Abstract
The use of antibiotic resistance genes in plasmids causes potential biosafety and clinical hazards, such as the possibility of horizontal spread of resistance genes or the rapid emergence of multidrug-resistant pathogens. This paper introduces a novel auxotrophy complementation system that allowed plasmids and host cells to be effectively selected and maintained without the use of antibiotics. An Escherichia coli strain carrying a defect in NAD de novo biosynthesis was constructed by knocking out the chromosomal quinolinic acid phosphoribosyltransferase (QAPRTase) gene. The resistance gene in the plasmids was replaced by the QAPRTase gene of E. coli or the mouse. As a result, only expression of the QAPRTase gene from plasmids can complement and rescue E. coli host cells in minimal medium. This is the first time that a vertebrate gene has been used to construct a nonantibiotic selection system, and it can be widely applied in DNA vaccine and gene therapy. As the QAPRTase gene is ubiquitous in species ranging from bacteria to mammals, the potential environmental biosafety problems caused by horizontal gene transfer can be eliminated.
Antibiotic resistance genes are the most commonly used markers for selecting and maintaining recombinant plasmids in hosts, such as Escherichia coli. However, the use of these genes has several drawbacks. For example, horizontal transfer of the antibiotic resistance gene can potentially contribute to the rapid emergence of multidrug-resistant organisms (e.g., superbacteria) (11, 29). Another significant concern is that the antibiotic resistance genes in DNA vaccines may become integrated into human chromosomes (23). The possibility arises, although the probability is low, that once the antibiotic resistance gene is translated into a functional protein, the vaccinee might be resistant to the corresponding antibiotic. This would add to the difficulty of curing diseases caused by infectious pathogens. Accordingly, the use of antibiotic resistance genes is undesirable in many areas of biotechnology, especially in gene therapy products and genetically engineered microorganisms (17, 23, 28). Furthermore, the addition of antibiotics is costly in large-scale cultivation, and there are risks of contamination of the final product with antibiotics (2, 3). Finally, the constitutively expressed antibiotic resistance genes impose a metabolic burden on the host cells, resulting in reduced growth rate and cell density (4, 27). An alternative strategy is to utilize antibiotic-free host-plasmid balanced lethal systems to select and maintain the recombinant plasmids.
To date, several such systems have been developed to replace traditional antibiotic selection systems. They include auxotrophy complementation (AC), postsegregational killing (PSK), and operator-repressor titration (ORT) (8). The AC system is based on a strain auxotrophic for an essential metabolite, obtained by mutating or knocking out the corresponding chromosomal gene, which can be complemented with the plasmid-borne selection gene. The choice of the essential gene used for complementation of host auxotrophy is critical, and it is mainly involved in DNA precursor, amino acid, or cell wall biosynthetic pathways. Various essential genes, such as asd, thyA, and glnA, have been utilized to construct AC systems (5, 9, 21, 22, 24, 26, 28). However, all of these systems require extra nutrients or expensive reagents. The PSK system relies on the balance between toxin and antitoxin, expressed from genome and plasmid, respectively. If a cell loses the plasmid, the corresponding antitoxin is degraded and the toxin then kills the cell. Unfortunately, this system has proven ineffective for plasmid maintenance during prolonged culture (6, 14). The ORT system utilizes plasmids with the lac operator to derepress a modified essential chromosomal gene. Loss of these types of plasmids no longer titrates the repressor and leads to the death of the bacterium. This system requires short, nonexpressed lac operator functions as the vector-borne selection marker and enables the selection and maintenance of plasmids free from expressed selectable marker genes (7, 8, 15, 30). Additionally, several other nonantibiotic selection systems (e.g., the fabI-triclosan system) have recently been developed (12, 17, 18).
Among the antibiotic-free selection systems that have been developed, the AC system has drawn much attention and has now been applied in numerous bacterial species, such as Lactococcus lactis, Salmonella spp., Vibrio cholerae, Mycobacterium bovis, and E. coli (5, 16, 21, 22, 24). However, all of the AC systems utilize plasmid-borne bacterial-origin genes to complement the auxotrophy. These systems may suffer from a potential risk that the bacterial-origin genes may be integrated into human chromosome when they are used in transgenic products, such as DNA vaccines. Therefore, a better strategy would be to use the genes of the vaccinees themselves to construct an AC system. Not only would this type of approach select and maintain plasmids in bacteria, but it could also be widely applied in the production of safer DNA vaccines.
In the present study, we successfully developed a novel antibiotic-free plasmid selection system based on complementation of host auxotrophy in the NAD synthesis pathway. The NAD synthesis pathway, including de novo and salvage pathways, differs among species. However, by comparison of NAD metabolism in different species, quinolinic acid phosphoribosyltransferase (QAPRTase) appears to be a common enzyme for de novo NAD biosynthesis in both prokaryotes and eukaryotes (13). Therefore, the QAPRTase gene was viewed as a favorable candidate that could potentially be utilized to construct a new AC system.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and reagents.
The bacterial strains and plasmids used in this study are described in Table 1. The bacterial strains were grown in Luria-Bertani (LB) broth, M9 broth, LB agar, and M9 agar. When required, the LB or M9 medium was supplemented with ampicillin (Amp) (100 μg/ml), kanamycin (Km) (50 μg/ml), nicotinic acid (NA) (10 μg/ml) (M9+NA), and l-arabinose (1 mmol/liter). M9 media were comprised of 17.1 mg/ml Na2HPO4·12H2O, 3 mg/ml KH2PO4, 0.5 mg/ml NaCl, 1 mg/ml NH4Cl, 2 mmol/liter MgSO4, 0.1 mmol/liter CaCl2, and 0.4% (wt/vol) glucose. All chemicals were purchased from Sigma. DpnI was from Toyobo (Japan); Taq polymerase, TaKaRa Ex Taq, and agarose were from Takara (Japan).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and comments | Source |
|---|---|---|
| Strains | ||
| DH5α | Routine cloning host | Department collection |
| BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | CGSC |
| BW25113ΔQAPRTase | Like BW25113 but with an entire chromosomal QAPRTase gene deletion | This study |
| Plasmids | ||
| pKD13 | Used in gene knockout procedure | CGSC |
| pKD46 | Used in gene knockout procedure | CGSC |
| pCP20 | Used in gene knockout procedure | CGSC |
| pUC19 | bla+ | Department collection |
| pZJU11 | Like pUC19 but with the bla gene replaced by the QAPRTase gene of E. coli | This study |
| pZJU12 | Like pUC19 but with the bla gene replaced by the QAPRTase gene of mouse | This study |
| pBAD-hisA | bla+ | Invitrogen |
| pZJU21 | Like pBAD-hisA but with the bla gene replaced by the QAPRTase gene of E. coli | This study |
| pZJU22 | Like pBAD-hisA but with the bla gene replaced by the QAPRTase gene of mouse | This study |
| pBAD-hisA-EGFP | EGFP gene was inserted into the MCS of pBAD-hisA | This study |
| pZJU21-EGFP | EGFP gene was inserted into the MCS of pZJU21 | This study |
| pZJU22-EGFP | EGFP gene was inserted into the MCS of pZJU22 | This study |
| pCDNA3.1+ | bla+ | Invitrogen |
| pZJU31 | Like pCDNA3.1+ but with the bla gene replaced by the QAPRTase gene of E. coli | This study |
| pZJU32 | Like pCDNA3.1+ but with the bla gene replaced by the QAPRTase gene of mouse | This study |
| pCDNA-EGFP | EGFP gene was inserted into the MCS of pCDNA3.1+ | This study |
| pZJU31-EGFP | EGFP gene was inserted into the MCS of pZJU31 | This study |
| pZJU32-EGFP | EGFP gene was inserted into the MCS of pZJU32 | This study |
| pEGFP-N2 | Containing the EGFP gene | BD Biosciences Clontech |
Cloning of QAPRTase genes.
Web searches for QAPRTase gene homology in different species were done using the BLAST or BLAT program from the NCBI (http://www.ncbi.nlm.nih.gov/) and the University of California—Santa Cruz (UCSC) genome bioinformatics website (http://genome.ucsc.edu/). Multiple-sequence alignments were generated using the CLUSTALW program (http://align.genome.jp/). The E. coli and mouse QAPRTase genes were selectively cloned in our study. The genomic DNA template of strain BW25113 was extracted with the QIAamp DNA Mini Kit (Qiagen, Germany), and the total liver RNA from BALB/c mice was extracted using TRIzol reagent (Gibco BRL) following the manufacturer's instructions. The concentration of total RNA was measured by spectrophotometry, and 1 μg was reverse transcribed into a cDNA template using a TaKaRa RNA PCR Kit (AMV) (TaKaRa, Japan). PCR amplification for gene cloning was performed in 50-μl reaction mixtures containing the following components: 5 μl of Pyrobest buffer (10×), 4 μl of deoxynucleotide triphosphate (dNTP) mixture (2.5 mM each), 0.5 μl of Pyrobest DNA polymerase (5 units/μl) (TaKaRa, Japan), 2 μl of forward and reverse primers (E-QAPRTase-F/E-QAPRTase-R for the E. coli QAPRTase gene and M-QAPRTase-F/M-QAPRTase-R for the mouse QAPRTase gene) (10 μM each), as listed in Table S1 in the supplemental material, and 1 μl of template and 35.5 μl of sterile distilled H2O. The cycling protocol was 1 cycle at 94°C for 4 min and 33 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, followed by 1 cycle at 72°C for 5 min. The PCR products were separated on 1.2% (wt/vol) agarose gels. After ethidium bromide staining, the gel was analyzed using 1D Image Analysis Software with the Kodak Gel Logic 200 Imaging System (Eastman Kodak Company). All products amplified by PCR were sequenced on an ABI 3730 automated sequencer (PE Applied Biosystems).
Construction of ΔQAPRTase E. coli.
The chromosomal QAPRTase gene in E. coli BW25113 was deleted using a λ Red-mediated recombination system, as previously described (10). Chromosomal QAPRTase gene deletion was confirmed by PCR and sequencing on an ABI 3730 automated sequencer (PE Applied Biosystems). The primers (QAPRTase-F/QAPRTase-R for gene knockout and QAPRTase-U/QAPRTase-D for knockout verification) used in the experiment are listed in Table S1 in the supplemental material.
Construction of ΔAmp::QAPRTase plasmids.
Three plasmids, pUC19, pBAD-hisA, and pCDNA3.1+, were selectively used as basic plasmids to construct the antibiotic resistance gene-free vector system. pUC19 and pBAD-hisA are commonly used cloning and prokaryotic expression vectors, respectively, in E. coli. pCDNA3.1+ is a commonly used eukaryotic expression vector that is also widely used as a DNA vaccine vector. The antibiotic resistance gene in the basic plasmids was replaced by the QAPRTase gene of E. coli and the mouse. For example, for the creation of pUC19ΔAmp from pUC19, the entire pUC19 plasmid DNA except for the ampicillin resistance gene (bla) was amplified by PCR with a pair of primers (pUC19-F/pUC19-R) listed in Table S1 in the supplemental material. The amplicons of the QAPRTase genes, as described above, and pUC19ΔAmp were then double digested with MluI and XhoI. After gel purification, the cleaved fragments were ligated with T4 DNA ligase and transformed into BW25113ΔQAPRTase to select the positive recombinant. The constructed vectors were verified by double digestion with MluI and XhoI and further confirmed by DNA sequencing. The bla genes of pBAD-hisA and pCDNA3.1+ were replaced by both E. coli and mouse QAPRTase genes using a similar method. The constructed plasmids were designated the pZJU series, as listed in Table 1.
Complementation test.
BW25113ΔQAPRTase transformed with different plasmids was dotted onto different plates, including M9 plates and M9 plates containing NA, with or without ampicillin, at 37°C for 12 h to test the validity of the newly constructed system. BW25113 suspension (0.2 μl; optical density at 600 nm [OD600] = 0.1) for each dot was also applied to the plates as a positive control.
Determination of transformation efficiency.
The transformation efficiencies of all the plasmids, including basic plasmids and constructed plasmids, were determined. Calcium-competent BW25113ΔQAPRTase cells were prepared as 0.1-ml aliquots (25) and transformed with 0.1 μg of each plasmid. Following a heat shock at 42°C for 60 s, 0.9 ml LB broth was added, and the transformation cultures were incubated at 37°C for 1 h. They were washed twice in fresh M9 medium, and a 10−3 dilution was plated on M9 plates (AC selection system) and M9 plates containing NA and ampicillin (antibiotic selection system). Experiments were performed in triplicate, and the colonies were counted to determine the transformation efficiency.
Determination of plasmid abundance.
Plasmid abundance was determined by relative quantitative PCR. The plasmid replicon (Rep) was used as the target gene and a single-copy chromosomal dxs as the reference gene (19, 20). BW25113ΔQAPRTase cells containing different plasmids were inoculated into cultures with relevant selection pressure, and total DNA was extracted from the cultures during the exponential phase, which was determined by periodic measurement of the OD600. The extraction was performed using the QIAamp DNA Mini Kit (Qiagen, Germany) as described in the instruction manual. Primers (Rep-F/Rep-R and dxs-F/dxs-R), listed in Table S1 in the supplemental material, amplifying the target gene and reference gene were validated for similar amplification efficiencies. Real-time data analyses were carried out by the 2−ΔΔCT method for quantitative real-time PCR. Each sample was run in three parallel reactions.
Determination of plasmid stability.
The stability of reconstructed plasmids was assayed in the presence (M9 medium) or absence (M9+NA) of selection pressure. A single colony was inoculated into M9 medium, and the culture was grown overnight at 37°C with shaking. The saturated culture was diluted to 10−3 cells per ml in defined medium, with or without selection pressure, and kept in continuous culture by similar dilutions every 24 h. Diluted culture samples were plated on M9+NA plates at various intervals and incubated overnight at 37°C. Colonies were replica plated onto M9 plates to confirm the presence of the plasmid. Meanwhile, BW25113ΔQAPRTase cells harboring the original plasmids, including pUC19, pBAD-hisA, and pCDNA3.1+, were tested as control groups, either in the presence (M9 medium containing NA and 200 μg/ml Amp) or absence (M9 medium containing NA) of selection pressure. Plasmid minipreparations were then performed on several independent colonies to confirm the presence of the plasmid.
Growth rate measurements.
Single colonies were inoculated into 5 ml of M9 minimal medium and grown overnight at 37°C in an orbital shaker at 200 rpm. These precultures were inoculated into 50 ml of the same medium in 250-ml Erlenmeyer flasks to a starting optical density at 600 nm of 0.005. The cultures were aerated with vigorous shaking at 37°C, and readings were taken hourly. All experiments were performed in triplicate.
Prokaryotic expression assay.
To confirm that the newly constructed AC system could be used to express foreign proteins in prokaryotic cells, the enhanced green fluorescent protein (EGFP) gene was inserted into the prokaryotic expression vectors pZJU21 and pZJU22 and the parent plasmid, pBAD-hisA. Using pEGFP-N2 plasmid DNA as a template, the EGFP gene was amplified with the primers EGFP-F1/EGFP-R for the construction of pBAD-hisA-EGFP and pZJU21-EGFP and with EGFP-F2/EGFP-R for pZJU22-EGFP construction, as listed in Table S1 in the supplemental material, and then inserted into the multiple cloning site (MCS) of the three plasmids, resulting in pZJU21-EGFP, pZJU22-EGFP, and pBAD-hisA-EGFP, respectively. Then, the plasmids were transformed into BW25113ΔQAPRTase, and the expression of EGFP was induced by adding 0.2% arabinose, according to the protocols provided by the manufacturer. The expression of EGFP in E. coli was visualized and photomicrographed under a phase-contrast microscope (MIC01631; Zeiss, Germany).
Eukaryotic expression assay.
To confirm that the newly constructed AC system could be used to express foreign proteins in eukaryotic cells, the EGFP gene was inserted into the eukaryotic expression vectors pZJU31 and pZJU32 and the parent plasmid, pCDNA3.1+. Using pEGFP-N2 plasmid DNA as a template, the EGFP gene was amplified with the primers EGFP-F3/EGFP-R for the construction of pCDNA-EGFP and pZJU31-EGFP and with EGFP-F4/EGFP-R for pZJU32-EGFP construction, as listed in Table S1 in the supplemental material, and then inserted into the MCS of the three plasmids, resulting in pZJU31-EGFP, pZJU32-EGFP, and pCDNA-EGFP, respectively. Human embryonic kidney cells (HEK293 cells) were incubated in RPMI 1640 medium (HyClone) supplemented with 10% fetal calf serum (HyClone) for 24 h at 37°C in the presence of 5% CO2. The cells were transfected with the three plasmids using the Lipofectamine 2000 Transfection Reagent (Invitrogen), as described by the manufacturer. Expression of EGFP was visualized and photomicrographed by fluorescence microscopy (MIC01631; Zeiss, Germany) at 488 nm 24 h after transfection.
RESULTS
Evaluation of the QAPRTase gene used for antibiotic-free selection.
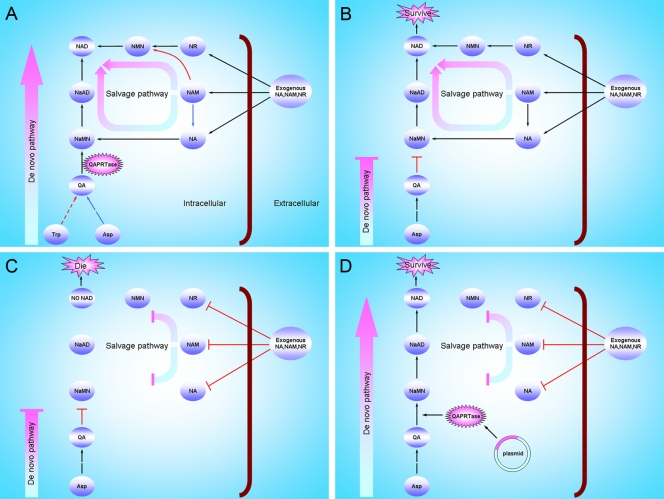
To evaluate the use of the QAPRTase gene in antibiotic-free selection, we initially analyzed the NAD metabolism pathways from prokaryotes to eukaryotes throughout evolution by bioinformatics. The result indicated that NAD in different species could be synthesized through two metabolic pathways referred to as the de novo pathway and the salvage pathway. As shown in Fig. 1A, NAD can be de novo synthesized from tryptophan or aspartate, also referred to as the kynurenine and aspartate pathways, respectively. Many prokaryotes and plants are capable of synthesizing NAD from aspartate, while most aerobes, including vertebrates and some plants, appear to utilize tryptophan for NAD biosynthesis. According to the different substrates, including NA, nicotinamide (NAM), and nicotinamide riboside (NR), the salvage pathway can be designated salvage pathway I, II, or III, respectively. However, the de novo pathway is critically important if the medium lacks any NAD precursor that can be utilized in the salvage pathway. One very interesting aspect of the de novo NAD biosynthetic pathways is that both pathways lead to the formation of a key intermediate, quinolinic acid (QA). Subsequently, QA is converted to nicotinic acid mononucleotide (NaMN) by means of the phosphoribosyl pyrophosphate-dependent enzyme quinolinic acid phosphoribosyltransferase (QAPRTase). Ultimately, NaMN is converted to NAD through the universal pathway, sequentially from NaMN to NaAD to NAD (Fig. 1A). Based on the fact that the QAPRTase gene exists in both prokaryotes and eukaryotes (see Table S2 in the supplemental material), the QAPRTase gene of vertebrates could potentially be utilized to construct an AC system in E. coli.
FIG. 1.
Schematic representations of NAD biosynthesis pathways and principles for the establishment of an antibiotic-free plasmid selection system based on complementation of host auxotrophy in the NAD de novo pathway. (A) NAD synthesis pathways in vertebrates and E. coli. The red arrows show the steps in vertebrates, the blue arrows show the steps in E. coli, and the black arrows show the common reactions. (B) A ΔQAPRTase E. coli strain was constructed, and E. coli was able to survive, even when the NAD de novo pathway was blocked, in a medium containing an NAD precursor. (C) ΔQAPRTase E. coli could not survive in media, such as M9 medium, that lacked an NAD precursor. (D) The ΔQAPRTase E. coli strain, which was auxotrophic in the NAD de novo pathway, could be complemented with the plasmid-borne selection gene.
Cloning and characterization of the QAPRTase gene.
The cloned E. coli and mouse QAPRTase genes were identical to the predicted sequences. Multiple-sequence alignment of QAPRTase in different species was examined. The deduced amino acid sequences of the QAPRTase genes exhibited different identities with each homologue in both prokaryotic and eukaryotic organisms (see Table S3 in the supplemental material). However, the active sites of QAPRTase in different species were well conserved (see Fig. S1 in the supplemental material), which implied a conserved function throughout its evolutionary history and indicated that the mammalian QAPRTase could function in prokaryotic cells.
Construction of an E. coli BW25113ΔQAPRTase strain.
BW25113, a strain with a well-defined pedigree that has not been subjected to mutagens, was selected to construct BW25113ΔQAPRTase (1). For this purpose, the chromosomal QAPRTase gene was replaced by a kanamycin resistance (Kmr) gene with 36-nucleotide (nt) homology extensions by λ Red-mediated recombination. After selection, 10 Kmr colonies were chosen, and the correct mutants were verified by PCR and sequencing. Subsequently, the Kmr gene was eliminated by pCP20, which expressed the FLP recombinase. Finally, all of the helper plasmids, which were temperature-sensitive replicons, were simply cured by growth at 37°C.
Construction of ΔAmp::QAPRTase plasmids.
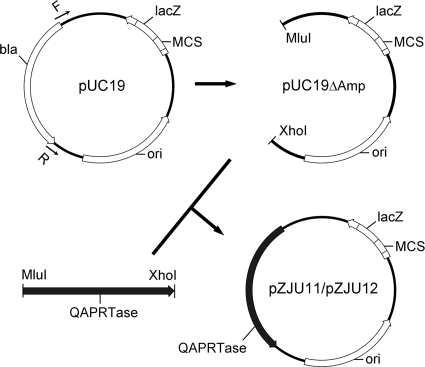
Three pairs of ΔAmp::QAPRTase plasmids were constructed. These plasmids were designated pZJU11/pZJU12, pZJU21/pZJU22, and pZJU31/pZJU32, and they represented the plasmid cloning vector, the prokaryotic expression vector, and the eukaryotic expression vector, respectively. A representative strategy for plasmid construction, in which pZJU11/pZJU12 was constructed, is shown in Fig. 2. pZJU21/pZJU22 and pZJU31/pZJU32 were constructed by similar strategies. The correct recombinant plasmids, confirmed by DNA sequencing, were used in the subsequent experiments.
FIG. 2.
Schematic representation of the strategy employed in this study for the construction of the pZJU11/pZJU12 vector series. The bla gene in the plasmid conferred ampicillin resistance on host cells. The E. coli and mouse QAPRTase genes replaced the bla gene in pUC19, resulting in pZJU11 and pZJU12, respectively.
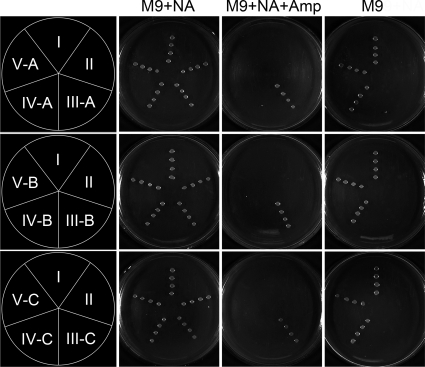
Complementation test.
The complementation test showed that all the strains, with or without plasmids, could propagate in M9 plates supplied with the NAD precursor NA, which was consistent with Fig. 1B. Meanwhile, the strains containing the reconstructed plasmids, excluding the bla gene, could not survive in M9 plates containing NA and Amp. As shown in Fig. 3, the E. coli strain deficient in the QAPRTase gene could not survive in M9 medium, indicating that the auxotroph for NAD de novo synthesis is a lethal phenotype in minimal medium. Besides the wild-type strain BW25113, only BW25113ΔQAPRTase with the E. coli or mouse QAPRTase gene was able to grow on M9 minimal medium in the absence of added NA, which was consistent with Fig. 1C and D. This indicated that, in addition to QAPRTase of E. coli, mouse QAPRTase was also able to confer the ability to convert QA to NaMN for NAD synthesis in E. coli. The experimental data showed that the bacteria deficient in the QAPRTase gene could be complemented by a plasmid-borne QAPRTase gene of either prokaryotic or eukaryotic organisms.
FIG. 3.
Complementation of the BW25113ΔQAPRTase E. coli strain with the QAPRTase gene from either E. coli or mouse. The BW25113ΔQAPRTase strain was transformed with plasmids carrying the E. coli QAPRTase gene, the mouse QAPRTase gene, or no insert. Culture plates were inoculated with equal amounts of each bacterial strain and incubated for 12 h at 37°C. Three kinds of culture media, M9 medium containing NA (M9+NA), M9 medium containing NA and ampicillin (M9+NA+Amp), and M9 medium without any supplement, were used. The bacterial strains were as follows: I, BW25113; II, BW25113ΔQAPRTase; III-A, BW25113ΔQAPRTase/pUC19; III-B, BW25113ΔQAPRTase/pBAD-hisA; III-C, BW25113ΔQAPRTase/pCDNA3.1+; IV-A, BW25113ΔQAPRTase/pZJU11; IV-B, BW25113ΔQAPRTase/pZJU21; IV-C, BW25113ΔQAPRTase/pZJU31; V-A, BW25113ΔQAPRTase/pZJU12; V-B, BW25113ΔQAPRTase/pZJU22; V-C, BW25113ΔQAPRTase/pZJU32.
Determination of transformation efficiency.
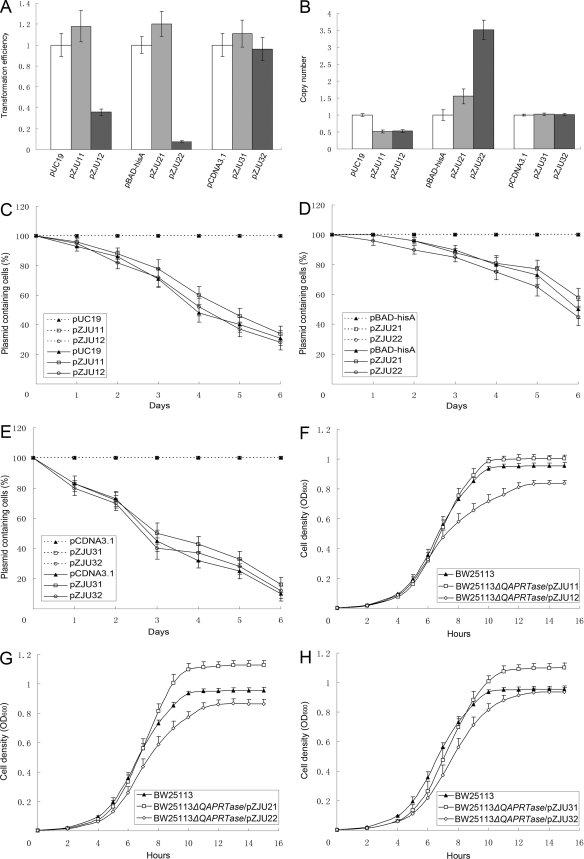
The transformation efficiencies of different plasmids in BW25113ΔQAPRTase are shown in Fig. 4A. The results showed that, when the bla gene in basic plasmids was replaced with the BW25113 origin QAPRTase gene, the transformation efficiencies of the constructed plasmids were equivalent to those of the parent plasmids, including pUC19, pBAD-hisA, and pCDNA3.1+. This indicated that the replacement of the bla gene with the E. coli QAPRTase gene did not influence the transformation efficiencies of the plasmids. However, when the bla gene in basic plasmids was replaced with the mouse QAPRTase gene, the transformation efficiencies of pZJU12 and pZJU22 were lower than those observed for the basic plasmids pUC19 and pBAD-hisA. The reason for this has not yet been clarified. Nevertheless, the transformation efficiency of the constructed plasmid pZJU32 was equivalent to that of the basic plasmid pCDNA3.1+.
FIG. 4.
Plasmid properties and determination of the growth rates of strains. (A) Transformation efficiencies of different plasmids in E. coli strain BW25113ΔQAPRTase. The transformation efficiencies of basic plasmids were arbitrarily set at 1.0, and the transformation efficiencies of constructed plasmids were adjusted accordingly. (B) Copy numbers of different plasmids in E. coli strain BW25113ΔQAPRTase. The copy numbers of basic plasmids were arbitrarily set at 1.0, and the copy numbers of constructed plasmids were adjusted accordingly. (C to E) Comparison of the growth rates of BW25113ΔQAPRTase strains transformed with different complement plasmids and BW25113. All strains were cultured in M9 minimal medium, and the cell density was measured as the increase in optical density. (F to H) Stability determination of different plasmids in E. coli strain BW25113ΔQAPRTase. Cells were grown continuously by subculturing them in the absence or presence of selection pressure. The frequency of plasmid-harboring cells was determined by replica plating onto selective plates. For basic plasmids, the selective plates were LB agar plates containing ampicillin. For the constructed plasmids, the selective plates were M9 agar plates. The data from three independent experiments were averaged. The error bars indicate standard errors of the means.
Determination of plasmid abundance.
The abundances of a different series of plasmids were examined by relative quantitative PCR. As shown in Table S4 in the supplemental material, the abundances of the various plasmids differed from each other. In general, the copy numbers of the plasmids were low in pBAD-hisA series plasmids, moderate in pUC19 series plasmids, and high in pCDNA3.1+ series plasmids. For pUC19 series plasmids, the abundances of the reconstructed plasmids were decreased by nearly 50% compared to that of the parent plasmid, pUC19 (Fig. 4B). In contrast, the copy numbers of the reconstructed pBAD-hisA series plasmids (pZJU21 and pZJU22) increased 55% and 250%, respectively, over that of the initial plasmid, pBAD-hisA. No significant difference in the plasmid abundances was noted among the pCDNA3.1+ series plasmids.
Determination of plasmid stability.
The stabilities of the reconstructed plasmids were evaluated in the presence and absence of selection pressure. As shown in Fig. 4C to E, when selection pressure was imposed, both the parent plasmids and the reconstructed plasmids were stable in the BW25113ΔQAPRTase strain. Even after continuous growth for 6 days, the stabilities of all plasmids were still 100% in the BW25113ΔQAPRTase strain. In addition, the stable maintenance of plasmids in host cells also demonstrated that the use of the QAPRTase gene for plasmid stabilization did not induce any cross-feeding effect in the BW25113ΔQAPRTase strains. However, plasmid loss occurred when the selection pressure was effectively removed. More than 60%, 40%, and 80% of the cultured parental strains had lost the plasmid after 6 days of culture without selection pressure (Fig. 4C to E). Although the loss rates of the three types of plasmids differed from each other, no significant difference was observed between the loss rates of the parent plasmids and the reconstructed plasmids.
Growth rate measurements.
Results from growth rate determinations showed that the BW25113ΔQAPRTase strain carrying QAPRTase genes with different origins could grow well in defined M9 minimal medium, although the growth rates differed slightly between strains (Fig. 4F to H). In general, when the auxotrophic strain was complemented by the constructed plasmids containing the BW25113 origin QAPRTase gene, such as pZJU11, pZJU21, and pZJU31, the growth status of the transformed strain was better than that of the prototrophic reference strain. The cell densities of the relevant transformed strains were 5%, 18%, and 15% higher than that of original strain. The intracellular overproduction of QAPRTase proteins did not lead to metabolic-burden effects, which was a further advantage compared with the traditional antibiotic selection systems. The detailed mechanism may be a result of the overexpression of QAPRTase, facilitating the synthesis of NAD and subsequently improving the growth of the transformed cells. When the auxotrophic strain was complemented by the constructed plasmids carrying the mouse origin QAPRTase gene, such as pZJU12, pZJU22, and pZJU32, the growth rates of the transformed strains were slightly lower than that of the prototrophic strain. This may have been due to the low identity between E. coli and mouse QAPRTase sequences.
Prokaryotic expression assay.
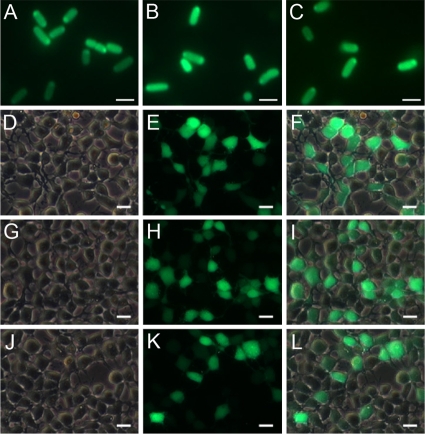
To confirm whether the newly constructed AC system could be used to express foreign proteins as the parent plasmid, the EGFP gene was inserted into the MCS of pBAD-hisA, pZJU21, and pZJU22, resulting in pBAD-hisA-EGFP, pZJU21-EGFP, and pZJU22-EGFP, respectively. After transformation, BW25113ΔQAPRTase cells containing the recombinant plasmids were selected and arabinose was added to the medium to induce the expression of EGFP in the host strain. As shown in Fig. 5A to C, both pZJU21 and pZJU22 could be used to effectively express the foreign protein, as well as the parent plasmid, pBAD-hisA, indicating that the replacement of the bla gene with the QAPRTase gene of E. coli or the mouse did not influence the expression cassette of the vector. This novel AC system can be widely and effectively applied in expressing foreign protein in prokaryotic expression systems.
FIG. 5.
Expression of EGFP protein in E. coli and HEK293 cells. (A to C) Confirmation of the expression of EGFP protein in E. coli strain BW25113ΔQAPRTase. BW25113ΔQAPRTase containing pBAD-hisA-EGFP (A), pZJU21-EGFP (B), or pZJU22-EGFP (C) was induced by arabinose, and EGFP protein was detected by fluorescence microscopy. (D to L) Confirmation of EGFP expression in HEK293 cells under a fluorescence microscope. The cells were transfected with pCDNA-EGFP (D to F), pZJU31-EGFP (G to I), or pZJU32-EGFP (J to L). The merged images (F, I, and L) indicate the merger between the light microscopy (D, G, and J) and EGFP fluorescence windows (E, H, and K). The images presented are in their original form without any modification. Scale bars, 2 μm (A to C) and 20 μm (D to L).
Eukaryotic expression assay.
To test whether the newly constructed AC systems were capable of expressing foreign proteins in eukaryotic cells, thus having potential use as DNA vaccines, the EGFP gene was inserted into the MCS of pCDNA3.1+, pZJU31, and pZJU32, resulting in pCDNA-EGFP, pZJU31-EGFP, and pZJU32-EGFP, respectively. The recombinant plasmids were transformed into HEK293 cells, and the protein expression was detected by fluorescence microscopy after 24 h. Both pZJU31 and pZJU32 effectively expressed the foreign protein as well as the parent plasmid, pCDNA3.1+ (Fig. 5D to L), indicating that the replacement of the bla gene with the QAPRTase gene of E. coli or the mouse did not influence the expression cassette of the vector. This novel AC system can be applied widely and effectively in expressing foreign proteins in eukaryotic expression systems. Furthermore, this type of AC system can also potentially be applied as a DNA vaccine. The replacement of antibiotic resistance genes in DNA vaccine vectors with the QAPRTase gene not only avoids the use of antibiotics, it also eliminates the transfer of resistance genes.
DISCUSSION
Previous research has shown that the AC system can effectively work and potentially replace the traditional antibiotic selection system, thus avoiding the use of antibiotics or antibiotic resistance genes from the outset. However, as the choice of the essential gene used for constructing the AC system is critical, the number of suitable selectable markers is limited at present. Additionally, for bacterial DNA with a mitogenic or immunostimulatory effect, if a cytokine whose expression cannot be terminated has been introduced in the DNA vaccine, the bacterial DNA may aggravate generalized immunosuppression or chronic inflammation (23). All of these factors reduce the generalization and application of the AC system. Accordingly, an alternative strategy is to use the vaccinee's own gene to construct the auxotroph complementation system. This novel approach would then not only be used to select and maintain plasmids in bacteria, but would also be widely applicable to the production of safer DNA vaccines.
The primary function of NAD and its phosphorylated form, NADP, is to serve as either electron donors or acceptors in over 300 enzymatically catalyzed oxidation-reduction reactions (13). Due to this important role of NAD in metabolism, the essential genes involved in NAD biosynthesis can be viewed as forming a new AC system. NAD is synthesized via the de novo pathway or the salvage pathway. Under conditions of nutritional deficiency in the culture medium, the de novo pathway is indispensable for NAD synthesis. Bioinformatics analysis showed that although the de novo pathways differed among species, the QAPRTase gene was the essential gene in both the kynurenine and aspartate pathways. Therefore, the QAPRTase gene was a logical candidate for construction of the AC system. Our experimental data demonstrated that although the identity of the E. coli and mouse QAPRTase genes was only 30%, the QAPRTase gene of the mouse could satisfactorily complement the function of the QAPRTase gene in E. coli strain BW25113ΔQAPRTase, so that it seems feasible to utilize QAPRTase genes of mammalian or even other species origin to construct an AC system.
After replacement of the resistance gene with the QAPRTase gene from E. coli or the mouse, the properties of the newly constructed plasmids were characterized, including transformation efficiency, copy number, stability, and growth rate. Most properties of the constructed plasmids were equivalent to those of the parent plasmids. Moreover, this type of AC system did not impose a metabolic burden on the host cells, which was a substantial improvement over antibiotic selection systems. Our newly developed AC systems were stable, and plasmid-free cells did not accumulate in the culture, indicating that the cross-feeding phenomenon was not a problem. Overall, this type of AC system appeared to be viable for the stable selection and maintenance of plasmids in hosts.
One novel feature of the new AC system was that host cells could be maintained in simple LB medium, which contains abundant NAD precursors, without the addition of any extra expensive reagents. In this respect, we can distinguish this system from other AC systems. Additionally, since most probiotic bacteria and engineering yeasts have the QAPRTase gene, the selection of this AC system can be easily achieved in minimal medium. This advantage could significantly reduce costs and readily facilitate large-scale industrial fermentation of microbiological cultures. The biosafety of DNA vaccines is one of the key questions that need to be addressed before clinical trials or animal testing. Due to the fact that almost all bacteria have a QAPRTase gene, the environmental bacteria cannot gain a selective advantage in a given environmental state even if horizontal gene transfer were to occur. Moreover, utilizing the vaccinee's own QAPRTase gene to replace the antibiotic resistance gene in the plasmid could also exclude the potential detrimental effects of introducing genes of bacterial or other species origin into the vaccinee. The replacement of the antibiotic resistance gene with the QAPRTase gene did not interfere with the expression cassette of either the prokaryotic or the eukaryotic expression plasmids. The expression of foreign protein using this system is therefore achievable. Consequently, the constructed AC system not only completely replaces the traditional antibiotic selection system, it can also be utilized to express foreign proteins. This is the first time that a vertebrate gene has been used to construct a nonantibiotic selection system. This type of system can be expected to produce a more defined and safer gene delivery product that should prove advantageous in all gene therapy applications and in DNA vaccination. Furthermore, our results indicated that an enzyme that is indispensable for the biosynthesis pathways of other important biomolecules (e.g., lipids, coenzyme A [CoA], and ATP) might also be a candidate for constructing chromosome-plasmid balanced lethal systems. It may broaden the choice of essential genes used for developing novel antibiotic-free plasmid selection systems.
Supplementary Material
Acknowledgments
This project was supported by the Hi-Tech Research and Development Program of China (863) (2008AA09Z409), the National Basic Research Program of China (973) (2006CB101805), the China Postdoctoral Science Foundation (20090461374), the National Natural Science Foundation of China (30871936 and 30571423), and the Science and Technology Foundation of Zhejiang Province (2006C12038, 2006C23045, 2006C12005, and 2007C12011).
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balbas, P. 2001. Understanding the art of producing protein and nonprotein molecules in Escherichia coli. Mol. Biotechnol. 19:251-267. [DOI] [PubMed] [Google Scholar]
- 3.Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10:411-421. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, W. E., N. Mirjalili, D. C. Andersen, R. H. Davis, and D. S. Kompala. 1990. Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol. Bioeng. 35:668-681. [DOI] [PubMed] [Google Scholar]
- 5.Borsuk, S., T. A. Mendum, M. Q. Fagundes, M. Michelon, C. W. Cunha, J. McFadden, and O. A. Dellagostin. 2007. Auxotrophic complementation as a selectable marker for stable expression of foreign antigens in Mycobacterium bovis BCG. Tuberculosis 87:474-480. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:12643-12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cranenburgh, R. M., J. A. Hanak, S. G. Williams, and D. J. Sherratt. 2001. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 29:E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cranenburgh, R. M., K. S. Lewis, and J. A. Hanak. 2004. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 7:197-203. [DOI] [PubMed] [Google Scholar]
- 9.Curtiss, R., III, K. Nakayama, and S. M. Kelly. 1989. Recombinant avirulent Salmonella vaccine strains with stable maintenance and high level expression of cloned genes in vivo. Immunol. Investig. 18:583-596. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 12.Fang, C. M., J. Y. Wang, M. Chinchilla, M. M. Levine, W. C. Blackwelder, and J. E. Galen. 2008. Use of mchI encoding immunity to the antimicrobial peptide microcin H47 as a plasmid selection marker in attenuated bacterial live vectors. Infect. Immun. 76:4422-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, J. W., and A. G. Moat. 1980. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol. Rev. 44:83-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galen, J. E., J. Nair, J. Y. Wang, S. S. Wasserman, M. K. Tanner, M. B. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garmory, H. S., M. W. Leckenby, K. F. Griffin, S. J. Elvin, R. R. Taylor, M. G. Hartley, J. A. Hanak, E. D. Williamson, and R. M. Cranenburgh. 2005. Antibiotic-free plasmid stabilization by operator-repressor titration for vaccine delivery by using live Salmonella enterica serovar typhimurium. Infect. Immun. 73:2005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glenting, J., S. M. Madsen, A. Vrang, A. Fomsgaard, and H. Israelsen. 2002. A plasmid selection system in Lactococcus lactis and its use for gene expression in L. lactis and human kidney fibroblasts. Appl. Environ. Microbiol. 68:5051-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goh, S., and L. Good. 2008. Plasmid selection in Escherichia coli using an endogenous essential gene marker. BMC Biotechnol. 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagg, P., J. W. de Pohl, F. Abdulkarim, and L. A. Isaksson. 2004. A host/plasmid system that is not dependent on antibiotics and antibiotic resistance genes for stable plasmid maintenance in Escherichia coli. J. Biotechnol. 111:17-30. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C., J. Kim, S. G. Shin, and S. Hwang. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123:273-280. [DOI] [PubMed] [Google Scholar]
- 20.Lee, C., S. Lee, S. G. Shin, and S. Hwang. 2008. Real-time PCR determination of rRNA gene copy number: absolute and relative quantification assays with Escherichia coli. Appl. Microbiol. Biotechnol. 78:371-376. [DOI] [PubMed] [Google Scholar]
- 21.Morona, R., J. Yeadon, A. Considine, J. K. Morona, and P. A. Manning. 1991. Construction of plasmid vectors with a non-antibiotic selection system based on the Escherichia coli thyA+ gene: application to cholera vaccine development. Gene 107:139-144. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama, K., S. M. Kelly, and R. Curtiss III. 1988. Construction of an ASD+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Biotechnology 6:693-697. [Google Scholar]
- 23.Robertson, J. S., and E. Griffiths. 2006. Assuring the quality, safety, and efficacy of DNA vaccines. Methods Mol. Med. 127:363-374. [DOI] [PubMed] [Google Scholar]
- 24.Ryan, E. T., T. I. Crean, S. K. Kochi, M. John, A. A. Luciano, K. P. Killeen, K. E. Klose, and S. B. Calderwood. 2000. Development of a DeltaglnA balanced lethal plasmid system for expression of heterologous antigens by attenuated vaccine vector strains of Vibrio cholerae. Infect. Immun. 68:221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Schneider, J. C., A. F. Jenings, D. M. Mun, P. M. McGovern, and L. C. Chew. 2005. Auxotrophic markers pyrF and proC can replace antibiotic markers on protein production plasmids in high-cell-density Pseudomonas fluorescens fermentation. Biotechnol. Prog. 21:343-348. [DOI] [PubMed] [Google Scholar]
- 27.Smith, M. A., and M. J. Bidochka. 1998. Bacterial fitness and plasmid loss: the importance of culture conditions and plasmid size. Can. J. Microbiol. 44:351-355. [PubMed] [Google Scholar]
- 28.Vidal, L., J. Pinsach, G. Striedner, G. Caminal, and P. Ferrer. 2008. Development of an antibiotic-free plasmid selection system based on glycine auxotrophy for recombinant protein overproduction in Escherichia coli. J. Biotechnol. 134:127-136. [DOI] [PubMed] [Google Scholar]
- 29.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 30.Williams, S. G., R. M. Cranenburgh, A. M. Weiss, C. J. Wrighton, D. J. Sherratt, and J. A. Hanak. 1998. Repressor titration: a novel system for selection and stable maintenance of recombinant plasmids. Nucleic Acids Res. 26:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.