies activation of the yeast pheromone response pathway. Genes and Development. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao CV, Wolf DM, Arkin AP. Control, exploitation, and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 39.Sanford C, Yip MLK, White C, Parkinson J. Cell++ – simulating biochemical pathways. Bioinformatics. 2006;22:2918–2925. doi: 10.1093/bioinformatics/btl497. [DOI] [PubMed] [Google Scholar]
- 40.Schneider P, Smith JM. Adsorption rate constants from chromatography. AIChE J. 1968;14:762–771. [Google Scholar]
- 41.Singer A, Schuss Z, Osipov A, Holcman D. Partially reflected diffusion SIAM. J. Appl. Math. 2008;68:844–868. [Google Scholar]
- 42.Stiles JR, Bartol TM. Monte Carlo methods for simulating realistic synaptic microphysiology using MCell. In: De Schutter E, editor. Computational Neuroscience: Realistic Modeling for Experimentalists. CRC Press; Boca Raton, FL: 2001. pp. 87–130. [Google Scholar]
- 43.Sundararaj S, et al. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 2004;32:D293–D295. doi: 10.1093/nar/gkh108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfram Research Inc. Mathematica 7.0. Champaign, IL: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.