Abstract
The impact of transgenic white spruce [Picea glauca (Moench) Voss] containing the endochitinase gene (ech42) on soil fungal biomass and on the ectendomycorrhizal fungi Wilcoxina spp. was tested using a greenhouse trial. The measured level of endochitinase in roots of transgenic white spruce was up to 10 times higher than that in roots of nontransformed white spruce. The level of endochitinase in root exudates of three of four ech42-transformed lines was significantly greater than that in controls. Analysis soil ergosterol showed that the amount of fungal biomass in soil samples from control white spruce was slightly larger than that in soil samples from ech42-transformed white spruce. Nevertheless, the difference was not statistically significant. The rates of mycorrhizal colonization of transformed lines and controls were similar. Sequencing the internal transcribed spacer rRNA region revealed that the root tips were colonized by the ectendomycorrhizal fungi Wilcoxina spp. and the dark septate endophyte Phialocephala fortinii. Colonization of root tips by Wilcoxina spp. was monitored by real-time PCR to quantify the fungus present during the development of ectendomycorrhizal symbiosis in ech42-transformed and control lines. The numbers of Wilcoxina molecules in the transformed lines and the controls were not significantly different (P > 0.05, as determined by analysis of covariance), indicating that in spite of higher levels of endochitinase expression, mycorrhization was not inhibited. Our results indicate that the higher levels of chitinolytic activity in root exudates and root tissues from ech42-transformed lines did not alter the soil fungal biomass or the development of ectendomycorrhizal symbiosis involving Wilcoxina spp.
White spruce [Picea glauca (Moench) Voss] is a tree species with an extensive distribution in boreal and subboreal forests and with significant ecological roles (37, 38). It is also an important commercial species for production of pulpwood and construction-grade lumber. However, in nurseries and plantations, white spruce is sensitive to multiple fungal diseases (23, 29, 42, 62, 76). Climate change scenarios suggest that diseases could result in increased mortality in conifer forests (22, 48). Genetic engineering offers a potential means to mitigate biotic and abiotic stresses.
During the last 2 decades, chitinase genes isolated from plants, fungi, or bacteria have been studied and used to transform crops or trees in order to increase their resistance to plant-pathogenic fungi. One potential goal is improving white spruce tolerance to fungal infection through insertion of a chitinase gene. Chitin is a biopolymer of β-(1-4)-linked molecules of N-acetylglucosamine (NAG), a derivative of glucose, and is the primary constituent of the fungal cell wall and arthropod exoskeleton (3, 51). Chitinases are plant defense pathogenesis-related proteins (6, 11) that break down the chitin chain either by cleavage of internal glycoside bonds (endochitinases), by hydrolysis of the nonreducing end of the chitin chain (exochitinases), or by hydrolysis of NAG oligomers and trimers into NAG monomers (chitobiases). Endo- and exochitinase genes have been well characterized using sugar beet (Beta vulgaris) (44) and the filamentous fungal genus Trichoderma (14, 24, 69). Chitinolytic genes have been inserted into the genomes of cultivated plants and trees in an attempt to boost plant chitinase activity. Among the different genes involved in the production of chitinolytic enzymes, the ech42 endochitinase gene from Trichoderma harzianum has been inserted into plant genomes to enhance their resistance against phytopathogenic fungi. In McIntosh apple cultivars transformed with the ech42 gene there was limited attack by the apple scab fungus Venturia inaequalis (5). Transgenic black spruce (Picea mariana) expressing the ech42 gene was more resistant to the root rot pathogen Cylindrocladium floridanum (45).
However, field deployment of crops and trees genetically transformed to improve nonspecific resistance against phytopathogenic fungi has raised concerns about the impact on nontarget fungi, including potentially beneficial symbionts. This is particularly worrisome when nonspecific constitutive promoters control expression of the resistance gene and the gene is expressed in all tissues from roots to leaves. As a consequence, the natural colonization of such transformed plants by endophytic or mycorrhizal fungi can be altered.
Mycorrhizal fungi play a key role in plant nutrition (55) by mobilizing and transferring nutrients to the host through an intimate and highly organized association with plant roots (52, 63). Furthermore, their involvement in soil nutrient recycling (56) makes mycorrhizal symbiosis a major ecological process that is important for the health of soil and forest ecosystems. Crops, fruits, and forest trees exhibit mycorrhizal colonization by arbuscular mycorrhizae, ectomycorrhizae, and ectendomycorrhizae (EEM). While numerous studies have addressed the impact of transgenic plants on arbuscular mycorrhizae (10, 26, 64, 68, 72, 73) and ectomycorrhizae (32, 43, 50, 60), no previous study focused on EEM.
Ectendomycorrhizal fungi can be distinguished from ectomycorrhizae by the presence of a thin or fragmented mantle and intracellular penetration into root cortical cells. All EEM fungi identified so far belong to the Ascomycetes, and these fungi are represented by several genera of Helotiales and Pezizales (77). EEM fungi are prevalent in conifer and deciduous tree nurseries (27, 39, 40, 70) and are also very common on seedling root tips at disturbed sites (15, 16, 19). The prevalence of EEM fungi on seedling roots, from which the genus Wilcoxina is frequently recovered (16, 67), suggests that they can play a significant role in establishment and growth of seedlings (77) and provide protection against root diseases (31, 61). Consequently, the potentially negative effects of chitinase-transformed trees on ectendomycorrhizal fungi could be detrimental to plant health.
The present study addressed the potential impact of ech42-transformed white spruce on soil fungal biomass and ectendomycorrhizal symbiosis. It was hypothesized that (i) the soil fungal biomass in a transgenic white spruce rhizosphere is less than the soil fungal biomass in a control tree rhizosphere and (ii) the development of Wilcoxina spp. on root tips of transgenic white spruce is less important than the development of Wilcoxina spp. on root tips of control trees. To test these hypotheses, 5-year-old white spruce trees transformed with the 35S promoter-ech42 construct were analyzed by performing a greenhouse trial. The amount of soil fungal biomass was estimated using measurements of ergosterol in soil. A real-time PCR method was developed to detect changes in the quantity of ectendomycorrhizal hyphae involved in colonization of transgenic white spruce root tips.
MATERIALS AND METHODS
Plant material and culture.
Embryogenic PG653 cell lines of white spruce were transformed by Noël et al. (45). The transformed lines were obtained by using Agrobacterium tumefaciens strain C58/pMP90 (35) and derivatives of the binary vector pB1N19ESR containing the complete coding sequence of the ech42 endochitinase gene, a duplicated enhancer 35S promoter from 35SCaMV, the alfalfa mosaic virus (AMV) leader sequence, and the neomycin phosphotransferase II (nptII) gene for kanamycin selection. Four transformed lines and control white spruce trees were grown in a greenhouse for 5 years with a photoperiod consisting of 16 h of light and 8 h of darkness in pots containing peat, perlite, and vermiculite (3:1:1, vol/vol/vol). The growth cycle was 17 weeks long. The plants were fertilized weekly with 11-41-8 (N-P-K; 50 ppm) for the first 3 weeks following dormancy and with a mixture of 20-8-20 and 20-20-20 (3:1, vol/vol; 100 ppm) for the remaining 14 weeks. The plants were then stored at 4°C for 8 weeks to induce dormancy. Two growth cycles were induced each year.
Endochitinase activity in root tissues and root exudates.
Root fragments were obtained from each tree, rinsed in distilled water to remove adherent soil particles, and crushed in liquid nitrogen using a pestle. To quantify the chitinolytic activity, 70 to 130 mg (fresh weight) of ground root tissues was vortexed for 10 min in 1.6 ml of sodium acetate buffer (96 mM sodium acetate, 0.1% SDS, 0.1% Triton X-100, 10 mM Na2EDTA) as described by Bolar et al. (4). Samples were centrifuged twice at 13,000 rpm for 5 min, and the supernatants were transferred into new tubes. One hundred microliters of the extraction solution was incubated for 30 min at 37°C with 50 μl of 0.2 mM 4-methylumbelliferyl-β-d-N,N′,N"-triacetylchitotrioside (Sigma-Aldrich Co., St. Louis, MO) dissolved in 100 mM sodium acetate. The reaction was stopped by adding 200 μl of 0.2 M sodium carbonate. Enzymatic activity was determined using an excitation wavelength of 365 nm and an emission wavelength of 450 nm with a Fluorolite 1000 microtiter plate reader (Dynatech Laboratories, Chantilly, VA). A standard curve for 4-methylumbelliferone (4-MU) (Sigma-Aldrich Co.) was used to convert levels of fluorescence into nanomoles of 4-MU released per minute per gram of root. Root exudates from three trees per transformed line or control were collected by soaking 3 to 5 living roots in 10 ml of Murashige and Skoog basal salt (MSS) mixture (Sigma-Aldrich Co.) for 7 days. The MSS solution was supplemented with sucrose (15 g liter−1), protease inhibitors (one Complete minitablet of protease inhibitors for every 50 ml; Roche, Indianapolis, IN), benomyl (5 ppm), and streptomycin (100 ppm). Root exudates were filtered on Acrodisc syringe filters with a polytetrafluoroethylene (PTFE) membrane (0.2 μm; Ultident Scientific, Saint-Laurent, Quebec, Canada) and concentrated 10-fold by ultrafiltration on Amicon PM-30 membranes (Millipore Corp., Bedford, MA). The endochitinase activity in root exudates was determined as previously described for root tissues. The dry weight of root segments soaked in MSS solution was determined after the preparation was dried at 70°C for 72 h.
Extraction of ergosterol and high-performance liquid chromatography.
For each tree, two soil samples per pot were collected using a metal punch and mixed with each other in a coffee mill. Five grams (wet weight) of soil was placed into 50-ml polypropylene Falcon tubes. The technique used for extraction of ergosterol was a modified version of the microwave-assisted extraction (MAE) technique of Montgomery et al. (41). Eight milliliters of methanol and 1 ml of 2 M NaOH were added to each sample. The internal standard 7-dehydrocholesterol (0.1 mg ml−1; Sigma-Aldrich Co.) was added to each soil sample. Samples were then homogenized using a vortex and heated until they boiled in a domestic microwave oven. Samples were cooled at room temperature for 15 min and heated a second time in a microwave oven. After cooling for 15 min, samples were neutralized with 1 ml of 1 M HCl and supplemented with 5 ml of methanol. Solutions were homogenized and supplemented with 3 ml of pentane. Samples were vortexed and centrifuged at 3,000 rpm for 5 min. Supernatants were transferred into 150-ml polypropylene Falcon tubes. The pentane extraction procedure was repeated three times. The organic phase was passed through 0.2-μm nylon syringe filters (Chromspec, Brockville, Ontario, Canada) and evaporated under an N2 atmosphere for 30 min. Each sample was then redissolved in 1 ml of methanol, incubated for 10 min, vortexed, and filtered through 0.2-μm nylon syringe filters. Samples were analyzed with a high-performance liquid chromatography (HPLC) system equipped with a Waters 1524 binary pump, a Waters 717 Plus autosampler, and a Waters 2487 dual-absorbance detector (Waters Corporation, Milford, MA). Ergosterol was separated from other organic soil compounds on a 4.6- by 250-mm Zorbax Rx C18 reverse-phase column packed with octyldecyl silane (ODS) (5 μm) preceded by a Zorbax guard column (Agilent Technologies, Palo Alto, CA). The mobile phase was methanol-acetonitrile (55:45, vol/vol) at a flow rate of 2 ml min−1. The experiment was performed at room temperature. Absorbance was read at 283 nm. The ergosterol content was determined using a standard curve based on the ergosterol/7-dehydrocholesterol (Sigma-Aldrich Co.) area ratio. Data were processed with Waters Breeze v.3.3 software (Waters Corporation, Milford, MA).
Mycorrhizal fungus identification and colonization.
The levels of mycorrhizal colonization of four trees of each transformed line and a control were estimated by visually scanning 1 g (fresh weight) of fine roots with a stereomicroscope, using the gridline intersection method (8, 21). Mycorrhizal root tips were then sorted based on their morphology and identified by sequencing internal transcribed spacer (ITS) regions of the rRNA. For genomic DNA (gDNA) isolation, root tips were crushed individually in liquid nitrogen using a polypropylene micropestle. The ground tissues were resuspended in 30 μl of ultrapure DNase- and RNase-free distilled water (Gibco, New York, NY) and 150 μl of a 15% Chelex 100 (Bio-Rad Laboratories, Richmond, CA) suspension with 1.6 μg μl−1 of proteinase K (Invitrogen, Carlsbad, CA). Samples were incubated for 2 h at 65°C and then for 20 min at 95°C to inactivate the proteinase K, and then they were centrifuged at 1,000 rpm for 5 min. One microliter of each supernatant was used as a template for PCR. The ITS regions were then amplified using primers ITS1-F (20) and ITS4 (75). Each PCR mixture (total volume, 25 μl) contained 1× PCR buffer, 1.6 mM MgCl2, each deoxynucleoside triphosphate (GE HealthCare Bio-Sciences) at a concentration of 1.25 mM, 25 μg of bovine serum albumin (BSA) (Sigma-Aldrich Co.), each primer at a concentration of 0.5 μM, and 1 U of Taq DNA polymerase (Invitrogen, Carlsbad, CA). The thermal cycling conditions were as follows: initial denaturation at 95°C for 2 min, followed by 37 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 45 s and a final elongation step consisting of 72°C for 10 min. PCRs were performed with an MJ Research PTC-200 (MJ Research Inc., Waltham, MA).
Real-time PCR assays.
To quantify root tip colonization by the ectendomycorrhizal fungi Wilcoxina spp., specific primers were designed to target the ITS2 region, beta-tubulin, and translation elongation factor 1-alpha genes. Wilcoxina-specific primers were designed with OligoAnalyzer v1.2 (Gene Link), based on sequence alignments constructed for the most closely related fungi as determined by preliminary root tip sequencing. A search of the GenBank public database using the BLASTN tool (2) was performed to ensure the specificity of all of the primers that were designed. The quantity of plant gDNA in each sample was also estimated by amplifying three housekeeping genes (Table 1). Real-time PCR conditions were optimized for each primer pair by using a temperature gradient with an Opticon-2 DNA engine (MJ Research, Cambridge, MA). Five root tips from each tree colonized by Wilcoxina spp. were pooled for gDNA extraction. Briefly, root tips were ground in liquid nitrogen with micropestles, incubated for 1 h at 65°C with 400 μl of Carlson lysis buffer and 2 μl of β-mercaptoethanol (9), and vortexed every 15 min. Four hundred microliters of phenol-chloroform-isoamyl alcohol (25:24:1) was added, and the aqueous phase was collected after centrifugation (13,000 rpm for 10 min). Nucleic acids were precipitated by incubating samples for 1 h at −20°C with 70 μl of 7.5 M sodium acetate and 600 μl of isopropanol. Pellets were obtained by centrifugation (13,000 rpm for 10 min), washed with 800 μl of 75% ethanol, dried using a thermoblock for 10 min at 55°C, and resuspended in 25 μl of Tris-EDTA buffer (pH 8). The DNA was diluted to obtain a concentration of 10 ng μl−1. Each real-time PCR mixture (total volume, 10 μl) contained 1× QuantiTect SYBR green mixture (Qiagen, Valencia, CA), each primer at a concentration of 600 nM, and 1 or 10 ng of gDNA depending on whether Wilcoxina sp. or P. glauca gDNA was targeted. The thermal cycling conditions were as follows: initial denaturation at 95°C for 15 min, followed by 40 cycles of 94°C for 5 s, the annealing temperature (see Table 1) for 30 s, and 65°C for 1 min 30 s. All reactions were performed with a Stratagene Mx3000P cycle engine (Stratagene, La Jolla, CA). Two fluorescence values were determined at the end of each cycle, and each run was followed by a melting curve analysis that confirmed the specificity of amplification, as well as the lack of primer dimer formation. The fluorescence threshold value was set at 500, and the filter gain setting used was 8×. Fluorescence and cycle threshold (CT) values were exported and analyzed using Excel spreadsheets (Microsoft Excel version 9.0.3821 SR-1; Microsoft, Redmond, WA). The amplification efficiency was determined by linear regression of efficiency analysis data (57). Gene stability and the normalization factor were determined using the geNorm VBA applet for Microsoft Excel (http://medgen.ugent.be/∼jvdesomp/genorm). To minimize sampling variations and to obtain normalized data, individual raw quantities (Q) were multiplied by the relative DNA quantification factor, (1+E)−CT.
TABLE 1.
Sequences of and organisms and loci targeted by the primer pairs used for real-time PCR analysis
| Target |
Primer | Sequence | Annealing temp (°C)b | Length (bp) | Reference | |
|---|---|---|---|---|---|---|
| Organism | Locusa | |||||
| Wilcoxina spp. | ITS2 | ITSWil F2 | TCATGGAAGATGAGTATGGTTGCAT | 60.4 | 120 | This study |
| ITSWil R2 | GTCAACGGCAGGACAATAACACACA | 61.4 | ||||
| Phialocephala fortinii | ITS1 | ITSPForti F | GTCAACGGCAGGACAATAACAC | 60.5 | 130 | This study |
| ITSPForti R | CTCTGGCGGGCACACACGAGCAGA | 62.4 | ||||
| Wilcoxina spp. | TEF1-α | tef F353 | GGAGGGTGGCAAGTCTAGC | 65 | 155 | This study |
| tef R488 | CCAGTCTCGACACGTCCGACA | 65 | ||||
| Wilcoxina spp. | Beta-tubulin | btub F340 | AGGAGTTGTTCAAGCGTGTCGGA | 64.8 | 119 | This study |
| btub R434 | ATCATAGCACAAGTTGGGAAACTCAC | 62.8 | ||||
| Picea glauca | PTSR | Pg PTSR K1 | TGGGAATTGATATAAGTGTTCTTGTGGAGGGTCT | 70.2 | 371 | 18 |
| Pg PTSR K2 | ACACCAAACCAGTAACCTGAGAAGGAAACA | 70.1 | ||||
| Picea glauca | TEF-α | Pg EXP 46630 F1 | GCTAGTCTGTCACAAGGTGCTTTCAAGT | 69.7 | 234 | 13 |
| Pg EXP 46630 R1 | TCCGAGTTTCTTTTCACAAGGAGTTGGC | 70.1 | ||||
| Picea glauca | Actin 2 | Pg ACT2 F1 | GTTTCCTGGTATTGCTGACCGTATGAGC | 70.4 | 450 | 13 |
| Pg ACT2 R1 | GTGCTGAGAGATGCCAAAATAGAACCTCC | 70.1 | ||||
TEF1-α, translation elongation factor 1-α; PTSR, peroxisomal targeting signal receptor.
The annealing temperature was determined by a gradient PCR assay using an Opticon II DNA engine (MJ Research, Cambridge, MA).
Microscopy.
For light microscopy, root tips were fixed in 4% paraformaldehyde in 0.1 M cacodylate (pH 7.3) at room temperature for 24 h and subsequently washed and dehydrated using a graded ethanol series. Root tips were transferred into toluene before infiltration in paraffin. Five-micron sections were cut with a sliding microtome, mounted on slides, and soaked in toluene to remove the paraffin. These sections were rehydrated using a descending ethanol series, stained with hematoxylin and eosin, dehydrated gradually using ethanol, and mounted permanently in Eukitt. Microscope observations were made with an Olympus BX-51 microscope.
Statistical analyses.
The experimental setup consisted of three blocks with one nontransformed line and four transformed lines per block and four trees per treatment. Data for the endochitinase activity, colonization rate, and ergosterol level were separated by using the Waller-Duncan k-ratio t test (P ≤ 0.05). All the data were checked for normality with the Shapiro-Wilk test and for homoscedasticity with Levene's test. Data for enzyme activity in root tissues were square root transformed, the logarithmic transformation log(1 + x) was applied to the values for endochitinase activity obtained for root exudates, and ergosterol data were reciprocal transformed to meet normality and homoscedasticity assumptions. The experiment was arranged in a randomized complete block manner. The potential effect of ech42-transformed white spruce on the ectendomycorrhizal fungi Wilcoxina spp. was evaluated by using covariance analysis (ANCOVA). The ANCOVA was performed using a linear mixed-effects model (lme). The variables tested in the ANCOVA were the five treatments (four transformed lines plus the control) and the three blocks and their interactions. The absolute number of white spruce molecules was considered a covariate. Log(1 + x) transformation was used for the absolute number of Wilcoxina sp. and P. glauca molecules to meet the assumption of homogeneity of variance. Statistical analyses were performed using the R statistical language (54) and the SAS software (58).
RESULTS
Endochitinase activity.
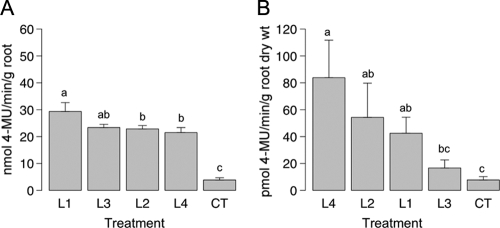
Endochitinase activity was first measured for root tissues isolated from eight trees per treatment. The means of the endochitinase activities obtained showed that there was a significant difference between the control and transformed spruce trees (Fig. 1A). The endochitinase activities in the control roots were 3.8 ± 0.8 nmol min−1 g−1 (average ± standard error of the mean), while they were 24.3 ± 1.1 nmol min−1 g−1 in the roots of the transformed lines. The highest endochitinase activity was obtained for roots from transformed spruce line 1, whose activity was 29.3 ± 3.3 nmol min−1 g−1. This value was significantly higher than the values obtained for roots of the other transformed lines.
FIG. 1.
Levels of endochitinase activity (A) in root tissues and (B) in root exudates for the four ech42-transformed lines and the control white spruce trees. The values for bars labeled with the same letter are not significantly different as determined by Waller-Duncan's multiple-range test (P ≤ 0.05). The error bars indicate standard errors of the means. CT, control.
The endochitinase activities in concentrated root exudates from four trees per treatment were also determined. The level of endochitinase activity was significantly increased in all transformed lines except line 3 (Fig. 1B). The endochitinase activities obtained for concentrated root exudates from the transformed lines were 2- to 10-fold higher than the endochitinase activities obtained for concentrated root exudates from control spruce trees. The highest enzyme activities were obtained for exudates from trees belonging to line 4 (83.8 ± 27.9 pmol min−1 g−1 [dry weight] of root).
Fungal biomass in the rhizosphere.
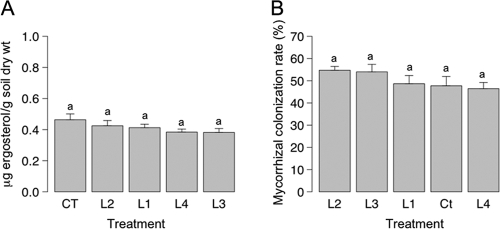
The amounts of fungal biomass present in the rhizospheres of control and transgenic white spruce trees were measured using a standard curve based on the ratio of ergosterol to the internal standard 7-dehydrocholesterol. The ergosterol concentration ranged from 0.38 ± 0.002 to 0.46 ± 0.037 μg g−1 (dry weight) of soil. The highest level of soil fungal biomass was obtained for soil samples from the rhizosphere of control trees; nevertheless, the differences in the mean soil ergosterol contents for pots subjected to different treatments were not significant (Fig. 2A).
FIG. 2.
(A) Levels of fungal biomass in pots based on measurement of ergosterol for the controls and the four transformed lines. (B) Levels of mycorrhizal colonization for the four ech42-transformed lines and the control white spruce trees. The values for bars labeled with the same letter are not significantly different as determined by Waller-Duncan's multiple-range test (P ≤ 0.05). The error bars indicate standard errors of the means. CT, control.
Characterization of fungal and mycorrhizal communities.
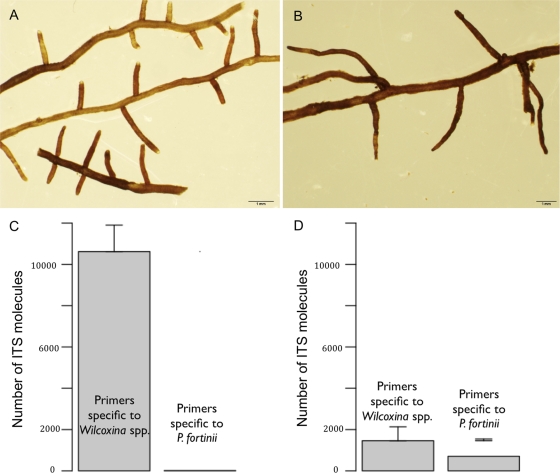
The mycorrhizal colonization rate (Fig. 2B) ranged from 46.4% ± 2.8% (line 4) to 54.7% ± 1.7% (line 2). Control roots had a mycorrhizal colonization rate of 47.7% ± 4.2%. No significant differences in mycorrhizal colonization intensity were found for the five treatments. Examination of root tips with a dissecting microscope revealed that two main morphological groups were present. The ITS sequence of the most abundant morphological group (MG1) (Fig. 3A) was similar to that of Wilcoxina mikolae var. mikolae (UAMH 6694) (92% similarity; 548 bp; 100% coverage), while the ITS sequence of the second most prevalent group (MG2) (Fig. 3B) was homologous to the ITS sequence of Phialocephala fortinii (UAMH 5524). Root tips colonized by Wilcoxina spp. were single to pinnate, 1 to 4 mm long, 0.2 to 0.3 mm in diameter, straight, smooth, glossy, and mainly yellowish brown with white to pale yellow tips. Root tips colonized by P. fortinii were 3 to 15 mm long, 0.2 to 0.3 mm in diameter, single, straight or slightly beaded, smooth, and brown or dark brown and sometimes had dark red tips.
FIG. 3.
(A and B) Stereomicroscope views of root tips colonized (A) by Wilcoxina spp. (MG1) and (B) by both Wilcoxina spp. and P. fortinii (MG2). (C and D) Variation in the numbers of Wilcoxina sp. and P. fortinii ITS molecules in root tips belonging to MG1 and MG2, respectively, as determined by amplification by real-time PCR with primer pairs specific for the two fungal taxa.
Inconsistent sequencing results were obtained for MG2. Fifty percent of the ITS sequences recovered corresponded to Wilcoxina sp. sequences instead of P. fortinii sequences, as expected. All ITS sequences obtained for MG1 corresponded to Wilcoxina sp. sequences, as expected.
The total DNA of 12 root tips characteristic of the two morphotypes was extracted and amplified with primer pairs specific for the ITS2 region of Wilcoxina spp. and the ITS1 region of P. fortinii using real-time PCR. The numbers of molecules at the cycle threshold obtained for the 12 samples characteristic of MG1 (Fig. 3C) were 10,621 ± 1,839 and 14 ± 5 (averages ± standard errors of the means) with primer pairs specific for Wilcoxina spp. and P. fortinii, respectively. Conversely, the numbers of molecules at the cycle threshold obtained for the 12 samples characteristic of MG2 (Fig. 3D) were 1,392 ± 935 and 689 ± 113 with primer pairs specific for Wilcoxina spp. and P. fortinii, respectively.
Quantification of Wilcoxina sp. symbiosis in root tips from control and ech42-transformed white spruce trees.
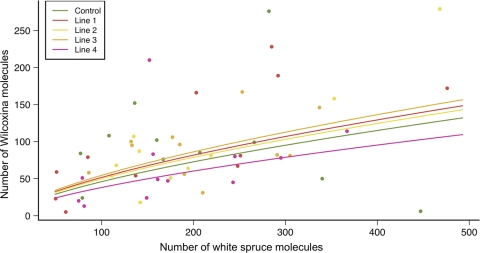
The absolute number of Wilcoxina sp. molecules was normalized based on data for beta-tubulin and translation elongation factor 1-alpha. The ITS data set was not included as it displayed the greatest gene instability. The three housekeeping genes were used to normalize the absolute number of plant molecules. The results of the ANCOVA (Fig. 4) showed that the absolute number of Wilcoxina molecules was significantly correlated with the absolute number of plant molecules (r = 0.6; F = 14.4; P = 0.0004). The mean absolute number of Wilcoxina molecules recovered was 66 (minimum, 42 molecules; maximum, 103 molecules) for controls. The number of these molecules was highest for line 3 (78 molecules; minimum, 50 molecules; maximum, 122 molecules) and lowest for line 4 (54 molecules; minimum, 35 molecules; maximum, 83 molecules). The results of the ANCOVA showed that variation in the absolute number of Wilcoxina molecules was not affected by the different levels of chitinase determined for the four transgenic lines (F = 0.44; P = 0.77).
FIG. 4.
Results of ANCOVA performed with the real-time PCR data. The circles indicate observed values, and the fitted lines indicate the predicted values based on the ANCOVA model.
Light microscopy.
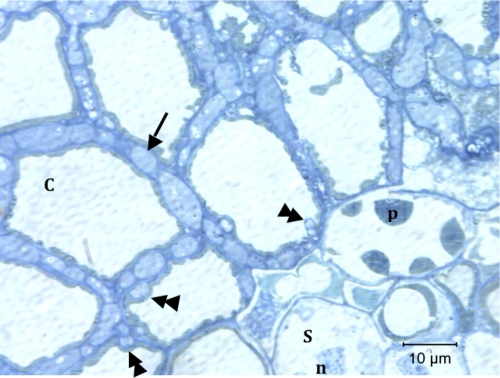
Figure 5 shows a transverse section of an MG1 root tip from a transformed line 1 tree. The entire cortex up to the stele is colonized by Wilcoxina sp. hyphae that form a Hartig net (arrow), while there were occasional intracellular structures (double arrowheads).
FIG. 5.
Transverse section of a root tip containing fungi belonging to MG1 collected from ech42-transformed line 1. The arrow indicates a Hartig net, and the double arrowheads indicate intracellular hyphae. C, cortical cell; p, tannin-filled cell; S, stele; n, nucleus.
DISCUSSION
Impact of chitinase overexpression on ectendomycorrhizal symbiosis.
The objective of this study was to determine if mycorrhizal colonization was inhibited in spruce trees that were genetically modified to overexpress endochitinase. Our analysis showed that an up-to-10-fold increase in chitinase activity in root tissues did not impede colonization of the inter- and intracellular spaces of root tips by ectendomycorrhizal hyphae. These results are in accordance with those of previous studies that investigated the effect of chitinase-transformed plants on symbiotic fungi. Vierheilig et al. (72) showed that a 5- to 16-fold increase in the level of chitinase in transgenic Nicotiana sylvestris enhanced its resistance against Rhizoctonia solani but did not alter its susceptibility to colonization by the endomycorrhizal fungus Glomus mosseae. In Betula pendula overexpressing the sugar beet chitinase, resistance against the leaf spot disease organism Pyrenopeziza betulicola and against Melampsoridium betulinum (birch rust) was improved compared with the control, as shown by greenhouse and field trials, respectively (46, 49). However, when the impact of the silver birch transformed lines on the ectomycorrhizal fungus Paxillus involutus was assessed in vitro, all but one transformed line showed similar levels of root mycorrhizal colonization (50). Pasonen et al. attributed the difference observed for one of the transformed lines to a pleiotropic effect due to transgene insertion. There was also a significant decrease in the root fresh weight for this B. pendula transformed line compared with the other transgenic lines and controls. Here, no evidence of a pleiotropic effect due to random insertion of the 35S-ech42 construct was found. All transformed lines were phenotypically similar to controls and were equally susceptible to mycorrhizal colonization.
The impact of chitinase-transformed plants has been monitored mainly using phytopathogenic fungi for which detrimental effects have been demonstrated. For example, attempts to increase tobacco and rice resistance by insertion of a chitinase gene were successful for Rhizoctonia solani (7, 34). Jayaraj and Punja (28) showed that for two foliar pathogens, Alternaria radicicola and Botrytis cinerea, the levels of infection were reduced up to 40 to 50% in transgenic carrots expressing a barley chitinase (chi-2) protein. Infection was reduced up to 90 to 95% when transgenic carrots coexpressed the chi-2 protein plus a wheat lipid transfer protein (ltp). Hybrid poplar trees (Populus nigra × P. maximowiczii) were transformed by insertion of the ech42 gene from T. harzianum controlled by the 35S promoter (45). The levels of endochitinase in foliar tissues increased 4- to 65-fold in the different transformed lines, which enhanced their resistance against the leaf rust pathogen Melampsora medusae f. sp. deltoidae. The same construct was used to transform black spruce (Picea mariana), and the endochitinase activity in embryogenic tissues was 2- to 8-fold higher than the activity in the controls. ech42-transformed black spruce trees were more resistant to the root rot pathogen Cylindrocladium floridanum than untransformed controls.
The release of more endochitinase into the rhizosphere from exudates of ech42-transformed white spruce roots did not affect the soil fungal biomass in pots, as determined by measurement of ergosterol using HPLC. Although efforts were made to select young and healthy-looking roots for each repetition, the variation in endochitinase activity observed was probably due to differences in the vigor of the roots soaking in the MSS mixture. Nevertheless, the level of and variation in the endochitinase activity observed for ech42-transformed white spruce root exudates were comparable to the level of and variation in the endochitinase activity observed by Tesfaye et al. (65). These workers transformed alfalfa with an acid phosphatase (APase) signal peptide region from white lupine fused to the N-terminal region of the ech42 protein controlled by the cassava vein mosaic virus (CsVMV) promoter to increase exudation of a transgenic protein.
Vauramo et al. (71) quantified fungal biomass by measuring the ergosterol in leaf litter from transgenic silver birch trees transformed with sugar beet chitinase. They found no significant difference in the fungal biomass between litter samples from transformed lines and litter samples from controls. The level of ergosterol that they obtained was 1,000- to 2000-fold higher than the level that we obtained. This can be explained by the potential limited production of extraradical mycelium of Wilcoxina spp. associated with very low mycorrhizal and nonmycorrhizal fungal diversity.
Real-time PCR to quantify colonization of the mycorrhizal root tip.
Real-time PCR provides useful tools for studying and quantifying mycorrhizal fungus interactions and biomass (33, 47, 53, 59). Unlike workers in previous studies, we did not use the ITS rRNA region to determine the number of Wilcoxina molecules. The expression of genes encoding beta-tubulin and translation elongation factor 1-alpha proved to be the most stable expression in repetitions and, consequently, more accurate for quantification than expression of the ITS rRNA region. This is not surprising considering that several dozen copies of rRNA genes are present in eukaryotic genomes (close to 150 copies) (17), while it is thought that there is one copy or a few copies of the genes encoding beta-tubulin and translation elongation factor 1-alpha (12, 66, 74). Also, we showed that determining the number of molecules for each root tip is a valuable approach for comparing the levels of root tip colonization without weighing root tips or a priori determining the relationship between fungal biomass and the number of molecules.
Interaction of Wilcoxina spp. and P. fortinii.
P. fortinii is part of the Mycelium radicis atrovirens (MRA) complex (25), which is also called the dark septate endophytes (DSE). The DSE group is a prevalent group of Ascomycetes that colonize a wide spectrum of hosts (30) either as root endophytes or as ectendomycorrhizal fungi (77). The nature of the association between P. fortinii and host plants is still unclear as there can be negative, neutral, or positive effects and it varies according to the experimental conditions and host plant (1, 31). Real-time PCR analysis of root tips containing fungi belonging to MG2 showed that there was colonization by both Wilcoxina spp. and P. fortinii. This may explain why direct ITS rRNA sequencing of root tips belonging to MG2 with the fungus-specific primer pair ITS1-F/ITS4 resulted in recovery of either Wilcoxina spp. or P. fortinii. Menkis et al. (39) characterized the fungal communities associated with conifer roots in nurseries and observed that, on average, 2.4 different morphotypes of an individual taxon were found. The detection of P. fortinii ITS molecules in MG2, while Wilcoxina spp. was dominant, suggests that the root tips containing fungi belonging to this morphological group were colonized initially by Wilcoxina spp. and later by P. fortinii. The underrepresentation of Wilcoxina sp. ITS molecules in MG2 compared with MG1 (7.5-fold less) might be explained by mycoparasitism of Wilcoxina spp. by P. fortinii. Krasowski et al. (36) observed the reverse relationship between the relative abundance of Wilcoxina spp. and the relative abundance of DSE in healthy and senescent root systems of white spruce seedlings. The presence of DSE, such as P. fortinii, has been observed in senescent roots of other conifers as well (1, 30).
Conclusion.
The level of endochitinase in roots of ech42-transformed white spruce did not alter establishment and development of the ectendomycorrhizal association. So far, there is no evidence that chitinase-transformed crops, fruits, and forest trees have a negative impact on mycorrhizal and nonpathogenic fungi. However, the number of studies that have assessed the impact of chitinase-transformed plants on nonpathogenic fungi is not sufficient to accurately predict the potential effects of such plants in nature. Moreover, greenhouse trial studies are only the first step in assessing whether chitinase transgenic trees are harmless, and the results of such studies would not necessarily reflect what would happen in the field. For example, Pasonen et al. (49) observed differences in the responses to leaf spot disease between sugar beet chitinase-transformed silver birch trees grown in the greenhouse and trees grown in the field. Long-term field deployment is required to assess the effect of chitinase-transformed trees on nonphytopathogenic fungi and to assess the impact of exudation of a higher level of chitinase by roots on soil mycorrhizal reserves. Finally, the real-time PCR technique is an accurate technique to obtain a better understanding of the intimate mycorrhizal association and the dynamics of root tip colonization by fungi.
Acknowledgments
We thank A. Séguin for production of transgenic white spruce trees and D. Stewart for providing primers for plant housekeeping genes. We also thank M. Bernier-Cardou for helping with statistical analyses and M.-J. Bergeron for a critical review of the manuscript.
This work was supported by a grant from the Canadian Regulatory System for Biotechnology Fund.
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Addy, H. D., M. M. Piercey, and R. S. Currah. 2005. Microfungal endophytes in roots. Can. J. Bot. 83:1-13. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell, J. 1988. Physical methods for the determination of chitin structure and conformation. Methods Enzymol. 161:435-442. [Google Scholar]
- 4.Bolar, J. P., J. L. Norelli, K.-W. Wong, C. K. Hayes, G. E. Harman, and H. S. Aldwinckle. 2000. Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and reduces vigor. Phytopathology 90:72-77. [DOI] [PubMed] [Google Scholar]
- 5.Bolar, J. P., J. L. Norelli, G. E. Harman, S. K. Brown, and H. S. Aldwinckle. 2001. Synergistic activity of endochitinase and exochitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Transgenic Res. 10:533-543. [DOI] [PubMed] [Google Scholar]
- 6.Boller, T. 1987. Hydrolytic enzymes in plant disease resistance. Mol. Genet. Perspect. 3:385-411. [Google Scholar]
- 7.Brogue, K., I. Chet, M. Holliday, R. Cressman, P. Biddle, S. Knowlton, C. J. Mauvais, and R. Broglie. 1991. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194-1197. [DOI] [PubMed] [Google Scholar]
- 8.Brundrett, M., N. Bougher, B. Dell, T. Grove, and N. Malajczuk. 1996. Working with mycorrhizas in forestry and agriculture. ACIAR monograph MNO 32. Australian Centre for International Agricultural Research, Canberra, Australia.
- 9.Carlson, J. E., L. K. Tulsieram, J. C. Glaubitz, V. W. K. Luk, C. Kauffeldt, and R. Rutledge. 1991. Segregation of random amplified DNA markers in F1 progeny of conifers. Theor. Appl. Genet. 83:194-200. [DOI] [PubMed] [Google Scholar]
- 10.Castaldini, M., A. Turrini, C. Sbrana, A. Benedetti, M. Marchionni, S. Mocali, A. Fabiani, S. Landi, F. Santomassimo, B. Pietrangeli, M. P. Nuti, N. Miclaus, and M. Giovannetti. 2005. Impact of Bt corn on rhizospheric and soil eubacterial communities and on beneficial mycorrhizal symbiosis in experimental microcosms. Appl. Environ. Microbiol. 71:6719-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinge, D. B., K. M. Kragh, J. D. Mikkelsen, K. K. Nielsen, U. Rasmussen, and K. Vad. 1993. Plant chitinases. Plant J. 3:31-40. [DOI] [PubMed] [Google Scholar]
- 12.Cottrelle, P., D. Thiele, V. L. Price, S. Memet, J.-Y. Micouin, C. Marck, J.-M. Buhler, A. Sentenac, and P. Fromageot. 1985. Cloning, nucleotide sequence, and expression of one of two genes coding for yeast elongation factor 1alpha. J. Biol. Chem. 260:3090-3096. [PubMed] [Google Scholar]
- 13.Czechowski, T., M. Stitt, T. Altmann, M. K. Udvardi, and W.-R. Scheible. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139:5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De la Cruz, J., A. Hidalgo-Gallego, J. M. Lora, T. Benitez, J. A. Pintor-Toro, and A. Llobell. 1992. Isolation and characterization of three chitinases from Trichoderma harzianum. FEBS J. 206:859-867. [DOI] [PubMed] [Google Scholar]
- 15.Egger, K. N., R. M. Danielson, and J. A. Fortin. 1991. Taxonomy and population structure of E-strain mycorrhizal fungi inferred from ribosomal and mitochondrial DNA polymorphisms. Mycol. Res. 95:866-872. [Google Scholar]
- 16.Egger, K. N. 1996. Molecular systematics of E-strain mycorrhizal fungi: Wilcoxina and its relationship to Tricharina (Pezizales). Can. J. Bot. 74:773-779. [Google Scholar]
- 17.Free, S. J., P. W. Rice, and R. L. Metzenberg. 1979. Arrangement of the genes coding for ribosomal ribonucleic acids in Neurospora crassa. J. Bacteriol. 137:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedmann, M., S. G. Ralph, D. Aeschliman, J. Zhuang, K. Ritland, B. E. Ellis, J. Bohlmann, and C. J. Douglas. 2007. Microarray gene expression profiling of developmental transitions in Sitka spruce (Picea sitchensis) apical shoots. J. Exp. Bot. 58:593-614. [DOI] [PubMed] [Google Scholar]
- 19.Fujimura, K. E., J. E. Smith, T. R. Horton, N. S. Weber, and J. W. Spatafora. 2005. Pezizalean mycorrhizas and sporocarps in ponderosa pine (Pinus ponderosa) after prescribed fires in eastern Oregon, U. S. A. Mycorrhiza 15:79-86. [DOI] [PubMed] [Google Scholar]
- 20.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 21.Giovannetti, M., and B. Mosse. 1980. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84:489-500. [Google Scholar]
- 22.Hamann, A., and T. Wang. 2006. Potential effects of climate change on ecosystem and tree species distribution in British Columbia. Ecology 87:2773-2786. [DOI] [PubMed] [Google Scholar]
- 23.Hamelin, R. C., P. Bérubé, M. Gignac, and M. Bourassa. 1996. Identification of root rot fungi in nursery seedlings by nested multiplex PCR. Appl. Environ. Microbiol. 62:4026-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harman, G. E., C. K. Hayes, M. Lorito, R. M. Broadway, A. Di Pietro, C. Peterbauer, and A. Tronsmo. 1993. Chitinolytic enzymes of Trichoderma harzianum: purification of chitobiosidase and endochitinase. Phytopathology 83:313-318. [Google Scholar]
- 25.Harney, S. K., S. O. Rogers, and C. J. K. Wang. 1997. Molecular characterization of dematiaceous root endophytes. Mycol. Res. 101:1397-1404. [Google Scholar]
- 26.Herrera Medina, M. J., H. Gagnon, Y. Piché, J. A. Ocampo, J. M. García Garrido, and H. Vierheilig. 2003. Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 164:993-998. [Google Scholar]
- 27.Iwanski, M., M. Rudawska, and T. Leski. 2006. Mycorrhizal associations of nursery grown Scots pine (Pinus sylvestris L.) seedlings in Poland. Ann. For. Sci. 63:715-723. [Google Scholar]
- 28.Jayaraj, J., and Z. K. Punja. 2007. Combined expression of chitinase and lipid transfer protein genes in transgenic carrot plants enhances resistance to foliar fungal pathogens. Plant Cell Rep. 26:1539-1546. [DOI] [PubMed] [Google Scholar]
- 29.Jeng, R. S., M. Dumas, F. H. Liu, C. L. Wang, and M. Hubbes. 1997. DNA analysis of Cylindrocladium floridanum isolates from selected forest nurseries. Mycol. Res. 101:285-291. [Google Scholar]
- 30.Jumpponen, A., and J. M. Trappe. 1998. Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol. 140:295-310. [DOI] [PubMed] [Google Scholar]
- 31.Jumpponen, A. 2001. Dark septate endophytes—are they mycorrhizal? Mycorrhiza 11:207-211. [Google Scholar]
- 32.Kaldorf, M., M. Fladung, H.-J. Muhs, and F. Buscot. 2002. Mycorrhizal colonization of transgenic aspen in a field trial. Planta 214:653-660. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy, P. G., S. E. Bergemann, S. Hortal, and T. D. Bruns. 2007. Determining the outcome of field-based competition between two Rhizopogon species using real-time PCR. Mol. Ecol. 16:881-890. [DOI] [PubMed] [Google Scholar]
- 34.Kim, J. K., I.-C. Jang, R. Wu, W.-N. Zuo, R. S. Boston, Y.-H. Lee, I.-P. Ann, and B. H. Nahm. 2003. Co-expression of a modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Transgenic Res. 12:475-484. [DOI] [PubMed] [Google Scholar]
- 35.Koncz, C., and J. Schell. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204:383-396. [Google Scholar]
- 36.Krasowski, M. J., J. N. Owens, L. E. Tackaberry, and H. B. Massicotte. 1999. Above- and below-ground growth of white spruce seedlings with roots divided into different substrates with or without controlled-release fertilizer. Plant Soil 217:131-143. [Google Scholar]
- 37.Labau, V. J., and W. S. van Hees. 1990. An inventory of Alaska's boreal forests: their extent, condition, and potential use, p. 30-99. In Condition, dynamics, anthropogenic effects. Proceedings of the International Symposium on Boreal Forests, 16-26 July 1990, Archangelsk, Russia. State Forest Committee of the USSR, Moscow, Russia.
- 38.McLeod, T. K., and G. MacDonald. 1997. Postglacial range expansion and population growth of Picea mariana, Picea glauca and Pinus banksiana in the western interior of Canada. J. Biogeogr. 24:865-881. [Google Scholar]
- 39.Menkis, A., R. Vasiliauskas, A. F. S. Taylor, J. Stenlid, and R. Finlay. 2005. Fungal communities in mycorrhizal roots of conifer seedlings in forest nurseries under different cultivation systems, assessed by morphotyping, direct sequencing and mycelial isolation. Mycorrhiza 16:33-41. [DOI] [PubMed] [Google Scholar]
- 40.Mikola, P. 1965. Studies on the ectendotrophic mycorrhiza of pine. Acta For. Fenn. 79:5-56. [Google Scholar]
- 41.Montgomery, H. J., C. M. Monreal, J. C. Young, and K. A. Seifert. 2000. Determination of soil fungal biomass from soil ergosterol analyses. Soil Biol. Biochem. 32:1207-1217. [Google Scholar]
- 42.Morrison, D. J., K. W. Pellow, D. J. Norris, and A. F. L. Nemec. 2000. Visible versus actual incidence of Armillaria root disease in juvenile coniferous stands in the southern interior of British Columbia. Can. J. For. Res. 30:405-414. [Google Scholar]
- 43.Newhouse, A. E., F. Schrodt, H. Liang, C. A. Maynard, and W. A. Powell. 2007. Transgenic American elm shows reduced Dutch elm disease symptoms and normal mycorrhizal colonization. Plant Cell Rep. 26:977-987. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen, K. K., P. Jørgensen, and J. D. Mikkelsen. 1994. Antifungal activity of sugar beet chitinase against Cercospora beticola: an autoradiographic study on cell wall degradation. Plant Pathol. 43:979-986. [Google Scholar]
- 45.Noël, A., C. Levasseur, V. Q. Le, and A. Séguin. 2005. Enhanced resistance to fungal pathogens in forest trees by genetic transformation of black spruce and hybrid poplar with a Trichoderma harzianum endochitinase gene. Physiol. Mol. Plant Pathol. 67:92-99. [Google Scholar]
- 46.Pappinen, A., Y. Degefu, L. Syrjälä, K. Keinonen, and K. von Weissenberg. 2002. Transgenic silver birch (Betula pendula) expressing sugarbeet chitinase 4 shows enhanced resistance to Pyrenopeziza betulicola. Plant Cell Rep. 20:1046-1051. [Google Scholar]
- 47.Parladé, J., S. Hortal, J. Pera, and L. Galipienso. 2007. Quantitative detection of Lactarius deliciosus extraradical soil mycelium by real-time PCR and its application in the study of fungal persistence and interspecific competition. J. Biotechnol. 128:14-23. [DOI] [PubMed] [Google Scholar]
- 48.Parmesan, C. 2006. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37:637-669. [Google Scholar]
- 49.Pasonen, H.-L., S. K. Seppänen, Y. Degefu, A. Rytkönen, K. von Weissenberg, and A. Pappinen. 2004. Field performance of chitinase transgenic silver birches (Betula pendula): resistance to fungal diseases. Theor. Appl. Genet. 109:562-570. [DOI] [PubMed] [Google Scholar]
- 50.Pasonen, H.-L., Y. Degefu, J. Brumós, K. Lohtander, A. Pappinen, S. Timonen, and S. K. Seppänen. 2005. Transgenic Betula pendula expressing sugar beet chitinase IV forms normal ectomycorrhizae with Paxillus involutus in vitro. Scand. J. For. Res. 20:385-392. [Google Scholar]
- 51.Peberdy, J. F. 1990. Fungal cell walls—a review, p. 5-30. In P. J. Kuhn, A. P. J. Trinci, M. J. Jung, and M. W. Goosey (ed.), Biochemistry of cell walls and membranes in fungi. Springer-Verlag, Berlin, Germany.
- 52.Perez-Moreno, J., and D. J. Read. 2000. Mobilization and transfer of nutrients from litter to tree seedlings via the vegetative mycelium of ectomycorrhizal plants. New Phytol. 145:301-309. [Google Scholar]
- 53.Raidl, S., R. Bonfigli, and R. Agerer. 2005. Calibration of quantitative real-time Taqman PCR by correlation with hyphal biomass and ITS copies in mycelia of Piloderma croceum. Plant Biol. 7:713-717. [DOI] [PubMed] [Google Scholar]
- 54.R Development Core Team. 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 55.Read, D. J. 1991. Mycorrhizas in ecosystems. Experientia 47:376-391. [Google Scholar]
- 56.Read, D. J., and J. Perez-Moreno. 2003. Mycorrhizas and nutrient cycling in ecosystems—a journey towards relevance? New Phytol. 157:475-492. [DOI] [PubMed] [Google Scholar]
- 57.Rutledge, R. G., and D. Stewart. 2008. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.SAS Institute Inc. 1999. SAS/STAT8 software. SAS Institute Inc., Cary, NC.
- 59.Schubert, R., S. Raidl, R. Funk, G. Bahnweg, G. Müller-Starck, and R. Agerer. 2003. Quantitative detection of agar-cultivated and rhizotron-grown Piloderma croceum Erikss. & Hjortst. by ITS1-based fluorescent PCR. Mycorrhiza 13:159-165. [DOI] [PubMed] [Google Scholar]
- 60.Seppänen, S.-K., H.-L. Pasonen, S. Vauramo, J. Vahala, M. Toikka, I. Kilpeläinen, H. Setälä, T. H. Teeri, S. Timonen, and A. Pappinen. 2007. Decomposition of the leaf litter and mycorrhiza forming ability of silver birch with a genetically modified lignin biosynthesis pathway. Appl. Soil Ecol. 36:100-106. [Google Scholar]
- 61.Sinclair, W. A., D. M. Sylvia, and A. O. Larsen. 1982. Disease suppression and growth promotion in Douglas-fir seedlings by the ectomycorrhizal fungus Laccaria laccata. For. Sci. 28:191-201. [Google Scholar]
- 62.Skilling, D. D. 1981. Scleroderris canker—the situation in 1980. J. For. 79:95-97. [Google Scholar]
- 63.Smith, S. E., and D. J. Read. 2008. Mycorrhizal symbiosis, 3rd ed. Academic Press, San Diego, CA.
- 64.Staehelin, C., C. Charon, T. Boller, M. Crespi, and A. Kondorosi. 2001. Medicago truncatula plants overexpressing the early nodulin gene enod40 exhibit accelerated mycorrhizal colonization and enhanced formation of arbuscules. Proc. Natl. Acad. Sci. U. S. A. 98:15366-15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tesfaye, M., M. D. Denton, D. A. Samac, and C. P. Vance. 2005. Transgenic alfalfa secretes a fungal endochitinase protein to the rhizosphere. Plant Soil 269:233-243. [Google Scholar]
- 66.Thon, M. R., and D. J. Royse. 1999. Partial β-tubulin gene sequences for evolutionary studies in the Basidiomycotina. Mycologia 91:468-474. [Google Scholar]
- 67.Trocha, L. K., M. Rudawska, T. Leski, and M. Dabert. 2006. Genetic diversity of naturally established ectomycorrhizal fungi on Norway spruce seedlings under nursery conditions. Microb. Ecol. 52:418-425. [DOI] [PubMed] [Google Scholar]
- 68.Turrini, A., C. Sbrana, L. Pitto, M. Ruffini Castiglione, L. Giorgetti, R. Briganti, T. Bracci, M. Evangelista, M. P. Nuti, and M. Giovannetti. 2004. The antifungal Dm-AMP1 protein from Dahlia merckii expressed in Solanum melongena is released in root exudates and differentially affects pathogenic fungi and mycorrhizal symbiosis. New Phytol. 163:393-403. [DOI] [PubMed] [Google Scholar]
- 69.Ulhoa, C. J., and J. F. Peberdy. 1991. Purification and characterization of an extracellular chitobiase from Trichoderma harzianum. Curr. Microbiol. 23:285-289. [Google Scholar]
- 70.Ursic, M., and R. L. Peterson. 1997. Morphological and anatomical characterization of ectomycorrhizas and ectendomycorrhizas on Pinus strobus seedlings in a southern Ontario nursery. Can. J. Bot. 75:2057-2072. [Google Scholar]
- 71.Vauramo, S., H.-L. Pasonen, A. Pappinen, and H. Setälä. 2006. Decomposition of leaf litter from chitinase transgenic silver birch (Betula pendula) and effects on decomposer populations in a field trial. Appl. Soil Ecol. 32:338-349. [Google Scholar]
- 72.Vierheilig, H., M. Alt, J.-M. Neuhaus, T. Boller, and A. Wiemken. 1993. Colonization of transgenic Nicotiana sylvestris plants, expressing different forms of Nicotiana tabacum chitinase, by the root pathogen Rhizoctonia solani and by the mycorrhizal symbiont Glomus mosseae. Mol. Plant-Microbe Interact. 6:261-264. [Google Scholar]
- 73.Vierheilig, H., M. Alt, J. Lange, M. Gut-Rella, A. Wiemken, and T. Boller. 1995. Colonization of transgenic tobacco constitutively expressing pathogenesis-related proteins by the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. Appl. Environ. Microbiol. 61:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wendland, J., and E. Kothe. 1997. Isolation of tef1 encoding translation elongation factor EF1[alpha] from the homobasidiomycete Schizophyllum commune. Mycol. Res. 101:798-802. [Google Scholar]
- 75.White, T. B. J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, NY.
- 76.Whitney, R. D., and R. A. Fleming. 2005. Quantifying relationships between root rot in a white spruce plantation and sporophores of Inonotus tomentosus. For. Pathol. 35:75-84. [Google Scholar]
- 77.Yu, T. E. J.-C., K. N. Egger, and R. L. Peterson. 2001. Ectendomycorrhizal associations—characteristics and functions. Mycorrhiza 11:167-177. [Google Scholar]