Abstract
Cellulose degradation, fermentation, sulfate reduction, and methanogenesis are microbial processes that coexist in a variety of natural and engineered anaerobic environments. Compared to the study of 16S rRNA genes, the study of the genes encoding the enzymes responsible for these phylogenetically diverse functions is advantageous because it provides direct functional information. However, no methods are available for the broad quantification of these genes from uncultured microbes characteristic of complex environments. In this study, consensus degenerate hybrid oligonucleotide primers were designed and validated to amplify both sequenced and unsequenced glycoside hydrolase genes of cellulose-degrading bacteria, hydA genes of fermentative bacteria, dsrA genes of sulfate-reducing bacteria, and mcrA genes of methanogenic archaea. Specificity was verified in silico and by cloning and sequencing of PCR products obtained from an environmental sample characterized by the target functions. The primer pairs were further adapted to quantitative PCR (Q-PCR), and the method was demonstrated on samples obtained from two sulfate-reducing bioreactors treating mine drainage, one lignocellulose based and the other ethanol fed. As expected, the Q-PCR analysis revealed that the lignocellulose-based bioreactor contained higher numbers of cellulose degraders, fermenters, and methanogens, while the ethanol-fed bioreactor was enriched in sulfate reducers. The suite of primers developed represents a significant advance over prior work, which, for the most part, has targeted only pure cultures or has suffered from low specificity. Furthermore, ensuring the suitability of the primers for Q-PCR provided broad quantitative access to genes that drive critical anaerobic catalytic processes.
The gene encoding the 16S small ribosomal subunit has served as a highly suitable target for studying bacterial species. When one obtains 16S rRNA gene sequence information, it is sometimes possible to infer function from an identical match to a well-characterized pure culture. More commonly, however, the similarity to pure cultures is low, and/or the highest similarities correspond to 16S rRNA gene sequences identified without isolation or phenotypic characterization. In either case, care must be taken, because distinct phenotypes [e.g., dissimilatory Fe(III) reduction, chlorate reduction] are found in microorganisms with highly similar (e.g., 99.5%) 16S rRNA gene sequences (1). In addition, 16S rRNA gene surveys of broad phylogenetic groups can be time-, labor-, and cost-intensive. For example, it is estimated that the 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing bacteria (SRB) would require approximately 132 16S rRNA gene-targeted microarray probes (32).
A more-direct approach for the study of microbes that span phylogenetic groups is to target them as a physiologically coherent guild by using specific genetic markers (functional genes) for the functions of interest. Functional genes have been successfully targeted in bioremediation studies to investigate microbial populations responsible for the degradation of various contaminants. Some examples include the use of the large alpha subunit of benzylsuccinate synthase to monitor anaerobic hydrocarbon-degrading bacteria (5), the monitoring of ars genes for the identification and quantification of arsenic-metabolizing bacteria (45), and the detection of catechol 1,2-dioxygenase in aromatic-hydrocarbon-degrading Rhodococcus spp. (48). In the field of mine drainage/metal remediation, functional genes have been used to target SRB (17, 26), but the methods have suffered both from a lack of broad specificity for SRB and from the inability to distinguish SRB from sulfur-oxidizing bacteria (SOB). A general challenge to the functional-gene approach has been the relative lack of characterization and unavailability of target sequences. As a consequence, the primer sets that are available tend to be more relevant to pure cultures than to complex environmental samples.
Microbial communities in natural and engineered anaerobic environments that utilize cellulose as the primary carbon source, such as those in rumina (56), termite guts (54), decomposing wood (7), sulfate-reducing and methanogenic sediments (9, 22), wetlands (28), and sulfate-reducing bioreactors (26), are particularly challenging to characterize. 16S rRNA gene-based studies have revealed the complexity of these microbial communities and their high levels of phylogenetic and functional diversity. In such anaerobic environments, mineralization of complex organic matter occurs through the concerted action of a variety of microorganisms. Primary fermenters, such as cellulose degraders, break down the complex molecules and ferment the hydrolysis products. Secondary fermenters also ferment the hydrolysis products. When sulfate is available, SRB utilize the fermentation products as carbon and energy sources. In addition, methanogens can also utilize some of the fermentation products. In many cases, functionally important members, such as SRB, are present only as a small fraction of the community (36, 38), making them difficult to detect by use of 16S rRNA gene-targeted fingerprinting methods. Furthermore, the phylogenetic diversity of cellulose degraders, fermenters, and SRB prevents their quantification using a small number of 16S rRNA gene-targeted probes.
In this study, degenerate PCR primers were developed, validated, and demonstrated for the amplification of key functional groups in anaerobic environments possessing genes encoding glycoside hydrolases of families 5 (collectively designated cel5 in this study) and 48 (collectively designated cel48 in this study) (cellulose degradation), the alpha subunit of iron hydrogenase (hydA) (fermentation), dissimilatory sulfite reductase (dsrA) (sulfate reduction), and methyl coenzyme M reductase (mcrA) (methanogenesis). This work is particularly novel considering that the vast majority of existing methods are suitable only for pure cultures, especially in the cases of cel5, cel48, and hydA (21, 44, 47). Thus, the approach provides access to uncultured and unsequenced markers, a critical feature for the study of key anaerobic processes in complex environments. Specificity was also enhanced where possible, notably in the case of dsrA, for which existing primers either do not distinguish SRB from SOB (14, 17) or have good alignment with only a narrow range of SRB (31, 52). Finally, all primers in this study were designed and validated for quantitative PCR (Q-PCR), in order to provide valuable quantitative functional information about complex anaerobic communities. The approach is demonstrated on mine drainage remediation systems and is expected to be of broad value to a variety of fields, including advancing the understanding of biohydrogen production, global carbon cycling, and other important biogeochemical processes.
MATERIALS AND METHODS
Primer design.
The consensus degenerate hybrid oligonucleotide primer (CODEHOP) strategy (43) was used to design all primer sets. CODEHOPs are hybrid primers that contain a relatively short 3′ degenerate core and a 5′ nondegenerate consensus clamp that stabilizes the hybridization of the 3′ end (43). The protein sequences used to design the primers were downloaded from the National Center for Biotechnology Information (NCBI) database (Table 1).
TABLE 1.
Sequences used for the design of the cel5, cel48, hydA, dsrA, and mcrA primer sets
| Primer set and target genea | Microorganism | NCBI accession no. |
|
|---|---|---|---|
| GenBank | GenPept | ||
| cel48 | |||
| celS | Clostridium thermocellum | L06942 | AAA23226 |
| celY | Clostridium thermocellum | AJ863163 | CAI06105 |
| celA | Bacteroides cellulosolvens | AY374129 | AAR23324 |
| celD | Clostridium josui | AB004845 | BAA32430 |
| celF | Clostridium cellulolyticum | U30321 | AAB41452 |
| celF | Clostridium acetobutylicum | AE001437 | AAK78887 |
| celA | Ruminococcus albus | AY422811 | AAR01217 |
| cel5 | |||
| celG | Fibrobacter succinogenes | U33887 | AAB38548 |
| endA | Ruminococcus flavefaciens | S55178 | AAB19708 |
| end1 | Butyrivibrio fibrisolvens | X17538 | CAA35574 |
| egl | Bacteroides ruminicola | M38216 | AAA22909 |
| engE | Clostridium cellulovorans | AF105331 | AAD39739 |
| celA | Clostridium longisporum | L02868 | AAC37035 |
| hydA | Clostridium acetobutylicum ATCC 824 | U15277 | AAB03723 |
| Clostridium pasteurianum ATCC 6013 | M81737 | AAA23248 | |
| Clostridium perfringens NCTC 8237 | AB016775 | BAA74726 | |
| Clostridium saccharobutylicum | U09760 | AAA85785 | |
| Clostridium difficile strain 630 | NC_009089 | YP_001089927 | |
| Clostridium novyi NT | NC_008593 | YP_877778 | |
| Clostridium thermocellum ATCC 27405 | AF148212 | AAD33071 | |
| dsrA | Desulfobacterium anilini | AF482455 | AAQ05939 |
| Desulfobacterium autotrophicum | AF418182 | AAL57437 | |
| Desulfococcus multivorans | U58126 | AAC24101 | |
| Desulfobacter curvatus | AF418199 | AAL57471 | |
| Desulfobacter postgatei | AF418198 | AAL57469 | |
| Desulfotomaculum ruminis | U58118 | AAC24103 | |
| Desulfobulbus propionicus | AF218452 | AAG28585 | |
| Desulfovibrio aerotolerans | AY749039 | AAU95684 | |
| Desulfovibrio alkalitolerans | AY864856 | AAW83205 | |
| Desulfovibrio burkinensis | AF418186 | AAL57445 | |
| Desulfovibrio desulfuricans | DQ092635 | AAZ04355 | |
| Desulfovibrio vulgaris | U16723 | AAA70107 | |
| mcrA | Methanococcus voltae | X07793 | CAA30633 |
| Methanococcus vannielii | M16893 | AAA72595 | |
| Methanothermus fervidus | J03375 | AAA72197 | |
| Methanosarcina barkeri | Y00158 | CAA68357 | |
| Methanocaldococcus jannaschii | L77117 | AAB98851 | |
| Methanothermobacter thermautotrophicus | NC_000916 | NP_276292 | |
| Methanopyrus kandleri | U57340 | 1E6VA | |
Target genes are listed only when the gene name differs from the name of the primer set.
Using the Gibbs algorithm of the Block Maker program (23), a total of 12, 5, 7, 5, and 8 blocks of conserved amino acids were found in the multiple alignments of the protein sequences of the cel5, cel48, hydA, dsrA, and mcrA genes presented in Table 1, respectively. These blocks were used as input to the CODEHOP program for the design of primers specific to each of the target genes. Primers were designed using all codon possibilities for the 3′ degenerate core and the most frequent nucleotide in each position for the 5′ consensus clamp. The CODEHOP default parameters were used for the design of all primer sets. For each block, the primer with the lowest degeneracy in the 3′ core was selected, and its sequence was compared in silico with the NCBI nucleotide database to confirm target specificity. Primer sequences that did not match the desired target were discarded, and the primer with the next lowest degeneracy was compared to the sequences in the NCBI database. The primers that were identified as specific were aligned with the DNA sequences of the proteins used for the design and were manually modified by increasing the degeneracy of the 3′ region to improve the match of the primer with the reference sequences or by decreasing the degeneracy when it was higher in the primer than in the alignment of reference sequences. The primers were arranged in pairs that would yield an amplicon of a size suitable for Q-PCR amplification (150 to 450 bp).
Primer validation. (i) Genomic DNA from pure cultures.
Genomic DNAs from the following microorganisms were used as positive or negative controls for PCR: Clostridium thermocellum (ATCC 27405D), Methanococcus maripaludis (ATCC 43000D), and Desulfobacterium autotrophicum (ATCC 43914D). Pure cultures of the following microorganisms were grown in the laboratory, and their genomic DNAs were used as positive or negative controls for PCR: Escherichia coli, Clostridium cellulovorans (ATCC 35296), Ruminococcus flavefaciens (ATCC 49949), and Fibrobacter succinogenes strain B1 (ATCC 51214).
(ii) Environmental samples.
For further primer validation, samples collected from two pilot-scale sulfate-reducing biochemical reactors that receive mine drainage from the National Tunnel in Black Hawk, CO (39), were selected. The sample sources differed primarily in the carbon substrate provided. The first reactor contained a complex lignocellulose (LC)-based substrate (wood chips [35%], limestone [20%], and corn stover [30%]). The second reactor received ethanol (Et) as its primary substrate and was packed with limestone and a zero-valent iron slag layer on the top. Both bioreactors were inoculated with horse manure. As a negative control, an aerated (dissolved oxygen [DO], >2.0 mg/liter) sequencing batch reactor (SBR) dominated by nontarget DNA was selected. The reactor was seeded with activated sludge from a local municipal wastewater treatment plant and was fed a mixture of Bacto peptone and acetate (OAc) at an influent chemical oxygen demand of 350 mg/liter. The hydraulic and solid residence times were 1 and 10 days, respectively.
DNA extraction.
DNA was extracted from portions (approximately 5 g each) of the environmental samples using the PowerMax soil DNA isolation kit and from pure cultures using the UltraClean microbial DNA isolation kit (Mo Bio, Carlsbad, CA) according to the manufacturer's protocols. The extracted DNA was stored at −80°C for subsequent studies.
Optimization of PCR conditions.
An annealing-temperature optimization was performed for all primer sets using pure-culture genomic DNA as the template at the following temperatures: 52.9°C, 54.3°C, 55.8°C, 59.2°C, and 60.7°C (cel48); 52.0°C, 52.8°C, 53.9°C, 55.3°C, 56.9°C, 58.5°C, 60.2°C, 61.7°C, 63.0°C, and 64.0°C (cel5); 56.0°C, 58.8°C, 62.8°C, and 65.2°C (hydA); 60.0°C and 63.0°C (dsrA); and 53.6°C, 54.7°C, 56.0°C, 58.8°C, and 60.1°C (mcrA). The optimized annealing temperature was selected based on the presence of one PCR product band of the expected size on a 1.2% agarose gel for the positive controls and the absence of any product in the pure-culture negative controls.
The optimized PCR master mix for the primer sets contained 1× reaction buffer with 2 mM magnesium (5 Prime, Gaithersburg, MD); 1× PCR enhancer (5 Prime); 0.05 mM each deoxynucleoside triphosphate; 1.0 mM (mcrA), 1.5 mM (dsrA), or 2.5 mM (cel5) Mg(OAc)2; 0.15 μM (cel48, hydA, dsrA), 0.2 μM (mcrA), or 0.25 μM (cel5) each primer; 0.125 μl of formamide; 0.875 U of Taq DNA polymerase (5 Prime); an additional 0.25 U (cel5) or 0.375 U (dsrA) of iTaq DNA polymerase (Bio-Rad, Hercules, CA); 1 μl (cel48, cel5, dsrA, and mcrA) or 0.5 μl (hydA) of the DNA template; and deionized water to a final volume of 12.5 μl. The additional iTaq added to the dsrA and cel5 PCR mixtures improved the yields of otherwise weak PCR products, presumably because the polymerase is more sensitive than Taq. The temperature program consisted of 3 min at 95°C, followed by 35 (hydA and mcrA), 40 (cel48), 45 (cel5), or 50 (dsrA) cycles of 40 s at 95°C, 30 s at the corresponding annealing temperature, 30 s at 72°C, and a final extension of 7 min at 72°C. The annealing temperatures were 56°C, 52°C, 59°C, 60°C, and 56°C for the cel48, cel5, hydA, dsrA, and mcrA primer sets, respectively.
Cloning.
The DNAs from the positive-control LC bioreactor and the negative-control SBR were used as the PCR templates for the primer combinations cel48_490F-cel48_920R, cel5_392F-cel5_754R, hydA_1290F-hydA_1538R, dsrA_290F-dsrA_660R, and mcrA_1035F-mcrA_1530R. In addition, the 16S rRNA gene was PCR amplified from DNA extracts of both the LC and Et bioreactors using primer sets 341F and 1492R (55). PCR products, if present, were cloned with the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Inserts were PCR amplified directly from colonies by using the vector-specific M13F and M13R primers. For the 16S rRNA gene clones, amplified rRNA gene restriction analysis (ARDRA) was performed visually with MspI restriction enzyme (Promega, Madison, WI)-digested inserts. The DNA corresponding to each unique ARDRA pattern was sequenced.
Selection of restriction enzymes.
The Restriction Enzyme Database (REBASE) (42) was used to select restriction endonucleases that could digest each functional gene PCR product. The NEBcutter (version 2.0) tool of REBASE was used to identify suitable restriction enzymes and the respective cleavage sites. The Hpy188III, Mbol, MnlI, NlaIII, and MboII restriction enzymes (New England Biolabs, Ipswich, MA) were chosen to digest the PCR products from the primer sets targeting the cel5, cel48, hydA, dsrA, and mcrA genes, respectively.
Restriction enzyme digestion and DNA sequencing.
M13 PCR products of cloned gene inserts were digested with the corresponding restriction enzymes according to the manufacturer's recommendations, with incubation conditions of 37°C for 1 h (mcrA), 1.5 h (cel5 and hydA), or 3 h (cel48 and dsrA), followed by an inactivation step of 20 min at 65°C. For each unique restriction pattern identified on 3% agarose gels, at least one clone was sequenced by the Proteomics and Metabolomics Facility (Colorado State University, Fort Collins, CO). The closest matches to microorganisms available in the NCBI database were determined using Basic Local Alignment Search Tool X (BLASTX), which searches the protein database using a translated nucleotide query.
Q-PCR.
The primer sets were adapted to Q-PCR and were further validated by quantifying the functional genes in samples collected from the LC and Et sulfate-reducing bioreactors. In addition, the total bacterial 16S rRNA gene was quantified using the conditions, universal primers (BACT1369F and PROK1492R), and probe (TM1389F) described by Suzuki et al. (46). For the functional-gene primers, Q-PCR amplification was performed with a 7300 real-time PCR system (Applied Biosystems, Foster City, CA) using a 12.5-μl reaction mixture consisting of 1× Power SYBR green PCR master mix (Applied Biosystems); 0.15 μM (cel48, hydA, and dsrA), 0.2 μM (mcrA), or 0.25 μM (cel5) each primer; 1 mM (mcrA and hydA), 1.5 mM (dsrA), or 2.5 mM (cel5) Mg(OAc)2; an additional 0.25 U (dsrA) or 0.375 U (cel5) of iTaq DNA polymerase (Bio-Rad); and 1 μl of the template. The temperature program was 10 min at 95°C, followed by 40 (cel48 and mcrA), 45 (hydA and cel5), or 50 (dsrA) cycles of 40 s at 95°C, 30 s at the corresponding annealing temperature, and 30 s at 72°C. DNA extracts were diluted 1:10 and were analyzed in five replicates.
For each functional gene primer set, a triplicate four- to six-point calibration curve was constructed using a standard mixture of purified PCR products obtained with the vector-specific primers T3 and T7 from sequenced clones. For the 16S rRNA gene primer set, a purified PCR product amplified from an environmental sample with primers 8F and 1492R (13, 55) was used as the standard. The calibration curves were used for absolute quantification of the target genes under the assumption that the PCR amplification efficiencies for standards and samples were the same. To ensure that only samples for which this assumption was valid were analyzed, the procedure proposed by Chervoneva et al. (10) was applied. Briefly, LinRegPCR (41) was used to calculate the amplification efficiency from the PCR amplification curve of each replicate sample and standard. A box plot outlier detection rule was applied to the amplification efficiency of the standards in order to identify calibration standards that amplified with dissimilar efficiencies, which were not used in data analysis. The Kinetic Outlier Detection method (3) was used to compare the average efficiency of the remaining standards and the efficiency of each replicate in order to detect samples with efficiencies dissimilar from those of the standards.
Nucleotide sequence accession numbers.
The cel5, cel48, hydA, dsrA, and mcrA sequence data from the LC bioreactor have been submitted to GenBank under accession numbers GQ184058 to GQ184072, GQ184073 to GQ184082, GQ184083 to GQ184095, GQ184096 to GQ184113, and GQ184114 to GQ184128, respectively. The cel48 and dsrA sequence data generated from the SBR have been submitted under accession numbers GU563804 to GU563811 and GU563812 to GU563821, respectively.
RESULTS
Selection of genetic markers for anaerobic cellulose degradation, fermentation, sulfate reduction, and methanogenesis.
The selection of appropriate genetic markers for the functions of interest was the first step in designing suitable primers for quantifying cellulose degraders, fermenters, sulfate reducers, and methanogens in anaerobic environments. The goal was to capture as much diversity among these groups as possible, including potentially uncultured members, through degenerate primer design. It was also desired that the primers should be appropriate for Q-PCR.
Of the more than 70 glycoside hydrolase families defined on the basis of amino acid sequence identity in their catalytic domains (18), cellulases are found in 15 families (families 5, 6, 7, 8, 9, 10, 11, 12, 26, 44, 45, 48, 51, 60, and 61) (24, 40). The cellulase type does not correlate closely with phylogeny: most cellulolytic microorganisms produce cellulases that belong to different families, and cellulases of the same family occur in widely different microorganisms (4). However, the majority of the glycoside hydrolases of anaerobes belong to just three families: families 5, 9, and 48 (24, 25). Families 5 and 48 were selected in this study as targets that were particularly well represented among anaerobic cellulose degraders. The majority of cel48 genes are found in Clostridium spp., although Bacteroides cellulosolvens (58) and Ruminococcus albus (12) are also known to have at least one cel48 gene each. cel48 genes have also been found in Bacillus, Streptomyces, and Cellulomonas, as well as in Eukarya; however, they are highly divergent and therefore were not considered as primary targets in the primer design. The cel5 target was selected in order to provide broader coverage of anaerobic cellulose degraders, since it is found in a wider range of phylogenetic groups (24).
Fermentation is carried out by a broad range of species using a variety of enzymes, some of which are also found in nonfermenters. In nearly all fermentative bacteria, the electrons from the reduced electron carriers are channeled to H2 via iron hydrogenases (2). All iron hydrogenases contain a highly conserved region of 350 amino acids known as the H-cluster domain in their alpha subunit (34, 50); this region was therefore selected as highly suitable for primer design. Specifically, the hydA primers were designed from the two conserved protein regions: GAGVIFGA and MACPGGCING. The second region is one of the characteristic sequence signatures of the H-cluster domain of iron hydrogenases (50).
The dissimilatory reduction of sulfate to hydrogen sulfide in SRB is performed in three steps (51). First, sulfate is activated to adenosine 5′-phosphosulfate by ATP sulfurylase. This is followed by the partial reduction to sulfite by adenosine 5′-phosphosulfate reductase (APSR) and the final reduction to sulfide by dissimilatory sulfite reductase (DSR). The genes encoding APSR and DSR are evolutionarily conserved among SRB, including deltaproteobacteria, clostridia, thermodesulfobacteria, nitrospirae, and certain archaea (27, 52). However, ATP sulfurylase, APSR, and DSR appear to be involved in both the oxidative and the reductive mode of the dissimilatory sulfur metabolism (27). This is a major challenge for the study of SRB in systems where SOB and SRB might coexist. Since the dsr genes of SRB and SOB are more distantly related than the corresponding aps genes (27), the dsr gene was selected as the preferred marker for SRB. Special attention was given in subsequent primer design to achieving broad specificity for SRB among the deltaproteobacteria and clostridia while avoiding matches for SOB.
Methanogens are a metabolically versatile group of microorganisms, capable of forming methane from acetate, CO2, H2, and C1 compounds (16). A variety of enzymes are involved in the methanogenic pathway, and many are not exclusive to methanogenesis. Except for coenzyme M, methyl coenzyme M, and methyl coenzyme M reductase, all the enzymes and coenzymes involved in methanogenesis are also present in sulfate-reducing archaea (49). Similarly, methylene tetrahydromethanopterin dehydrogenase and methenyl tetrahydromethanopterin cyclohydrolase are also present in methanotrophs (11). Thus, the genes encoding methyl coenzyme M reductase (mcrA) were selected as the ideal genetic marker for methanogens, since they appear to be unique to methanogens and are evolutionarily conserved (15).
Primer design, PCR optimization, and in silico analysis.
Based on in silico specificity with minimum degeneracy using the CODEHOP strategy, a total of three mcrA primer sets, two primer sets each for cel5, cel48, and dsrA, and one hydA primer set were identified as most promising (Table 2). One of the dsrA primer sets contained a modified version of the reverse primer RH3-dsr-R designed by Ben-Dov et al. (6) and an in-house-designed forward primer. One of the mcrA primer sets (mcrA_1035F-mcrA_1530R) was designed by Luton et al. (33). The other two mcrA primer pairs each combined one of the Luton primers with one in-house-designed primer.
TABLE 2.
Primer combinations designed by the CODEHOP strategy and selected for further validation PCR
| Target and primer | Sequence (5′-3′) | Primer combination(s) (approximate amplicon size [bp])a |
|---|---|---|
| Family 48 glycoside hydrolases | ||
| cel48_490F | TNATGGTTGAAGCTCCDGAYTAYGG | cel48_490F-cel48_920R (430), cel48_880F-cel48_980R (100) |
| cel48_880F | CAYTGGHTNNTGGAYGTTGAYAACTGGTA | |
| cel48_920R | CCAAANCCRTACCAGTTRTCAACRTC | |
| cel48_980R | CCTGTTCACCTCTYTGRWARGTRTT | |
| Family 5 glycoside hydrolases | ||
| cel5_392F | GAGCATGGGCTGGAAYHTNGGNAA | cel5_392F-cel5_525R (133), cel5_392F-cel5_754R (362) |
| cel5_525R | GAAAGGAATACGGACGGYNTTRAAHCC | |
| cel5_754R | CATCATAATCTTTGAAGTGGTTTGCAATYTGDKTCCA | |
| Alpha subunit iron hydrogenases | ||
| hydA_1290F | GGTGGAGTTATGGAAGCWGCHHT | hydA_1290F-hydA_1538R (248) |
| hydA_1538R | CATCCACCWGGRCAHGCCAT | |
| Alpha subunit dissimilatory sulfite reductase | ||
| dsrA_290F | CGGCGTTGCGCATTTYCAYACVVT | drsA_290F-RH3-dsr-R′ (140), dsrA_290F-dsrA_660R (370) |
| RH3-dsr-R′ | GTGGMGCCGTGCATGTT | |
| dsrA_660R | GCCGGACGATGCAGHTCRTCCTGRWA | |
| Alpha subunit of methyl coenzyme M reductase | ||
| mcrA_1035F | GGTGGTGTMGGATTCACACARTAYGCWACAGC | mcrA_1035F-mcrA_1530R (495), mcrA_1035F-mcrA_1450R (415), mcrA_1430F-mcrA_1530R (100) |
| mcrA_1530R | TTCATTGCRTAGTTWGGRTAGTT | |
| mcrA_1450R | TTTGAAGCWCCRCAYTGGTCYT | |
| mcrA_1430F | TTCTATGGTTACGACTTVCAGGACCARTGYGG |
The annealing temperature and magnesium concentration were optimized for all primer sets (Table 2), which were then subjected to specificity tests with positive and negative controls (Table 3). Suitability for a complex environmental sample known to contain all the target (as well as nontarget) groups and PCR inhibitors was also tested. On the basis of the results, the following primer sets were selected for further validation: cel48_490F-cel48_920R, cel5_392F-cel5_754R, hydA_1290F-hydA_1538R, dsrA_290F-dsrA_660R, and mcrA_1035F-mcrA_1530R.
TABLE 3.
Results of PCR specificity tests with positive and negative controls
| Primer combination | Result with the following positive/negative controla: |
Result with an environmental sampleb | ||||||
|---|---|---|---|---|---|---|---|---|
| C. thermocellum | C. cellulovorans | R. flavefaciens | F. succinogenes | E. coli | D. autotrophicum | M. maripaludis | ||
| cel48_490F-cel48_920R | + | − | − | − | − | − | − | + |
| cel48_880F-cel48_980R | + | − | − | − | − | − | − | − |
| cel5_392F-cel5_525R | − | − | − | − | − | − | − | − |
| cel5_392F-cel5_754R | + | + | + | + | − | − | − | + |
| hydA_1290F-hydA_1538R | + | − | − | − | − | − | − | + |
| drsA_290F-RH3-dsr-R′ | + | − | − | − | − | + | + | + |
| dsrA_290F-dsrA_660R | − | − | − | − | − | + | − | + |
| mcrA_1035F-mcrA_1530R | − | − | − | − | − | − | + | + |
| mcrA_1035F-mcrA_1450R | − | − | − | − | − | − | + | + |
| mcrA_1430F-mcrA_1530R | − | − | − | − | + | − | + | + |
C. thermocellum possesses cel5, cel48, and hydA; C. cellulovorans, R. flavefaciens, and F. succinogenes possess cel5; D. autotrophicum possesses dsrA; and M. maripaludis possesses mcrA. E. coli does not possess any of the target genes.
Substrate from a lignocellulose-based sulfate-reducing bioreactor.
In order to determine the broader specificities of the primers beyond the reference organisms used for their design (Table 1), in silico analysis was performed. This was achieved by interrogating the NCBI Reference Sequence (RefSeq) collection (http://www.ncbi.nlm.nih.gov/RefSeq/) for each of the target genes except mcrA, since the mcrA primers had been described in detail previously (33). A total of 29, 397, 202, and 14 bacterial reference sequences were found for the cel48, cel5, hydA, and dsrA genes, respectively. Since the dsrA sequences from the RefSeq collection did not include sequences from all the different genera that contain this gene, additional sequences were included in the analysis (32 in total) in order to better represent the diversity. The results of the in silico analysis suggest that the hydA and dsrA primer sets have a broader phylogenetic coverage than that for which they were directly designed (Table 4). The cel48 forward and reverse primers matched mainly Clostridium sp. sequences. The reverse primer also matched Bacillus and Anaerocellum sp. sequences. The RefSeq collection did not include cel48 sequences from Ruminococcus or Bacteriodes spp. However, the Bacteroides cellulosolvens and Ruminococcus albus sequences used for primer design aligned with the forward primer with 1 and 4 mismatches and with the reverse primer with 4 and 1 mismatches, respectively. In all cases, the mismatches were not in the 3′ primer region. In the case of the cel5 primer set, all reference sequences had more than 2 mismatches with the primer sequence. This was not unexpected given the sequence diversity of the cel5 genes. Nonetheless, by cloning and DNA sequencing of cel5 PCR products, it was confirmed that these primers amplify cel5 sequences. The primers had either no mismatches or 1 mismatch with the cel5 sequences recovered from cloning.
TABLE 4.
Results of in silico analysis of primer specificity for NCBI RefSeq database sequences
| Primera | Representative species with 2 or fewer mismatches |
|---|---|
| cel48_490F | Clostridium spp. |
| cel48_920R | Anaerocellum, Bacillus, and Clostridium spp. |
| hydA_1290F | Clostridium, Coprococcus, Dehalococcoides, Desulfitobacterium, Geobacter, Marinitoga, Opitutus, Thermoanaerobacterium, Thermotoga, Tolumonas, and Veillonella spp. |
| hydA_1538R | Clostridium, Desulfovibrio, Fervidobacterium, Finegoldia, Fusobacterium, Marinitoga, Petrotoga, Thermodesulfovibrio, Thermosipho, and Thermotoga spp. |
| dsrA_290F | Desulfobotulus, Desulfomicrobium, Desulfomonile, Desulforhopalus, Desulfosalina, and Desulfovibrio spp. |
| dsrA_660R | Desulfobacter, Desulfobulbus, Desulfococcus, Desulfomicrobium, Desulfonema, Desulforhopalus, Desulfosalina, Desulfosarcina, Desulfotalea, and Desulfovibrio spp. |
In the case of the cel5 primer set (cel5_392F-cel5_754R), all reference sequences had more than 2 mismatches with the primer sequence.
Experimental validation of primer specificity.
PCR products obtained from the LC bioreactor sample were cloned, and DNA sequences were determined for 10, 15, 13, 18, and 15 clones (out of 20, 30, 26, 30, and 28 per library) resulting from the cel48, cel5, hydA, dsrA, and mcrA primer sets, respectively. The results confirmed that the primer sets amplified only the desired targets in all cases (Table 5). The cel48 and cel5 clones matched with exo- or endoglucanases belonging to families 48 and 5 of glycoside hydrolases, respectively. The sequences obtained with the hydA primers matched the C-terminal domain of the large subunit of iron hydrogenases, which contains the H-cluster sequence used to design the primers. All of the sequences obtained with the dsrA primers matched the alpha subunit of the dissimilatory sulfite reductase of uncultured SRB. Finally, all of the mcrA sequences matched the alpha subunit of methyl coenzyme M reductase, and the majority of the sequences matched an uncultured archaeon belonging to the Methanosarcinaceae. Good's coverage estimates (19), calculated from the results of the restriction digest analysis, were 96.6%, 90.0%, 84.6%, 93.3%, and 82.1% for the cel5, cel48, hydA, dsrA, and mcrA clones, respectively. This suggested that the number of clones screened was representative of the genetic diversity captured by each primer set.
TABLE 5.
DNA sequencing results of clones of PCR products obtained with the cel5, cel48, hydA, dsrA, and mcrA primer sets on a sample from the lignocellulose-based sulfate-reducing bioreactor
| Primer set and microorganism | BLASTX identity (% match)a | % of screened clones | GenPept accession no. |
|---|---|---|---|
| cel48 | |||
| Clostridium josui | Exoglucanase S, glycosyl hydrolase family 48 (75) | 14.3 | AAC38571 |
| Clostridium acetobutylicum ATCC 824 | Processive endoglucanase, glycosyl hydrolase family 48 (56) | 14.3 | NP_347547 |
| Clostridium cellulolyticum H10 | Glycoside hydrolase, family 48 (69) | 28.6 | ZP_01575465 |
| Clostridium cellulovorans | Exoglucanase S, glycosyl hydrolase family 48 (53-61) | 42.9 | AAC38571 |
| cel5 | |||
| Saccharophagus degradans 2-40 | Endoglucanase-like protein, glycoside hydrolase family 5 (45-59) | 46.7 | ABD81896 |
| Clostridium cellulovorans | Endoglucanase EngB, glycoside hydrolase family 5 (43) | 3.3 | AAA23231 |
| Flavobacterium johnsoniae UW101 | Hypothetical protein EUBSIR_0099, glycoside hydrolase family 5 (44) | 6.7 | ABQ03808 |
| Eubacterium siraeum DSM 15702 | Hypothetical protein EUBSIR_00991, glycoside hydrolase family 5 (52-53) | 43.3 | EDS01174 |
| hydA | |||
| Clostridium botulinum B1 strain Okra | [Fe] hydrogenase (72) | 3.7 | YP_001781395 |
| Clostridium cellulolyticum H10 | Hydrogenase, Fe only (75-82) | 7.4 | ZP_01573883 |
| Bacteroides capillosus ATCC 29799 | Hypothetical protein BACCAP 02746, iron hydrogenase large subunit, C-terminal domain (73) | 7.4 | ZP_02037133 |
| Thermosinus carboxydivorans Nor1 | Hydrogenase, Fe only (74) | 11.1 | EAX48145 |
| Pelotomaculum thermopropionicum SI | Hydrogenase subunit (63) | 11.1 | YP_001212560 |
| Clostridium bolteae ATCC BAA-613 | Hypothetical protein CLOBOL 01023, iron hydrogenase large subunit, C-terminal domain (60-74) | 22.2 | ZP_02083500 |
| Clostridium thermocellum | Hydrogenase-1(67-69) | 37.0 | AAD33071 |
| dsrA | |||
| Uncultured sulfate-reducing bacterium | Dissimilatory sulfite reductase alpha subunit (96) | 3.3 | AAQ7928 |
| Uncultured sulfate-reducing bacterium | Dissimilatory sulfite reductase alpha subunit (90-91) | 63.3 | ACD37519 |
| Uncultured sulfate-reducing bacterium | Dissimilatory sulfite reductase subunit A (82-88) | 13.3 | ABR45803 |
| Uncultured sulfate-reducing bacterium | Dissimilatory sulfite reductase alpha subunit (89-92) | 3.3 | ABR09856 |
| Uncultured sulfate-reducing bacterium | Dissimilatory sulfite reductase alpha subunit (96) | 16.7 | ABW69247 |
| mcrA | |||
| Uncultured Methanosarcinaceae archaeon | Methyl coenzyme M reductase alpha subunit (96) | 3.6 | AAN02199 |
| Uncultured euryarchaeote | Methyl coenzyme M reductase alpha subunit (90-91) | 3.6 | AAR22542 |
| Uncultured methanogenic archaeon | Methyl coenzyme M reductase alpha subunit (96) | 7.1 | CAN99766 |
| Uncultured archaeon | Methyl coenzyme M reductase alpha subunit (97-98) | 21.4 | ABX75060 |
| Uncultured euryarchaeote | Methyl coenzyme M reductase alpha subunit (96) | 25.0 | AAR24558 |
| Methanobacterium ivanovii | Methyl coenzyme M reductase alpha subunit (99) | 39.3 | ABO93183 |
Where several clones matched the same protein, the percentage of matching sequence is given as a range.
With respect to the aerated SBR dominated by nontemplate DNA, which served as a negative control, no PCR product was obtained from any of the primer sets, except for faint products yielded by the cel48 and dsrA primers. Cloning and sequencing revealed that the products did represent viable targets. dsrA clones (n = 10) were most similar to sequences present in the following microorganisms (with GenBank accession numbers and percentages of similarity given in parentheses): Desulfobulbus elongatus (AJ310430; 90%), Desulfobulbus propionicus (AF218452; 82%), Desulfobulbus rhabdoformis (AJ250473; 85%), Desulforhopalus singaporensis (AF418196; 77 to 82%), Desulfovibrio aerotolerans (AY749039; 96%), Desulfovibrio aminophilus (AY626029; 88%), and Desulfovibrio magneticus RS-1 (AP010904; 93 to 95%). cel48 clones (n = 19) were all most similar to a glycoside hydrolase present in Herpetosiphon aurantiacus (CP000875; 93 to 94%).
Adaptation to and validation of Q-PCR.
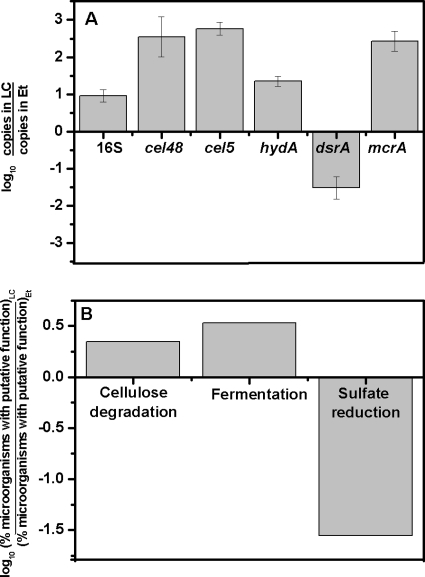
The selected primer sets were adapted to Q-PCR and were used for the quantification of the functional genes in the LC and Et bioreactors treating mine drainage. All functional genes were successfully quantified in both samples (Fig. 1A). Assuming that the numbers of copies of functional genes present per organism are distributed similarly between the two samples, the ratios represent how many more (or less) organisms with each function are present in the LC versus the Et bioreactor. In the LC bioreactor, the abundances of the cel48, cel5, and mcrA genes were approximately 350 ± 180, 580 ± 100, and 270 ± 70 times higher, respectively, than those in the ethanol-fed bioreactor. The concentration of hydA genes in the LC bioreactor was also higher (22 ± 3) than that in the Et reactor. On the other hand, the Et reactor contained approximately 33 ± 9 times more dsrA genes than the LC reactor. Based on the quantification of the 16S rRNA gene, the LC bioreactor contained a higher concentration of bacterial biomass than the Et bioreactor (about 10 times higher).
FIG. 1.
(A) Numbers of copies of the 16S rRNA, cel5, cel48, hydA, dsrA, and mcrA genes in the lignocellulose (LC)-based bioreactor relative to those in the ethanol (Et)-fed bioreactor as determined by Q-PCR. (B) Ratios of the percentages of putative cellulose degraders, fermenters, and sulfate-reducing bacteria in the LC bioreactor to those in the Et bioreactor as determined by ARDRA and 16S rRNA gene sequencing, as well as by a subsequent literature review.
Characterization by 16S rRNA gene cloning.
A total of 109 and 84 cloned 16S rRNA genes were analyzed by ARDRA for the LC and Et reactors, respectively. As expected, the LC reactor was found to have higher overall bacterial diversity, with a Shannon index of 2.97, compared to 1.03 for the Et reactor. Putative functions were assigned to the bacteria represented by the sequences based on information obtained from the highest matches in the GenBank database and a corresponding literature review (Table 6). Based on the putative functions assigned to the bacteria, the relative proportions of cellulose degraders, fermenters, and sulfate reducers were estimated, and these estimates were compared with the results obtained by Q-PCR targeting functional genes (Fig. 1B). The two approaches yielded similar overall trends, with higher concentrations of cellulose degraders and fermenters in the LC bioreactor than in the Et bioreactor, and higher concentrations of sulfate reducers in the ethanol bioreactor. However, the values of the ratios cannot be directly compared, because the ARDRA results were determined as ratios of the frequencies (expressed as percentages) in the clone library, and the Q-PCR results were determined as ratios of the numbers of gene copies.
TABLE 6.
Summary of 16S rRNA gene sequence analysis of the lignocellulose-based versus the ethanol-fed bioreactora
| Bioreactor and highest match (GenBank accession no.) | % Match | % of screened clones | Putative functionb |
|---|---|---|---|
| Lignocellulose-based sulfate-reducing bioreactor | |||
| Clostridium from anoxic bulk soil, strain XB90 (AJ229234) | 97 | 6.5 | CD |
| Uncultured bacterium, clone RB323 (AB240338) | 87-99 | 4.7 | CD |
| Uncultured bacterium, clone LCFA-B03 (AB244310) | 98-99 | 7.5 | F |
| Uncultured spirochete, clone C6_2 (EF562545) | 96 | 7.5 | F |
| Uncultured bacterium, clone Eb26 (EF063613) | 98 | 6.5 | F |
| Uncultured bacterium, FukuN63 (AJ290056) | 96 | 1.9 | F |
| Uncultured bacterium, clone RA13C8 (AF407407) | 99 | 2.8 | PH/F |
| Uncultured Bacteroidetes bacterium, clone CF_5 (EF562567) | 98 | 1.9 | PH/F |
| Uncultured Bacteroidetes bacterium, clone RBE2CI-81 (EF111172) | 98-99 | 8.4 | PH/F |
| Uncultured bacterium, clone 060F03_B_SD_P93 (CR933306) | 97 | 2.8 | PH/F |
| Uncultured bacterium, clone 1013-1-CG49 (AY532557) | 94-99 | 3.7 | PH/F |
| Uncultured sulfate-reducing bacterium, clone 39B (AB069774) | 99 | 1.9 | SR |
| Uncultured bacterium, clone ELME-7 (AB288666) | 96 | 1.9 | UNK |
| Failed sequencing | 3.7 | ||
| Ethanol-fed sulfate-reducing bioreactor | |||
| Uncultured eubacterium, AA05 (AF275916) | 97 | 2.3 | F |
| Unidentified eubacterium, clone BSV87 (AJ229231) | 99 | 2.3 | F |
| Uncultured Acetobacterium sp. (EF420226) | 99 | 2.3 | F |
| Uncultured betaproteobacterium, clone UCT N123 (AY064177) | 98 | 2.3 | AO |
| Iron-reducing enrichment clone Cl-A9 (DQ677001) | 99 | 6.9 | PH/F |
| Uncultured Bacteroidetes bacterium (EF111172) | 97 | 4.6 | PH/F |
| Iron-reducing enrichment clone Cl-A4 (DQ676996) | 99 | 2.3 | PH/F |
| Desulfomicrobium sp. BL (AY928663) | 99 | 14.9 | SR |
| Desulfovibrio sp. GWE2 (AF159119) | 99 | 14.9 | SR |
| Desulfomicrobium baculatum (AJ277896) | 99 | 10.3 | SR |
| Uncultured deltaproteobacterium (DQ676356) | 99 | 8.0 | SR |
| Desulfovibrio putealis (AY574979) | 99 | 5.7 | SR |
| Geobacter sp. Ply1 (EF527233) | 98 | 4.6 | SR |
| Uncultured bacterium (AB237688) | 98 | 4.6 | SR |
| Uncultured deltaproteobacterium (EF111173) | 98 | 4.6 | SR |
| Rhodopseudomonas sp. ORS 1416ri (AJ968691) | 98 | 2.3 | UNK |
| Uncultured bacterium (AY570636) | 98 | 6.9 | UNK |
The percentage of the total community was estimated by ARDRA.
CD, cellulose degradation; PH, polysaccharide hydrolysis; F, fermentation; AO, aromatics oxidation; SR, sulfate reduction; UNK, unknown.
DISCUSSION
Advantages of new primers relative to existing primers.
This study provides a useful new suite of tools for characterizing anaerobic sulfate-reducing and methanogenic communities that utilize cellulose as the primary carbon source. The approach is much less cumbersome than 16S rRNA gene profiling and provides direct functional information. Significant advances were also made over prior functional-gene methods, which have been developed primarily for pure cultures or do not discriminate well against nonspecific targets. Even when challenged with samples dominated by nontarget DNA, all primer sets still yielded 100% specificity. The ability to detect functional genes corresponding to both cultured and uncultured organisms in complex environments is of particular value. With this approach, ubiquitous microbial functions, such as cellulose degradation, fermentation, sulfate reduction, and methanogenesis, can be studied with a smaller number of primer sets than that required for the 16S rRNA gene-based approach. All of the primer sets were also confirmed to be suitable for Q-PCR, providing valuable quantitative information about important functional groups in complex anaerobic environments.
The CODEHOP strategy proved to be effective for degenerate primer design, particularly for the cel5, cel48, and hydA sequences, which, because of their lower degree of conservation, were more challenging than the mcrA and dsrA sequences. Although it was possible to identify blocks of conserved protein sequences and to design primers from them for all of the target genes, the divergence of the cel5, cel48, and hydA sequences limited the number of primer combinations that could be designed and, in general, increased the degeneracy of their 3′ degenerate cores. As was intended, all primers were capable of detecting an array of sequences even broader than that directly used to design the primers (Table 1). This was confirmed by in silico analysis and/or experimental validation. Interestingly, the similarity of the sequences from the LC bioreactor, obtained from experimental validation, to the sequences in the NCBI database was lower than what is typically observed for 16S rRNA gene sequences (>90%), particularly for the cel5, cel48, and hydA sequences. This is likely because the 16S rRNA gene has changed very slowly throughout the course of evolution relative to the rate of change of functional genes. Also, there is limited sequence information in the database relative to that for 16S rRNA.
In particular, the development of degenerate primers targeting cel5 and cel48 represents a significant advance over prior work. While several sets of primers targeting various genes encoding glycoside hydrolases have been identified in the literature, all were designed to target pure cultures, except for one study in which the primers were specific to the cellulases of red claw crayfish (8). Many existing primers may, in fact, amplify more cellulases than the single target for which they were designed; however, considering the broad sequence diversity among glycoside hydrolases, a direct degenerate design approach has been greatly needed. This is especially true for applications in complex anaerobic environments utilizing cellulose as the primary carbon substrate.
Prior efforts to develop primers for the hydA gene have also focused primarily on pure cultures (37, 44, 53). In this study, the hydA primers were designed using sequences obtained from several Clostridium species, and the DNA sequencing results demonstrated that an even broader range of genera was captured by the primer set (Table 5). Recently, broadly specific primers were also developed by Xing and colleagues (57) to amplify the hydA gene of H2-producing bacteria in acidophilic communities. The forward primer targeted the ADLTIMEE signature sequence of the H-cluster domain to produce a PCR product of 500 to 600 bp. However, this fragment length is not appropriate for Q-PCR. Therefore, in our study, a new forward primer was designed from a conserved region closer to MACPGGCING to produce a smaller product (∼248 bp), which is preferable for Q-PCR (35).
The major challenge in seeking universal primers for SRB remains the ability to distinguish SRB from SOB while maintaining broad coverage of SRB, which span phyla and are present in both bacteria and archaea. Primers targeting the genes encoding APSR have been particularly problematic in environmental surveys and have been observed to favor SOB over SRB when both are present (20, 26). The dsrA and dsrB genes appear to be superior in this regard, but an in silico analysis (data not shown) of published primers conducted during the design phase of this study still revealed limitations. Many published dsrA or dsrB gene primer sets suffered from either high specificity for SOB (14, 17, 31) or low specificity for various SRB phyla (14, 17, 31, 52). Based on our literature survey and in silico analysis, primer RH3-dsr-R, described by Ben-Dov et al. (6), appeared to be the most promising and therefore was modified for broader specificity in this study. An in silico analysis of the dsrA primer set developed and applied in this study indicated that there were several mismatches between known dsrA genes of Thiobacillus spp. and the primers (particularly at the 3′ end of the reverse primer) to prevent their amplification. In addition, no Thiobacillus spp. were detected by these primers in the aerobic SBR sample. Furthermore, DNA sequencing of clones of a PCR product obtained with the dsrA primer set from a sample of the Peerless Jenny King bioreactor, which was known to contain Thiobacillus sp. DNA (26), revealed that no Thiobacillus sp. dsrA sequences were amplified (data not shown).
Primers mcrA_1450R and mcrA_1430F were designed and tested in combination with the forward and reverse primers designed by Luton et al. (33). While the in-house-designed primers were confirmed experimentally to be specific to mcrA genes, the Luton combination provided better sequence diversity coverage (data not shown) and therefore was selected for adaptation to Q-PCR. In addition to the Luton primers, several other primer sets for mcrA gene amplification have been published. However, these were not found to be suitable for Q-PCR and/or did not provide reliable information regarding the proportion of different operational taxonomic units (OTUs) in a sample. For example, Juottonen et al. (30) concluded that the Luton primer combination provided better performance, in terms of quantitative characterization of methanogenic communities in peatland samples, than two other primer sets.
Application for quantifying key functional guilds in complex anaerobic environments.
Traditionally, the 16S rRNA gene has been used for the biomolecular characterization of microbial communities. The design of primers targeted to the 16S rRNA gene is generally effective for the study of microorganisms that form tight phylogenetic clusters but becomes impractical when primers are used to capture the diversity of microorganisms that span phylogenetic groups. Thus, the 16S rRNA gene-based approach is particularly problematic for characterizing complex anaerobic environments, in which cellulose degraders, fermenters, sulfate reducers, and methanogens are key populations with vast phylogenetic diversity. The processes driven by these groups are of growing interest in various arenas, including carbon cycling/climate studies, biofuel production, and bioremediation.
Mine drainage remediation is an important example of a microbially mediated process that is poorly understood, largely due to the complexity of the microbial communities involved. Although sulfate reduction was the primary engineered function, SRB were estimated to be present at only 1.9% of the total bacterial community of the LC bioreactor based on ARDRA (Table 6). This observation is supported by several other 16S rRNA gene surveys of similar systems (26, 29, 36, 38). This indicates that for every 100 clones screened, only 2 are likely to represent sulfate reducers. This is a frustrating approach to the study of the bacteria that drive the main function of interest in these bioreactors, especially if quantitative information is desired.
The mine drainage treatment systems investigated in this study were particularly suitable for validating the primers, because they differed only in the type of carbon substrate applied to support the microbial community. The presence of ethanol was expected to produce a microbial community enriched in SRB, because, as a product of fermentation, ethanol is not a favorable substrate for fermenters, and it can be directly utilized by most SRB. On the other hand, the complex lignocellulosic carbon source was expected to produce a more functionally diverse microbial community, as was confirmed by ARDRA. Q-PCR utilizing the primers designed in this study provided quantitative support for these assumptions and was also in good general agreement with the 16S rRNA gene-based characterization (Fig. 1). The LC bioreactor contained higher concentrations of the cel5, cel48, hydA, and mcrA genes than the Et bioreactor. In contrast, the Et bioreactor contained approximately 1.5 orders of magnitude more dsrA genes. While genetic markers for cellulose degradation and fermentation were detected in the Et bioreactor, these were likely a result of the residual horse manure used for inoculation.
Even though the overall trends in the results obtained from the 16S rRNA gene analysis and the functional gene analysis were similar (Fig. 1), it is important to keep in mind that assignment of bacteria to functional groups based on 16S rRNA gene sequence similarity can be highly dubious. While functional genes also are not perfect in this regard, they present a much higher likelihood of corresponding to the function of interest.
Analysis of the 16S rRNA genes present in the sulfate-reducing bioreactors also confirmed the dominance of uncultured organisms in these systems (Table 6). In fact, 100% of the sequences obtained from the LC bioreactor presented the highest similarities to uncultured bacteria. This underscores the lack of suitability of primers targeting pure cultures for the characterization of complex anaerobic environments. While there is no guarantee that the primers designed in this study provide 100% coverage of the targets, especially with respect to uncultured organisms, this is the case with all primers and probes published to date. Thus, the degenerate primers developed in this study represent an important first step to broad and quantitative characterization of key functional groups in anaerobic environments.
Acknowledgments
Financial support for this work was provided by NSF CBET award 0651947, by the EPA Office of Research and Development Engineering Technical Support Center (contract 68-C-00-136; David Reisman, Project Officer), and by the Rocky Mountain Hazardous Substance Research Center through the EPA Science to Achieve Results (STAR) program (contract R 8395101-0).
The results of this study do not necessarily represent the views of the EPA or the NSF.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Achenbach, L. A., and J. D. Coates. 2000. Disparity between bacterial phylogeny and physiology—comparing 16S rRNA sequences to assess relationships can be a powerful tool, but its limitations need to be considered. ASM News 66:714-715. [Google Scholar]
- 2.Adams, M. W. W., L. E. Mortenson, and J. S. Chen. 1980. Hydrogenase. Biochim. Biophys. Acta 594:105-176. [DOI] [PubMed] [Google Scholar]
- 3.Bar, T., A. Stahlberg, A. Muszta, and M. Kubista. 2003. Kinetic outlier detection (KOD) in real-time PCR. Nucleic Acids Res. 31:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Béguin, P. 1990. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 44:219-248. [DOI] [PubMed] [Google Scholar]
- 5.Beller, H. R., S. R. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Dov, E., A. Brenner, and A. Kushmaro. 2007. Quantification of sulfate-reducing bacteria in industrial wastewater by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microb. Ecol. 54:439-451. [DOI] [PubMed] [Google Scholar]
- 7.Borsodi, A. K., A. Micsinai, A. Rusznyak, P. Vladar, G. Kovacs, E. M. Toth, and K. Marialigeti. 2005. Diversity of alkaliphilic and alkalitolerant bacteria cultivated from decomposing reed rhizomes in a Hungarian soda lake. Microb. Ecol. 50:9-18. [DOI] [PubMed] [Google Scholar]
- 8.Byrne, K. A., S. A. Lehnert, S. E. Johnson, and S. S. Moore. 1999. Isolation of a cDNA encoding a putative cellulase in the red claw crayfish Cherax quadricarinatus. Gene 239:317-324. [DOI] [PubMed] [Google Scholar]
- 9.Cardenas, E., W. M. Wu, M. B. Leigh, J. Carley, S. Carroll, T. Gentry, J. Luo, D. Watson, B. Gu, M. Ginder-Vogel, P. K. Kitanidis, P. M. Jardine, J. Zhou, C. S. Criddle, T. L. Marsh, and J. A. Tiedje. 2008. Microbial communities in contaminated sediments, associated with bioremediation of uranium to submicromolar levels. Appl. Environ. Microbiol. 74:3718-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chervoneva, I., T. Hyslop, B. Iglewicz, L. Johns, H. R. Wolfe, S. Schulz, E. Leong, and S. Waldman. 2006. Statistical algorithm for assuring similar efficiency in standards and samples for absolute quantification by real-time reverse transcription polymerase chain reaction. Anal. Biochem. 348:198-208. [DOI] [PubMed] [Google Scholar]
- 11.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C-1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 12.Devillard, E., D. B. Goodheart, S. K. R. Karnati, E. A. Bayer, R. Lamed, J. Miron, K. E. Nelson, and M. Morrison. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eden, P. A., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 14.Foti, M., D. Y. Sorokin, B. Lomans, M. Mussman, E. E. Zacharova, N. V. Pimenov, J. G. Kuenen, and G. Muyzer. 2007. Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl. Environ. Microbiol. 73:2093-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich, M. W. 2005. Methyl-coenzyme M reductase genes: unique functional markers for methanogenic and anaerobic methane-oxidizing Archaea. Methods Enzymol. 397:428-442. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 17.Geets, J., K. Vanbroekhoven, B. Borremans, J. Vangronsveld, L. Diels, and D. Van der Lelie. 2006. Column experiments to assess the effects of electron donors on the efficiency of in situ precipitation of Zn, Cd, Co and Ni in contaminated groundwater applying the biological sulfate removal technology. Environ. Sci. Pollut. Res. 13:362-378. [DOI] [PubMed] [Google Scholar]
- 18.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, and R. A. J. Warren. 1991. Domains in microbial beta-1,4-glycanases—sequence conservation, function, and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 20.Hallberg, K. B., and D. B. Johnson. 2005. Microbiology of a wetland ecosystem constructed to remediate mine drainage from a heavy metal mine. Sci. Total Environ. 338:53-66. [DOI] [PubMed] [Google Scholar]
- 21.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heijs, S. K., R. R. Haese, P. van der Wielen, L. J. Forney, and J. D. van Elsas. 2007. Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a Mediterranean cold seep. Microb. Ecol. 53:384-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henikoff, S., J. G. Henikoff, W. J. Alford, and S. Pietrokovski. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163:GC17-GC26. [DOI] [PubMed] [Google Scholar]
- 24.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiibel, S. R., L. P. Pereyra, L. Y. Inman, A. Tischer, D. J. Reisman, K. F. Reardon, and A. Pruden. 2008. Microbial community analysis of two field-scale sulfate-reducing bioreactors treating mine drainage. Environ. Microbiol. 10:2087-2097. [DOI] [PubMed] [Google Scholar]
- 27.Hipp, W. M., A. S. Pott, N. Thum-Schmitz, I. Faath, C. Dahl, and H. G. Trüper. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891-2902. [DOI] [PubMed] [Google Scholar]
- 28.Ibekwe, A. M., C. M. Grieve, and S. R. Lyon. 2003. Characterization of microbial communities and composition in constructed dairy wetland wastewater effluent. Appl. Environ. Microbiol. 69:5060-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, D. B., and K. B. Hallberg. 2005. Biogeochemistry of the compost bioreactor components of a composite acid mine drainage passive remediation system. Sci. Total Environ. 338:81-93. [DOI] [PubMed] [Google Scholar]
- 30.Juottonen, H., P. E. Galand, and K. Yrjala. 2006. Detection of methanogenic Archaea in peat: comparison of PCR primers targeting the mcrA gene. Res. Microbiol. 157:914-921. [DOI] [PubMed] [Google Scholar]
- 31.Karr, E. A., W. M. Sattley, M. R. Rice, D. O. Jung, M. T. Madigan, and L. A. Achenbach. 2005. Diversity and distribution of sulfate-reducing bacteria in permanently frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 71:6353-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 34.Meyer, J. 2007. [FeFe] hydrogenases and their evolution: a genomic perspective. Cell. Mol. Life Sci. 64:1063-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammadi, M., and P. J. R. Day. 2004. Oligonucleotides used as template calibrators for general application in quantitative polymerase chain reaction. Anal. Biochem. 335:299-304. [DOI] [PubMed] [Google Scholar]
- 36.Morales, T. A., M. Dopson, R. Athar, and R. B. Herbert. 2005. Analysis of bacterial diversity in acidic pond water and compost after treatment of artificial acid mine drainage for metal removal. Biotechnol. Bioeng. 90:543-551. [DOI] [PubMed] [Google Scholar]
- 37.Ozkan, M., S. G. Desai, Y. Zhang, D. M. Stevenson, J. Beane, E. A. White, M. L. Guerinot, and L. R. Lynd. 2001. Characterization of 13 newly isolated strains of anaerobic, cellulolytic, thermophilic bacteria. J. Ind. Microbiol. Biotechnol. 27:275-280. [DOI] [PubMed] [Google Scholar]
- 38.Pereyra, L. P., S. R. Hiibel, A. Pruden, and K. F. Reardon. 2008. Comparison of microbial community composition and activity in sulfate-reducing batch systems remediating mine drainage. Biotechnol. Bioeng. 101:702-713. [DOI] [PubMed] [Google Scholar]
- 39.Prieto, M. V., S. R. Hiibel, L. P. Pereyra, A. Pruden, K. F. Reardon, and D. Reisman. 2008. Effect of organic substrate composition on microbial community structure of pilot-scale biochemical reactors treating mining influenced water, p. 878-891. In R. Barnhisel (ed.), Proceedings of the 2008 National Meeting of the American Society of Mining and Reclamation: new opportunities to apply our science. American Society for Mining Reclamation, Richmond, VA.
- 40.Rabinovich, M. L., M. S. Melnik, and A. V. Bolobova. 2002. Microbial cellulases (review). Appl. Biochem. Microbiol. 38:305-321. [Google Scholar]
- 41.Ramakers, C., J. M. Ruijter, R. H. L. Deprez, and A. F. M. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, R. J., T. Vincze, J. Posfai, and D. Macelis. 2007. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 35:D269-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson, D. M., and P. J. Weimer. 2005. Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl. Environ. Microbiol. 71:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, Y. M., E. A. Polishchuk, U. Radoja, and W. R. Cullen. 2004. Identification and quantification of arsC genes in environmental samples by using real-time PCR. J. Microbiol. Methods 58:335-349. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Syutsubo, K., Y. Nagaya, S. Sakai, and A. Miya. 2005. Behavior of cellulose-degrading bacteria in thermophilic anaerobic digestion process. Water Sci. Technol. 52:79-84. [PubMed] [Google Scholar]
- 48.Táncsics, A., S. Szoboszlay, B. Kriszt, J. Kukolya, E. Baka, K. Marialigeti, and S. Revesz. 2008. Applicability of the functional gene catechol 1,2-dioxygenase as a biomarker in the detection of BTEX-degrading Rhodococcus species. J. Appl. Microbiol. 105:1026-1033. [DOI] [PubMed] [Google Scholar]
- 49.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 50.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, M., A. Loy, M. Klein, N. Lee, N. B. Ramsing, D. A. Stahl, and M. W. Friedrich. 2005. Functional marker genes for identification of sulfate-reducing prokaryotes. Methods Enzymol. 397:469-489. [DOI] [PubMed] [Google Scholar]
- 52.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, M. Y., B. H. Olson, and J. S. Chang. 2007. Improving PCR and qPCR detection of hydrogenase A (hydA) associated with Clostridia in pure cultures and environmental sludges using bovine serum albumin. Appl. Microbiol. Biotechnol. 77:645-656. [DOI] [PubMed] [Google Scholar]
- 54.Warnecke, F., P. Luginbuhl, N. Ivanova, M. Ghassemian, T. H. Richardson, J. T. Stege, M. Cayouette, A. C. McHardy, G. Djordjevic, N. Aboushadi, R. Sorek, S. G. Tringe, M. Podar, H. G. Martin, V. Kunin, D. Dalevi, J. Madejska, E. Kirton, D. Platt, E. Szeto, A. Salamov, K. Barry, N. Mikhailova, N. C. Kyrpides, E. G. Matson, E. A. Ottesen, X. N. Zhang, M. Hernandez, C. Murillo, L. G. Acosta, I. Rigoutsos, G. Tamayo, B. D. Green, C. Chang, E. M. Rubin, E. J. Mathur, D. E. Robertson, P. Hugenholtz, and J. R. Leadbetter. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560-565. [DOI] [PubMed] [Google Scholar]
- 55.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitford, M. F., R. J. Forster, C. E. Beard, J. H. Gong, and R. M. Teather. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153-163. [DOI] [PubMed] [Google Scholar]
- 57.Xing, D. F., N. Q. Ren, and B. E. Rittmann. 2008. Genetic diversity of hydrogen-producing bacteria in an acidophilic ethanol-H2-coproducing system, analyzed using the [Fe]-hydrogenase gene. Appl. Environ. Microbiol. 74:1232-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Q., E. A. Bayer, M. Goldman, R. Kenig, Y. Shoham, and R. Lamed. 2004. Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase. J. Bacteriol. 186:968-977. [DOI] [PMC free article] [PubMed] [Google Scholar]