One of the most promising new chemotherapeutic strategies is the RNA interference (RNAi)-based approach, wherein small double-stranded RNA molecules can sequence-specifically inhibit the expression of targeted oncogenes.[1] In principle, this method has high specificity and broad applicability for chemotherapy. For example, the small interfering RNA (siRNA) strategy enables manipulation of key oncogenes that modulate signaling pathways and thereby regulate the behavior of malignant tumor cells. To harness the full potential of this approach, the prime requirements are to deliver the siRNA molecules with high selectivity and efficiency into tumor cells and to monitor both siRNA delivery and the resulting knock-down effects at the single cell level. Although several approaches such as polymer- and nanomaterial-based methods[2] have been attempted, limited success has been achieved for delivering siRNA into the target tumor cells. Moreover, these types of approaches mainly focus on the enhancement of transfection efficiency, knock-down of non-oncogenes (e.g. green fluorescent protein (GFP)), and the use of different nanomaterials such as quantum dots (QDs), iron oxide nanoparticles, and gold nanoparticles.[3,4] Therefore, to narrow the gap between current nanomaterial-based siRNA delivery and chemotherapies, there is a clear need to develop methods for target-oriented delivery of siRNA [5], for further monitoring the effects of siRNA-mediated target gene silencing via molecular imaging probes[4], and for investigating the corresponding up/down regulation of signaling cascades.[6] Perhaps most importantly, to begin the development of the necessary treatment modalities, the nanomaterial-based siRNA delivery strategies must be demonstrated on oncogenes involved in cancer pathogenesis.
Herein, we describe the synthesis and target-specific delivery of multifunctional siRNA-QD constructs for selectively inhibiting the expression of epidermal growth factor receptor variant III (EGFRvIII) in target human U87 glioblastoma cells, and subsequently monitoring the resulting down-regulated signaling pathway with high efficiency.[7]
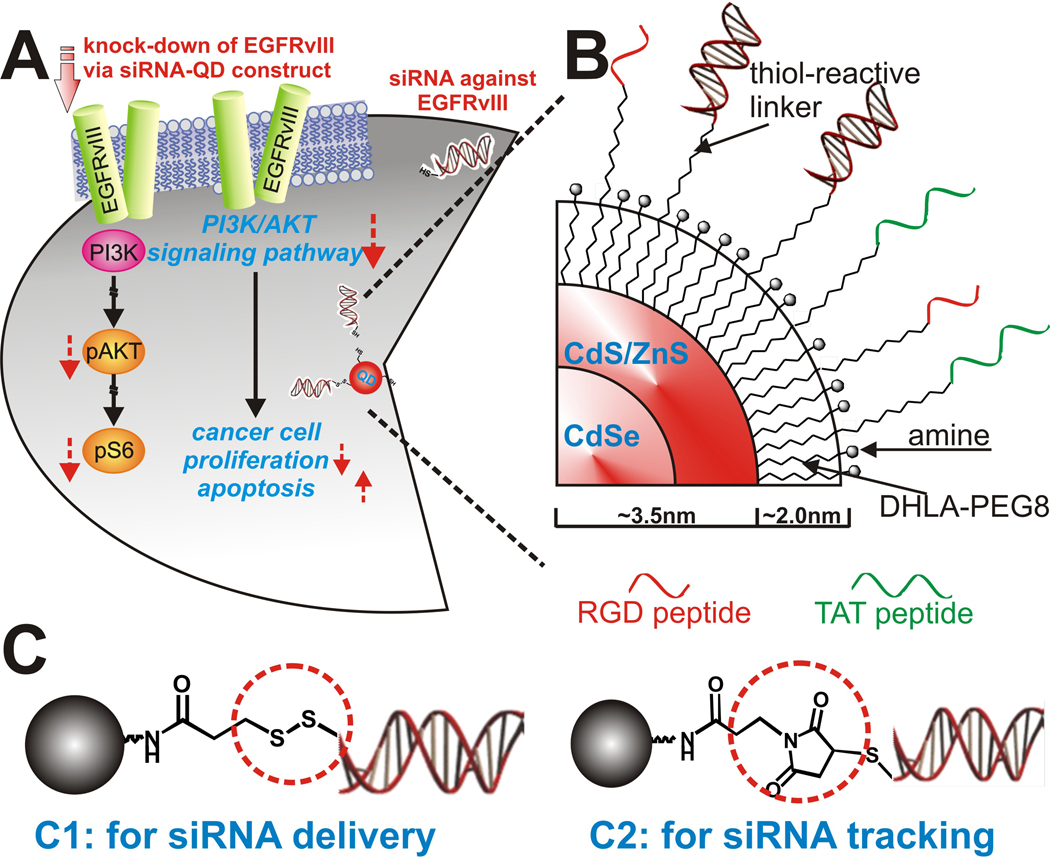
Glioblastoma multiforme (GBM) is the most malignant, invasive, and difficult-to-treat primary brain tumor. Successful treatment of GBM is rare with a mean survival of only 10–12 months.[8] EGFRvIII, the key growth factor receptor triggering cancer cell proliferation in many cancer diseases such as brain tumors and breast cancer, is a constitutively active mutant of EGFR which is expressed in only human GBM and several other malignant cancers, but not in normal healthy cells (Figure 1A).[9] We targeted EGFRvIII, since it has been known that knock-down of this gene is one of the most effective ways to down-regulate the PI3K/Akt signaling pathway, a key signal cascade for cancer cell proliferation and apoptosis. [6,10] Hence by targeting EGFRvIII, our multi-functional nanoparticle-based siRNA delivery strategy could potentially minimize the side effects caused by conventional chemotherapies, specifically immune suppression, while significantly improving the efficacy of chemotherapy against GBM.
Figure 1.
(A): Quantum dots as a multi-functional nanoplatform to deliver siRNA and to elucidate of EGFRvIII- knockdown effect of PI3K signaling pathway in U87-EGFRvIII (B): Detailed structural information of multifunctional siRNA-QDs (C): Two different strategies for the siRNA-QD conjugate. (C1) Linker for attaching siRNA to QDs through a disulfide linkage which was easily reduced within the cells to release the siRNA. (C2) Linker for covalently conjugating siRNA to QDs which enabled tracking of siRNA-QDs within the cells.
We prepared two types of siRNA-QD conjugates, one for siRNA delivery and the other for siRNA tracking (Figure 1B and C). Core-shell CdSe/CdS/ZnS QDs with a 7 nm diameter were synthesized.[11] The QDs were coated with trioctyl-phosphine oxide (TOPO) or hexadecylamine (HDA). In order to make the QD constructs water-soluble and suitable for conjugating with siRNA, we displaced these hydrophobic ligands with a dihydrolipoic acid (DHLA) derivatized with an amine terminated poly [ethylene glycol] (PEG) spacer. The expectation was that the dithiol moiety would provide strong coordination to the QD surface and increase stability in aqueous media, the PEG spacer would increase water solubility and reduce non-specific binding, and the amine group would enable conjugation to the siRNA element.[12] Two bifunctional linkers were synthesized and evaluated for siRNA conjugation. Linker C1, PTPPf [3-(2-pyridyl)-dithiopropionic acid pentafluorophenyl ester], was designed to release siRNA upon cellular entry through cleavage of the disulfide linkage, through enzymatic reduction or ligand-exchange (e.g. glutathione).[13] Linker C2, MPPF (3-maleimidopropionic acid pentafluorophenyl ester), was designed to be more robust, thereby enabling evaluation of cellular uptake and localization of the siRNA construct within the cellular compartments.[14] [See supporting information for details of synthesis, characterization and conjugation protocols]
The final design component was to functionalize the construct for tumor cell-selective transfection. For this purpose two functional peptides, thiol-modified RGD peptide and thiol-modified HIV-Tat derived peptide, were attached to the siRNA-QDs via the conjugation methods described above. Brain tumor cells (U87 and U87-EGFRvIII) overexpress the integrin receptor protein αvβ3, which strongly binds to the RGD binding domain.[15] RGD functionalized siRNA-QDs selectively accumulate in brain tumor cells in vitro, and can be tracked by fluorescence microscopy.[16] In addition, the HIV-Tat peptide enables efficient transfection of siRNA-QDs in cells when directly attached to the QD surface.[17] The density of siRNA on the QDs and the ratio between siRNA strands and peptides were optimized for gene knockdown. It was found that the density of 10 siRNAs/per nanoparticle and the ratio of 1:10 (siRNA: each peptide) was in close agreement with literature values[4], and was optimal for knocking down the target genes (EGFP and EGFRvIII) overexpressed in our U87 cell lines. To optimize gene silencing with our siRNA-QD constructs and to assess the transfection efficiency and RNA interference (RNAi) activity, we examined the suppression of EGFP expressed in U87 cell lines that were genetically modified to express EGFP. Cytotoxicity of the constructs was tested by serial dilution studies. The range of concentration causing minimal/negligible cytotoxicity was identified and the subsequent experiments employed the concentrations within this range (See Supporting information, Figure S1).[18] Importantly, the EGFP cell line has been widely used to investigate siRNA-based silencing of EGFP, since the suppression of EGFP expression does not compromise cell viability. The transfection efficiency of three different kinds of constructs were evaluated; constructs modified with the RGD peptide only, those modified with the HIV Tat peptide only, and those with both HIV-Tat and RGD peptide. Although the siRNA-QDs modified with only RGD showed considerable selective internalization within U87-EGFP cells, siRNA-QDs modified with a combination of RGD and HIV-tat peptides (the ratio of siRNA: RGD: HIV-Tat being 1:10:10 per QD) showed maximum internalization within U87-EGFP cells, in close agreement with previous studies.[4] This optimal condition was used for subsequent siRNA-QD experiments.
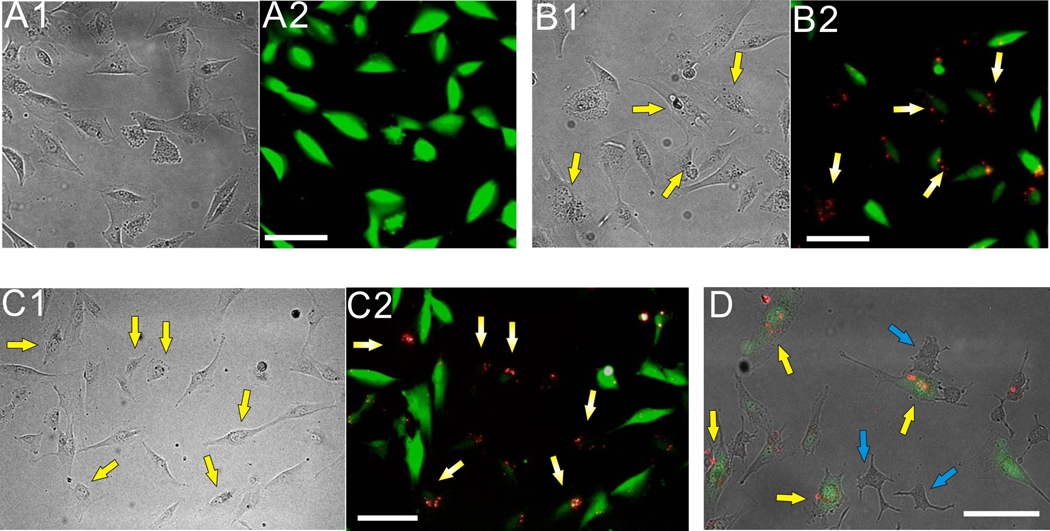
The U87-EGFP cell line was then treated with siRNA-QDs (siRNA:QDs = 0.12 µM:0.011 µM), modified with HIV-Tat [~1.2 µM] and RGD [~1.2 µM], and simultaneously imaged using fluorescence microscopy (Figure 2). Cationic lipids (X-tremeGENE, Roche) were used to further enhance cellular uptake and prevent degradation of the siRNA within the endosomal compartment of the cells. The siRNA-QDs showed significant internalization into the cells. Knockdown of the EGFP signal was observed after 48–72 hrs (Figure 2B). Fluorescence intensity was influenced by other factors such as exposure time, media condition, and cell shrinkage. To minimize the influence from these external factors, the control U87-EGFP cells (without siRNA) were trypsinized and co-cultured with U87-EGFP cells transfected with siRNA-QDs in the same well. The U87 cells containing siRNA-QDs were easily distinguishable from the control cells due to the bright fluorescent property of the QDs (Figure 2C2). Cells with internalized siRNA-QDs showed considerable knockdown of the EGFP protein when compared with the surrounding control U87-EGFP cells (Figure 2C).
Figure 2.
Knockdown of EGFP in U87 cells using siRNA-QD modified with RGD and HIV-Tat peptides (note that yellow arrows represent U87- EGFP cells transfected with the siRNA-QDs and the blue arrows indicate PC-12 cells) (A): Control U87-EGFP cells without siRNA-QDs; (A1) represents the phase contrast image and (A2) is the corresponding fluorescence image. (B): EGFP knockdown using multifunctional siRNA-QDs; (B1) Phase contrast image showing that the morphology of U87-EGFP cells has not changed as compared to the control cells in (A). (B2) Fluorescence image clearly shows the knockdown of EGFP in cells (marked by yellow arrows) which have internalized the siRNA-QDs (red) after 48 hrs. (C): Co-cultures of U87-EGFP control cells (without siRNA-QDs) and U87-EGFP cells transfected with siRNA-QDs so as to investigate them under the same conditions; (C1) Phase contrast image clearly shows no difference in the morphology of the U87-EGFP control cells and the siRNA-QDs transfected cells. (C2) Fluorescence image clearly shows the decrease in the EGFP signal in the U87-EGFP cells transfected with siRNA-QD as compared to the surrounding U87-EGFP control cells. (D): Phase contrast image showing the target-oriented delivery of siRNA-QDs in co-cultures of the malignant U87-EGFP cells, overexpressing the αvβ3 integrin receptors, and the less tumorigenic PC-12 cells(blue arrows) incubated with the siRNA-QDs. It can be clearly seen that most of the siRNA-QDs, due to the presence of RGD and HIV-tat peptides, were taken up by the U87-EGFP cells and not by the PC-12 cells. (Scale bar: 100 µm)
To further demonstrate the target-specific delivery of the siRNA-QDs, we incubated the siRNA-QDs modified with Tat and RGD against EGFP in co-cultures of the U87-EGFP cell line with other less-tumorigenic cell lines, such as PC-12 and SK-N-BE(2)C (See Supporting information, Figure S2), having a considerably small number of integrin receptors.[19] The presence of RGD tripeptide molecules on the surface of the siRNA-QDs led to specific binding with integrin receptors overexpressed in the U87 cells, resulting in higher cellular uptake by the malignant U87 cells as compared to the less tumorigenic PC-12 cells as seen by the selective accumulation of the QDs within the U87-EGFP cells (Figure 2D). These results confirmed our hypothesis that the target-specific delivery of the siRNA-QDs into brain cancer cells can be significantly enhanced by functionalizing the QDs with targeting moieties like RGD tripeptide.
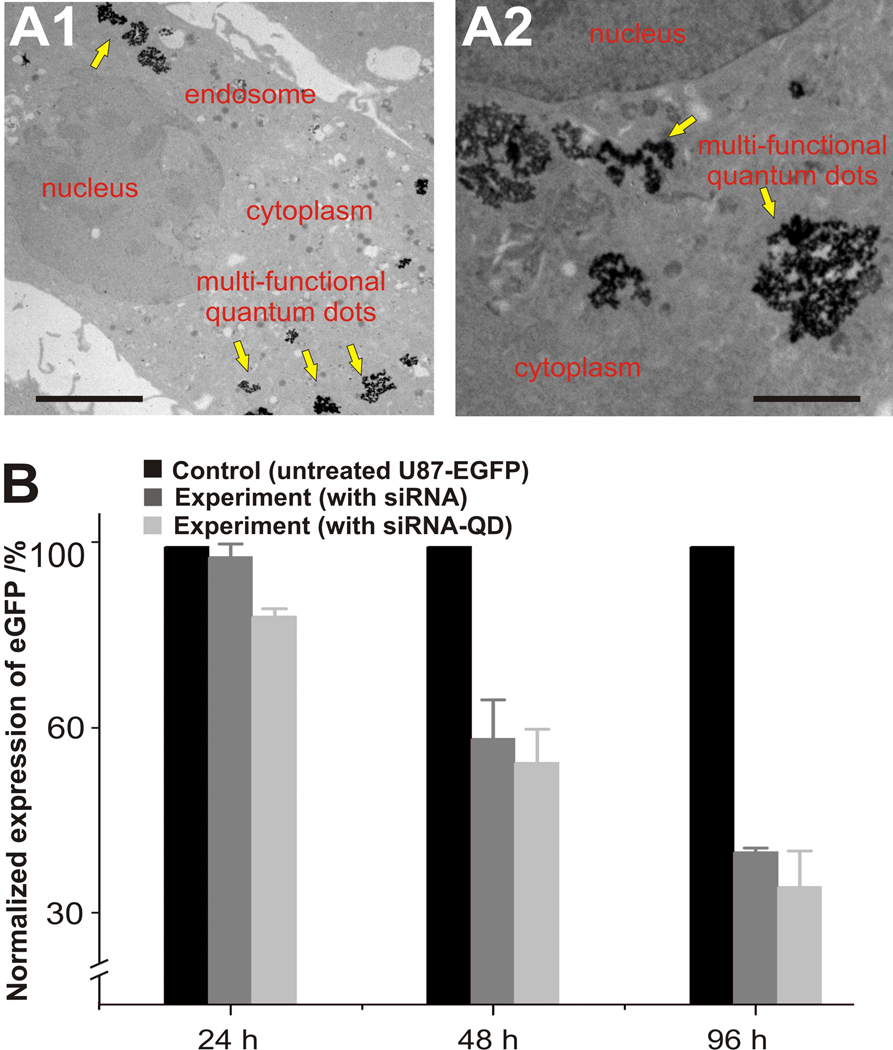
The intracellular delivery of the siRNA-QDs within the U87-EGFP cells was also confirmed by transmission electron microscopy (TEM), which clearly shows the presence of QDs in the cytoplasm of the cells (Figure 3A). The knockdown efficiency of the siRNA-QDs was similar to or slightly better than that of the positive control consisting of U87-EGFP cells transfected with only siRNA using X-tremeGENE (See Supporting information, Figure S3). This high transfection efficiency appears to be due to synergistic effects of the two transfection peptides. Decrease in fluorescence intensities (EGFP signal, green fluorescence) within cells treated with the above mentioned systems were then compared with the intensity of U87-EGFP without siRNA. As shown in (Figure 3B), the decrease in fluorescence intensity of U87-EGFP incubated with siRNA-QDs and siRNA alone was comparable, but drastically lower than that observed for the control without siRNA. Cells with internalized siRNA-QDs show decreased green fluorescence (EGFP signal) when compared with the control. This data strongly suggests that siRNA-QDs can be simultaneously used as delivery and imaging probes.
Figure 3.
Knockdown efficiency of EGFP within U87-EGFP cells and internalization of multifunctional siRNA-QDs. (A): TEM analysis of the internalization of the multifunctional siRNA-QDs into the U87-EGFP cells; (A1) Presence of multi-functional siRNA-QDs (yellow arrows) within the cytoplasm and the endosome (Scale bar: 5 µm). (A2) Zoomed-in image showing individual siRNA-QDs within the cytoplasm (Scale bar: 2.5 µm). (B): The bar graph represents the knockdown of EGFP over 24h, 48h, and 96h in U87-EGFP cells treated with siRNA [0.12µM] only (dark grey), and siRNA-QD [siRNA:QD = 0.12µM:0.011µM] (light grey). The EGFP knockdown data was normalized with the expression levels of EGFP in the control U87-EGFP cells (black).
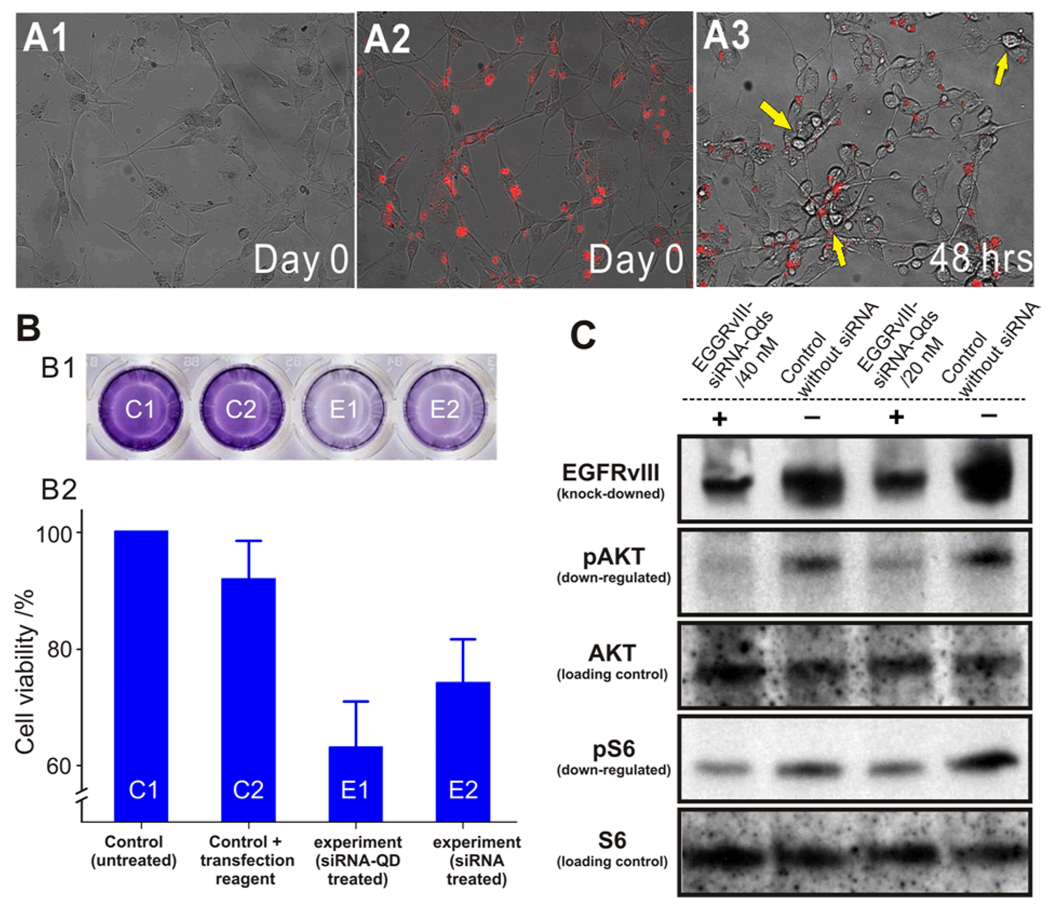
Having demonstrated the selective manipulation of the U87-EGFP cell line, we then focused on the knockdown of EGFRvIII with our siRNA-QD constructs. U87-EGFRvIII cells were genetically modified to overexpress EGFRvIII, a mutant-type epidermal growth factor receptor (EGFR) only expressed within cancer cells.[20] This cell type was incubated with our siRNA-QDs modified with Tat and RGD peptides and armed with EGFRvIII-targeting siRNA. The cells were simultaneously imaged for the internalization of siRNA-QDs using fluorescence microscopy. Significant cell death was observed in the wells loaded with siRNA-QDs against EGFRvIII after 48h (Figure 4A.) Quantitative analysis revealed that the number of viable U87-EGFRvIII cells, as observed via fluorescence microscopy, decreased with increased incubation time. When compared with the control (U87-EGFRvIII without siRNA-QDs), there was a significant decrease in the number of viable cells, thus demonstrating the effectiveness of our nanoparticle-based siRNA delivery to knock-down the oncogene. The result was confirmed using the MTT assay which showed a decrease in the number of viable cells in the well incubated with siRNA-QDs against EGFRvIII (Figure 4B). This assay further confirmed that the QDs themselves were noncytotoxic when used alone as they did not result in any appreciable cell death (See Supporting information, Figure S1). The knockdown of EGFRvIII and the inhibition of the downstream proteins in the PI3K signaling pathway were confirmed using Western Immunoblotting. The results (Figure 4C) confirm a considerable decrease in the expression of EGFRvIII, and down-regulation of phospho-Akt and phospho-S6 as compared to the control. Thus, these results demonstrate the specificity of the siRNA against EGFRvIII, the inherent noncytotoxicity of the QDs, and the facile evaluation and manipulation of cancer cell proliferation with multifunctional-QD constructs.
Figure 4.
Knockdown of EGFRvIII in U87 EGFRvIII using multifunctional siRNA-QDs (A): Phase contrast images showing the internalization of siRNA-QDs into the U87-EGFRvIII cells. (A1) Morphology of U87-EGFRvIII cells before incubation with siRNA-QDs on Day 0. (A2) U87-EGFRvIII cells after incubation with siRNA-QDs (red) on Day 0. (A3) Morphology of U87-EGFRvIII 48hrs after incubation with siRNA-QDs. Note that effect of the EGFRvIII knockdown by the siRNA-QDs can be clearly seen as they have clearly shrunk (yellow arrows) and look collapsed as compared to Day 0, marking the onset of apoptosis (Scale bar = 100 µm). (B): Cell viability assay using MTT assay. (B1) Optical image of cell viability (MTT) assay in a well plate. Dark blue color represents high number of viable cells and pale blue indicates low viable cell population. (B2) MTT assayed wells were quantified with UV absorbance and converted to cell viability. Untreated control C1 and C2 represent control cell population and viable cell population in presence of a cationic lipid based transfection reagent respectively. siRNA-QD transfected cells in experiment (E1) and siRNA treated cells in E2 show low numbers of the viable cells due to knockdown of EGFRvIII gene (C) Western Immunoblotting to show silencing effect of EGFRvIII gene. Protein expression level of EGFRvIII is dramatically decreased, and phosphorylation levels of key proteins in PI3K signaling pathway are reduced significantly. The upstream protein (AKT) and the downstream protein (S6), which play an important role in cell proliferation are selected to investigate the gene-knockdown effect on the PI3K signaling pathway.
In summary, this work is a demonstration of the multi-functional siRNA-QD strategy focusing on targeted delivery, high transfection efficiency, and multi-modal imaging/tracking. Our siRNA-QDs could be used for the development of novel chemotherapies and diagnostics relevant to brain cancer research. These novel methods and applications complement recent advances in nanomaterial-based siRNA delivery, nanomaterial-based molecular imaging, as well as siRNA-based chemotherapeutic strategies reported recently. While the ability to functionalize as well as control the surface of quantum dots with specific linkers and multi-functional molecules (siRNA and peptides) is critical for nanoparticle-based drug delivery, this method could also provide highly useful information regarding bio-surface chemistry of nanomaterials. In addition, the application of multi-functional siRNA-QDs to modulate the key cancer signaling pathways is important not only for selective chemotherapeutic strategy but also for dissecting signaling cascades triggered by inhibiting specific proteins. Collectively, our multi-functional QD-based siRNA delivery strategy has significant potential for simultaneous prognosis, diagnosis, and therapy.
Footnotes
We especially appreciate Mr. V. Starovoytov and Dr. Y. Horibe for helping us with TEM, and we would like to thank NJ Biomaterial Center (Prof. Kohn) for allowing us to use the cell culture facilities. KBLee acknowledges the NIH Director’s Innovator Award (1DP20D006462-01) and is also grateful to the N.J. Commission on Spinal Cord grant (09-3085-SCR-E-0).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the authors.
Contributor Information
Jongjin Jung, Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA).
Aniruddh Solanki, Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA).
Kevin A. Memoli, Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA)
Ken-ichiro Kamei, Dr., Department of Molecular and Medical Pharmacology, University of California, Los Angeles. Los Angeles, CA 90095 (USA)
Hiyun Kim, Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA).
Michael A. Drahl, Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA)
Lawrence J. Williams, Prof., Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA)
Hsian-Rong Tseng, Prof., Department of Molecular and Medical Pharmacology, University of California, Los Angeles. Los Angeles, CA 90095 (USA)
Ki-Bum Lee, Prof., Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854 (USA)
References
- 1.a) Medarova Z, Pham W, Farrar C, Petkova V, Moore A. Nat. Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]; b) Dykxhoorn DM, Palliser D, Lieberman J. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 2.a) Wasungu L, Hoekstra D. J.Contr.Release. 2006;116:255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]; b) Jeong JH, Mok H, Oh YK, Park TG. Bioconjugate Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]; c) Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Bioconjugate Chem. 2008;19:1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]; d) Zintchenko A, Philipp A, Dehshahri A, Wagner E. Bioconjugate Chem. 2008;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- 3.a) Lee JH, Lee K, Moon SH, Lee Y, Park TG, Cheon J. Angew.Chem.Int.Ed. 2009;48:4174–4179. doi: 10.1002/anie.200805998. [DOI] [PubMed] [Google Scholar]; b) Qi LF, Gao XH. Acs Nano. 2008;2:1403–1410. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Nano Lett. 2009;9:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yezhelyev MV, Qi LF, O'Regan RM, Nie S, Gao XH. J. Am. Chem. Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J. Am. Chem. Soc. 2009;131:2072. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Bioconjugate Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 5.a) Kikkeri R, Lepenies B, Adibekian A, Laurino P, Seeberger PH. J. Am. Chem. Soc. 2009;131:2110. doi: 10.1021/ja807711w. [DOI] [PubMed] [Google Scholar]; b) Bhirde AA, Patel V, Gavard J, Zhang GF, Sousa AA, Masedunskas A, Leapman RD, Weigert R, Gutkind JS, Rusling JF. Acs Nano. 2009;3:307–316. doi: 10.1021/nn800551s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan QW, Weiss WA. Oncogene. 2005;24:829–837. doi: 10.1038/sj.onc.1208227. [DOI] [PubMed] [Google Scholar]
- 7.Arwert E, Hingtgen S, Figueiredo JL, Bergquist H, Mahmood U, Weissleder R, Shah K. Cancer Res. 2007;67:7335–7342. doi: 10.1158/0008-5472.CAN-07-0077. [DOI] [PubMed] [Google Scholar]
- 8.a) Veeravagu A, Liu ZF, Niu G, Chen K, Jia B, Cai WB, Jin CJ, Hsu AR, Connolly AJ, Tse V, Wang F, Chen XY. Clin. Cancer Res. 2008;14:7330–7339. doi: 10.1158/1078-0432.CCR-08-0797. [DOI] [PubMed] [Google Scholar]; b) Lesniak MS, Brem H. Nature Reviews Drug Discovery. 2004;3:499–508. doi: 10.1038/nrd1414. [DOI] [PubMed] [Google Scholar]
- 9.a) Ekstrand AJ, Sugawa N, James CD, Collins VP. Proc. Natl. Acad. Sci. USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Humphrey PA, Wong AJ, Vogelstein B, Zalutsky MR, Fuller GN, Archer GE, Friedman HS, Kwatra MM, Bigner SH, Bigner DD. Proc. Natl. Acad. Sci. USA. 1990;87:4207–4211. doi: 10.1073/pnas.87.11.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Vivanco I, Sawyers CL. Nature Reviews Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]; b) Yamoutpour F, Bodernpudi V, Park SE, Pan WH, Mauzy MJ, Kratzke RA, Dudek A, Potter DA, Woo RA, O'Rourke DM, Tindall DJ, Farassati F. Mol. Cancer Ther. 2008;7:3586–3597. doi: 10.1158/1535-7163.MCT-08-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Li JJ, Wang YA, Guo WZ, Keay JC, Mishima TD, Johnson MB, Peng XG. J. Am. Chem. Soc. 2003;125:12567–12575. doi: 10.1021/ja0363563. [DOI] [PubMed] [Google Scholar]; b) Xie RG, Kolb U, Li JX, Basche T, Mews A. J. Am. Chem. Soc. 2005;127:7480–7488. doi: 10.1021/ja042939g. [DOI] [PubMed] [Google Scholar]
- 12.a) Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. J. Am. Chem. Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Susumu K, Uyeda HT, Medintz IL, Pons T, Delehanty JB, Mattoussi H. J. Am. Chem. Soc. 2007;129:13987–13996. doi: 10.1021/ja0749744. [DOI] [PubMed] [Google Scholar]
- 13.a) Kida S, Maeda M, Hojo K, Eto Y, Nakagawa S, Kawasaki K. Chem.Pharmaceut.Bulletin. 2007;55:685–687. doi: 10.1248/cpb.55.685. [DOI] [PubMed] [Google Scholar]; b) Kamruzzahan ASM, Ebner A, Wildling L, Kienberger F, Riener CK, Hahn CD, Pollheimer PD, Winklehner P, Holzl M, Lackner B, Schorkl DM, Hinterdorfer P, Gruber HJ. Bioconjugate Chem. 2006;17:1473–1481. doi: 10.1021/bc060252a. [DOI] [PubMed] [Google Scholar]; c) Bauhuber S, Hozsa C, Breunig M, Göpferich A. Adv. Mater. 2009;21:3286–3306. doi: 10.1002/adma.200802453. [DOI] [PubMed] [Google Scholar]
- 14.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Chemistry & Biology. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Xie J, Chen K, Lee HY, Xu CJ, Hsu AR, Peng S, Chen XY, Sun SH. J. Am. Chem. Soc. 2008;130:7542. doi: 10.1021/ja802003h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai WB, Shin DW, Chen K, Gheysens O, Cao QZ, Wang SX, Gambhir SS, Chen XY. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 17.a) Berry CC. Nanomedicine. 2008;3:357–365. doi: 10.2217/17435889.3.3.357. [DOI] [PubMed] [Google Scholar]; b) Ruan G, Agrawal A, Marcus AI, Nie S. J. Am. Chem. Soc. 2007;129:14759–14766. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]
- 18.a) Derfus AM, Chan WCW, Bhatia SN. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kirchner C, Liedl T, Kudera S, Pellegrino T, Javier AM, Gaub HE, Stolzle S, Fertig N, Parak WJ. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 19.a) Meyer A, van Golen CM, Kim B, van Golen KL, Feldman EL. Neoplasia. 2004;6:332–342. doi: 10.1593/neo.03445. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Martiniova L, Lai EW, Elkahloun AG, Abu-Asab M, Wickremasinghe A, Solis DC, Perera SM, Huynh TT, Lubensky IA, Tischler AS, Kvetnansky R, Alesci S, Morris JC, Pacak K. Clin. Exp. Metastasis. 2009;26:239–250. doi: 10.1007/s10585-009-9236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Li ZB, Cai WB, Cao QZ, Chen K, Wu ZH, He LN, Chen XY. J. Nucl.Med. 2007;48:1162–1171. doi: 10.2967/jnumed.107.039859. [DOI] [PubMed] [Google Scholar]
- 20.Wang MY, Lu KV, Zhu SJ, Dia EQ, Vivanco I, Shackleford GM, Cavenee WK, Mellinghoff IK, Cloughesy TF, Sawyers CL, Mischel PS. Cancer Res. 2006;66:7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]