Abstract
Clamp loader proteins catalyze assembly of circular sliding clamps on DNA to enable processive DNA replication. During the reaction, the clamp loader binds primer-template DNA and positions it in the center of a clamp to form a topological link between the two. Clamp loaders are multi-protein complexes, such as the five protein Escherichia coli, Saccharomyces cerevisiae, and human clamp loaders, and the two protein Pyrococcus furiosus and Methanobacterium thermoautotrophicum clamp loaders, and thus far the site(s) responsible for binding and selecting primer-template DNA as the target for clamp assembly remain unknown. To address this issue, we analyzed the interaction between the E. coli γ complex clamp loader and DNA using UV-induced protein–DNA cross-linking and mass spectrometry. The results show that the δ subunit in the γ complex makes close contact with the primer-template junction. Tryptophan 279 in the δ C-terminal domain lies near the 3′-OH primer end and may play a key role in primer-template recognition. Previous studies have shown that δ also binds and opens the β clamp (hydrophobic residues in the N-terminal domain of δ contact β. The clamp-binding and DNA-binding sites on δ appear positioned for facile entry of primer-template into the center of the clamp and exit of the template strand from the complex. A similar analysis of the S. cerevisiae RFC complex suggests that the dual functionality observed for δ in the γ complex may be true also for clamp loaders from other organisms.
Keywords: DNA replication, circular sliding clamp, clamp loader, UV-cross-linking, mass spectrometry
Introduction
Sliding clamp proteins are used by DNA polymerases as mobile tethers for highly processive replication of DNA. By themselves, the polymerases can incorporate a few nucleotides into the growing DNA polymer in a single binding event; in association with clamps, however, their processivity can increase to several thousand nucleotides. DNA polymerases in viruses, bacteria and archaebacteria, as well as eukaryotes use such clamps for rapid and efficient replication of chromosomal DNA.1,2 Sliding clamps are composed of two (Escherichia coli β clamp) or three subunits (bacteriophage T4 gp45, Sacharomyces cerevisiae PCNA, human PCNA), arranged in the form of a ring with a central cavity wide enough to accommodate double-stranded DNA (dsDNA).3–8 Upon encircling the duplex, clamps are linked topologically to DNA, and yet free to move on it; therefore, they can serve effectively as mobile tethers for polymerases during DNA synthesis. Several recent reports indicate that circular sliding clamps also play important roles in other cellular processes, including DNA repair and recombination, DNA methylation, chromatin remodeling, and cell-cycle control, perhaps by helping target key proteins in these processes to their sites of action on DNA.9,10
Circular sliding clamps must be loaded onto primed sites on template DNA by multi-protein complexes known as clamp loaders.1 These proteins use ATP to fuel their actions, which include binding the clamp, opening it, binding the DNA, and facilitating closure of the clamp around the duplex portion of the primer-template.11–15 Consistent with their essential role in DNA metabolism and possibly other cellular processes, clamp loader proteins appear to be conserved across evolution.16–18 Numerous studies of clamp loaders, including the E. coli γ complex, S. cerevisiae, Pyrococcus furiosus, and human RFC complexes, have revealed detailed information about their structure and mechanisms of action. For example, γ complex, the clamp loader of E. coli DNA polymerase III holoenzyme, is composed of five different proteins, γ/τ, δ, δ′, χ, and ψ, with three copies of γ/τ and one each of δ and δ′ forming the minimal functional body of the loader.15,19,20 (χ and ψ serve accessory functions, such as coordinating clamp assembly with primase and single-stranded DNA (ssDNA) binding protein activity at the replication fork.)21–23 The γ, δ, and δ′ subunits are arranged in a pentameric ring in the shape of a claw, with the β clamp binding sites at the tips of the fingers (see model in Discussion).19,24 The γ/τ subunits bind and hydrolyze ATP and serve as the motors of the clamp-loading machine (γ/τ belong to the AAA+ ATPase family).16,25,26 The δ subunit is the main contact between γ complex and the β clamp, and can open the clamp by itself.11 The δ′ and γ subunits modulate interaction between δ and β.11,27,28 ATP binding to the γ/τ subunits triggers conformational changes in γ complex that allow δ to bind β with high affinity and open the ring.12,24,29 The ATP-bound γ complex-β complex binds primer-template DNA with high affinity, presumably positioning it within the central cavity of the opened ring.12,30 The DNA-binding event triggers rapid ATP hydrolysis at the γ subunits, which is accompanied by a reduction in γ complex affinity for both β and DNA.13,14,31–33 Release of γ complex from β and DNA, and closure of the clamp around DNA complete the assembly process, following which DNA polymerase (or other proteins) can bind the clamp and commence work on DNA.
Clamp loaders from other organisms are composed of multiple subunits: the bacteriophage T4 clamp loader has four copies of the gp44 subunit and one copy of gp62;34 the S. cerevisiae and human RFC clamp loaders contain one copy each of five different proteins, RFC1, RFC2, RFC3, RFC4 and RFC5;35–38 archaebacterial clamp loaders contain two proteins, RFC-l and RFC-s.39–41 The gp44 and the RFC proteins share sequence similarities with γ and δ′, and are members of the AAA+ family; thus, like γ complex, these clamp loaders utilize multiple ATPase-active subunits for clamp assembly.16,17 A new report on S. cerevisiae RFC structure from the Kuriyan research group shows that the five RFC subunits adopt a claw-like arrangement, reminiscent of γ complex.42 Electron microscopy images of human RFC and P. furiosus RFC43 show the five subunits in a pentameric ring arrangement, and indicate ATP-dependent changes in clamp loader conformation.44 The conservation of many elements of clamp loader structure and biochemical activity across different species suggests a common overall mechanism of clamp assembly.
A key piece of information missing from our knowledge of the clamp loader mechanism is how this multi-protein complex binds DNA and recognizes primer-template as the target site for clamp assembly. Thus far, no discrete DNA-binding site(s), or even subunit(s) in the clamp loader has been identified as having DNA-binding activity that is essential for clamp assembly. Prior reports indicate that an N-terminal domain in RFC1 that shares homology with DNA ligase can bind DNA;45–47 however, removal of this domain from RFC1 appears to have no effect on RFC clamp-loading activity.48–50 The γ complex and S. cerevisiae RFC can interact with ssDNA and primer-template DNA, and RFC can even bind dsDNA with high affinity.12,51 Nonetheless, γ complex ATPase kinetics reveal that only primer-template DNA triggers rapid ATP hydrolysis, which in turn results in release of the DNA and clamp.30,33 In the case of RFC as well, primer-template DNA stimulates the greatest increase in steady-state ATPase activity.52 These data indicate that clamp loaders have a refined DNA-binding activity that is capable of distinguishing primer-template DNA from other DNA structures, and is coupled to both clamp-binding/opening and ATPase activities to facilitate efficient assembly of clamps on primed DNA during DNA replication.
In this study, we probed the identity of the DNA-binding subunit(s) on E. coli γ complex and S. cerevisiae RFC clamp loaders. Using UV-induced protein–DNA cross-linking at specific locations on primer-template DNA, we found that δ, the clamp-binding/opening subunit, also contacts DNA at the 3′-OH primer-template junction. In the case of RFC, the RFC1 subunit appears to be the major contact between the clamp loader and DNA. Further characterization of the cross-linked complexes by mass spectrometry revealed the DNA-binding site on δ, and these findings, in conjunction with the known crystal structure of γ complex, suggest a possible mechanism by which clamp loaders can recognize primed DNA as the substrate for clamp assembly.
Results and Discussion
The following sections describe: (a) preparation of DNA substrates with photo-reactive and radioactive nucleotides at desired locations, necessary for identification of proteins bound at the primer-template junction; (b) SDS-PAGE used to identify the DNA-binding subunit in the γ complex and S. cerevisiae RFC; (c) mass spectrometric analysis of the protein–DNA complex to identify the DNA-binding site; and (d) a model for primer-template DNA binding and selection by the clamp loader for clamp assembly.
Primer–template DNA with 5-bromodeoxyuridine (BrdU) and 32P inserted at specific locations
We have used site-specific photo-affinity labeling to map interactions between protein(s) in the multi-subunit clamp loader and primed DNA.53 The schematic in Figure 1 shows polymerase-catalyzed synthesis and purification of a DNA substrate containing BrdU and 32P-labeled deoxyadenosine monophosphate (dA). During biosynthesis, the polymerase incorporates BrdU, [32P]dA, and then the other nucleotides into a DNA primer as defined by the template strand sequence. The duplex DNA product is bound to streptavidin beads (via the biotin-tagged template) and the extended primer strand is separated by alkali denaturation and purified. This strand now serves as the template for primers of different lengths that position BrdU at various distances from the primer-template junction (primer-template DNA length is greater than the minimal length required for interaction with γ complex and RFC).54,55 BrdU allows zero-length cross-linking between protein and DNA, and is therefore a good probe for close protein–DNA contacts.56 The 32P-label marks the protein linked covalently to DNA and, most importantly, proximity of the 32P label to BrdU permits nearly complete degradation of DNA in the protein–DNA complex without loss of the radioactive marker, allowing identification of the cross-linked protein simply by size.
Figure 1.
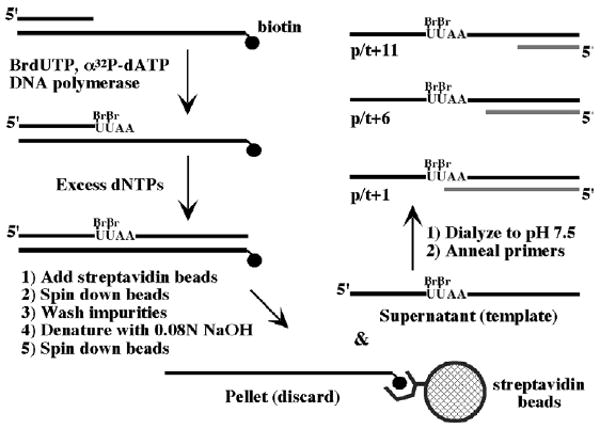
Design and synthesis of primed DNA templates containing 5-bromodeoxyuridine and 32P at specific locations. BrdU and [32P]dATP were incorporated into indicated positions within a 54 nucleotide DNA strand by T7 DNA polymerase. The 54-mer was purified and a 33 nucleotide primer annealed to it to form p/t+1, a primed DNA substrate with two [32P]dA in the double-stranded region and two BrdU in the single-stranded region immediately flanking the primer-template junction. Primers composed of 28 nucleotides and 23 nucleotides were used to generate p/t+6 and p/t+11 DNA substrates, respectively.
A C-terminal domain of the δ subunit in γ complex cross-links DNA at the primer-template junction
Binding of γ complex to p/t+1 primer-template DNA was examined first; p/t+1 contains two BrdU nucleotides on the single-stranded region immediately flanking the 3′-OH primer end (Figure 1). The reactions were performed in the presence of ATPγS, as ATP binding to γ complex facilitates its interactions with both DNA and the β clamp and, in this regard, ATPγS is a good non-hydrolyzable mimic of ATP.12 Exposure of the protein and DNA to UV light resulted in a single predominant complex that was separated from free DNA by SDS-PAGE (Figure 2(A), lane 3; 3% yield); a control experiment showed no such complex formation with DNA alone (Figure 2(A), lane 1). The protein–DNA complex was treated with DNaseI and S1 nuclease, but the 32P label adjacent to BrdU was protected by the protein and the complex remained radiolabeled (Figure 2(A), lane 4; in the corresponding control in lane 2 no free DNA remains after treatment with nuclease). Since the mass of the protein–DNA complex is reduced to nearly that of the free protein following treatment with nuclease, comparison of the radioactive image and Coommassie brilliant blue stain of the same gel reveals the identity of the cross-linked protein/s (Figure 2(A) and (B), lane 4). Because of their similar sizes, both δ (39 kDa) and δ′ (37 kDa) subunits of γ complex were implicated in DNA binding by this analysis. Further investigation of the interaction showed that the extent of DNA cross-linking to δ and/or δ′ increased from 3% to 10% when β was added to the reaction (Figure 2(A), compare lanes 3/4 and 7/8), even though β itself does not form any covalent links with DNA (Figure 2(A), lanes 5 and 6). The stimulatory effect of the clamp implies that the protein–DNA complex trapped by UV cross-linking is likely a relevant intermediate in the clamp assembly process. This effect is consistent with prior reports that γ complex-β has higher affinity for primed DNA, and rapidly assembles clamps on it, as compared with free γ complex.12,31,32
Figure 2.

The δ subunit in γ complex cross-links primer-template DNA. Protein–DNA complexes formed by UV-induced cross-linking were analyzed by SDS-PAGE. (A) Phosphorimage and (B) Coommassie brilliant blue-stained gel showing cross-linking of γ complex to p/t+1 DNA: lanes 1 and 2 contain p/t+1 alone, loaded onto the gel before and after treatment with DNaseI+S1 nuclease, respectively; lanes 3 and 4 show similar analysis of p/t+1 DNA with γ complex in the reaction; lanes 5 and 6 show p/t+1 with β clamp; lanes 7 and 8 show p/t+1 DNA with γ complex and β. (C) Phosphorimage and (D) Coommassie brilliant blue-stained gel showing similar analysis of p/t+1 cross-linking to γ complex subunits: lanes 1 and 2 show γ, lanes 3 and 4 show γ+β, lanes 5 and 6 show δ, and lanes 7 and 8 show δ+β loaded onto the gel before (lanes 1, 3, 5 and 7) and after (lanes 2, 4, 6 and 8) treatment with nucleases. (E) Phosphorimage and (F) Coommassie brilliant blue-stained gel of a similar analysis as in (C and D), except with δ′ and χ/ψ (−/+β).
Next we attempted to resolve whether δ or δ′, or both proteins, bind primer-template DNA. The cross-linking experiment was performed with individual γ, δ, and δ′ subunits, as well as a complex of χ/ψ (ψ alone is insoluble). Only δ exhibits strong cross-linking to p/t+1 DNA (Figure 2(C) and (D), lanes 5 and 6; 11% cross-linking yield). A trace amount of cross-linked product detected with the γ subunit, although not reproducible in all experiments, may indicate contact between γ and DNA as well (Figure 2(C) and (D), lanes 1/2 and 3/4; 0.4% yield −/+β). In the presence of β, the yield of δ-DNA increases from 11% to 20%, suggesting that β binding to δ (and β opening?) may stabilize the interaction between δ and DNA and/or help position δ to favor cross-linking BrdU in the pt+1 DNA (Figure 2C, compare lanes 5/6 and 7/8). Thus, the δ subunit in γ complex appears to play a dominant role in DNA binding during clamp assembly.
We questioned whether the interaction detected between δ and DNA is specific to a primed DNA template. The BrdU and 32P-containing template was used alone (ssDNA), annealed to its complement (dsDNA), annealed to a 33 nt primer (p/t+1), a 28 nt primer (p/t+6), or a 23 nt primer (p/t+11), in cross-linking reactions with γ complex in the presence of ATPγS (−/+β) and with δ (−/+β). The γ complex does cross-link to ssDNA (Figure 3(A), lane 1) but, unlike the single predominant species observed with p/t+1 DNA (Figure 3(A), lane 5), ssDNA yields multiple cross-linked species (the origin of non-specific species is not entirely clear). Following treatment with nuclease, however, the 32P label again marks δ as the subunit cross-linked to BrdU in ssDNA (Figure 3(A) and (C), lanes 1 and 2). Interestingly, both p/t+6 and p/t+11 DNA, which have BrdU positioned in the single-stranded region at six and 11 nucleotides from the primer-template junction, respectively, show reduced levels of cross-linking to γ complex compared with p/t+1 DNA (Figure 3(A), lanes 5/6, 7/8, and 9/10). The effect is more striking in the presence of β (p/t+1: 10%; p/t+6: 2%; p/t+11: 2%; Figure 3(B), lanes 5/6, 7/8, and 9/10); according to nitrocellulose membrane filtration assays, γ complex can bind all these DNAs (data not shown). Barely any cross-linking is detectable with the dsDNA substrate, which is consistent with prior reports of low or no interaction between γ complex and duplex DNA12 but may reflect inaccessibility of BrdU within the double helix. Cross-linking experiments with δ indicate that it can distinguish the primer-template junction by itself (Figure 3(C) (−β) and D (+β), compare lanes 5/6, 7/8, and 9/10). Thus, it appears that the amino acid residue(s) that can cross-link DNA are located in a region of δ that binds the primer-template junction specifically.
Figure 3.
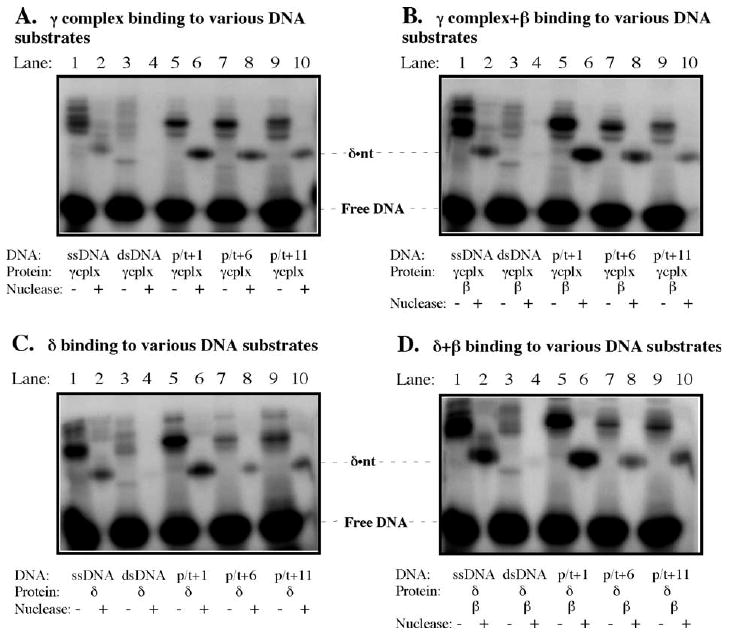
The δ-DNA cross-link is specific to the primer-template junction. (A) Lanes 1 and 2 show cross-linking of γ complex to single-stranded DNA (ssDNA), loaded onto the gel before and after treatment with nuclease, respectively; lanes 3 and 4 show similar analysis with double-stranded DNA; lanes 5 and 6 show p/t+1 DNA; lanes 7 and 8 show p/t+6 DNA and lanes 9 and 10 show p/t+11 DNA (containing BrdU 5 and 10 bases from the junction, respectively). (B) Similar analyses with γ complex+β, (C) with the δ subunit, (D) with δ+β.
Further investigation of the DNA binding/recognition properties of δ and γ complex was facilitated by knowledge of the crystal structure of δ, which shows three distinct domains: N-terminal domain I (1–140), domain II (141–210), and C-terminal domain III (211–343) (Figure 7(A)).19 The δ domains I (δI) and III (δIII) can be expressed at high levels and in soluble form,24,57 and δI has in fact been co-crystallized with the β clamp and is similar in structure to domain I within δ in the γ complex (except for elements that change upon contact with β).24 Experiments performed with the δ domains show that δIII forms a cross-link with p/t+1 DNA, but δI does not (Figure 4(A) and (B), lanes 3/4 and 5/6, respectively). These data suggest that both clamp-binding and DNA-binding activities of the clamp loader are located on the same protein, in separate domains, I and III, respectively. Moreover, δIII exhibits some preference for p/t+1 DNA over the other DNAs, although the difference is not as striking as that seen for full-length δ (Figure 4(C), compare lanes 1, 3, 5, and 7). However, δIII in complex with γ and δ′ (γ3δIIIδ′) can apparently distinguish a primer-template junction as it cross-links BrdU in p/t+1 DNA but not in p/t+6, p/t+11 DNAs (χ and ψ were not included in the complex because they are not absolutely necessary for clamp assembly and because similarity in their sizes with δIII would have complicated analysis). These results indicate that domain III is necessary and sufficient for interaction between δ and DNA, although specific placement of δ at the primer-template junction may be most effective when full-length δ protein or δ domain III bind DNA within the context of the γ complex.
Figure 7.
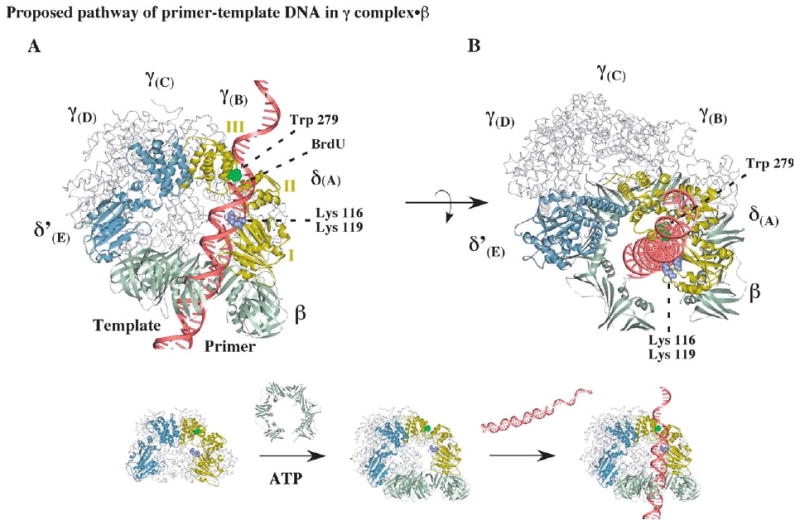
Possible positioning of primed DNA within the clamp loader-open clamp complex during clamp assembly. (A) A model of the open β clamp docked onto γ complex, illustrating a possible path for primer-template DNA with the duplex portion passing through the β ring and the single-stranded portion exiting the complex near the C-terminal domain III of δ. The structure of δ is shown with the DNA-cross-linking residue, tryptophan 279, colored green and lysine 116 and lysine 119 colored purple (the alphabet nomenclature for γ complex subunits corresponds to that used for the S. cerevisiae RFC crystal structure).11 (B) Another view of the complex, illustrating possible stacking of tryptophan 279 against the primer-template junction.
Figure 4.
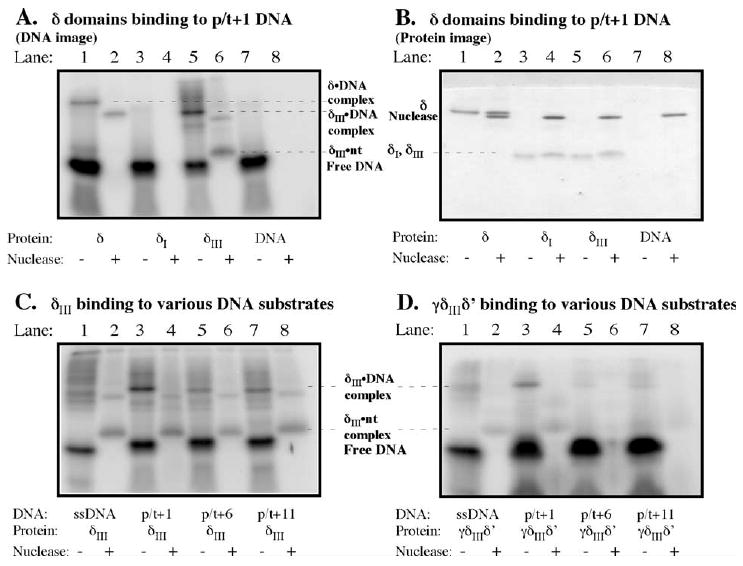
A C-terminal domain of δ (δIII) is responsible for interaction with the primer-template junction. (A) Phosphorimage and (B) Coommassie brilliant blue-stained gel with p/t+1 and full-length δ in lanes 1 and 2 (before and after treatment with nuclease, respectively), N-terminal δI domain in lanes 3 and 4, and C-terminal δIII domain in lanes 5 and 6. (C) Analysis of δIII and (D) γ3δIIIδ′ cross-linking to single-stranded DNA, p/t+1, p/t+6, and p/t+11 DNAs.
Identification of the cross-linked amino acid residue in δIII and δ by mass spectrometry
The UV cross-linking experiments yielded a fairly substantial amount of δIII-DNA complex (10–15% yield), enabling further analysis by mass spectrometry (MS) and identification of the amino acid residue(s) in close proximity to the 3′-OH primer end. p/t+1 DNA was synthesized on a large scale, and cross-linking of 1 nmol of DNA to δIII followed by SDS-PAGE and electro-elution from the gel yielded 30–50 pmol of pure δIII-DNA complex (Figure 5(A)). The complex was subjected to proteolysis by immobilized trypsin. Because of the high molecular mass of the DNA attached to the protein, it proved convenient to treat the resulting tryptic peptides with nuclease to digest the majority of the DNA.58 This treatment reduces the m/z of the cross-linked peptide ions to a convenient range for analysis (m/z<4000) and improves their MS response. Direct comparison of the MS profile before and after treatment with nuclease enabled us to distinguish and identify peptides modified by DNA cross-links among the other unmodified δIII tryptic peptides (Figure 5(B)). Six peaks emerged reproducibly in the MS spectrum after exhaustive treatment with nuclease (Figure 5(B) and Table 1). These six peaks were analyzed by multi-stage MS in order to identify the modified peptide, the modified residue, and the nature of the modification (Figure 5(C)). For example, the peak at m/z 1301.6 was present in the MS spectrum only after exhaustive treatment with both DNaseI and S1 nuclease, but not in the MS spectrum of a sample treated with only DNaseI, suggesting it as a potential candidate containing the DNA cross-linked site. MS2 analysis on 1301.6 gave characteristic neutral losses of 98 Da and 196 Da, which correspond to H3PO4 and phosphodeoxyribose, respectively. MS3 on the peak at m/z 1105.5 yielded a dominant product ion at m/z 949.5, corresponding to loss of the C-terminal arginine residue. Further MS4 analysis on m/z 949.5 gave a series of b and y ions that indicated the original peptide sequence was HRVWQNR, and that the tryptophan residue in this peptide was modified with a uracilyl group. These results agreed well with the photo-cross-linking chemistry of the BrdU reagent used in the experiment.59 Similar analysis of the other five modified peptide peaks revealed that they all contained the same cross-linked tryptophan 279 residue, and originated from partial cleavage by trypsin or nuclease (Table 1). A similar approach was applied to δ-p/t+1 DNA complex, but because MS peaks from the cross-linked tryptic peptides of full-length δ were masked by background noise, we resorted to analysis by hypothesis-driven multistage MS as described.60 As shown in Figure 5(D), we found that even in the full-length δ protein, the same tryptophan 279 residue cross-links with the uracil base at the primer-template junction.
Figure 5.

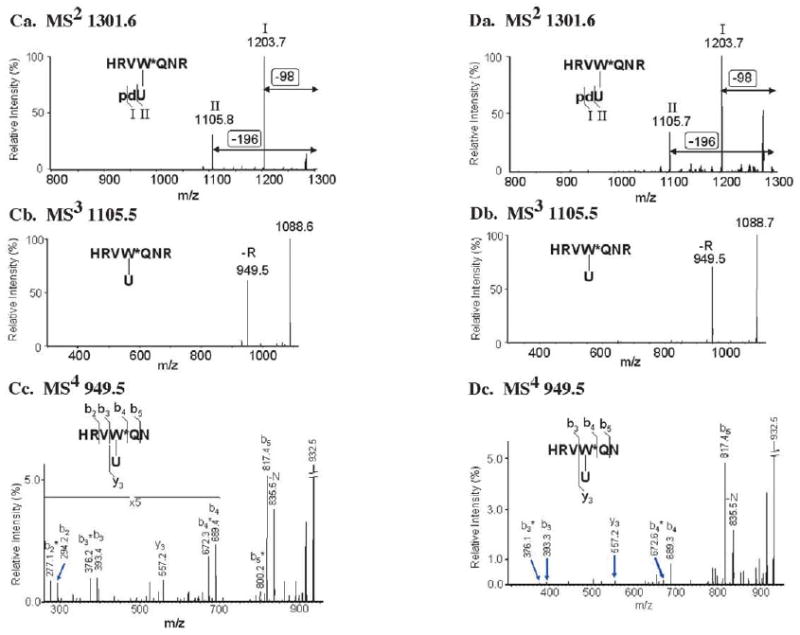
Differential peptide mass mapping and multiple stage mass spectrometry analysis of δIII·p/t+1 DNA and δ·p/t+1 DNA complexes reveal tryptophan-279 as the site of cross-linking to DNA. (A) Phosphorimage and Coommassie brilliant blue-stained gel showing separation of cross-linked protein–DNA complex from impurities, including unreacted protein and DNA; lanes 1, 2 and 3 have δIII alone, δIII+p/t+1 DNA, and protein size marker, respectively. (B) Comparison of MS spectra of δIII−p/t+1 DNA complex tryptic digest treated with (top) only DNaseI and (bottom) both DNaseI and S1 nuclease. Labeled peaks emerged after treatment with S1 nuclease, indicating that they contained the DNA cross-linked site. (C) Identification of m/z 1301.6 cross-linked peptide from δIII-p/t+1 DNA and site of cross-linking using multiple stage MS. (D) Identification of m/z 1301.6 cross-linked peptide from δ-DNA p/t+1 and site of cross-linking using multiple stage MS.
Table I.
Tryptic peptides containing DNA cross-linked at Tryptophan-279
| No. | Chemical Formula [M+H]+ | m/z [M+H]+ Calc. (Da) | m/za [M+H]+ Expt.(Da) | Peptide Sequence b | Modification b |
|---|---|---|---|---|---|
| 1 | C35H50N13O10 | 812.380 | 812.387 | VW*QNR | U |
| 2 | C40H59N13O16P | 1008.394 | 1008.408 | VW*QNR | pdU |
| 3 | C47H69N20O12 | 1105.540 | 1105.542 | HRVW*QNR | U |
| 4 | C46H71N17O17P | 1164.495 | 1164.522 | VW*QNRR | pdU |
| 5 | C52H78N20O18P | 1301.554 | 1301.560 | HRVW*QNR | pdU |
| 6 | C61H89N22O25P5 | 1591.584 | 1591.607 | HRVW*QNR | pdUpdU |
RFC1 subunit in the S. cerevisiae clamp loader cross-links primer-template DNA
The five subunits of the S. cerevisiae clamp loader, RFC (RFC1-5), are homologous to the γ and δ′ subunits of γ complex; however, as there is no clear sequence similarity between any of these proteins and δ, it is not immediately obvious which subunit(s) in RFC binds and opens the PCNA clamp. RFC1 is proposed to be the functional equivalent of δ due to certain common structural features, including a pair of conserved hydrophobic residues that contact PCNA.24,61 However, the RFC3 subunit also appears to bind PCNA, and it is not known which subunit actually opens the clamp for assembly on DNA.61,62 In the recently solved structure of S. cerevisiae RFC-PCNA, RFC1 and RFC3 (and RFC 4 to some extent) contact PCNA.42 Given our finding that δ possesses both clamp-binding/opening and DNA-binding activities, we wondered if RFC1 and/or RFC3 might also bind primer-template DNA.
In UV cross-linking experiments, the large RFC1 subunit was found linked to p/t+1 DNA (Figure 6, lanes 1 and 2). Less prominent products of the size of RFC2-5 were visible, but these were further reduced in the presence of PCNA (Figure 6(A), compare lanes 2 and 4). Since reports in the literature indicate that an N-terminal domain in RFC1, which is unnecessary for clamp loading, binds DNA, we tested an RFC complex with RFC1 replaced by RFC1S, which has the 282 amino acid residue domain deleted from the N terminus.37 RFC1S also cross-links p/t+1 DNA and weaker cross-links between the small RFC subunits and DNA are lost when PCNA is included in the reaction, highlighting the relevance of the contact between RFC1 and DNA for clamp assembly (Figure 6(A), lanes 5/6 and 7/8).
Figure 6.
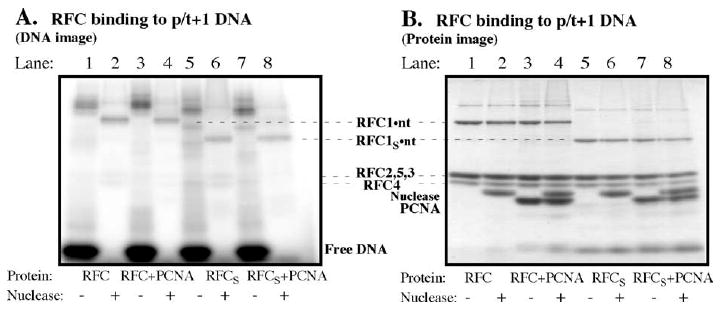
The RFC1 subunit in S. cerevisiae RFC clamp loader, which is analogous to the δ subunit, cross-links primed DNA. (A) Phosphorimage and (B) Coommassie brilliant blue-stained gel showing cross-linking of RFC with p/t+1 DNA in the absence (lanes 1 and 2) and in the presence (lanes 3 and 4) of PCNA; lanes 5–8 show similar analysis of RFC containing an N-terminal-truncated RFC1 subunit (RFC1s).
Implications for the mechanism of primer-template recognition and clamp assembly by clamp loader proteins
Of the five subunits in the γ complex, δ is the only one capable of opening the β clamp by itself. While essential, this action of δ is not sufficient for clamp assembly, as γ and δ′ are required for placement of the clamp around a primed DNA template.11 Since the identity of the DNA-binding subunit was unknown, it was considered possible that the γ and/or δ′ subunits were required for interaction between γ complex and DNA, and therefore δ alone could not function as a clamp loader. Here, we show that the δ protein has both clamp-binding/opening and primer-template binding/recognition activities, located on two separate domains.
Tryptophan 279 found cross-linked at the primer-template junction is in a cationic amino acid-rich region in the C-terminal domain of δ (δIII; Figure 7).42 The tryptophan flanks a narrow pass in δ, with two lysine residues located on the other side of the pass (lysine 113 and lysine 116 in domain I). The average width of the pass appears to be about 12 Å, which is wide enough to accommodate ssDNA, but not dsDNA. The correspondence between the cross-linking data and the protein structure in proximity to the cross-link suggests a pathway for entry of DNA into the γ complex as well as a means for selection of primer-template as the substrate for clamp assembly (Figure 7(A)). Once the clamp loader-open clamp complex is formed, primed DNA can be positioned in it with part of the double-stranded region in the central cavity of the clamp, and the rest, including the primer-template junction and single-stranded region, located within the dome-shaped chamber of the γ complex. As the template strand goes through the pass, the lysine residues may help position the DNA backbone,42 while the tryptophan residue stacks against a base at or near the primer-template junction, and perhaps helps lock the DNA substrate into place for clamp assembly (Figure 7(B)). Viewed from the top of the γ complex, the template appears to exit from the gap between the δ and δ′ subunits (consistent with proposed paths for DNA in structural analyses of the S. cerevisiae and P. furiosus RFC clamp loaders).42,43 In this model pathway for DNA, the tryptophan residue is in the right location to form a covalent link with a UV-induced uracilyl radical in the template strand flanking the primer-template junction. Tryptophan 279 function may be analogous to the “bracketing” action of tryptophan residues in the structure of p19 protein bound to its siRNA substrate, a 19-bp duplex with 3′-dinucleotide overhangs at both ends.63,64 In this case, tryptophan residues stack over the terminal base-pairs of the substrate, facilitating sequence-independent selection of duplex RNAs of specific lengths.
The defined position of BrdU in our DNA substrates allowed us to identify an amino acid present near that location. However, BrdU tends to couple most easily to tryptophan residues, particularly when excited at shorter wavelengths (254 nm in this case),56 so it is highly likely that there are other interactions between γ complex/RFC along the length of the DNA substrate that are not detected by this particular analysis. For example, the lysine residues in δ domain I, which also appear to be in close proximity to the DNA, were not picked up in the cross-linking analysis with DNA. In fact, the RFC structure reported by the Kuriyan research group shows a track of positive electrostatic potential along all the RFC subunits that matches the minor groove of a double helix placed within the central cavity in the complex.42 The cross-link formed between tryptophan 279 and BrdU at the 3′-OH end of the primer does, however, highlight a special role for δ versus the other clamp loader subunits in primer-template selection. We do not know whether the observed cross-link between RFC1 and DNA also reflects interaction between a tryptophan residue (or another base-stacking residue) and the primer-template junction. However, previous studies have noted several similarities between RFC1 and δ, including the fact that both RFC1 and δ contact their clamps; both lack the SRC amino acid motif essential for ATPase activity; both do not contribute ATPase activity towards clamp assembly; both are most divergent in sequence from other subunits in their respective complexes; both appear to be positioned similarly within their respective complexes, relative to the other subunits, and these observations fuel the hypothesis that RFC1 functions like δ in clamp assembly. Here, we add to this list of similarities by demonstrating that both proteins bind primer-template DNA. Further analysis is underway to identify the amino acid(s) in RFC1 that contacts DNA (note: the RFC1 DNA-binding site detected in this study is not located in the N-terminal ligase homology domain, which is known to bind DNA but is unnecessary for clamp assembly).48–50
The δ sequence is poorly conserved, making it difficult to predict even its functional homologue in other clamp loaders, let alone the location of the DNA-binding site in the said δ homologue. Recently, however, the McHenry research group used an iterative ψ-blast approach to identify δ sequences in known bacterial genomes and noted six conserved regions in these proteins.57 Of the amino acid residues we have identified as potential contributors to the DNA-binding site in δ, lysine 116 and lysine 119 are conserved in Region 3. Tryptophan 279 in δ does not lie in a conserved region; however, most of the proteins contain tryptophan, tyrosine, or phenylalanine residues at or within two to three residues of that location.57 Thus, it is possible that the DNA-binding site and mechanism for recognition of primer-template DNA might be conserved among bacterial clamp loaders.
An interesting corollary to the above discussion is the growing evidence that alternate RFC complexes function in DNA metabolic processes other than genomic DNA replication. These eukaryotic RFC-like complexes are pentamers of RFC2, RFC3, RFC4, and RFC5 subunits plus an RFC1 homologue, such as Rad17 (S. cerevisiae Rad24), Ctf18/Chl12, or Egl1, and are known to clamp proteins other than PCNA onto DNA (e.g. human Rad1-Hus1·Rad9 and S. cerevisiae Ddc1-Mec3-Rad17).65,66 A recent report on the human Rad17-RFC2-5 complex revealed that it catalyzes assembly of Rad1-Hus1-Rad9 preferentially on primer-template DNA with a recessed 5′ primer end, in marked contrast to the RFC complex, which loads PCNA onto primer-template DNA with a recessed 3′ primer end.67 The protein-loading activity of alternate RFC complexes is proposed to play a role in recognition/resolution of DNA structures containing recessed 5′ ends, such as are formed during stalled DNA replication, nucleotide excision repair, recombination-coupled DNA repair, and telomere maintenance.68 These observations are strikingly consistent with our finding that the RFC1 subunit in RFC complex binds primer-template DNA with a recessed 3′ end. Perhaps RFC1-like proteins perform analogous functions with other DNA structures.
An exact understanding of the mechanism of clamp loader action awaits further structural and biochemical studies; in the meantime, however, it is tempting to speculate the existence of a primordial clamp loader comprising a single protein that could bind DNA, bind and open the clamp, and load the clamp on DNA, and perhaps even had ATPase activity to drive needed conformational changes. Multiplication of this primordial clamp loader possibly yielded better clamp-loading efficiency and regulation of activity; however, the key activities of clamp binding/opening and DNA substrate recognition were retained on one subunit.
Experimental Procedures
Preparation of protein and nucleotide reagents
The γ, δ, δ′, χ, and ψ proteins were purified and the γ complex reconstituted,11 S. cerevisiae RFC (containing RFC1 or RFC1S) was over-expressed and purified from E. coli,37 δI was cloned and purified,24 and E. coli β3 and S. cerevisiae PCNA51 were purified as described in earlier reports. δIII, a 212–343 amino acid residue fragment of δ, was cloned into pET11a, over-expressed in E. coli, and purified over an SP Sepharose column (Amersham Biosciences; 100–700 mM NaCl gradient). Bacteriophage T7 DNA polymerase (exo− mutant) was a gift from Dr Smita Patel (Robert Wood Johnson Medical School), thioredoxin was purchased from Sigma-Aldrich, and Deoxyribonuclease I and S1 nuclease were purchased from Life Technologies.
The 5-BrdUTP was purchased from Sigma-Aldrich, other dNTPs were purchased from Amersham Biosciences, and [α-32P]dATP was purchased from Perkin Elmer. Short synthetic DNAs were purchased from Integrated DNA Technologies:
ST-5′ biotin-TAGCAGGGTGGAGGTTGTCAGGGTAGGGGGCTTAACGGCGCGGTCTTAGTCAGC 3′
SP-5′ GCTGACTAAGACCGCGCCG 3′
P+1-5′ TAGCAGGGTGGAGGTTGTCAGGGTAGG GGGCTT 3′
P+6-5′ TAGCAGGGTGGAGGTTGTCAGGGTAGGG 3′
P+11-5′ TAGCAGGGTGGAGGTTGTCAGGG 3′
For synthesis of BrdU and 32P-containing template DNA, first the ST and SP DNAs were annealed at a concentration of 5–25 μM (90 °C for five minutes and slow cooling to 25 °C in 20 mM Tris–HCl (pH 7.5), 150 mM NaCl). SP/ST at a concentration of 0.2 μM (or 2.5 μM for large-scale synthesis) was extended by 0.2 μM phage T7 DNA polymerase (pre-incubated for 20 minutes with 1 μM thioredoxin, 3 mM DTT) at 37 °C in 40 mM Tris–HCl (pH 7.5), 12.5 mM MgCl2, 50 mM NaCl, 0.1 mg/ml of bovine serum albumin (BSA), 1 mM DTT, 1 mM EDTA. Fifty micromolar BrdUTP and trace amounts of [α-32P]dATP were added to the reaction and incubated for ten minutes (25 minutes for large scale), followed by all four dNTPs at 1 mM each and further incubation for 30 minutes. The reaction was mixed with MagnaBind Streptavidin beads (Pierce) for 30 minutes at 37 °C to bind the DNA. The beads were collected and washed three times with 10 mM Tris–HCl (pH 7.5), 2 M NaCl, 1 mM EDTA, then incubated with 0.08 M NaOH for one minute to denature the duplex, and then centrifuged to separate ST (bound to the beads via biotin–streptavidin interaction) from the extended SP DNA (released into the supernatant). The supernatant was collected and dialyzed against 20 mM Hepes–NaOH (pH 7.5), 30 mM NaCl. 0.2 μM (or 2 μM for large scale) of the DNA was annealed with P+1, P+6, or P+11 DNAs to prepare the primer-templates.
UV-induced protein–DNA cross-linking and SDS-PAGE analysis
The γ complex, γ3δδ′χψ, or γ3δIIIδ′ (1 μM), δ (2.5 μM), δIII (5 μM), or RFC (1 μM) was incubated with 0.05 μM p/t DNA (or 1 μM DNA for large scale) and 200 μM ATPγS (in the case of γ complexes) in 30 μl of 30 mM Hepes–NaOH (pH 7.5), 4 mM MgCl2 for ten minutes at 25 °C. The reactions were placed on ice and exposed to UV light (254 nm) in a StrataLinker (Stratagene) for 25 minutes. Following cross-linking, each reaction was split into two: (a) 15 μl was quenched with 6 μl of SDS dye (1.5 M Tris base, 3% (w/v) SDS, 35% (v/v) glycerol, 150 mM DTT, bromophenol blue) + 6 μl of 0.5 M EDTA; (b) 15 μl was treated with 0.2 μl of DNaseI (200 units/μl) for ten minutes at 25 °C, then with 0.5 μl of 10% SDS and boiling for three minutes to destroy DNaseI, followed by addition of 2 μl of 10×S1 nuclease buffer and 1 μl of S1 nuclease (20 units/μl) and incubation at 37 °C for five minutes, after which the reaction was quenched with 0.5 μl of 0.5 M Tris base and 15 μl of SDS/dye. The reactions were boiled and analyzed by SDS-PAGE. The gels were scanned by Phosphorimager (Molecular Dynamics) and then stained by GelCode Blue stain (Pierce). For mass spectrometry analysis, the protein–DNA band was cut out of the gel and eluted using the BioRad Electro-Eluter (50 mM NH4HCO3 with 0.1% SDS for six hours and without SDS for four hours more). The eluate was dialyzed against 20 mM Tris–HCl (pH 7.5), 30 mM NaCl and dried under vacuum. The pellet was washed twice with 50 μl of water, 0.5 μl of 1 M HCl, 450 μl of acetone (12 hours at −20 °C) and twice with 50 μl of water/150 μl of acetone to remove SDS.
Protease digestion and mass spectrometry analysis
δIII-DNA and δ-DNA were dissolved in 20 μl of 50 mM NH4HCO3/acetonitrile (9/1, v/v), and digested with 1.5 μl of immobilized trypsin (Pierce) at 37 °C, shaking at 900 rpm for five hours. Next, 125 pg DN-EP (Sigma-Aldrich) was added to digest duplex DNA at room temperature for two hours; then 2 μl of the reaction product was removed for MS analysis. The rest of the sample was neutralized to pH 5 with 10% acetic acid and then zinc acetate (to 1 mM) and 0.5 unit of S1 nuclease added to digest ssDNA at room temperature for eight hours. MS spectra were collected on an in-house modified prototype Sciex QqTOF mass spectrometer. Tandem mass spectrometry experiments were performed on an in-house assembled matrix-assisted laser desorption/ionization (MALDI)-ion trap mass spectrometer that incorporates a Finnigan LCQ DecaXP (Thermo Electron) mass analyzer. 2,5-Dihydroxybenzoic acid (DHB) (Sigma-Aldrich) was prepared as a saturated solution in methanol/acetonitrile/water/trifluoroacetic acid (TFA) (50/20/30/0.1, by vol.) and diluted threefold in the same solution. Portions (2 μl) of the samples (before and after treatment with S1 nuclease) were mixed with 6 μl of diluted DHB solution and deposited onto MALDI target CD at 4 μl per spot, and air-dried before the mass spectrometry analysis. For the multi-stage MS experiments, samples were further concentrated using reversed phase oligoR3 beads (Applied Biosystems) and eluted with 4 μl of 1/4 saturated DHB solution.
Acknowledgments
This work was supported by grants GM64514-01 (to M.M.H), RR00862 (to B.T.C), and GM38839 (to M.O'D). We thank Smita S. Patel for the generous gift of T7 DNA polymerase (exo− mutant), Ishita Mukerji and Kelly Knee for tests with UV laser cross-linking, David Jeruzalmi and Gregory Bowman for helpful ideas and discussions, and Sanchaita Das for artistic input.
Abbreviations used
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- BrdU
bromodeoxyuridine
- MS
mass spectrometry
- BSA
bovine serum albumin
- DHB
2,5-dihydroxybenzoic acid
- TFA
trifluoroacetic acid
References
- 1.Jeruzalmi D, O'Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr Opin Struct Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 2.Kelman Z, O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 3.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 4.Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 5.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 7.Matsumiya S, Ishino Y, Morikawa K. Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 2001;10:17–23. doi: 10.1110/ps.36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hingorani MM, O'Donnell M. Sliding clamps: a (tail)ored fit. Curr Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 9.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 10.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 11.Turner J, Hingorani MM, Kelman Z, O'Donnell M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hingorani MM, O'Donnell M. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 13.Williams CR, Snyder AK, Kuzmic P, O'Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: I. Two distinct activities for individual ATP sites in the γ complex. J Biol Chem. 2004;279:4376–4385. doi: 10.1074/jbc.M310429200. [DOI] [PubMed] [Google Scholar]
- 14.Snyder AK, Williams CR, Johnson A, O'Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: II. Uncoupling the β and DNA binding activities of the γ complex. J Biol Chem. 2004;279:4386–4393. doi: 10.1074/jbc.M310430200. [DOI] [PubMed] [Google Scholar]
- 15.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J Biol Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 16.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Davey MJ, Jeruzalmi D, Kuriyan J, O'Donnell M. Motors and switches: AAA+ machines within the replisome. Nature Rev Mol Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- 18.O'Donnell M, Onrust R, Dean FB, Chen M, Hurwitz J. Homology in accessory proteins of replicative polymerases–E. coli to humans. Nucl Acids Res. 1993;21:1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader γ complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ′ with DnaX(4) forms DnaX(3)δδ′. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glover BP, McHenry CS. The χ ψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 23.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 24.Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, et al. Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 25.Johnson A, O'Donnell M. Ordered ATP hydrolysis in the γ complex clamp loader AAA+ machine. J Biol Chem. 2003;278:14406–14413. doi: 10.1074/jbc.M212708200. [DOI] [PubMed] [Google Scholar]
- 26.Podobnik M, Weitze TF, O'Donnell M, Kuriyan J. Nucleotide-induced conformational changes in an isolated Escherichia coli DNA polymerase III clamp loader subunit. Structure (Camb) 2003;11:253–263. doi: 10.1016/s0969-2126(03)00027-3. [DOI] [PubMed] [Google Scholar]
- 27.Indiani C, O'Donnell M. Mechanism of the δ wrench in opening the β sliding clamp. J Biol Chem. 2003;278:40272–40281. doi: 10.1074/jbc.M305828200. [DOI] [PubMed] [Google Scholar]
- 28.Leu FP, O'Donnell M. Interplay of clamp loader subunits in opening the β sliding clamp of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 2001;276:47185–47194. doi: 10.1074/jbc.M106780200. [DOI] [PubMed] [Google Scholar]
- 29.Goedken ER, Levitus M, Johnson A, Bustamante C, O'Donnell M, Kuriyan J. Fluorescence measurements on the E. coli DNA polymerase clamp loader: implications for conformational changes during ATP and clamp binding. J Mol Biol. 2004;336:1047–1059. doi: 10.1016/j.jmb.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 30.Ason B, Bertram JG, Hingorani MM, Beechem JM, O'Donnell M, Goodman MF, Bloom LB. A model for Escherichia coli DNA polymerase III holoenzyme assembly at primer/template ends. DNA triggers a change in binding specificity of the γ complex clamp loader. J Biol Chem. 2000;275:3006–3015. doi: 10.1074/jbc.275.4.3006. [DOI] [PubMed] [Google Scholar]
- 31.Bloom LB, Turner J, Kelman Z, Beechem JM, O'Donnell M, Goodman MF. Dynamics of loading the β sliding clamp of DNA polymerase III onto DNA. J Biol Chem. 1996;271:30699–30708. doi: 10.1074/jbc.271.48.30699. [DOI] [PubMed] [Google Scholar]
- 32.Bertram JG, Bloom LB, Hingorani MM, Beechem JM, O'Donnell M, Goodman MF. Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when γ complex loads β on DNA. J Biol Chem. 2000;275:28413–28420. doi: 10.1074/jbc.M910441199. [DOI] [PubMed] [Google Scholar]
- 33.Ason B, Handayani R, Williams CR, Bertram JG, Hingorani MM, O'Donnell M, et al. Mechanism of loading the Escherichia coli DNA polymerase III β sliding clamp on DNA. Bona fide primer/templates preferentially trigger the γ complex to hydrolyze ATP and load the clamp. J Biol Chem. 2003;278:10033–10040. doi: 10.1074/jbc.M211741200. [DOI] [PubMed] [Google Scholar]
- 34.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 35.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgers PM. Overexpression of multisubunit replication factors in yeast. Methods. 1999;18:349–355. doi: 10.1006/meth.1999.0796. [DOI] [PubMed] [Google Scholar]
- 37.Finkelstein J, Antony E, Hingorani MM, O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 38.Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee CG, Phillips B, et al. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc Natl Acad Sci USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelman Z, Hurwitz J. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum deltaH. J Biol Chem. 2000;275:7327–7336. doi: 10.1074/jbc.275.10.7327. [DOI] [PubMed] [Google Scholar]
- 40.Oyama T, Ishino Y, Cann IK, Ishino S, Morikawa K. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol Cell. 2001;8:455–463. doi: 10.1016/s1097-2765(01)00328-8. [DOI] [PubMed] [Google Scholar]
- 41.Seybert A, Scott DJ, Scaife S, Singleton MR, Wigley DB. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucl Acids Res. 2002;30:4329–4338. doi: 10.1093/nar/gkf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 43.Miyata T, Oyama T, Mayanagi K, Ishino S, Ishino Y, Morikawa K. The clamp-loading complex for processive DNA replication. Nature Struct Mol Biol. 2004;11:632–636. doi: 10.1038/nsmb788. [DOI] [PubMed] [Google Scholar]
- 44.Shiomi Y, Usukura J, Masamura Y, Takeyasu K, Nakayama Y, Obuse C, et al. ATP-dependent structural change of the eukaryotic clamp-loader protein, replication factor C. Proc Natl Acad Sci USA. 2000;97:14127–14132. doi: 10.1073/pnas.97.26.14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen BL, Uhlmann F, Gaur LK, Mulder BA, Posey KL, Jones LB, Hardin SH. DNA recognition properties of the N-terminal DNA binding domain within the large subunit of replication factor C. Nucl Acids Res. 1998;26:3877–3882. doi: 10.1093/nar/26.17.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burbelo PD, Utani A, Pan ZQ, Yamada Y. Cloning of the large subunit of activator 1 (replication factor C) reveals homology with bacterial DNA ligases. Proc Natl Acad Sci USA. 1993;90:11543–11547. doi: 10.1073/pnas.90.24.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fotedar R, Mossi R, Fitzgerald P, Rousselle T, Maga G, Brickner H, et al. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells. EMBO J. 1996;15:4423–4433. [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlmann F, Cai J, Gibbs E, O'Donnell M, Hurwitz J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- 49.Podust VN, Tiwari N, Stephan S, Fanning E. Replication factor C disengages from proliferating cell nuclear antigen (PCNA) upon sliding clamp formation, and PCNA itself tethers DNA polymerase δ to DNA. J Biol Chem. 1998;273:31992–31999. doi: 10.1074/jbc.273.48.31992. [DOI] [PubMed] [Google Scholar]
- 50.Gomes XV, Gary SL, Burgers PM. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J Biol Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
- 51.Hingorani MM, Coman MM. On the specificity of interaction between the Saccharomyces cerevisiae clamp loader replication factor C and primed DNA templates during DNA replication. J Biol Chem. 2002;277:47213–47224. doi: 10.1074/jbc.M206764200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomes XV, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 53.Persinger J, Bartholomew B. Site-directed DNA photoaffinity labeling of RNA polymerase III transcription complexes. DNA-Protein interactions: Principles and Protocols. In: Moss T, editor. Methods in Molecular Biology. 2nd. Vol. 148. Human Press; Totowa, NJ: 2001. p. 363. [DOI] [PubMed] [Google Scholar]
- 54.Yao N, Leu FP, Anjelkovic J, Turner J, O'Donnell M. DNA structure requirements for the Escherichia coli γ complex clamp loader and DNA polymerase III holoenzyme. J Biol Chem. 2000;275:11440–11450. doi: 10.1074/jbc.275.15.11440. [DOI] [PubMed] [Google Scholar]
- 55.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 56.Dietz TM, Koch TH. Photochemical coupling of 5-bromouracil to tryptophan, tyrosine and histidine, peptide-like derivatives in aqueous fluid solution. Photochem Photobiol. 1987;46:971–978. doi: 10.1111/j.1751-1097.1987.tb04879.x. [DOI] [PubMed] [Google Scholar]
- 57.Bullard JM, Pritchard AE, Song MS, Glover BP, Wieczorek A, Chen J, et al. A three-domain structure for the δ subunit of the DNA polymerase III holoenzyme δ domain III binds δ′ and assembles into the DnaX complex. J Biol Chem. 2002;277:13246–13256. doi: 10.1074/jbc.M108708200. [DOI] [PubMed] [Google Scholar]
- 58.Qin J, Chait BT. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal Chem. 1997;69:4002–4009. doi: 10.1021/ac970489n. [DOI] [PubMed] [Google Scholar]
- 59.Steen H, Petersen J, Mann M, Jensen ON. Mass spectrometric analysis of a UV-cross-linked protein–DNA complex: tryptophans 54 and 88 of E. coli SSB cross-link to DNA. Protein Sci. 2001;10:1989–2001. doi: 10.1110/ps.07601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalkum M, Lyon GJ, Chait BT. Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc Natl Acad Sci USA. 2003;100:2795–2800. doi: 10.1073/pnas.0436605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, et al. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J Biol Chem. 2003;278:50744–50753. doi: 10.1074/jbc.M309206200. [DOI] [PubMed] [Google Scholar]
- 62.Venclovas C, Colvin ME, Thelen MP. Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 2002;11:2403–2416. doi: 10.1110/ps.0214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 65.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J, MacNeill SA. Genome stability: a new member of the RFC family. Curr Biol. 2003;13:R873–R875. doi: 10.1016/j.cub.2003.10.048. [DOI] [PubMed] [Google Scholar]


