Abstract
Evidence is presented here that in vivo bacteriophage T3 DNA packaging includes capsid hyper-expansion that is triggered by lengthening of incompletely packaged DNA (ipDNA). This evidence includes observation that some of the longer ipDNAs in T3-infected cells are packaged in ipDNA-containing capsids with hyper-expanded outer shells (HE ipDNA-capsids). In addition, artificially induced hyper-expansion is observed for the outer shell of a DNA-free capsid. Detection and characterization of HE ipDNA-capsids is based on non-denaturing two-dimensional agarose gel electrophoresis, followed by structure determination with electron microscopy and protein identification with SDSPAGE/mass spectrometry. After expulsion from HE ipDNA-capsids, ipDNA forms sharp bands during gel electrophoresis. The hypotheses are presented that (1) T3 has evolved feedback-initiated, ATP-driven capsid contraction/hyper-expansion cycles that accelerate DNA packaging when packaging is slowed by increase in the packaging-resisting force of the ipDNA and (2) each gel electrophoretic ipDNA band reflects a contraction/hyper-expansion cycle.
Keywords: Agarose gel electrophoresis, two-dimensional, Bacteriophage DNA packaging motor, Biological energy transduction, Electron microscopy, Mass spectrometry
Introduction
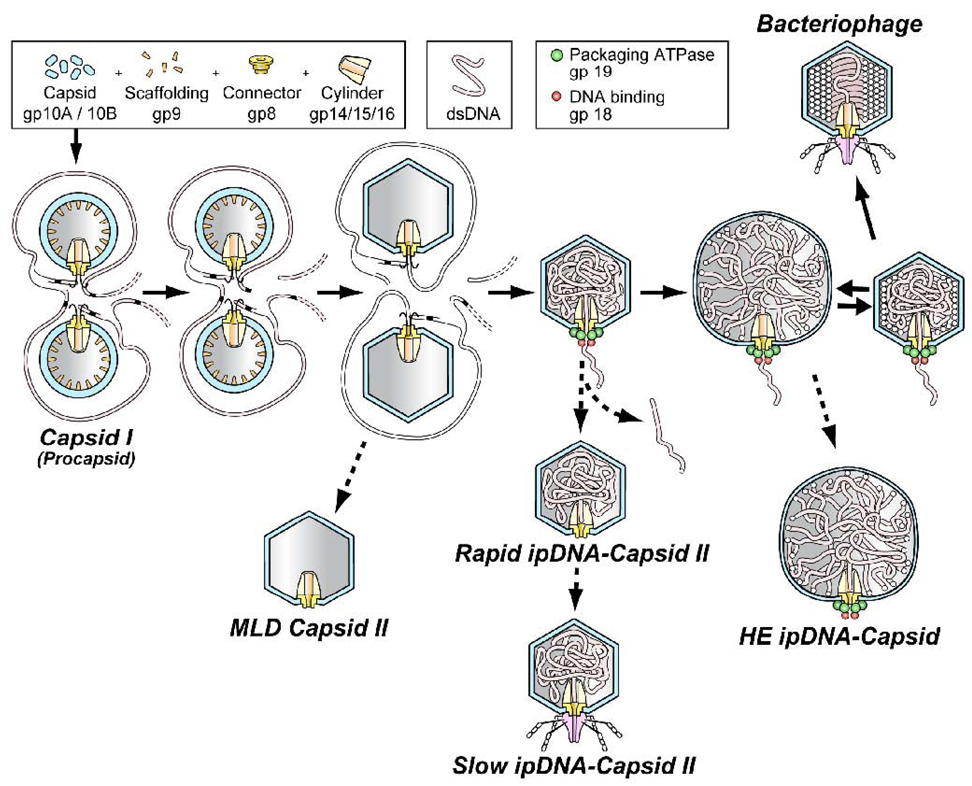
Bacteriophage double-stranded DNA packaging motors produce ATP-fueled DNA motion and are studied because they are comparatively accessible to investigation, among the various nucleoside triphosphate fueled biological motors. A DNA packaging motor drives a double-stranded DNA molecule into a protective protein shell (capsid). The source of energy is ATP cleavage thought to be catalyzed entirely by a DNA packaging ATPase that has been identified for all studied double-stranded DNA bacteriophages.1 Several copies of the ATPase are attached to a ring-shaped oligomer called the connector or portal. The ATPase/connector structure has a central channel through which the DNA molecule enters the capsid. Most entry occurs after the initially assembled capsid (procapsid; also called capsid I for the related bacteriophages, T3 and T7) converts to a more mature capsid, also called capsid II for T3 and T7 (illustrated in Figure 1).2,3 The DNA packaging ATPase is gp19 in the case of T3 and T7 (proteins are labeled by gp, followed by gene number 2,3).
Figure 1.
The DNA packaging pathway of the related bacteriophages, T3 and T7. The pathway indicated by solid arrows is the deduced in vivo DNA packaging pathway. The pathways indicated by the dashed arrows are proposed abortive branches that produce the various particles that have been detected, isolated and characterized.11 A legend for protein components and DNA is at the top. The HE ipDNA-capsid component of the figure is the contribution of the current study.
To understand mechanisms of a biological motor, the most direct strategy is to characterize the motor as it progresses through its various states. Visible light-based single-molecule procedures have recently been used for this purpose in the case of several motors, including F1-ATPase 4,5 and bacteriophage DNA packaging 6,7,8,9 motors. These single-molecule procedures have the advantage that information about the dynamics is not lost via motor asynchrony during analysis. Much of this information is lost via motor asynchrony during use of most (not all) ensemble averaging procedures of analysis. However, thus far, visible light-based single-molecule procedures have the disadvantage that they cannot be used simply and directly (without probes) for analysis of structure and structural dynamics (reference 6, for example, in the case of bacteriophage DNA packaging).
Alternatively, the needed dynamics of motor structure can be obtained by use of a fractionation procedure that retains information about the temporal order of structural transitions. The final analysis is multidimensional in character, in that the contents of each fraction are subsequently either again fractionated or structurally analyzed. Optimally, the first fractionation is based on a motor characteristic that is a measure of the progression of the motor; the second dimension is based on structure-based characteristics of the motor.10 This analysis works comparatively well with bacteriophage DNA packaging motors because fractionation for the first dimension can be based on the length of incompletely packaged DNA (ipDNA).10 To facilitate comparisons, the ratio (F) of ipDNA length to mature DNA length is used here. In practice, buoyant density centrifugation fractionates bacteriophage T3 ipDNA-containing capsids (called ipDNA-capsids) by F. In initial studies, the progression of ipDNA conformations during packaging in capsid II was determined vs. F (F < 0.58) by use of cryo-electron microscopy of particles in fractions of a cesium chloride density gradient.11
To initially detect and characterize ipDNA-capsids after buoyant density centrifugation, the following high-throughput second dimension can be used: one dimensional, non-denaturing agarose gel electrophoresis (1d-AGE) of each fraction of a density gradient. In the past, all T3 ipDNA-capsids detected by 1d-AGE had a capsid indistinguishable from capsid II.11 As recently reviewed1, the assumption has usually been made that the radius of the capsid does not change after a procapsid expands to a bacteriophage-like capsid, e.g., the capsid I to capsid II transition in Figure 1 (constant capsid assumption). The constant capsid assumption is made in most models of DNA packaging motors.1 It is also a fundamental aspect of both analytical 12,13,14 and molecular dynamics simulations 15,16,17 of the conformation and energetics of ipDNA during packaging.
However, the constant capsid assumption is problematic because slowing of packaging near the end is a consequence unless the capsid ruptures.7 Capsid rupture is the explanation that was used for a dramatic, unexpected decrease in the in vitro nanometry determined packaging force when F reached 0.9–1.0 in the case of bacteriophage lambda.7 However, an alternative explanation is that this decrease in force was caused by capsid hyper-expansion; hyper-expansion would better account for the observed preservation of motor activity after the decrease of packaging force.2,18 In addition, capsid hyper-expansion in vivo would increase the chance that packaging is completed before an infected cell either lyses or becomes incompatible with packaging for other reasons, including decrease in ATP concentration. In the case of T3, ATP binding to gp10 (major outer shell protein of the capsid; Figure 1) has been observed 19 and this binding potentially drives changes in capsid size via cleavage of the gp10-bound ATP.
In the present study, the constant capsid hypothesis has been tested by probing for hyper-expanded (HE) ipDNA-capsids in lysates of bacteriophage T3-infected cells. To conduct this study, a procedure was needed for rapidly and sensitively detecting HE ipDNA-capsids. Thus, the present study adds use of a capsid radius detecting, two-dimensional, non-denaturing agarose gel electrophoresis (2d-AGE) and also a more sensitive dye for in-gel detection. The results provide an alternative perspective for understanding data obtained to understand the biochemistry, biophysics and evolution of bacteriophage DNA packaging motors.
Results
Initial fractionation of ipDNA-capsids
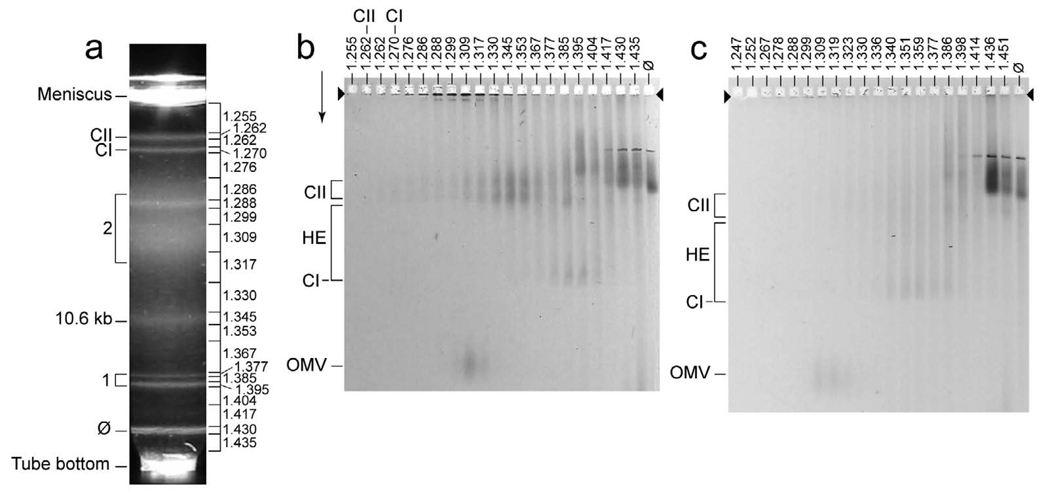
To probe for HE ipDNA-capsids in a T3-infected cell lysate, we began by concentrating and enriching all ipDNA-capsids via centrifugation in a cesium chloride step gradient, followed by buoyant density centrifugation in a cesium chloride density gradient of the entire ipDNA-capsid region of the step gradient.11 Figure 2a has a representative example of the profile of the light scattering from particles fractionated by the buoyant density centrifugation. Zones of light scattering were formed by the following: bacteriophage particles (ϕ), DNA-free capsid I (CI), DNA-free capsid II (CII), an ipDNA-capsid II with an ipDNA length of 10.6 kb (10.6 kb), two capsid-bacteriophage dimers (bracket 1 at the left) and cellular debris (bracket 2 at the left), as previously found.11 Density (g/ml) is indicated at the right in Figure 2a. The length of ipDNA in kb will sometimes be used to label ipDNA-capsids, as done for the 10.6 kb ipDNA in Figure 2a.
Figure 2.
Detection of new ipDNA-capsids. A lysate of wild type T3-infected E. coli was prepared, concentrated and fractionated by centrifugation, as described in the Materials and Methods Section. The same was done with a lysate of T3-LG114-infected E. coli. Then, each fraction of the buoyant density centrifugation was subjected to 1d-AGE with GelStar staining. (a) The light-scattering profile of the buoyant density centrifugation for wild type T3. (b) The profile of the agarose gel electrophoresis for wild type T3. (c) The profile of the agarose gel electrophoresis for T3-LG114. In (a), density (g/ml) of fractions is indicated at the right; in (b) and (c), density (g/ml) is indicated above a lane. The arrowheads indicate the origins of electrophoresis; the arrows indicate the direction of electrophoresis. The significance of other markings is indicated in the text. The contrast of (c) was enhanced more than the contrast of (b) to make more clearly visible the stained particles other than bacteriophages.
Higher sensitivity detection
To improve upon past detection, isolation and characterization of ipDNA-capsids, we made two changes to the previous post-centrifugation analysis by 1d-AGE.11 (1) We used a more sensitive nucleic acid-specific stain, GelStar, instead of the previously used stain, ethidium, to improve detection of ipDNA-capsids after fractionating them in agarose gels. (2) We introduced 2d-AGE (in addition to 1d-AGE), the details of which are discussed below.
The increased fluorescence staining sensitivity revealed new particles, i.e., particles not previously observed, although the highest fluorescence was, as expected, associated with previously observed ipDNA-capsid II. For the 1.262–1.377 g/ml fractions in Figure 2a, 1d-AGE revealed that the ipDNA-capsid II particles formed two bands (CII at the left in Figure 2b); density (g/ml) is indicated at the top of a lane in Figure 2b. The more slowly migrating ipDNA-capsid II particles (slow ipDNA-capsid II) have the bacteriophage tail; the more rapidly migrating ipDNA-capsid II particles (rapid ipDNA-capsid II) do not (see Figure 1).11 In Figure 2b, new particles were observed in the 1.309 g/ml and neighboring samples. The position in the agarose gel of these 1.309 g/ml new particles is indicated by OMV at the left. Additional new particles were observed at 1.345–1.417 g/ml. These latter new particles are seen in Figure 2b between the position of capsid I (indicated at the left by CI) and the position of capsid II (indicated at the left by CII). HE at the left indicates the position of these new particles in the agarose gel. Neither the OMV particles nor the HE particles were observed for the samples from the same gradient fractions when ethidium was the agarose gel stain, because of the lower detection sensitivity of ethidium.
Nucleic acid-free versions of both capsid I and capsid II were also present in the agarose gel of Figure 2b, but were not revealed by the nucleic acid-specific GelStar stain. These two particles were observed via Coomassie staining (not shown) of the gel in Figure 2b, with a peak in the fractions marked CI and CII, respectively, at the top of Figure 2b. CI and CII, respectively, at the left in Figure 2b indicate the positions in the agarose gel.
Our preliminary conjecture was that OMV particles were outer membrane vesicles because the OMV particles formed a band at approximately the position in the agarose gel of previously identified 20 host outer membrane vesicles. We knew that the OMV particles had protein because they stained with Coomassie blue (not shown). The OMV particles were confirmed to be host be outer membrane vesicles by mass spectral analysis of their proteins after trypsin digestion. The most abundant proteins observed were chain A ompF porin, TolB translocation protein, ompA and LppC lipoprotein. As previously described21,22, the first three of these proteins have all been observed in host outer membrane vesicles. Some OMVs are known to have nucleic acid21, as do the OMVs indicated in Figure 2b. The presence of nucleic acid is the likely explanation for a density higher in Figure 2b than the density of OMVs previously observed.20 The OMVs were not further investigated in this study.
HE particles: Analysis by 2d-AGE
Potentially, HE particles were novel ipDNA-capsids and, therefore, we further characterized the HE particles, beginning with 2d-AGE. As illustrated in the diagram of Figure 3a, the 2d-AGE used here fractionates through a comparatively dilute gel in the first dimension (illustrated by the rectangle with diagonal lines in Figure 3a). The primary, but not the only, source of fractionation is electrostatic force that is proportional to the average electrical surface charge density (σ), assuming that particles are accurately modeled by fluid flow-impermeability (hard particles).23,24 The first dimensional gel is dilute enough so that size-dependent sieving effects are small, though not zero, for T3 capsid-sized particles. If particles cannot be modeled by fluid flow impermeability (soft particles), then electrical charges beneath the surface are also a source of electrostatic force during fractionation.23 Here, we use σ to characterize the electrostatic force-derived fractionation because, thus far, all ipDNA-capsids are hard particles, based on independence of electrophoretic mobility from the amount of DNA packaged.11,25
Figure 3.
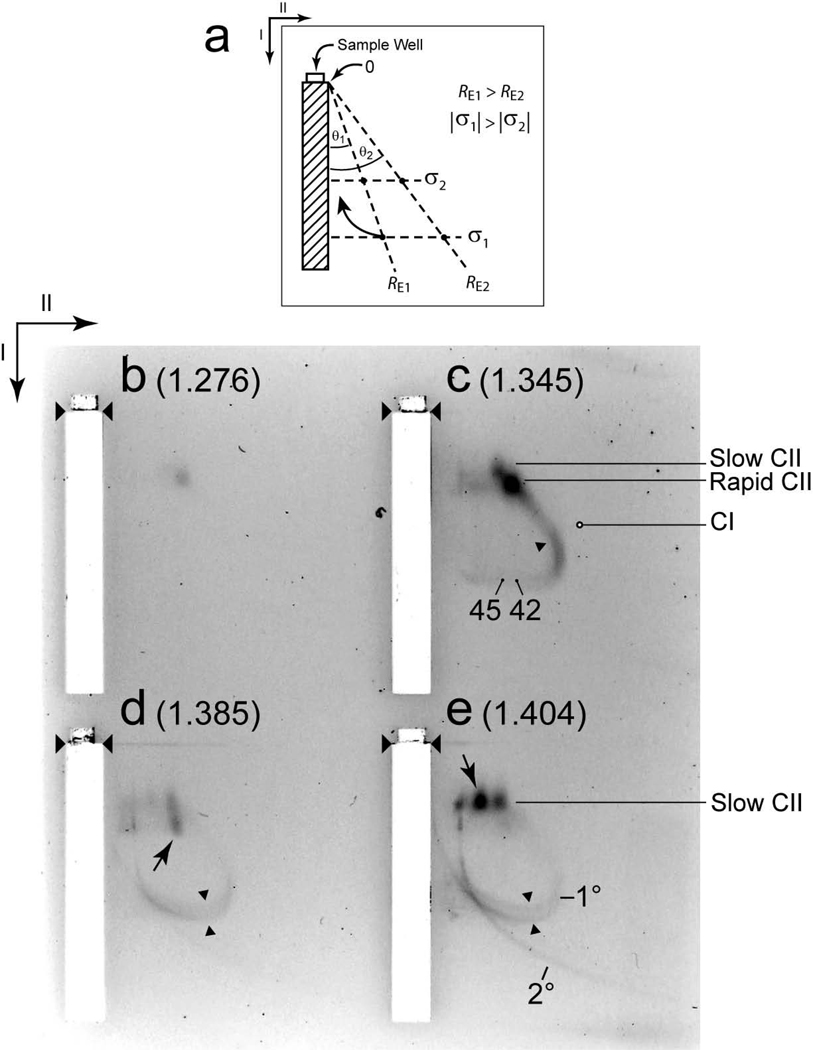
Two-dimensional, non-denaturing agarose gel electrophoresis (2d-AGE). (a) A diagram illustrates 2d-AGE. Wild type samples with the following densities (g/ml) were subjected to 2d-AGE with staining by GelStar: (b) 1.276, (c) 1.345, (d) 1.385, (e) 1.404. The first and second dimension gels were all embedded in one agarose slab. Thus, all four samples were co-fractionated, thereby assuring the same field and temperature among them. The samples come from the gradient in Figure 2a. The arrows indicate the directions of the first (I) and second (II) electrophoresis. The unnumbered arrowheads indicate the origins of electrophoresis. The purposes of both the numbered arrowheads and other markings are described in the text.
In the second dimension, 2d-AGE fractionates by both size (i.e., effective radius, RE) and σ. In the ideal condition of no sieving in the first dimension, the result is conceptualized as a series of straight size lines (dashed, diagonal lines in the second dimension gel drawn in Figure 3a) that extend from the origin of electrophoresis. Each size line connects all points on the gel that have particles of equal RE. The value of RE is a decreasing function of the angle (θ) between a size line and the direction of the first dimension electrophoresis (illustrated in Figure 3a). In the present study, we used a first dimension gel concentration (0.3%) that was high enough to produce significant (about 22% for a particle with a RE of 30 nm) dependence of mobility on RE in the first dimension. The 0.3% gel was more concentrated than the most dilute gel possible (0.075–0.2%) in order to both sharpen patterns and avoid gel breakage that sometimes occurs with more dilute gels. Thus, the size lines are no longer straight and, although change in size is still directly observed via θ (independently of either σ or mass), quantification of RE is accurate only ± 5%.
The use of both 2d-AGE and GelStar staining succeeded in providing additional characterization of HE particles. When the1.345 g/ml fraction was the sample for 2d-AGE, bands were observed at the positions of both slow ipDNA-capsid II (tail-containing; labeled slow CII in Figure 3c) and rapid ipDNA-capsid II (without a tail; labeled rapid CII in Figure 3c), as expected from Figure 2b. No band was seen either at or near the position of capsid I (position indicated by CI in Figure 3c). The remaining stained particles in Figure 3c formed an extension of the ipDNA-capsid II bands. This extension continued in a straight, diagonal line (an indication of no change in RE) until a bend formed in the direction of an increase in RE, (single arrowhead in Figure 3c), thereby forming an arc. The linear extension and the arc-forming particles are the only non-band-forming particles that migrate more rapidly than capsids II in the second dimension and, therefore, are assumed to be the HE particles from the 1d-AGE. As expected from Figure 2b, the HE particles were not observed in a 1.276 g/ml (less dense) fraction, although slow capsid II was observed (Figure 3b).
HE particles were also found in higher density fractions of the same density gradient. The 1.385 g/ml and 1.404 g/ml fractions yielded arcs (1.385 g/ml, Figure 3d; 1.404 g/ml, Figure 3e) that were continuations of the arc of the 1.345 g/ml fraction of Figure 3c. As density increased (and DNA/protein ratio increased, assuming that the HE particles contain only DNA and protein), the average RE also increased. In addition, the arc of Figure 3d appeared to split to two, as indicated by the oppositely pointing two arrowheads, and this splitting became more pronounced in Figure 3e (oppositely pointing two arrowheads). In addition, a second arc, that was comparatively faint in Figure 3d, was clearly observed in Figure 3e. The most intense arc (seen in Figures 3c–3e) will be called the primary arc (indicated by 1° in Figure 3e). The second arc, clearly seen in only Figure 3e, will be called the secondary arc (2° in Figure 3e). The following observation indicates that neither of the two arcs derives curvature from change in electrophoretic mobility with time. When the 2d-AGE was repeated with 1.8% agarose gels used for both the first and the second dimensions, all particles migrating more rapidly than rapid ipDNA-capsid II were on the same straight (diagonal) line, as though the separation was a 1d-AGE (not shown; to be called the 2d-AGE control).
To complete the description of the 2d-AGE patterns, we note that bands (without arcs) were observed for particles larger than capsid II, in both Figure 3d and Figure 3e (arrows). The density gradient fractions that produced these agarose gel bands were centered on the fractions with the light scattering bands indicated by bracket 1 in Figure 2a. Electron microscopy of gradient fractions from another experiment (not shown) has revealed that the positions of the light scattering bands of Figure 2a correspond to the positions of bacteriophage-capsid dimers in the density gradient. These dimers are assumed to form the agarose gel bands marked with arrows in Figures 3d and 3e. The band marked by an arrow in Figure 3d was formed by a capsid I-bacteriophage dimer; the band marked by an arrow in Figure 3e was formed by a capsid II-bacteriophage dimer. These two dimers were not further investigated.
The secondary arc had a shape that has been observed before. Specifically, in the first dimension, the mobility of secondary arc-forming particles monotonically increased and approached a limit, as θ increased. An arc of this shape is expected of particles that all have the same σ, but vary in RE and sometimes have RE large enough to be significantly sieved in the first dimension (i.e., particles with the smaller values of θ).26 The curved arrow in Figure 3a illustrates an arc of this type; examples of this behavior have been previously described.26,27.
For the following reasons, the above results indicate that the secondary arc-forming particles were molecules of DNA that were not packaged in either a protein or any other container. The limiting mobility at high θ (low RE) revealed a σ that was ~ 4.5× the σ of slow capsid II, in agreement with previous observations for unpackaged nucleic acid.25 The GelStar-staining molecule was DNA for both primary and secondary arcs, based on sensitivity to DNase, and not RNase, as described below. Although proteins may have been bound to the DNA molecules of the secondary arc, the σ of the DNA molecules was retained and, therefore, the DNA molecules were not packaged. The source of the secondary arc-associated DNA is likely to be aggregated bacteriophage particles that expelled DNA that was later partially digested by environmental nucleases. In support, bacteriophage-capsid aggregates became more numerous as density increased.11 Although some particles of the secondary arc migrated in the HE region during 1d-AGE (Figure 2b), these particles were not considered to be HE particles and were not investigated further.
The primary arc-forming particles are new and have not previously been reported; we consider them to be unique, in part because of the shape of the primary arc. Specifically, the distance migrated in the first dimension decreased as θ increased in the large-θ range for particles that formed the primary arc. This pattern of migration indicates that the dominant molecule on the surface of the primary arc-forming particles is not DNA because (1) the distance migrated in the first dimension gel is determined primarily by σ and the σ of DNA is invariant in any given solution28 and (2) the σ of DNA is higher (over 1.5×)25 in magnitude than the apparent σ of particles in the primary arc. The comparatively high magnitude of the σ of DNA is illustrated by the position of secondary arc in the high θ region of the gel. Thus, the DNA of primary arc-forming particles is at least partially packaged.
Additional observations from the 1d-AGE and 2d-AGE
The most plausible interpretation of the primary arc in Figures 3c–e is that the primary arc-forming particles are ipDNA-capsids that are conversion products of ipDNA-capsid II and that have not previously been detected because of limitations of methods. Precursor/product relationship of ipDNA-capsid II to primary arc-forming particles is suggested by the fact that the linear extension of the primary arc merges precisely with the bands of ipDNA-capsid II after 2d-AGE (Figure 3c). At the point of merger, the linear extension-forming particles have the RE of capsid II (± 5%) and the σ of rapid capsid II (± 5%). The ipDNA-capsid II would be the precursor because it is the only ipDNA-capsid observed when packaging begins, i.e., at the lower F values (and densities) in Figure 2a,b. If, as this observation suggests, the primary arc-forming particles are ipDNA-capsids that are derived from a functional DNA packaging motor, then their amount should not progressively increase as F increases to 1.0 because they eventually become bacteriophage particles. Therefore, the primary arc-forming particles are expected to have maximal staining intensity at densities lower than the density of bacteriophage particles in Figure 2b. This pattern is, in fact, observed in Figure 2b.
The RE (in nm) along the primary arc in Figure 3c is indicated for RE = 45 and RE = 42. The approach used for determining these numbers is described in the Materials and Methods Section. Most detected primary arc-forming particles are either larger than capsid II or appear larger than capsid II, perhaps because they have a DNA segment protruding from a capsid. The latter possibility also qualitatively explains the variation of σ and RE if the protruding DNA segment varies in length. However, electron microscopy, presented below, indicates that the primary arc-forming particles are, in fact, larger than capsid II.
In summary, the above observations indicate that the primary arc-forming particles are larger than capsid II and have the potential to be HE ipDNA-capsids. Further tests of HE ipDNA-capsid status are in subsequent sections.
Proteins
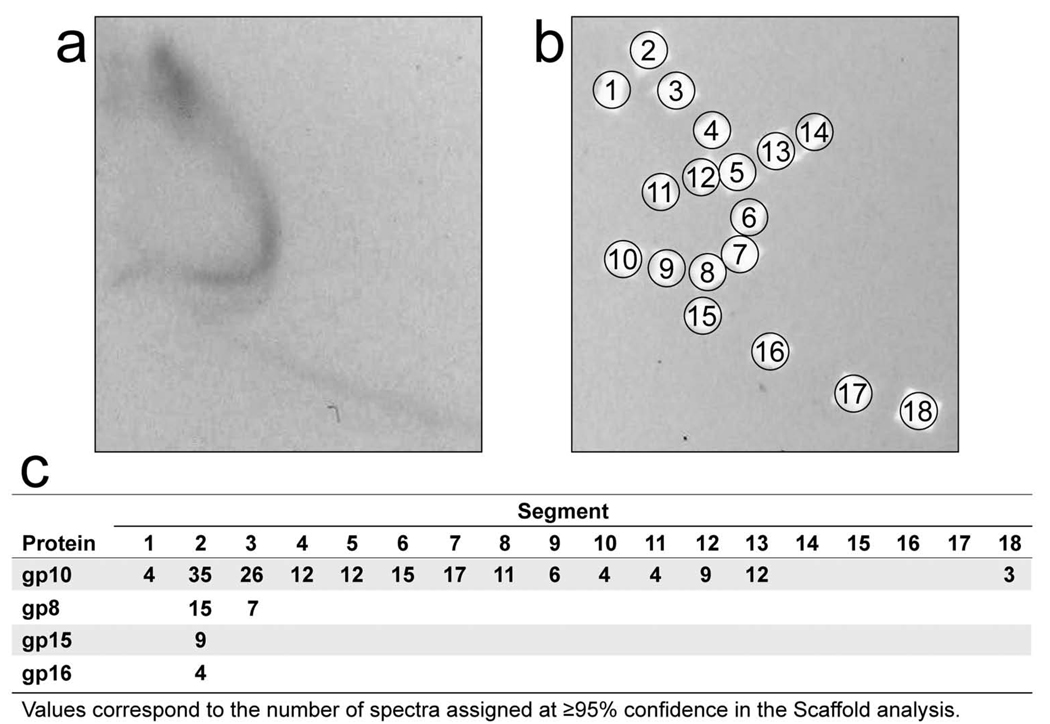
To identify the proteins of primary arc-forming particles, segments of agarose gel were excised after 2d-AGE and their contents were analyzed by SDSPAGE, followed by trypsin digestion and analysis by HPLC/ESI-MS/MS. To reduce the potential for contamination with proteins from ipDNA-capsid II, the source of the particles was a T3 mutant (described and discussed, below) that, for reasons not known, produced a comparatively small amount of slow and rapid ipDNA-capsid II (Figure 2c). The 2d-AGE shown in Figure 4a is for a 1.340 g/ml fraction. The result from database searching of the tandem mass spectra indicated that the major T3 capsid protein, gp10, was detected in all gel segments tested from the primary arc; no host proteins (and, therefore, no OMVs) were detected in any of these gel segments. The absence of host OMVs from the primary arc is supported by the higher σ of the OMVs and the absence of detected variation in σ for OMVs (no sub-arcs).20 For each gel segment (positions indicated by numbered circles in Figure 4b), a measure of the relative quantity of each protein was obtained by comparison of the number of mass spectra assigned with > 95% confidence (Figure 4c). In the segments with the highest protein content (gel segments 2 and 3), minor T3 capsid proteins were also detected in addition to gp10, including gp8, gp15 and gp16 (see Figure 1 for other information about these proteins). The quantities of protein for primary arc-forming particles were too low to detect T3 capsid proteins other than gp10. The primary arc-forming particles were present in less than 1 per 10 T3-infected cells. The gp10 was concentrated in the arc-containing region of the gel, as seen by analysis of gel segments from background (non-arc) regions in Figure 4b (Figure 4c). Thus, the arc is formed by a capsid of some type.
Figure 4.
Analysis of proteins. (a) A sample of a cesium chloride density gradient fractionation of a T3-LG114 lysate (density = 1.340 g/ml) was subjected to 2d-AGE with staining by GelStar; the first dimension gel is not shown. (b) The gel in (a) was then dissected and analyzed by SDS-PAGE, followed by trypsin digestion and mass spectrometry. (c) Spectrum counts of T3 gp10 are displayed as a function of both gel segment numbers (columns) and gp number (rows).
Further analysis supported the conclusion that the primary arc is formed by HE ipDNA-capsids. The only known alternatives to ipDNA-capsids are (1) DNA-containing aberrant capsids, such as the tubular polycapsids observed for T729 and (2) aggregates of either ipDNA-capsids or bacteriophages and capsids. As shown above and in reference 11, bacteriophage-capsid aggregates were present in the fractions analyzed by 2d-AGE; polycapsids were seen by electron microscopy of these fractions (not shown). However, polycapsids have an outer shell like that of capsid II and are, therefore, expected to have an invariable σ equal to that the σ of capsid II. Thus, alternative-1 is contradicted by the data (Figure 3) that show a σ that is both higher in magnitude than that of capsid II and variable. Capsid multimers (alternative-2) form discrete bands30, as illustrated by the dimer bands in Figures 3d,e (arrows), if they enter an agarose gel. Thus, the primary arc, which is continuous, is not formed by capsid multimers. Thus, among the known possibilities, we are left with the conclusion that the primary arc is formed by HE ipDNA-capsids.
Structure-based evidence
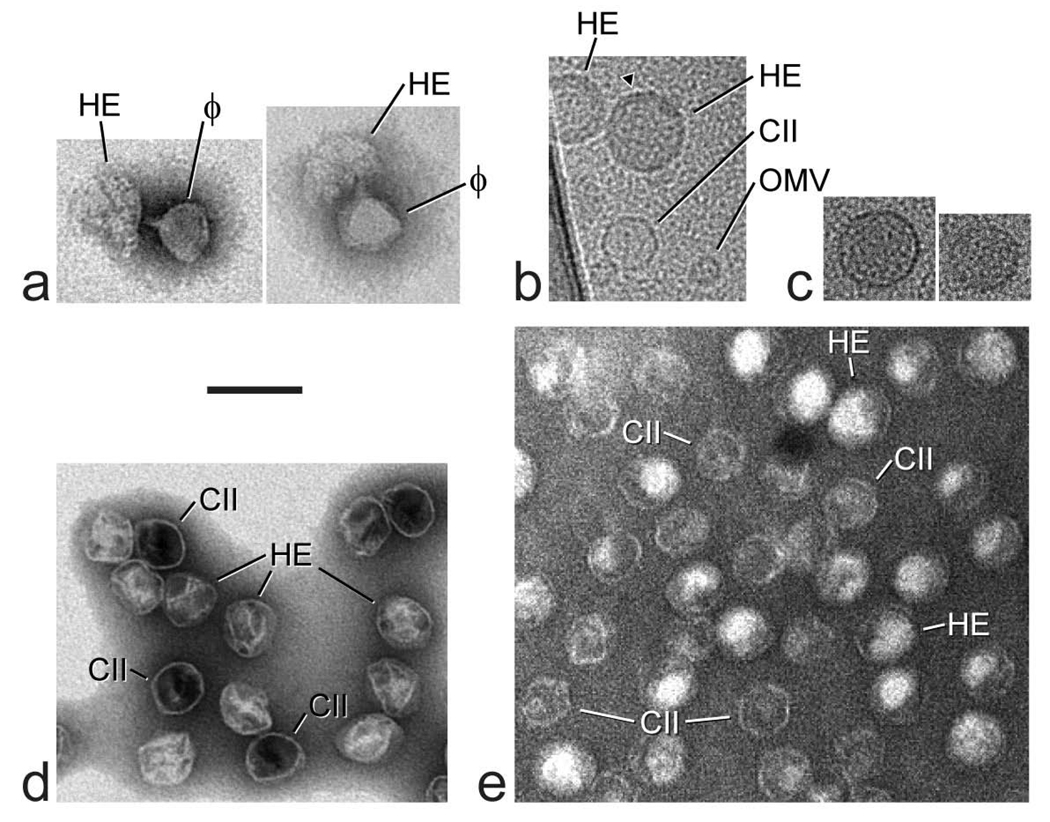
Electron microscopy of the density gradient fractions (like those in Figure 2a) with HE intensity after 1d-AGE (Figure 2b) confirmed the existence of “large” HE capsid-like particles that are never observed in samples of purified DNA-free capsid II. Negatively stained specimens taken from a 1.394 g/ml fraction had collapsed particles, some of which were associated with a bacteriophage particle to form a dimer. The collapsed particle of a dimer retained enough of its original structure to see that it was larger than a bacteriophage particle (HE particles in Figure 5a). Although the bacteriophage particle of the two dimers in Figure 5a had its tail pointing toward the HE particle, the tail did not always point in this direction. Unlike capsid II, the subunits within the HE particles were separated by a space large enough to see the subunits. The visible separation of subunits after negative staining is probably caused by the process of particle disintegration and distinguishes the HE particles from OMVs, polycapsids and, indeed, any other particle observed in T3 lysates. In addition, OMVs and polycapsids have stain-penetrated central cavities not seen in the disintegrating HE particles.
Figure 5.
Electron microscopy. (a) Electron microscopy of negatively stained dimers that consist of one T3 HE particle (labeled HE) and one T3 bacteriophage particle (labeled ϕ) in a 1.394 g/ml fraction of a density gradient like the one in Figure 2a. (b) Cryo-EM of a 1.340 g/ml fraction of a density gradient like the one in Figure 2a. (c) Cryo-EM of two particles that were eluted from an agarose gel like the one in Figure 2b. The density of sample for the agarose gel was 1.395 g/ml; the distance migrated in the agarose gel was 1.7× the distance migrated by rapid ipDNA-capsid II. (d) Electron microscopy of particles of undialyzed T3 MLD capsid II that had been negatively stained while still in the Metrizamide solution in which they had been previously purified. (e) Electron microscopy of particles of undialyzed T7 MLD capsid II that had been negatively stained while still in the Metrizamide solution in which they had been previously purified.
Cryo-EM revealed large HE capsid-like particles in more intact form. HE capsid-like particles were observed in a 1.340 g/ml fraction of the T3 mutant discussed below (marked HE in Figure 5b), together with particles of capsid II (identified by comparison with previous images of purified capsid II; marked CII in Figure 5b) and smaller, apparent OMVs (marked OMV in Figure 5b). The HE capsid-like particles sometimes had a DNA-like fiber next to the inner surface of the outer shell (arrowhead in Figure 5b). This fiber is presumably the primary arc DNA detected by GelStar staining after 2d-AGE and did not usually coat the entire inner surface of the outer shell. The apparent OMV particles were assumed to be contaminating OMVs that were not sufficiently stained with GelStar to be seen in Figure 2b.
To investigate the apparent HE ipDNA-capsids at higher levels of purity, cryo-EM was performed of 1.395 g/ml particles eluted by multi-day soaking of agarose gel segments from the HE region after 1d-AGE (Materials and Methods Section). Over 40 fields were imaged at random and two particles were found (38 fields had no particles). Both particles observed by cryo-EM were HE ipDNA-capsid-like spheres, larger than capsid II (Figure 5c). The agarose gel fractionation had removed both the OMVs and the capsid II particles. The radii of the HE ipDNA-capsids in Figures 5b and 5c are 1.2–1.4× as large as the radius of capsid II. More detailed analysis of structure, including 3-D reconstruction, requires the production and isolation of ipDNA-capsids in amount at least two orders of magnitude larger than the amounts obtained here.
Artificial capsid hyper-expansion
If ipDNA-capsid II particles undergo conversion to HE ipDNA-capsids in vivo, then one should be able to mimic this conversion by artificially providing the energy for hyper-expansion of capsid II. This was achieved with MLD (Metrizamide low density) capsid II, a DNA-free capsid II isolated via its low density during fractionation by buoyant density centrifugation in a Metrizamide density gradient. The low density is caused by impermeability to Metrizamide, a characteristic that suggests the possibility of providing energy for capsid expansion by transiently lowering capsid permeability and allowing entry of molecules that subsequently generate expansion-promoting osmotic pressure. Both T3- and T7-infected cells produce MLD capsid II, a particle that has an outer shell that is uniform in size and structure.11, 31 When negative staining was performed without dialyzing the Metrizamide present in a solution of MLD capsid II, the result was the apparent hyper-expansion of 20–50% of the MLD capsid II particles in the case of both T3 (Figure 5d) and T7 (Figure 5e). Some apparent hyper-expanded particles are indicated by the symbol, HE. The interior of these particles also had lowered electron density (less negative stain) as though they had internal content that the other, smaller particles did not have. The results for T7 were more dramatic in that the T7 hyper-expanded MLD capsid II was about 20% larger than the original, in contrast to about 10% larger for T3, on average.
Based on the following evidence, the apparent hyper-expanded particles in Figure 5d and 5e are true hyper-expanded MLD capsid II particles that are not produced only by flattening during specimen preparation. (1) At least in the case of T7, the smaller particles of Figure 5d are already flattened to an extent that mathematically precludes enough further flattening to cause the apparent increase in size without breaking the capsid’s outer shell32 and the outer shells are not broken based on the retention of internal contents. (2) Apparent hyper-expanded MLD capsid II particles are intermixed with the smaller MLD capsid II particles, all of which were presumably exposed to the same surface tension during specimen drying. Surface tension is the cause of the flattening. Furthermore, the apparent hyper-expanded MLD capsid II also had outer shells that generally appeared thinner than the outer shells of the more traditional MLD capsid II particles.
We propose the following to explain the MLD capsid II hyper-expansion in Figures 5d and 5e. (1) At the beginning of specimen preparation (Methods Section), adsorption of MLD capsid II particles to a carbon support film transiently opened a hole in MLD capsid II and allowed diffusion of Metrizamide (present in the specimen) into MLD capsid II particles. (2) The hole closed and trapped some Metrizamide in MLD capsid II particles. (3) Next, washing during specimen preparation removed the Metrizamide outside of the capsid, but left Metrizamide inside of the capsid because of the hole closing. (4) The higher Metrizamide concentration inside of the capsid produced an outward (expansionary) osmotic force on Metrizamide-loaded capsids. (5) This force expanded the capsid’s outer shell. In support of this idea, the HE particles of Figures 5d and 5e appear to have internal material, presumably Metrizamide, which is not present in the other, smaller particles.
In contrast to the low permeability of MLD capsid II, the HE ipDNA-capsids of Figure 2–Figure 4 have at least one hole that is not present in most ipDNA-capsid II particles. This conclusion is based on the result of DNase I digesting HE-ipDNA-capsids in fractions with densities of 1.353 – 1.396 g/ml, as described in the Methods Section. The result was the complete elimination of both the secondary arc (as expected) and the primary arc. The DNase digestion did not alter the GelStar staining of ~70% of the ipDNA-capsid II particles, although the GelStar staining of the remaining ipDNA-capsid II particles was lost (data not shown). No change in the gel profile was caused by RNase A digestion (not shown).
The ipDNA of HE ipDNA-capsids
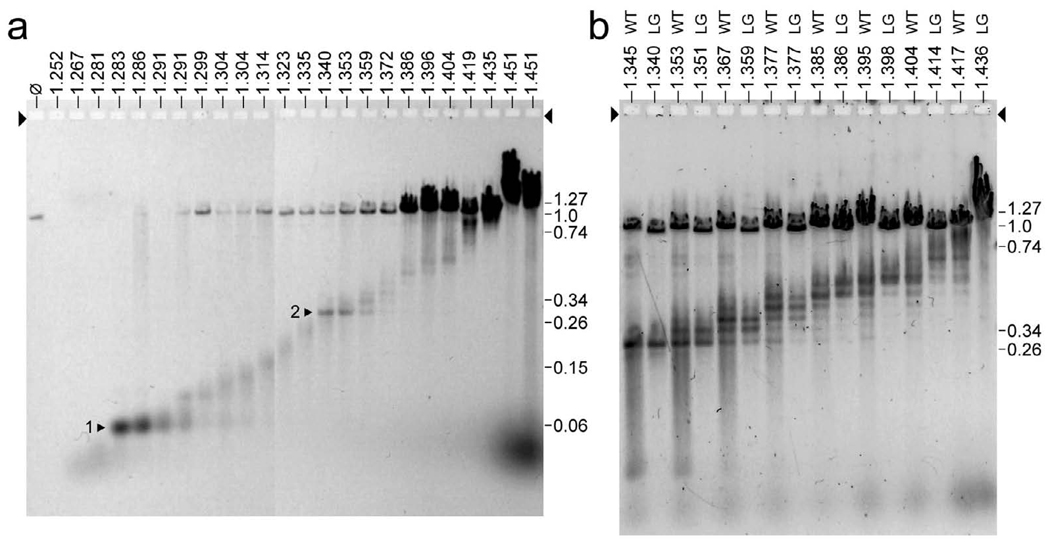
Surprisingly, most ipDNAs from fractions with T3 HE ipDNA-capsids had quantized lengths, based on the results of expelling ipDNAs from their capsids and then gel electrophoretically separating the ipDNAs. The gel electrophoretic conditions were optimized for resolution of linear DNA molecules with lengths between 1 and 40 kb.11 The first result was the finding, as expected, that the length of the ipDNA increased (distance from the origin decreased) as the density of the fraction increased [Figure 6a; density (g/ml) is indicated at the top of each lane]. With one exception, the ipDNAs at the lower densities (density < 1.340 g/ml; F < 0.28) had a distribution of lengths that was continuous, i.e., without formation of bands any sharper than what is expected from the density fractionation that had been performed before analysis of ipDNA. The exception was ipDNA from a fraction with a density of 1.283–1.286 g/ ml, which formed a dominant, sharp band at 2.3 kb, indicating that production of an ipDNA-capsid was more efficient at this ipDNA length than it was for either shorter or longer ipDNAs (arrowhead 1 in Figure 6a). A second dominant, sharp ipDNA band occurred at a density of 1.340 g/ml and an ipDNA length of 10.6 kb (arrowhead 2 in Figure 6). The value of F is indicated at the right in Figure 6a, as determined from co-fractionated DNA length standards that are not shown.
Figure 6.
The ipDNAs. After a fractionation like the fractionation in Figure 2a, DNA was expelled from the ipDNA-capsids in each fraction. Then, the ipDNAs were separated by gel electrophoresis. (a) ipDNAs from an entire density gradient of wild type T3, with the density of a fraction indicated at the top (g/ml), (b) ipDNAs from the 1.345–1.436 g/ml region of the density gradients of both wild type T3 (lanes marked WT) and the T3-LG114 mutant (lanes marked LG), with the density of a fraction indicated at the top. An arrow indicates the direction of electrophoresis; arrowheads indicate the origins of electrophoresis. In order to have enough lanes, results from two ethidium stained gels run in parallel are combined in (a); the gel in (b) was stained with GelStar.
As sample density increased beyond 1.340 g/ml, additional sharp ipDNA bands were visible in Figure 6a, although with intensity lower than the intensity of the 10.6 kb band. The spacing between ipDNA bands was ~1.100 kb for densities between 1.335 and 1.396 g/ml, and progressively increased to 3.1 kb as the density increased to 1.419 g/ml. Mature length T3 DNA (F = 1.0), derived from bacteriophage-capsid aggregates, was present in some fractions and increased in quantity as density increased. The 1.340 g/ml fraction (F = 0.28) was the low-density border for the presence of both HE ipDNA-capsids and, within experimental error, all sharp bands formed by ipDNA longer than 2.3 kb. This joint border suggests the possibility that the longer-than-2.3 kb, sharp band-forming ipDNAs were expelled from HE ipDNA-capsids, not from ipDNA-capsid II.
This conclusion was supported by the results of repeating the experiment of Figure 2a, 2b and Figure 6a with T3 deletion mutant LG114 (Materials and Methods). We found that, in relation to the staining intensity of HE ipDNA-capsids, the staining intensity of ipDNA-capsid II was reduced by over 90% (Figure 2c). As expected, the amount of ipDNA between 2.3 and 10.6 kb Figure 6a was reduced by over 90% relative to the amount of the band-forming ipDNAs with length either equal to or greater than 10.6 kb (F = 0.28; not shown). Besides the HE ipDNA-capsids, ipDNA-capsid II is the only other detected possible source of the longer-than-2.3 kb, sharp band-forming ipDNAs. However, the deficiency of ipDNA-capsid II in the T3-LG114 lysate means that ipDNA-capsid II cannot be the primary source. The HE ipDNA-capsids are the only known possible source for most of the longer-than-2.3 kb, sharp band-forming ipDNAs in the case of the T3-LG114 lysate.
A test of the relationship between the wild type and mutant band-forming ipDNAs can be seen in Figure 6b where ipDNA lengths in the F > 0.28 region of a cesium chloride density gradient are compared. Sample density (g/ml) and source [either wild type (WT) or LG114 (LG)] are shown above each lane in Figure 6b. The ipDNAs for T3-WT and T3-LG-114 are the same length (±1 %), based on band alignment when a LG lane is compared to a WT lane of comparable density in Figure 6b. As expected, the mature length T3-LG114 DNA migrates more rapidly than the mature T3-LG114 DNA in Figure 6b. The alignment of the T3-WT and T3-LG-114 sharp band-forming ipDNAs indicates that the mechanism for producing these ipDNAs is the same for the wild type lysate as it is for the mutant lysate. A hypothesis for the biochemistry of sharp band-forming ipDNA production is presented below.
Discussion
The research presented here was based on leads provided by both 1d-AGE and 2d-AGE. These leads were followed by rigorous characterization by both mass spectrometry and cryo-EM. However, we have not yet explained the split in some regions of the primary arc (oppositely pointing two arrowheads in Figure 3d, e). A possible explanation arises from the conclusion drawn here that particles of ipDNA-capsid II are likely to convert to HE ipDNA-capsids in vivo. Since some of the ipDNA-capsid II particles have a tail (slow ipDNA-capsid II) and others do not (rapid ipDNA-capsid II), some hyper-expanded ipDNA-capsids should have a tail and others should not. Thus, a possible explanation of the split in the primary arc is that HE ipDNA-capsids of the more origin-distal primary arc branch do not have a tail and HE ipDNA-capsids of the more origin-proximal primary arc branch do have a tail. Testing of this explanation requires the isolation of HE ipDNA-capsids in greater amount.
The unusual shape of the primary arc has additional points of interest. At the higher values of θ along the arc and its linear extension in Figure 3,c-e, the variation of σ has two possible explanations. (1) The particles involved are hard and have an outer shell that increases its surface exposure of negatively charged amino acids as RE increases. This change can be caused by movement of peptide segments to and from the capsid surface, a phenomenon already described for the procapsid-to-mature capsid transition of bacteriophage T4.33 Similarly, T3 and T7 capsid I each has a negative σ value higher in magnitude than that of the corresponding capsid II, even though the surface protein of both capsid I and capsid II is gp10.31 (2) An alternative possible explanation is that the primary arc-forming particles become soft particles during expansion and, therefore, the packaged, internalized DNA contributes progressively more to the electrophoretic mobility. Either explanation can be correct, according to current data.
At the lower values of θ along the primary arc in Figure 3, c–e, the magnitude of σ appears to decrease as θ decreases, as most dramatically seen in Figure 3e. However, sieving in the first dimension is so large for these comparatively large particles that the apparent decrease in the magnitude of σ might, in fact, be primarily the result of an increase in sieving in the first dimension. Discriminating effects of σ from effects of RE at these low θ’s (high RE’s) is not possible with the current experiments.
Role and significance of HE ipDNA-capsids
The data presented here are best interpreted by the assumptions that the HE ipDNA-capsids were produced (1) during DNA packaging that would have produced an infective bacteriophage if packaging had not been interrupted and (2) by a DNA packaging motor that was in a hyper-expanded state during packaging. The alternative to (1) is that the HE ipDNA-capsids were generated during packaging that had become abortive before the infection was terminated by lysis. The alternative to (2) is that HE ipDNA-capsids were generated not during DNA packaging, but after either productive or abortive packaging, during cellular lysis, for example. In the case of either alternative, the hyper-expansion would probably not be informative for understanding DNA packaging. However, hyper-expansion while maintaining capsid integrity is so complicated that the most reasonable assumption is that hyper-expansion is evolutionarily selected. That is to say, the HE ipDNA-capsids represent states of the DNA packaging motor during productive DNA packaging. This conclusion is supported by the observation that the HE ipDNA-capsids do not accumulate at the highest F values (less than one).
The following are implications of both a hyper-expanded capsid state during T3 DNA packaging and a quantization of T3 DNA packaging, the latter as observed via sharp ipDNA bands in Figure 6. First of all, in the case of at least T3, the constant capsid assumption can be strictly made only for F values between 0.0 and 0.28; the accuracy of this assumption decreases as F increases. Accounting for changes in capsid size would produce more useful analytical and computer simulation-based prediction of packaged ipDNA conformations and energetics. Second, a single DNA packaging event is the product of motors of more than one type, given that hyper-expansion occurs only for the higher F values. Furthermore, we speculate that the bands marked by arrows 1 and 2 in Figure 2b are comparatively intense because a transition occurred from one motor type to another. That is to say, such transitions are accompanied by increased probability of DNA cleavage during packaging. A more detailed hypothesis is presented in the next section.
If packaging motors of more than one type function during a single DNA packaging event, one questions whether the ATP cleaved per base pair packaged is constant throughout packaging. Assumption of constant ATP utilization per base pair packaged (constant ATP assumption) is implicit in almost all models for bacteriophage DNA packaging motors (recently reviewed in reference 1). Nanometry has detected progression of in vitro bacteriophage ϕ29 DNA packaging in equal, quantized steps34, an observation that can be interpreted by making the constant ATP assumption, although ATP consumption was not monitored during nanometry. However, packaging in equal, quantized steps can also occur if the constant ATP assumption is incorrect, for example, via ATP cleavage-derived potential energy stored within the connector. In addition, in vitro packaging used for nanometry may, in some cases, occur without the feedback, discussed below, necessary for capsid hyper-expansion. Empirically, one ATP molecule has been found cleaved, on average, per two base pairs packaged in purified in vitro systems for ϕ2935 and T3.36 This number is about 6× higher for λ.37 However, these numbers were obtained in experiments that average ATP usage for all F values. No data exist, to the authors’ knowledge, that measure the dependence of the ATP usage on F. Given the highly evolved character of bacteriophage DNA packaging motors and a very high dependence of force-of-packaging on F7, 8, 9, the constant ATP assumption is questionable even without the data presented here.
Details of the T3 DNA packaging motor
The existence of HE ipDNA-capsids implies that some energy transduction occurs via the outer shell of the capsid. For example, if RE increases from 30 nm to 43 nm at the end of packaging, F decreases from 1.0 to 0.33 and the force-of-packaging decreases from ~110 pN to less than 7 pN, based on nanometry of in vitro ϕ29 packaging.38 Thus, ATP cleavage-driven hyper-expansion would transfer to the outer shell some, if not most, of the work that the gp19 DNA packaging ATPase is usually assumed to do. The major T3 outer shell protein, gp10, does bind ATP.19 Packaging in a protein outer shell at DNA density below that of T3 is reasonably assumed to have evolved before a complex DNA packaging motor and accompanying high forces needed for packaging. Adaptation of the outer shell for ATP fueled energy transduction is simpler than production of an entirely new protein and its assembly on the connector. Thus, the use of only the outer shell for energy transduction may have predated the use of DNA packaging ATPases and may not have been retained for all bacteriophages. If so, the time involved is before the prokaryote/eukaryote splits, because Herpes viruses have DNA packaging ATPases that have detectable similarity to those of double-stranded DNA bacteriophages.39
Capsid hyper-expansion as F increases implies a signal that triggers hyper-expansion. Signaling during DNA packaging has previously been demonstrated for filling-of-the-capsid (headful) triggering of the second of two DNA cleavages that are used by some bacteriophages to produce a mature genome from a concatemeric precursor40, 41. In the case of bacteriophages P2241 and SPP142, detailed cryo-EM analysis of the connector (homolog of gp8 in Figure 1) indicates that the connector is part of the signaling pathway.
A previously presented hypothesis18 goes further in proposing that the DNA packaging motor engages in signal-driven, multiple contraction/expansion (including contraction/hyper-expansion) cycles in the later stages of DNA packaging. By this hypothesis, the contraction/expansion cycles are coupled with cycles of permeability change that, together, cause non-DNA molecules to be expelled from the capsid. In reference 18, the capsid of the contracted state was proposed to be smaller than the mature capsid. In the present study, an ipDNA-capsid smaller than ipDNA-capsid II was not detected. No other direct evidence for cycling of hyper-expansion was found.
However, several observations are best, but not uniquely, explained by the hypothesis that some T3 HE ipDNA-capsids are produced during a contraction/expansion cycle that includes terminase-derived premature DNA cleavage that produces the observed T3 ipDNAs. A key observation is that slowing of in vitro T3 DNA packaging stimulates the terminase-induced second cleavage associated with the packaging of T3 concatemers.43 Slowing of packaging after each contraction/expansion cycle might, therefore, with low probability, signal premature terminase-induced DNA cleavage that produces the ipDNA bands of Figure 6.
The following additional observations support the idea that the ipDNAs observed here are, in fact, produced by premature terminase cleavage. (1) Some ipDNA-capsid II has a tail, which is not added until after a terminase cut is made.44 (2) The sharp ipDNA bands of Figure 6 are not produced during in vitro T3 DNA packaging in unfractionated T3-infected cell extracts that have all T3 gene products and are used for high efficiency (10–20%) packaging. Instead a broad, more continuous distribution of ipDNAs was observed (P. Serwer and S. J. Hayes, unpublished observation). The absence of the sharp bands of Figure 6 in vitro indicates that no restriction endonuclease-like activity is present in the T3-infected cell extracts and suggests that the presence of these bands depends on the influence on DNA packaging of cellular conditions that are not mimicked in vitro. Terminase cleavage is the only known alternative for producing the band-forming ipDNAs.
Future integration of genetics and ipDNA-capsid characterization
Greater production of the various known ipDNA-capsids is needed to have sufficient amounts to determine structure with high-resolution 3-D reconstructions. One can presumably reach this goal by either selecting or screening for T3 mutants that accumulate the known ipDNA-capsids in amount higher than achieved here with wild type T3. The mutants obtained might also help to reveal other aspects of the DNA packaging motor, possibly through accumulating ipDNA-capsids that have not yet been detected because of low amount. Previous searches for DNA packaging-altered mutants 37, 44–47 have been disadvantaged by the absence of such a screenable, packaging-related phenotype. The work presented here suggests that non-denaturing agarose gel electrophoresis can be used for screening for HE ipDNA-capsid accumulation in lysates of cells infected with T3 mutants.
Materials and Methods
Production and initial fractionation of lysates of bacteriophage-infected cells
Bacteriophage-related particles were prepared and concentrated by the following procedure. Lysates of bacteriophage-infected Escherichia coli BB/1 were prepared after infection at a multiplicity of 0.01, by use of procedures previously described.11 The medium was 2×LB broth: 20 g Bacto tryptone, 10 g yeast extract, 5 g NaCl in 1 liter of water. After addition of NaCl to a lysate (final concentration of 0.5 M), debris was immediately removed from the lysed culture by pelleting in six 1.0 liter bottles at 4,000 rpm, for 15 min. in a Beckman J-6B centrifuge (JS4.2 rotor). The lysate was then brought to 9% polyethylene glycol 8,000 by the addition of solid polyethylene glycol 8,000. The capsids, ipDNA-capsids and bacteriophage particles were precipitated by leaving this mixture at 4 °C for 2–4 days. These procedures were used for wild type bacteriophages T3 and T7 and a T3 deletion mutant LG114 (received from Dr. Ian Molineux), a mutant that deleted parts of genes 0.3 (SAMase) and 0.7 (protein kinase) in the left half of T3 DNA.49
The concentrates were then prepared for buoyant density-based fractionation. The supernatant of the polyethylene glycol precipitate was decanted and the precipitate was collected by centrifugation at 4,000 rpm for 45 min in the Beckman JS4.2 rotor. Precipitated particles were resuspended in 7 ml per liter of lysate of resuspension buffer: 0.5 M NaCl, 0.01 M Tris-Cl, pH 7.4, 0.001 M MgCl2. The resuspended lysate was clarified by centrifugation for 10 min. at 10,000 rpm, 4 °C in a Beckman Avanti J-25 centrifuge (JLA16.250 rotor). The supernatant and two resuspension buffer washes of the pellet were combined and brought to a volume of 160 ml in resuspension buffer. Then, the resuspended particles were again brought to 9% polyethylene glycol 8,000 and precipitated at 4 °C, this time for ~ 16 hr. After resuspension (final volume = ~17 ml) and clarification, concentrated particles were then digested with DNase I (2.5 µg/ml) mg/ml, final concentration) for 60 min at 30 °C. This concentration of DNase reduces the viscosity, but does not completely degrade DNA.
Density-based fractionation was then performed. First, the above mixture was fractionated by centrifugation through a cesium chloride step density gradient (5.6 ml per gradient; 6 gradients).11 Particles in the entire ipDNA-capsid region of the step gradient were further purified by clarification, followed by buoyant density centrifugation in a cesium chloride density gradient, by use of procedures previously described.11 Identical fractions of three step gradients were pooled for the buoyant density centrifugation. Light scattering from particles in the density gradient was photographed and the gradient was collected by pipeting from the top. To prepare MLD capsid II from either T3 or T7, the particles in the CII band (see Figure 2a) were subjected to buoyant density centrifugation in a Metrizamide density gradient.31
Nondenaturing agarose gel electrophoresis
The ipDNA-capsids were initially detected by use of 1d-AGE. A sample of a density gradient fraction was added to a 0.35× amount of the following buffer (electrophoresis buffer), without dialysis: 0.09 M Tris-acetate, pH 8.4, 0.001 M MgCl2. A 0.11× amount of 50% sucrose in electrophoresis buffer was then added. The fractions were then layered in the wells of a horizontal, submerged agarose slab gel. The gel had been cast in and submerged under electrophoresis buffer. The layered fractions were allowed to remain in place for 60 min pre-electrophoresis in order to dialyze enough cesium chloride out of the sample so that increased ionic strength at the origin did not interfere with the initial movement of particles during electrophoresis. Electrophoresis was then conducted at 2.0 V/cm, 25 °C for 10 hr. After electrophoresis, the gel was stained for DNA with either 1 µg/ml ethidium bromide (final concentration) or 1/10,000 diluted GelStar (Lonza, catalogue #50535) in electrophoresis buffer, visualized with an ultraviolet trans-illuminator and photographed. The gel was then stained for protein with Coomassie blue.11
Some particles in density gradients were also characterized in more detail by use of 2d-AGE. The sample preparation for 2d-AGE was the same as it was for 1d-AGE, except that time in the sample well was increased for enzyme digestions (below) and 100 µg/ml bovine serum albumen was added to reduce adherence to the gel origin. The first dimensional gel was 0.3% agarose (Seakem LE, Lonza, Rockland Maine) that was embedded within a 1.8% second dimensional Seakem LE agarose gel, submerged beneath electrophoresis buffer and cast in an apparatus in which the direction of electrophoresis is changed by 90° without manipulating the gel.26,27 The first dimensional electrophoresis was performed at 2.0 V/cm for 4.0 hr. The second dimensional electrophoresis was performed at 1.8 V/cm for 10.0 hr, after changing the direction of electrophoresis by 90°. Extrapolations by use of previously described procedures50 were made to determine RE values in Figure 3c. Tests of enzyme susceptibility were performed by adding to the sample either (1) 200 µg/ml (final concentration) DNase I (Worthington) or (2) 100 µg/ml (final concentration) boiled RNase A (Sigma). A control without enzyme was also run. After layering in the sample well, the time of incubation before starting the first dimensional electrophoresis was raised to 1.5 hr in order to promote digestion by the added enzymes, after the in-sample well dialysis of the cesium chloride.
Gel electrophoresis of DNA
The lengths of ipDNAs were measured by gel electrophoresis of DNA expelled from a capsid in which the DNA had been packaged. To expel DNA from capsids, the following was sequentially added to 20 µl of a fraction from a density gradient: 5 µl of 0.1 M NaCl, 0.01 M Tris-Cl, pH 7.4, 0.001 M EDTA and then 5 µl of 30% sucrose, 0.6 M NaCl, 0.06 M Tris-Cl, pH 7.4, 0.06 M EDTA, 6% Sarkosyl. Next, the temperature was raised to 85 °C for 10 min. The DNA was then subjected to electrophoresis through a 0.25 % horizontal agarose gel (Seakem Gold agarose, Lonza) cast in and submerged under the following buffer: 0.05 M sodium phosphate, pH 7.4, 0.001 M EDTA. The electrophoresis was conducted at 0.34 V/cm for 26 hr at 20 °C. These conditions were chosen for high resolution in the 5–40 kb range.
SDS-PAGE and Mass spectrometry
To determine the proteins in GelStar-stained particles fractionated by 2d-AGE, we began by removing segments of the agarose gel. The locations were selected by viewing the gel while it was illuminated by an ultraviolet trans-illuminator. Proteins in each agarose gel segment were prepared for SDS-PAGE by soaking in the following buffer (three changes) for 1.5 hr and then boiling for 3 min: 20.5% glycerol, 4.8% SDS, 0.23 M Tris-Cl, pH 6.8, 0.008 M EDTA, 4.8% β- mercaptoethanol. The boiled mixture was maintained at 70 °C to avoid re-gelation of the agarose and was then loaded for SDS-PAGE with pipets pre-warmed to 37 °C. Proteins were fractionated by SDS-PAGE (12%, Precast Criterion XT, Bio-Rad) for a time sufficient to run the tracking dye (bromphenol blue) about 2.5 cm into the separating polyacrylamide gel. The gels were stained with Coomassie blue and the protein-containing region of each lane was sliced into four segments, each of which was digested in situ with trypsin (Promega modified) in 40 mM NH4HCO3 overnight at 37 °C. The digests for each gel lane were combined and analyzed by capillary HPLC-electrospray ionization-tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher LTQ mass spectrometer as previously described.11 The uninterpreted collision-induced dissociation spectra were searched against the NCBInr database using Mascot (Matrix Science; London, UK). Methionine oxidation was considered as a variable modification for all searches. Cross correlation of the Mascot results with X! Tandem and determination of protein identity probabilities were accomplished by Scaffold (Proteome Software).
Electron microscopy
Particle size and shape were characterized by electron microscopy. Negatively stained specimens were made by (1) placing a drop of specimen on a specially prepared25 carbon film and allowing particles to adhere to the film for 2–3 min, (2) washing the film with 3 drops of water, (3) negative staining with 3 drops of 1.0 % sodium phosphotungstate, pH 7.6 and (4) wicking dry with filter paper. The ipDNA-capsids examined were obtained from a fraction of a cesium chloride density gradient, such as the gradient in Figure 2a; i.e., no dialysis was performed. T7 MLD capsid II was also not dialyzed after preparation by buoyant density centrifugation in a Metrizamide density gradient. Negatively stained specimens were observed in either a Philips 208S or a JEOL100CX electron microscope in The Department of Pathology at The University of Texas Health Science Center at San Antonio.
Cryo-EM was also performed to minimize specimen preparation-induced changes. The procedures have been previously described.11 Samples were taken either directly from a cesium chloride density gradient, such as the one in Figure 2b, or from a segment of an agarose gel, such as the gels in Figures 2b and 2c. For extraction of particles from a segment of agarose gel, the segment was placed in a roughly equal volume of saturated cesium chloride in 0.01 M Tris-Cl, pH 7.4, 0.001 M MgCl2. The contents of the gel were allowed to diffuse into solution for at least 4 days at 4 °C. Cesium chloride solution was used for extraction because of the previously demonstrated stabilizing effect of cesium chloride solutions on ipDNA-capsid II particles.11 After elution, the liquid phase was shipped at ambient temperature from San Antonio to Purdue for cryo-EM.11
Acknowledgements
This work was supported by grants from the National Institutes of Health to P.S. (GM24365 and GM069757) and W.J. (AI072035) and from the Welch Foundation (AQ-764) and The Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, to P.S. The cryo-EM images were taken in the Purdue Biological Electron Microscopy Facility. Protein identifications were conducted in the UTHSCSA Institutional Mass Spectrometry Laboratory.
Glossary
- ipDNA-capsid
A capsid that contains an incompletely packaged molecule of DNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Philip Serwer, Email: serwer@uthscsa.edu.
Elena T. Wright, Email: wrighte@uthscsa.edu.
Kevin Hakala, Email: hakala@uthscsa.edu.
Susan T. Weintraub, Email: weintraub@uthscsa.edu.
Min Su, Email: su5@purdue.edu.
Wen Jiang, Email: jiang12@purdue.edu.
References
- 1.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 2.Serwer P. T3/T7 DNA packaging. In: Catalano CE, editor. Viral genome packaging machines: genetics, structure, and mechanism. Georgetown, TX: Landes Publishing; 2004. pp. 59–79. [Google Scholar]
- 3.Fujisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 4.Okuno D, Fujisawa R, Iino R, Hirono-Hara Y, Imamura H, Noji H. Correlation between the conformational states of F1-ATPase as determined from its crystal structure and single-molecule rotation. Proc Natl Acad Sci USA. 2008;105:20722–20727. doi: 10.1073/pnas.0805828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masaike T, Koyama-Horibe F, Oiwa K, Yoshida M, Nishizaka T. Cooperative three-step motions in catalytic subunits of F(1)-ATPase correlate with 80 degrees and 40 degrees substep rotations. Nature Struct Mol Biol. 2008;15:1326–1333. doi: 10.1038/nsmb.1510. [DOI] [PubMed] [Google Scholar]
- 6.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage ϕ29 capsids. Plos Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage ϕ29. Proc Natl Acad Sci, USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 10.Serwer P. Analysis of biological motors via multidimensional fractionation: a strategy. Electrophoresis. 2005;26:494–499. doi: 10.1002/elps.200406166. [DOI] [PubMed] [Google Scholar]
- 11.Fang PA, Wright ET, Weintraub ST, Hakala K, Wu W, Serwer P, Jiang W. Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J Mol Biol. 2008;384:1384–1399. doi: 10.1016/j.jmb.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci U S A. 2001;98:13671–13274. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrov AS, Harvey SC. Structural and thermodynamic principles of viral packaging. Structure. 2007;15:21–27. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spakowitz AJ, Wang ZG. DNA packaging in bacteriophage: is twist important? Biophys J. 2005;88:3912–3923. doi: 10.1529/biophysj.104.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serwer P. Models of bacteriophage DNA packaging motors. J Struct Biol. 2003;141:179–188. doi: 10.1016/s1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 19.Hamada K, Fujisawa H, Minagawa T. Characterization of ATPase activity of a defined in vitro system for packaging of bacteriophage T3 DNA. Virology. 1987;159:244–249. doi: 10.1016/0042-6822(87)90461-2. [DOI] [PubMed] [Google Scholar]
- 20.Serwer P, Wright ET, Hakala KW, Weintraub ST. Evidence for bacteriophage T7 tail extension during DNA injection. BMC Res Notes. 2008;1:36. doi: 10.1186/1756-0500-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiner JH, Li L. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim. Biophys. Acta. 2008;1778:1698–1713. doi: 10.1016/j.bbamem.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, Kim KP, Gho YS. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 23.Langlet J, Gaboriaud F, Gantzer C, Duval JF. Impact of chemical and structural anisotropy on the electrophoretic mobility of spherical soft multilayer particles: the case of bacteriophage MS2. Biophys J. 2008;94:3293–3312. doi: 10.1529/biophysj.107.115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw DJ. Electrophoresis. London: Academc Press; 1972. pp. 4–36. [Google Scholar]
- 25.Serwer P, Pichler ME. Electrophoresis of bacteriophage T7 and T7 capsids in agarose gels. J Virol. 1978;28:917–928. doi: 10.1128/jvi.28.3.917-928.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serwer P, Griess GA. Advances in the separation of bacteriophages and related particles. J Chromatogr B, Biomed Sci Appl. 1999;722:179–190. doi: 10.1016/s0378-4347(98)00404-6. [DOI] [PubMed] [Google Scholar]
- 27.Tietz D. Computer-assisted 2-D agarose electrophoresis of Haemophilus influenzae type B meningitis vaccines and analysis of polydisperse particle populations in the size range of viruses: a review. Electrophoresis. 2007;28:512–524. doi: 10.1002/elps.200600532. [DOI] [PubMed] [Google Scholar]
- 28.Allison S, Chen C, Stigter D. The length dependence of translational diffusion, free solution electrophoretic mobility, and electrophoretic tether force of rigid rod-like model duplex DNA. Biophys J. 2001;81:2558–2568. doi: 10.1016/S0006-3495(01)75900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serwer P. Fibrous projections form the core of a bacteriophage T7 procapsid. J Supramol Struct. 1979;11:321–326. doi: 10.1002/jss.400110307. [DOI] [PubMed] [Google Scholar]
- 30.Serwer P. A technique for electrophoresis in multiple-concentration agarose gels. Anal Biochem. 1980;101:154–159. doi: 10.1016/0003-2697(80)90054-8. [DOI] [PubMed] [Google Scholar]
- 31.Serwer P, Watson RH, Hayes SJ, Allen JL. Comparison of the physical properties and assembly pathways of the related bacteriophages T7, T3 and ϕII. J Mol Biol. 1983;170:447–469. doi: 10.1016/s0022-2836(83)80157-0. [DOI] [PubMed] [Google Scholar]
- 32.Serwer P. Flattening and shrinkage of bacteriophage T7 after preparation for electron microscopy by negative staining. J Ultrastruct Res. 1977;58:245–243. doi: 10.1016/s0022-5320(77)90015-6. [DOI] [PubMed] [Google Scholar]
- 33.Steven AC, Bauer AC, Bisher ME, Robey FA, Black LW. The maturation-dependent conformational change of phage T4 capsid involves the translocation of specific epitopes between the inner and the outer capsid surfaces. J Struct Biol. 1991;106:221–236. doi: 10.1016/1047-8477(91)90072-5. [DOI] [PubMed] [Google Scholar]
- 34.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–451. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 36.Hamada K, Fujisawa H, Minagawa T. Characterization of ATPase activity of a defined in vitro system for packaging of bacteriophage T3 DNA. Virology. 1987;159:244–249. doi: 10.1016/0042-6822(87)90461-2. [DOI] [PubMed] [Google Scholar]
- 37.Dhar A, Feiss M. Bacteriophage lambda terminase: alterations of the high-affinity ATPase affect viral DNA packaging. J Mol Biol. 2005;347:71–80. doi: 10.1016/j.jmb.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 38.Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage ϕ29. Biophys J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serwer P, Hayes SJ, Zaman S, Lieman K, Rolando M, Hardies SC. Improved isolation of undersampled bacteriophages: finding of distant terminase genes. Virology. 2004;329:412–424. doi: 10.1016/j.virol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- 41.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 42.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujisawa H, Kimura M, Hashimoto C. In vitro cleavage of the concatemer joint of bacteriophage T3 DNA. Virology. 1990;174:26–34. doi: 10.1016/0042-6822(90)90050-2. [DOI] [PubMed] [Google Scholar]
- 44.Catalano CE. Viral genome packaging machines: an overview. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Georgetown, TX: Landes Bioscience; 2005. pp. 1–4. [Google Scholar]
- 45.Duffy C, Feiss M. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 46.Morita M, Tasaka M, Fujisawa H. Analysis of functional domains of the packaging proteins of bacteriophage T3 by site-directed mutagenesis. J Mol Biol. 1994;235:248–259. doi: 10.1016/s0022-2836(05)80031-2. [DOI] [PubMed] [Google Scholar]
- 47.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J Mol Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 48.Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 49.Studier FW, Movva NR. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J Virol. 1976;19:136–135. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griess GA, Moreno ET, Easom RA, Serwer P. The sieving of spheres during agarose gel electrophoresis: Quantitation and modeling. Biopolymers. 1989;28:1475–1484. doi: 10.1002/bip.360280811. [DOI] [PubMed] [Google Scholar]