Abstract
Many genes are toxic when overexpressed, but general mechanisms for this toxicity have proven elusive. Vavouri et al. (2009) find that intrinsic protein disorder and promiscuous molecular interactions are strong determinants of dosage sensitivity, explaining in part the toxicity of dosage-sensitive oncogenes in mice and humans.
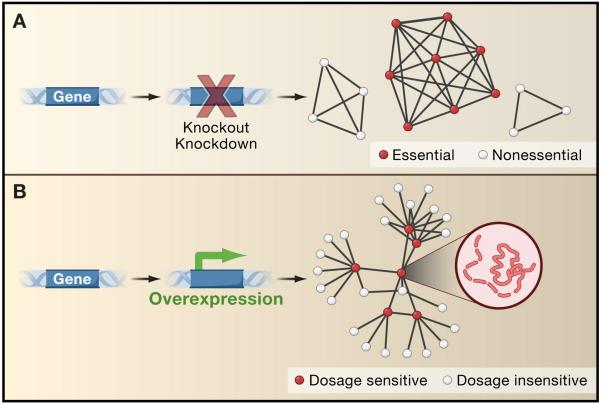
In yeast, approx. 15-20% of genes are toxic when overexpressed (Gelperin et al., 2005; Sopko et al., 2006), a phenomenon known as dosage sensitivity. A similar fraction of genes are known to be essential for growth in standard laboratory conditions, and can lead to cell death upon deletion or down-regulation (Giaever et al., 2002). Too much expression of a gene is not the same as too little, and it is not surprising that the mechanisms of toxicity appear quite different. What has been surprising is that a general understanding is emerging for the causes of cell death stemming from loss-of-function, but not from overexpression. In the case of loss-of-function, essential genes tend to preferentially interact with each other, both as hubs in gene networks and as components of essential protein complexes (Figure 1). If any one component of an essential complex is deleted, the whole system fails. This trend explains a large fraction of the cases in which reducing or abolishing expression of a particular gene results in lethality (Hart et al., 2007; Wang et al., 2009). In contrast, dosage sensitivity has thus far eluded any such general explanation. One is now provided, at least in part, by Vavouri et al. in this issue (Vavouri et al., 2009).
Figure 1. Too much gene expression is not the same as too little.
(A) Genes that lead to cell death or growth defects upon knockout or knockdown tend to preferentially associate with each other in the same protein complexes, shown here by representing proteins as circles and protein-protein associations by lines forming a network of protein interactions. Complexes of essential proteins tend to contain more protein constituents than nonessential complexes. These trends explain a large fraction (>30%) of the essential genes in yeast cells (Hart et al., 2007; Wang et al., 2009).
(B) In contrast to genes that are essential upon knockout or knockdown, a different mechanism appears to underlie the toxicity observed upon gene overexpression (Vavouri et al., 2009). Such dosage sensitive genes tend to participate in larger numbers of direct pairwise protein interactions, shown here with dosage sensitive genes forming hubs in a network of yeast two-hybrid protein interactions. Dosage sensitive genes also tend to be intrinsically disordered, as illustrated by the dashed line for the protein structure pictured in the inset. Overexpression of an intrinsically disordered protein may increase its already high degree of interaction promiscuity, leading to toxic partnerships and detrimental effects on the cell.
This notable lack of a general mechanism for dosage sensitivity has been a source of considerable frustration given its contribution to many diseases, including a wide variety of amyloid, prion, and other diseases of protein misfolding and aggregation. For example, one of the first mouse models to develop the predominant features of Alzheimer’s disease entails overexpressing a variant of the human amyloid precursor protein (Games et al., 1995). Overexpression of dosage sensitive oncogenes also leads to a variety of human cancers. For example, it has recently been shown that overexpression (up to 100-fold) of the angiotensin II receptor type I enables a highly invasive phenotype in mammalian epithelial cells, thereby defining a subpopulation of breast cancers (10-20%) that might respond to treatment with angiotensin receptor antagonists (Rhodes et al., 2009). A better notion of why some genes are toxic when overexpressed and others are not might therefore help in understanding mechanisms of disease progression and even in designing new therapeutics.
In order to search for general mechanisms of dosage sensitivity, Vavouri and colleagues tested a variety of likely factors that failed to provide a reliable prediction, including protein abundance, aggregation potential, half-life, and the degree to which codon usage is optimized for protein translation (Vavouri et al., 2009). Instead the strongest predictors of dosage sensitivity are intrinsic protein disorder and the tendency to participate directly in large numbers of pairwise protein-protein interactions. The latter is especially predictive when either partner contains functional motifs completely contained in a single linear stretch of amino acids. These factors do not explain all (or even most) of the trend—this is clearly only part of the story. Nonetheless these factors provide successful predictions of dosage sensitive genes in yeast and metazoans, to the extent that a computer algorithm trained to identify such proteins correctly predicts the dosage toxicity of 6 of 8 genes tested in worms.
One interpretation, offered by Vavouri et al., is that intrinsically disordered proteins are more likely to participate in promiscuous protein interactions when overexpressed, simply as a consequence of mass action. This explanation draws support from two intriguing sets of contrasting observations: First, dosage sensitive genes show higher numbers of direct protein interactions (measured by the yeast two-hybrid assay) than dosage insensitive genes, but interestingly do not share more interaction partners in protein complexes (as measured by affinity purification or co-immunoprecipitation). This is in strong contrast to genes essential upon knockout or knockdown (Figure 1) and suggests that there is something special about the excess participation specifically in pairwise interactions. Conceivably, the yeast two-hybrid assay might preferentially detect interactions that result from non-native expression of highly interactive (or even just sticky) proteins; the assay requires constitutive nuclear expression of proteins whose native expression is neither constitutive nor nuclear, and in fact, hub proteins in yeast two-hybrid interaction networks are known to exhibit increased disorder (Haynes et al., 2006). Nonetheless, the signal for dosage sensitivity appears stronger than this trend alone. Second, dosage sensitive proteins show increased intrinsic disorder, but not increased tendencies to self-aggregate, at least as predicted computationally. Thus, dosage sensitivity does not appear to be predominantly driven by self-aggregation, although it is possible that this trend might be overturned with direct experimental data. Nonetheless, such aggregation would still be consistent with the notion of mass-action driven interaction promiscuity being a major determinant of dosage sensitivity.
This model in which increased dosage drives toxic interactions by mass action—tested in yeast, flies, and worms—might also explain a fraction of dosage sensitive oncogenes in humans and mice. These cases abound in the literature, especially for oncogenic transcription factors, such as the gene Oct-4. Oct-4 is a homeobox gene responsible for maintaining pluripotency in embryonic stem cells. Strikingly, as the expression level of Oct-4 is varied from 0% to 150% relative abundance, the incidence of tumors formed following injection into syngeneic mice varies from 4% to 64-83%, an effect possibly stemming from promiscuous activation of growth factors (Gidekel et al., 2003). Vavouri et al. systematically analyze a set of dosage-sensitive oncogenes. They show that genes known to be amplified and causally linked to cancer tend to encode proteins that are enriched with intrinsically-disordered segments, contain or bind linear functional motifs, and make many direct pairwise physical interactions with other proteins. Thus, at least a substantial portion of these genes are consistent with the model.
Intrinsic disorder and protein-protein interaction promiscuity account only for a portion of dosage sensitivity cases. This begs the question of what accounts for the rest of the cases? A test of the dosage sensitive yeast genes assembled by Vavouri et al. shows that the 839 dosage sensitive genes are strongly statistically enriched for proteins with transcription factor and DNA binding activities (p ≤ 10−10; hypergeometric probability), with 110 of the proteins involved in some aspect of transcriptional regulation. The set is enriched for many other regulatory functions as well, including 50 genes known to participate in phosphorylation, dephosphorylation, or autophosphorylation, and 30 in RNA transport. Thus, the notion of mass action-driven protein interaction promiscuity can probably be generalized to mass action-driven promiscuity of many types of macromolecular interactions, including transcription factor-target interactions, kinase/phosphatase-target interactions, and so on. Vavouri et al. point out a prime candidate for overexpression-induced promiscuity leading to toxicity, overexpression of microRNAs, which might lead to many off-target gene regulatory interactions. Therefore, although the current data primarily implicate promiscuous protein interactions and disordered proteins, a broader exploration of regulatory promiscuity may yet reveal mechanisms of dosage sensitivity with even greater predictive value
REFERENCES
- Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, Carr T, Clemens J, Donaldson T, Gillespie F, et al. Nature. 1995;373:523–527. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gidekel S, Pizov G, Bergman Y, Pikarsky E. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Hart GT, Lee I, Marcotte ER. BMC Bioinformatics. 2007;8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. PLoS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Ateeq B, Cao Q, Tomlins S, Mehra R, Laxman B, Kalyana-Sundaram S, Lonigro R, Helgeson B, Bhojani M, et al. Proc Natl Acad Sci U S A PNAS. 2009 doi: 10.1073/pnas.0900351106. early edition, published online June 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple J, Garci-Verdugo R, Lehner B. Cell. 2009;(this issue) doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Wang H, Kakaradov B, Collins SR, Karotki L, Fiedler D, Shales M, Shokat KM, Walther T, Krogan NJ, Koller D. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]