Abstract
Proteus mirabilis is naturally resistant to polymyxin B (PB). To investigate the underlying mechanisms, Tn5 mutagenesis was performed, and a mutant exhibiting increased PB susceptibility was isolated. The mutant was found to have Tn5 inserted into the PpmrI (Proteus pmrI) gene, a gene which may encode a UDP-glucuronic acid decarboxylase. In other bacteria, pmrI belongs to the seven-gene pmrF operon, which is involved in lipopolysaccharide (LPS) modification. While the PpmrI knockout mutant had a wild-type LPS profile and produced amounts of LPS similar to those produced by the wild type, LPS of the knockout mutant had higher PB-binding activity than that of the wild type. PB could induce alterations of LPS in the wild type but not in the PpmrI knockout mutant. Moreover, the PpmrI knockout mutant exhibited decreased abilities in biofilm formation and urothelial cell invasion. Complementation of the PpmrI mutant with the full-length PpmrI gene led to restoration of the wild-type phenotypic traits. Previously we identified RppA, a response regulator of the bacterial two-component system, as a regulator of PB susceptibility and virulence factor expression in P. mirabilis. Here we showed that RppA could mediate the induction of PpmrI expression by PB. An electrophoretic mobility shift assay further demonstrated that RppA could bind directly to the putative PpmrI promoter. Together, these results provide a new insight into the regulatory mechanism underlying PB resistance and virulence expression in P. mirabilis.
Cationic antimicrobial polypeptides (CAP) play an important role in host defense against microbial infection by affecting the membrane integrity of the pathogen and are key effectors of the host innate immune response (20). CAP, with antimicrobial and immunomodulatory activities, have the potential to be developed into antimicrobial drugs (12). Microbial pathogens have evolved distinct mechanisms to resist killing by CAP, including expelling CAP through pumps and cleaving CAP with proteases (39). One of the important mechanisms of resistance to CAP in Gram-negative bacteria involves remodeling the composition of the outer membrane through modifications of lipopolysaccharide (LPS) with positively charged substituents, which leads to the repulsion of CAP (39). In Salmonella enterica serovar Typhimurium, a seven-gene polycistronic unit (pmrHFIJKLM, called the pmrF operon for short) is involved in LPS modification (17, 18). The gene products of the pmrF operon, except that of pmrM, are necessary for the biosynthesis and addition of 4-aminoarabinose (Ara4N) to the 4′ phosphate of lipid A, a modification which contributes to a reduction in the net negative charge of LPS and consequently decreases attraction and binding of CAP to the outer membrane (17, 18).
In a large number of bacterial species, the genes conferring resistance to CAP are regulated by the bacterial two-component system (24, 28, 29, 31, 32, 37). In Salmonella, transcriptional activation of the pmrF operon requires the PmrA-PmrB two-component regulatory system, where PmrB is the sensor kinase and PmrA is the cognate response regulator that controls pmrF operon expression directly (16). A decrease in the Mg2+ concentration promotes PmrA-dependent upregulation of the pmrF operon. This process additionally requires the PhoP (regulator)-PhoQ (sensor kinase) two-component system, a master regulator of S. enterica virulence functions (14). PhoP positively controls the pmrF operon at the transcriptional level by increasing production of PmrD (24), which then activates the PmrA protein, resulting in modification of LPS. More recently, studies have shown that the transcription of PhoP-activated genes is upregulated by sublethal concentrations of CAP (3, 6) and that CAP can bind to and activate the PhoQ sensor directly (4).
Proteus mirabilis exhibits a form of multicellular behavior known as swarming migration (23) and is highly resistant to polymyxin B (PB) (30, 42), a kind of CAP (48). It has been shown that LPS plays a critical role in swarming (30, 43) and that an LPS modification affects both swarming and PB resistance in P. mirabilis (30). Previously we identified a gene, rppA, which may encode a response regulator of the two-component system (48). Our study showed that RppA plays an essential role in regulating PB susceptibility, swarming, and virulence functions in P. mirabilis (48).
Though the detailed mechanisms underlying resistance to PB are not known, an Ara4N LPS modification has been shown to be involved in PB resistance in P. mirabilis (30, 42). In this study, by a Tn5 transposon mutagenesis approach, we identified a gene, PpmrI (pmrI in Proteus), which was shown to be involved in PB resistance, biofilm formation, and urothelial cell invasion in P. mirabilis. We also found that expression of PpmrI is under the control of RppA. These studies provide a new insight into the regulation of PB resistance and virulence in P. mirabilis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB) medium. The LSW− agar, a medium that can inhibit swarming motility of P. mirabilis, was used for selecting Tn5-mutagenized clones (48).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant phenotype | Source or reference |
|---|---|---|
| Strains | ||
| P. mirabilis | ||
| N2 | Wild type; Tcr | Clinical isolate |
| I2 | N2 derivative; PpmrI Tn5-mutagenized mutant; PBs Kmr | This study |
| dIp | N2 derivative; PpmrI knockout mutant; PBs Kmr | This study |
| dIpc | dIp containing pACYC184-PpmrI; PPmrI -complemented strain; Cmr | This study |
| dA10 | N2 derivative; rppA knockout mutant; PBs Kmr | 48 |
| dA10c | dA10 containing pGEM-T Easy-rppA; RppA-complemented strain; Ampr | This study |
| E. coli | ||
| TOP10 | F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| S17-1 λ pir | λ pir lysogen of S17-1 [thi pro hsdR hsdM+recA RP4 2-Tc::Mu-Km::Tn7 (Tpr Smr)]; permissive host able to transfer suicide plasmids requiring the Pir protein by conjugation to recipient cells | 9 |
| Plasmids | ||
| pGEM-T Easy | High-copy TA cloning vector; Ampr | Promega |
| pGEM-4Z | High-copy cloning vector; Ampr | Promega |
| yT&A-xylE | High-copy TA cloning vector containg xylE coding region; Ampr | Provided by T. C. Yang (21) |
| pUT/mini-Tn5(Km) | Suicide plasmid requiring Pir protein for replication and containing mini-Tn5 cassette containing Kmr gene | 9 |
| pACYC184 | Low-copy cloning vector, P15A replicon; Cmr Tetr | 7 |
| pACYC184-PpmrI | pACYC184 containing intact PpmrI sequence, including its ribosome binding site (rbs); Cmr | This study |
| pGEM-T Easy-rppA | pGEM-T Easy containing intact rppA sequence, including its rbs; Ampr | This study |
Transposon mutagenesis and identification of the mutated gene.
P. mirabilis mini-Tn5 (Km)-mutagenized mutants were isolated as described previously (5). PB-susceptible mutants were identified by inoculating Tn5-mutagenized clones onto LSW− plates with or without 400 μg/ml PB. Chromosomal DNA was extracted from the mutants and partially digested with EcoRV, and fragments of more than 4 kb in size were cloned into EcoRV-digested pGEM-4Z (Promega). Following transformation of Escherichia coli TOP10, Km-resistant Tn5 Km-containing clones were selected. The nucleotide sequence of the cloned DNA fragments was determined and subjected to blast analysis (http://www.ncbi.nlm.nih.gov/). We then searched the sequence in the released genome sequence of P. mirabilis (http://www.sanger.ac.uk/) and cloned the full PpmrI gene by PCR-TA cloning with the primer pair pmrI-F1 and pmrI-R (Table 2). The nucleotide sequence was determined step by step using a 373A DNA sequencer (Applied Biosystems). PPmrI alignment with other PmrI proteins was performed using the DNAMAN software program (version 4.15).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Description |
|---|---|---|
| Km1-I out | GAGCTCGAATTCGGCCTAG | For identification of transposon-inserted gene |
| Km1-O out | CCTGCAGGCATGCAAGCTTC | For identification of transposon-inserted gene |
| Real-time-pmrIF | CACGTCCAGTGGCAGATAAC | For real-time PCR; paired with Real-time-pmrIR |
| Real-time-pmrIR | CCCCATTTCAGTGGCTAAAC | Paired with Real-time-pmrIF |
| pmrI-F1 | GCGTCCTCGGTATTTTATCC | For complementation and PpmrI full-length sequencing; paired with pmrI-R |
| pmrI-R | ACATCGACTCTCAAGCCAAC | Paired with pmrI-F1 |
| pmrI-R1 | CACATACCGTATACTTCTGATG | For PpmrI full-length sequencing |
| pmr-promoter-F | CAACAATCGTTAGCTTTGCC | For reporter assay; paired with pmr-promoter-R |
| pmr-promoter-R | GGAGAATGGAAGAAAGTGAC | Paired with pmr-promoter-F |
| pmr-promoter-F2 | TTGTCTACAAGGTGGCAGTA | For EMSA; paired with pmr-promoter-R |
| pmrI-upF | ATCGCGGTAGTGCAATTAAG | For PpmrI knockout; paired with Xba-pmrI-upR |
| Xba-pmrI-upR | TCTAGACCAACACAGCCAATATCATG | Paired with pmrI-upF |
| Xba-pmrI-downF | TCTAGATATCGAAACACGCCAAACGG | For PpmrI knockout; paired with pmrI-downR1 |
| pmrI-downR1 | TTCCCCGACAATCACTATTG | Paired with Xba-pmrI-downF |
| RppA-F EcoRI | CGGAATTCATGAATATTTTATTAGTTGA | For His-tagged RppA expression; paired with RppA-R XhoI |
| RppA-R XhoI | CCGCTCGAGAAGAGCGGATCTTCATTTAG | Paired with RppA-F EcoRI |
Gene knockout by homologous recombination.
Sequences flanking the PpmrI gene were amplified by PCR using the primer pairs pmrI-upF/Xba-pmrI-upR and Xba-pmrI-downF/pmrI-downR1 (Table 2), respectively, and cloned into pGEM-T Easy (Promega) to generate pGPpmrI-up and pGPpmrI-dn. pGPpmrI-up was digested with SalI/XbaI, and the PpmrI upstream sequence-containing fragment was ligated to SalI/XbaI-digested pGPpmrI-dn to produce the pGPpmrI-updn plasmid, which contains both upstream and downstream sequences of PpmrI. A Kmr cassette was inserted in the XbaI-digested pGPpmrI-updn plasmid to generate pGPpmrI-updn-Km, a plasmid containing the Kmr-cassette-disrupted combined upstream and downstream sequence of PpmrI. The DNA fragment containing the Kmr-cassette-disrupted combined upstream and downstream sequence of PpmrI was cleaved by SalI/SphI from pGPpmrI-updn-Km and ligated into SalI/SphI-cleaved pUT/mini-Tn5(Km) (9) to generate pUTPpmrI-Km. Gene inactivation mutagenesis by homologous recombination and confirmation of mutants with double-crossover events were performed as described previously (48).
Construction of PPmrI-complemented strains.
Full-length PpmrI was amplified by PCR using the primer pair pmrI-F1 and pmrI-R (Table 2) and cloned into pGEM-T Easy (Promega) to generate pGPpmrI. The DNA fragment containing full-length PpmrI was excised from pGPpmrI with SalI and SphI. The DNA fragment was ligated into a SalI/SphI-digested low-copy-number plasmid, pACYC184, to generate the PpmrI complementation plasmid, pACYC184-PpmrI. PpmrI is thus driven by the promoter of the tetracycline-resistant gene in the pACYC184 plasmid. pACYC184-PpmrI was then transformed into the PpmrI knockout mutant to generate the PPmrI-complemented strain.
Real-time RT-PCR.
To study the effect of rppA mutation and PB on the expression of PpmrI mRNA, overnight LB cultures of the wild-type, rppA knockout, and RppA-complemented strains were washed with N minimal medium (5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.2% glucose, 0.01% Casamino Acids, 0.1 mM Tris-HCl, pH 7.4), diluted to an optical density at 600 nm (OD600) of 0.3 to 0.4 in N minimal medium without or with 1 μg/ml PB, and grown for 3 to 6 h at 37°C. Total RNA was extracted, and real-time reverse transcriptase PCR (RT-PCR) was carried out as described previously (48) to monitor the expression of PpmrI mRNA. The levels of PpmrI mRNA were normalized against those of 16S rRNA.
Reporter assay.
The 0.674-kb promoter region of the putative pmrF operon, amplified by the primer pair pmr-promoter-F and pmr-promoter-R, was cloned into pGEM-T Easy to create pGEMPpmrI. xylE-containing yT&A plasmid (21) was cut by SacI/PstI, and the xylE-containing fragment was ligated to SacI/PstI-cleaved pGEMPpmrI to create pGEMPpmrI-xylE. pGEMPpmrI-xylE was then digested with SmaI/SphI, and the pmrF promoter-xylE fusion DNA fragment was ligated to EcoRV/SphI-cleaved pACYC184 to produce the PpmrI-xylE reporter plasmid. The PpmrI-xylE reporter plasmid-transformed wild type, rppA knockout, and RppA-complemented strains were grown overnight in LB broth with 20 μg/ml chloramphenicol. The cultures were diluted 100-fold in the same medium with or without PB and incubated for 3 to 6 h before XylE activity was measured as described previously (21). Briefly, a 1-ml cell suspension was centrifuged, resuspended in assay buffer, and subjected to kinetic assay of the enzyme activity after addition of the substrate (catechol). Meanwhile, the optical density of the cell suspension at 600 nm was recorded. One unit of enzyme activity was defined as the amount of enzyme that converted 1 nmol substrate per minute. The specific activity of the enzyme was defined in terms of units per OD600 unit. Each experiment was repeated three times. Specific activity of XylE (nmol catechol/min/OD600) was calculated as ΔA375 × Δmin−1 × ɛ−1 × OD600−1 × dilution factor (ɛ = 22,000 M−1 cm−1).
Purification of His-tagged recombinant protein.
The rppA gene was cloned into the plasmid pET32a (Novagen) to generate the pET32a-RppA plasmid. An overnight culture of E. coli strain BL21 containing pET32a-RppA was diluted 1:100 into 200 ml of LB broth supplemented with ampicillin and grown at 37°C with vigorous shaking to an optical density at 600 nm of 0.5 to 0.6. The culture was then incubated for 7 h at 25°C after addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (0.25 mM) to induce expression of the His-tagged RppA protein. After sonication, the E. coli cell lysate was centrifuged and the supernatant was applied to a Ni2+-nitrilotriacetic acid column (Invitrogen). After washing three times with wash buffer (500 mM NaCl, 50 mM or 75 mM imidazole, 50 mM sodium phosphate, pH 8.0), the His-tagged RppA protein was eluted with the same buffer containing an increased concentration of imidazole (500 mM). Finally, the His-tagged RppA protein was concentrated using a centrifugal filter (Amicon Ultra-4; Millipore) against binding buffer (150 mM KCl, 10 mM MgCl2, 30 mM Tris-HCl, pH 9.0). The protein concentration was determined using the Bradford protein assay (Bio-Rad). The purity of the RppA preparation was confirmed by SDS-PAGE.
EMSA.
Amplified DNA fragments (372 bp putative PpmrI promoter DNA or 330 bp irrelevant DNA fragments, 0.1 μg each) were incubated with 0, 1, or 2 μM His-tagged RppA in a 10-μl binding buffer (150 mM KCl, 10 mM MgCl2, 30 mM Tris-HCl, pH 9.0). For the supershift assay, the amplified promoter fragments (0.1 μg) were incubated with 1 μM His-tagged RppA and 0.3 μl antipolyhistidine antibody (1:33) in a 10-μl reaction mixture. After incubation for 30 min at room temperature, the reaction mixtures were loaded onto 5% nondenaturing polyacrylamide gels, and then the gel was electrophoresed at 100 V for 1.5 to 2 h before staining with ethidium bromide. An electrophoretic mobility shift assay (EMSA) with purified His-tagged RppA and digoxigenin (DIG)-labeled PpmrI promoter DNA was also performed, and unlabeled PpmrI promoter DNA was added as competitors to demonstrate the binding specificity of the PpmrI promoter DNA and His-tagged RppA.
MIC assay.
In vitro determination of the MIC for PB was performed by the broth microdilution method according to the guidelines proposed by the Clinical and Laboratory Standards Institute (8). A stock solution of PB (40,960 μg/ml) prepared in sterile water was added to 96-well microtiter plates in 2-fold serial dilutions. Aliquots of bacterial culture (5 × 104 CFU) were then dispensed into each well and incubated for 16 to 18 h. The MIC was defined as the lowest PB concentration at which no visible growth occurred. MICs of ampicillin, gentamicin, ciprofloxacin, tetracycline, and imipenem for wild-type P. mirabilis, the PpmrI knockout mutant, and the PPmrI-complemented strain were also determined according to the guidelines proposed by the Clinical and Laboratory Standards Institute (8).
Preparation and analysis of LPS.
LPS extraction and analysis were performed as described previously (48).
Binding of PB by LPS.
The experiments for determining the binding of PB by LPS were performed as described previously (48), except that a final concentration of 15 μg/ml of PB was used in the PB-binding reaction.
Biofilm formation assay.
Biofilm formation was assayed by measuring the ability of cells to adhere to the wells of 96-well microtiter dishes made of polyvinylchloride (Becton Dickinson), as described previously (27), with some modifications. Overnight LB cultures (10 μl) were diluted to 1 ml with LB broth, and 100 μl was transferred to the microtiter well. The microtiter dishes were sealed with parafilm and incubated at 37°C for 16 h. After incubation, the wells were rinsed with phosphate-buffered saline (PBS) and air dried at room temperature for 15 min. Two hundred microliters of crystal violet (1%) solution was added to each well, and the dishes were incubated for 20 min. The crystal violet-stained biofilms were then extracted twice with 200 μl of 95% ethanol, and the absorbance at 550 nm was determined.
Cell invasion assay.
Overnight culture was diluted 100-fold and incubated for 3 h before the cell invasion assay according to the protocol of Liaw et al. (26), with some modifications. Briefly, human urothelial NTUB1 cells were grown and then infected at 37°C with 1 ml of a bacterial suspension containing ca. 5 × 107 bacteria for 1.5 h. Urothelial cells were then washed twice with PBS and incubated at 37°C in 1 ml of RPMI 1640 medium containing streptomycin (250 μg/ml) for another 1.5 h. Cells were washed twice again with PBS and then lysed by incubation with 1 ml of lysis solution at 37°C for 30 min. Cell lysates were diluted serially in saline, and viable bacteria were counted by plating on LSW− agar plates. Cell invasion ability was expressed as the percentage of viable bacteria that survived the streptomycin treatment versus the total inoculum.
Nucleotide sequence accession number.
The nucleotide sequence of P. mirabilis N2 PpmrI has been deposited in the DDBJ/EMBL/GenBank databases under accession no. GQ457568.
RESULTS
Isolation of PB-sensitive P. mirabilis mutant.
In order to investigate the underlying mechanisms of natural PB resistance in P. mirabilis, we undertook a mini-Tn5 transposon mutagenesis approach to isolate the PB-sensitive mutants of P. mirabilis. Through characterizing these mutants, we identified a mutant, I2, which was more than 10,240-fold more susceptible to PB than wild-type P. mirabilis N2 (MIC of 4 versus >40,960 μg/ml). The Tn5-disrupted nucleotide sequence was obtained from cloning the mini-Tn5-containing DNA fragment in the mutant I2. Through searching the P. mirabilis released genome sequence (http://www.sanger.ac.uk/) using the sequence we obtained, we found that Tn5 was inserted into a gene which we named PpmrI (pmrI in Proteus). The location of the Tn5 insertion in PpmrI was 88 bp downstream from the initiation codon. PpmrI was cloned and sequenced. The nucleotide sequence of PpmrI was found to be 97% identical to the corresponding sequence of the published P. mirabilis strain HI4320 (GenBank accession no. AM942759).
PpmrI is within a putative seven-gene pmrF operon.
The sequence of PpmrI was similar to that of the third gene of the pmrF operon in Salmonella and E. coli. Analysis of the amino acid sequence indicated that PpmrI may encode a bacterial UDP-glucuronic acid decarboxylase. PPmrI is homologous to S. enterica serovar Typhimurium PmrI (66% identity, 80% similarity), E. coli PmrI (66% identity, 81% similarity), and Pseudomonas aeruginosa PmrI (64% identity, 80% similarity). Further analysis of the deduced amino acid sequences revealed that this Tn5-inserted P. mirabilis locus may encode seven putative proteins with 81%, 87%, 72%, 68%, 63%, and 55% similarities to Salmonella PmrH, PmrF, PmrJ, PmrK, PmrL, and PmrM, respectively (GenBank accession no. AE006468). Immediately upstream and downstream of the operon are the btuD and nlpC genes, respectively, in P. mirabilis N2.
The P. mirabilis PpmrI knockout mutant exhibited increased susceptibility to PB.
To demonstrate the role of PPmrI in regulating PB susceptibility, we sought to construct the PpmrI mutant through allelic-exchange mutagenesis. A PpmrI knockout mutant (dIp) was constructed by homologous recombination using the plasmid pUTPpmrI-Km, harboring a Kmr-cassette-disrupted combined upstream and downstream sequence of PpmrI (see Materials and Methods). Southern blot analysis indicated that the dIp mutant contained a single disrupted PpmrI gene and no wild-type PpmrI allele (data not shown). The MICs of PB for wild-type P. mirabilis N2 and the dIp mutant were determined. While the MIC of PB for the wild-type strain was >40,960 μg/ml, that for the dIp mutant was 4 μg/ml. To further confirm that PPmrI can affect PB susceptibility, we constructed a PPmrI-complemented strain of dIp, dIpc, by transforming pACYC184-PpmrI (a low-copy-number plasmid carrying a wild-type PpmrI gene) into the dIp mutant. We found that the MIC of PB for the dIpc strain was >40,960 μg/ml. Together, these data suggest that PPmrI is a determinant of PB susceptibility in P. mirabilis.
P. mirabilis PpmrI knockout mutant exhibited changes in LPS.
LPS modifications play an important role in PB susceptibility in many Gram-negative bacteria, including Salmonella, Yersinia, Pseudomonas, E. coli, and P. mirabilis (30, 31, 41, 44). To investigate the underlying cause of PB sensitivity in the PpmrI knockout mutant, we compared the LPS profile of the PpmrI knockout mutant (dIp) with that of the wild-type strain (N2). The LPS was extracted from equal amounts of the wild-type or mutant cells and was subjected to SDS-PAGE analysis. As shown in Fig. 1A, the PpmrI knockout mutant had an LPS profile similar to that of the wild type. In addition, the concentration of LPS was determined from equal amounts of the wild-type or mutant cells. Table 3 showed that the PpmrI knockout mutant synthesized an amount of LPS similar to that of the wild-type strain (13.1 versus 13.2 mg/ml).
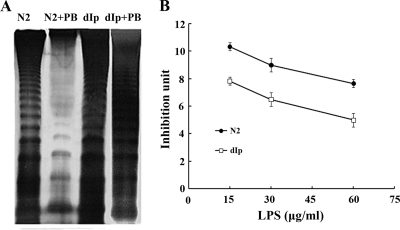
FIG. 1.
(A) LPS profiles of wild-type P. mirabilis (N2) and the PpmrI knockout mutant (dIp) in the presence or absence of 1 μg/ml PB. Six microliters of LPS purified from the same cell number (OD600 × volume [in ml] = 100) of the respective strains was subjected to SDS-PAGE analysis. (B) PB-binding ability of LPS purified from wild-type P. mirabilis (N2) and the PpmrI knockout mutant (dIp). Various amounts of purified LPS were subjected to the PB-binding assay. The unbound PB was then subjected to the E. coli inhibition assay (see Materials and Methods). The data are the averages of results from three independent experiments with standard deviations.
TABLE 3.
Amounts of LPS in wild-type P. mirabilis (N2), the PpmrI knockout mutant (dIp), and the PPmrI-complemented strain (dIpc) in the absence and presence of PB
| Strain | LPS concn (mg/ml)a |
|
|---|---|---|
| PB (−) | PB (+) | |
| N2 | 13.2 ± 0.7 | 6.8 ± 0.6 |
| dIp | 13.1 ± 1.1 | 12.9 ± 0.9 |
| dIpc | 13.0 ± 0.8 | 7.0 ± 0.6 |
LPS was quantitated as described in Materials and Methods in the absence [(−)] or presence [(+)] of 1 μg/ml polymyxin B.
To explain the reason why the PpmrI knockout mutant has increased PB susceptibility, we tested whether the LPS purified from the PpmrI knockout mutant and the wild-type strain had a different binding affinity for PB. Equal amounts of LPS from the PpmrI knockout mutant (dIp) and the wild-type strain (N2) were incubated with PB, and the unbound fraction was subjected to the E. coli inhibition assay. As shown in Fig. 1B, LPS from the PpmrI knockout mutant bound a larger amount of PB than LPS from the wild-type strain. Since identical concentrations of LPS were used in the binding assay, these data indicate that there was a qualitative change in the LPS of the PpmrI knockout mutant and that this change caused it to have a higher binding affinity for PB. This increased PB-binding activity of the PpmrI knockout mutant may contribute to its susceptibility to PB.
In addition, it is noteworthy that when the wild-type P. mirabilis strain was incubated in the medium containing a low concentration of PB (1 μg/ml), its LPS profile changed dramatically: the intensity of the LPS ladder decreased, and the band shift was observed (Fig. 1A, compare lane 1 with lane 2). In contrast, the LPS profile of the PpmrI knockout mutant did not change after incubation in the medium containing 1 μg/ml PB (Fig. 1A, compare lane 3 with lane 4). Moreover, while the amounts of LPS in the wild-type strain were reduced upon addition of 1 μg/ml PB, the amounts of LPS in the PpmrI knockout mutant were not affected by PB (Table 3). Together, these data suggest that PB can affect the modification of LPS in P. mirabilis and that this effect is dependent on the presence of PPmrI.
Knowing the alterations of LPS in the PpmrI knockout mutant, we also performed in vitro susceptibility testing using ampicillin, gentamicin, ciprofloxacin, tetracycline, and imipenem in wild-type P. mirabilis (N2), the PpmrI knockout mutant (dIp), and the PPmrI-complemented strain (dIpc). The PpmrI knockout mutant exhibited the same susceptibilities to all drugs except imipenem (Table 4). The increased susceptibility of the PpmrI knockout mutant to imipenem indicated that LPS may also play some roles in imipenem resistance.
TABLE 4.
MICs of ampicillin, gentamicin, ciprofloxacin, tetracycline, and imipenem for wild-type P. mirabilis (N2), PpmrI knockout mutant (dIp), and PPmrI-complemented strain (dIpc)
| Strain | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Ampicillin | Gentamicin | Ciprofloxacin | Tetracycline | Imipenem | |
| N2 | 16 | 4 | 2−6 | 32 | 2 |
| dIp | 16 | 4 | 2−6 | 32 | 1/4 |
| dIpc | 16 | 4 | 2−6 | 32 | 2 |
Expression of PPmrI is modulated by PB through the RppA-dependent pathway.
Knowing that PPmrI is required for resistance to PB and for PB-mediated changes in LPS and that pmrI expression (i.e., the pmrF operon) is controlled by a two-component systems in Salmonella, we wondered whether the expression of PPmrI was regulated by RppA, a response regulator of the two-component system that was shown by us to be able to regulate PB susceptibility (48). We thus investigated the expression of PpmrI mRNA in the wild type, the rppA knockout mutant, and the RppA-complemented strain in the presence or absence of 1 μg/ml PB by real-time RT-PCR. In the absence of PB, the expression of PpmrI mRNA in the rppA knockout mutant was about the same as that in the wild-type and RppA-complemented strains. While PB can induce the expression of PpmrI mRNA in the wild-type and RppA-complemented strains, it cannot do so in the rppA knockout mutant (Fig. 2A shows the results after a 3.5-h incubation). These data indicate that PB can regulate the expression of PPmrI in the RppA-dependent pathway.
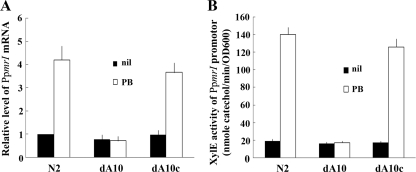
FIG. 2.
(A) Effect of rppA mutation on the expression of PpmrI mRNA in the presence or absence of PB (1 μg/ml). The amounts of PpmrI mRNA for the wild-type, rppA mutant, and RppA-complemented strains were quantified by real-time PCR after a 3.5-h incubation. The value obtained with the wild-type cells in the absence of PB was set at 1. The data represent the averages of results of four independent experiments with standard deviations. (B) The activity of XylE in the PpmrI-xylE reporter plasmid-transformed wild-type, rppA mutant, and RppA-complemented strains in the presence or absence of 1 μg/ml PB after a 3.5-h incubation. The data represent the averages of results of three independent experiments with standard deviations. N2, wild type; dA10, rppA mutant; dA10c, RppA-complemented strain.
To confirm this, a promoter-reporter assay using a xylE transcriptional fusion was used. By performing RT-PCR using primers spanning the putative pmrI and pmrH genes in the N2 strain, we demonstrated that the pmrH, pmrF, and pmrI genes are cotranscribed and share the promoter upstream of the putative pmrH gene (data not shown). Therefore, the promoter region upstream of the putative pmrH gene was used to construct the xylE transcriptional fusion. The activity of XylE in the rppA knockout mutant was about the same as that in the wild type and the RppA-complemented strain after incubation for 3 to 6 h in the absence of PB (only the results after 3.5 h of incubation are shown in Fig. 2B). In consistence with the results of real-time RT-PCR, PB can induce XylE activity in the wild type and the RppA-complemented strain but not in the rppA knockout mutant (only the results after 3.5 h of incubation are shown in Fig. 2B).
To study further whether RppA could stimulate the transcription of PpmrI directly by binding to the putative promoter of the pmrF operon in P. mirabilis, an EMSA was performed. The data in Fig. 3 indicated that the purified His-tagged RppA proteins could bind specifically to the putative PpmrI (i.e., the pmrF operon) promoter DNA but not to the irrelevant DNA fragment (compare lanes 1 to 3 with lanes 4 to 6 in Fig. 3), and the supershift band was visible when the antihistidine antibody was included in the binding reaction (Fig. 3, lane 7). Another EMSA with purified His-tagged RppA and DIG-labeled PpmrI promoter DNA showed unlabeled PpmrI promoter DNA can act as a competitor to reduce the binding of DIG-labeled PpmrI promoter DNA with His-tagged RppA (data not shown). Therefore, RppA can activate the expression of PPmrI through binding to the promoter of the latter. Taken together, the above data suggest that PB can modulate the expression of PPmrI through the RppA-dependent pathway.
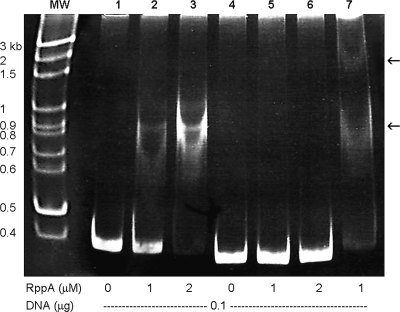
FIG. 3.
Electrophoretic mobility shift assay with purified His-tagged RppA and PpmrI promoter DNA. DNA fragments (0.1 μg) containing either the PpmrI promoter region (372 bp) or unrelated DNA (330 bp) were incubated with the indicated concentrations (0, 1, or 2 μM) of the His-tagged RppA protein. After protein-DNA complex formation, the fragments were resolved on a 5% nondenaturing polyacrylamide gel. The supershift assay (lane 7) was performed using 0.3 μl antihistidine antibody. The lower arrow indicates the position of the protein-DNA complex, and the upper arrow indicates the position of the supershifted band. MW, marker; lanes 1 to 3, PpmrI promoter DNA with His-tagged RppA; lanes 4 to 6, negative control DNA with His-tagged RppA.
Biofilm formation and urothelial cell invasion ability of PpmrI knockout P. mirabilis.
It is well known that surface structures, such as alterations of LPS, have a significant impact on biofilm formation of Gram-negative bacteria (1, 46) and that LPS modifications or defects can affect bacterial invasion ability and bacterial viability after invasion of eucaryotic cells (10, 49). In this study, we found that the PpmrI knockout mutant of P. mirabilis underwent qualitative changes in LPS (see above). We thus investigated whether the PpmrI knockout mutant had biofilm-forming and cell invasion abilities different from those of the wild-type and PPmrI-complemented strains. As shown in Fig. 4A, the biofilm-forming ability of the wild-type and PPmrI-complemented strains was significantly higher than that of the PpmrI knockout mutant. Moreover, the wild-type and PPmrI-complemented strains also had greater urothelial cell invasion ability than the PpmrI knockout mutant (Fig. 4B). Together, these data indicate that PPmrI is a positive determinant of biofilm formation and cell invasion behavior in P. mirabilis.
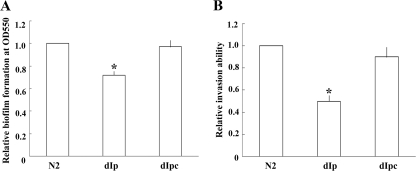
FIG. 4.
(A) Effect of PpmrI mutation on the biofilm-forming ability of P. mirabilis. Biofilm-forming abilities of the wild-type (N2), the PpmrI knockout mutant (dIp), and the PPmrI-complemented strain (dIpc) of P. mirabilis were determined as described in Materials and Methods. The value obtained with wild-type cells was set at 1. The data represent the averages of results of three independent experiments with standard deviations. A significant difference between the biofilm-forming ability of the wild-type strain and that of the mutant was observed by Student's t-test analysis (*, P < 0.05). (B) Effect of PpmrI mutation on the cell invasion ability of P. mirabilis. Invasion abilities of the wild-type (N2), the PpmrI knockout mutant (dIp), and the PPmrI-complemented strain (dIpc) of P. mirabilis were determined as described in Materials and Methods. The cell invasion ability of wild-type cells was set at 1. The data represent the averages of results from three independent experiments with standard deviations. A significant difference between the invasion ability of the wild-type strain and that of the mutant was observed by Student's t-test analysis (*, P < 0.001).
DISCUSSION
P. mirabilis is known to be naturally highly resistant to PB, a kind of CAP often used for treatment of multidrug-resistant Gram-negative bacterial infections (11, 12). The ability of P. mirabilis to survive the killing action of CAP is clearly important in the pathogenesis of urinary tract infections. Though studies have suggested that an LPS modification appears to be critical for PB resistance in P. mirabilis (15, 22, 30), the molecular mechanism underlying PB resistance remains elusive. In this study, we took a Tn5 mutagenesis approach to isolate PB-sensitive P. mirabilis mutants. Several mutants were identified as being more sensitive to PB than the wild-type strain. Among them, one mutant was found to have Tn5 inserted in the PpmrI gene, a homologue of the pmrI gene located in the pmrF operon of S. enterica and E. coli (17, 18). The S. enterica and E. coli pmrF operons are flanked at the 5′ end by the homologous genes pmrG and ais, respectively (17). pmrG was shown to be regulated by PmrA-PmrB (38). Located at the 3′ end of this operon in both S. enterica and E. coli is pmrD, which is regulated by PhoP-PhoQ (14). These upstream and downstream genes of the pmrF operon in S. enterica and E. coli may be related to LPS modification (14, 38). In contrast, the putative pmrF operon in P. mirabilis is flanked by the btuD (encoding a transporter of vitamin B12) and nlpC (encoding a lipoprotein hydrolase) genes. It is not known whether the gene products of btuD and nlpC are involved in PB resistance.
Other PB-sensitive mutants we isolated had a Tn5 insertion in the Proteus galU (PgalU) (may encode UDP-glucose pyrophosphorylase), Proteus ugd (Pugd) (may encode UDP-glucose dehydrogenase), Proteus pmrJ (PpmrJ), or Proteus pmrK (PpmrK) gene (our unpublished data). Both UDP-glucose pyrophosphorylase and UDP-glucose dehydrogenase are necessary for synthesis of bacterial surface structures (such as LPS) and of Ara4N, which is involved in LPS modification (25, 36, 40, 47). The PpmrJ and PpmrK mutants had wild-type LPS profiles, while the PgalU and Pugd mutants had a defective LPS profile which lacks an O-antigen ladder (data not shown). Knowing the PpmrJ and PpmrK genes are members of the putative pmrF operon, it seems reasonable that the PpmrI mutant also had a wild-type LPS profile but exhibited increased sensitivity to PB, as did the PpmrJ and PpmrK mutants. Our data from PB binding assays suggest that a subtle change in LPS exists in the PpmrI mutant and this change confers higher PB-binding activity on the PpmrI mutant. In addition, by RT-PCR using primers annealing to the beginning of the PpmrJ gene and the end of the PpmrK gene, we detected an amplicon of the expected size (ca. 2.7 kb) from RNA samples of both the wild-type strain and the PpmrI knockout mutant (unpublished results). This implies the transcript containing the PpmrJ and PpmrK genes was produced in the PpmrI knockout mutant. The transcriptional readthrough may explain the restoration of wild-type PB susceptibility in the complemented strain with PpmrI alone. Taken together, these findings demonstrated the function of the putative pmrF operon in PB resistance.
LPS of the Gram-negative bacteria is highly modified in response to environmental conditions (13-15, 29, 40). In S. enterica and E. coli, the products of the pmrF operon are required for the synthesis of Ara4N and the addition of Ara4N to lipid A and thus are involved in controlling PB resistance (17, 45). Transcription of the Salmonella pmrF operon is induced by Fe3+ via the PmrA-PmrB two-component system and by low Mg2+ in a process that requires the PhoP-PhoQ two-component system, the PmrD protein, and the PmrA-PmrB system (33, 34). In this regard, we found that the transcription of the pmrF (PpmrI) operon was induced by PB via RppA, a response regulator of the two-component system, in P. mirabilis (48). This increased expression of the pmrF (PpmrI) operon may somehow cause alterations in LPS composition in P. mirabilis, as suggested by previous studies in Salmonella enterica serovar Typhimurium (17) and the results that PB could affect the LPS profile in wild-type P. mirabilis but not in the PpmrI mutant (Fig. 1A). Therefore, PB may serve as a signal to induce the expression of RppA (48), which in turn binds to the PpmrI promoter (Fig. 3) and induces the expression of PPmrI, leading to modification of LPS and PB resistance.
Previous studies indicated that surface structures such as LPS are crucial determinants of biofilm formation (1, 2, 35, 46). In Pseudomonas, LPS is known to undergo structural modifications (primarily alterations in O antigen) in response to changes in the mode of life (from biofilm to planktonic) (2). Rhizobium deficient in 27-hydroxyoctacosanoate-modified LPS is impaired in biofilm formation (46). In the present study, we also found that the PpmrI mutant of P. mirabilis, which exhibited changes in LPS (had higher binding affinity for PB), had decreased biofilm-forming ability (Fig. 4A). LPS also is involved in bacterial adherence and subsequent invasion into host cells (10, 19, 49, 50). P. aeruginosa LPS cores and terminal glucose are required for binding and ingestion by the corneal cells (50), and mutants with defects in LPS cores and O antigen exhibited reduced viability after internalization by corneal epithelial cells (10). LPS glucosylation in Shigella promotes bacterial invasion of host cells and evasion of innate immunity (49). Here we also found that the PpmrI mutant of P. mirabilis had a lesser ability to invade urothelial cells (Fig. 4B). Although the detailed mechanism underlying the low biofilm-forming and cell invasion ability of the PpmrI mutant is not known, it is possible that these phenotypes are associated with subtle changes in LPS which cause it to have higher binding affinity for PB (Fig. 1B).
To disclose the subtle difference in the LPS structures of the wild type and the PpmrI mutant, analysis of LPS composition by liquid chromatography-mass spectrometry (LC-MS) needs to be performed. This approach not only will reveal the LPS structures associated with PB resistance but also may highlight the importance of LPS structures in the biofilm formation and cell invasion ability of P. mirabilis.
In conclusion, by using Tn5 transposon mutagenesis, we have identified a gene, PpmrI, which plays important roles in controlling LPS modification, PB susceptibility, biofilm formation, and cell invasion in P. mirabilis. The expression of PpmrI can be induced by PB via RppA, a response regulator of the bacterial two-component system. Together, these results reveal a novel mechanism by which P. mirabilis regulates PB resistance and virulence.
Acknowledgments
This work was supported by grants from the National Science Council and National Taiwan University Hospital, Taipei, Taiwan.
We thank Yeong-Shiau Pu (National Taiwan University Hospital) for providing the NTUB1 cell line and Yang Tsuey-Ching for giving us the yT&A-xylE plasmid.
Footnotes
Published ahead of print on 1 February 2010.
REFERENCES
- 1.Amini, S., H. Goodarzi, and S. Tavazoie. 2009. Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5:e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augustin, D. K., Y. Song, M. S. Baek, Y. Sawa, G. Singh, B. Taylor, A. Rubio-Mills, J. L. Flanagan, J. P. Wiener-Kronish, and S. V. Lynch. 2007. Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa. J. Bacteriol. 189:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 4.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 5.Belas, R., D. Erskine, and D. Flaherty. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodsky, I. E., R. K. Ernst, S. I. Miller, and S. Falkow. 2002. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J. Bacteriol. 184:3203-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D., T. Kuo, M. Kwong, R. Van, and S. Fleiszig. 2002. Pseudomonas aeruginosa strains with lipopolysaccharide defects exhibit reduced intracellular viability after invasion of corneal epithelial cells. Exp. Eye Res. 75:635-643. [DOI] [PubMed] [Google Scholar]
- 11.Evans, M. E., D. J. Feola, and R. P. Rapp. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 33:960-967. [DOI] [PubMed] [Google Scholar]
- 12.Falagas, M. E., and S. K. Kasiakou. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333-1341. [DOI] [PubMed] [Google Scholar]
- 13.Geurtsen, J., M. Dzieciatkowska, L. Steeghs, H. J. Hamstra, J. Boleij, K. Broen, G. Akkerman, H. El Hassan, J. Li, J. C. Richards, J. Tommassen, and P. van der Ley. 2009. Identification of a novel lipopolysaccharide core biosynthesis gene cluster in Bordetella pertussis, and influence of core structure and lipid A glucosamine substitution on endotoxic activity. Infect. Immun. 77:2602-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunn, J. S. 2001. Bacterial modification of LPS and resistance to antimicrobial peptides. J. Endotoxin Res. 7:57-62. [PubMed] [Google Scholar]
- 16.Gunn, J. S. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16:284-290. [DOI] [PubMed] [Google Scholar]
- 17.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 18.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta, S. K., S. Masinick, M. Garrett, and L. D. Hazlett. 1997. Pseudomonas aeruginosa lipopolysaccharide binds galectin-3 and other human corneal epithelial proteins. Infect. Immun. 65:2747-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U. S. A. 97:8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, R. M., K. J. Huang, L. T. Wu, Y. J. Hsiao, and T. C. Yang. 2008. Induction of L1 and L2 beta-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 52:1198-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaca, W., J. Radziejewska-Lebrecht, and U. R. Bhat. 1990. Effect of polymyxins on the lipopolysaccharide-defective mutants of Proteus mirabilis. Microbios 61:23-32. [PubMed] [Google Scholar]
- 23.Kaiser, D. 2007. Bacterial swarming: a re-examination of cell-movement patterns. Curr. Biol. 17:R561-R570. [DOI] [PubMed] [Google Scholar]
- 24.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacour, S., E. Bechet, A. J. Cozzone, I. Mijakovic, and C. Grangeasse. 2008. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS One 3:e3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liaw, S. J., H. C. Lai, S. W. Ho, K. T. Luh, and W. B. Wang. 2001. Characterisation of p-nitrophenylglycerol-resistant Proteus mirabilis super-swarming mutants. J. Med. Microbiol. 50:1039-1048. [DOI] [PubMed] [Google Scholar]
- 27.Liaw, S. J., H. C. Lai, and W. B. Wang. 2004. Modulation of swarming and virulence by fatty acids through the RsbA protein in Proteus mirabilis. Infect. Immun. 72:6836-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 29.Marceau, M., F. Sebbane, F. Ewann, F. Collyn, B. Lindner, M. A. Campos, J. A. Bengoechea, and M. Simonet. 2004. The pmrF polymyxin-resistance operon of Yersinia pseudotuberculosis is upregulated by the PhoP-PhoQ two-component system but not by PmrA-PmrB, and is not required for virulence. Microbiology 150:3947-3957. [DOI] [PubMed] [Google Scholar]
- 30.McCoy, A. J., H. Liu, T. J. Falla, and J. S. Gunn. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moskowitz, S. M., R. K. Ernst, and S. I. Miller. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moss, J. E., P. E. Fisher, B. Vick, E. A. Groisman, and A. Zychlinsky. 2000. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell. Microbiol. 2:443-452. [DOI] [PubMed] [Google Scholar]
- 33.Mouslim, C., and E. A. Groisman. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 34.Mouslim, C., T. Latifi, and E. A. Groisman. 2003. Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J. Biol. Chem. 278:50588-50595. [DOI] [PubMed] [Google Scholar]
- 35.Nakao, R., H. Senpuku, and H. Watanabe. 2006. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect. Immun. 74:6145-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcombe, J., J. C. Jeynes, E. Mendoza, J. Hinds, G. L. Marsden, R. A. Stabler, M. Marti, and J. J. McFadden. 2005. Phenotypic and transcriptional characterization of the meningococcal PhoPQ system, a magnesium-sensing two-component regulatory system that controls genes involved in remodeling the meningococcal cell surface. J. Bacteriol. 187:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino, K., F. F. Hsu, J. Turk, M. J. Cromie, M. M. Wosten, and E. A. Groisman. 2006. Identification of the lipopolysaccharide modifications controlled by the Salmonella PmrA/PmrB system mediating resistance to Fe(III) and Al(III). Mol. Microbiol. 61:645-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 40.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76:295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 42.Rozalski, A., Z. Sidorczyk, and K. Kotelko. 1997. Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 61:65-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran, A. X., M. E. Lester, C. M. Stead, C. R. Raetz, D. J. Maskell, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186-28194. [DOI] [PubMed] [Google Scholar]
- 45.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 46.Vanderlinde, E. M., A. Muszynski, J. J. Harrison, S. F. Koval, D. L. Foreman, H. Ceri, E. L. Kannenberg, R. W. Carlson, and C. K. Yost. 2009. Rhizobium leguminosarum biovar viciae 3841, deficient in 27-hydroxyoctacosanoate-modified lipopolysaccharide, is impaired in desiccation tolerance, biofilm formation and motility. Microbiology 155:3055-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vilches, S., R. Canals, M. Wilhelms, M. T. Salo, Y. A. Knirel, E. Vinogradov, S. Merino, and J. M. Tomas. 2007. Mesophilic Aeromonas UDP-glucose pyrophosphorylase (GalU) mutants show two types of lipopolysaccharide structures and reduced virulence. Microbiology 153:2393-2404. [DOI] [PubMed] [Google Scholar]
- 48.Wang, W. B., I. C. Chen, S. S. Jiang, H. R. Chen, C. Y. Hsu, P. R. Hsueh, W. B. Hsu, and S. J. Liaw. 2008. Role of RppA in the regulation of polymyxin b susceptibility, swarming, and virulence factor expression in Proteus mirabilis. Infect. Immun. 76:2051-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 50.Zaidi, T. S., S. M. Fleiszig, M. J. Preston, J. B. Goldberg, and G. B. Pier. 1996. Lipopolysaccharide outer core is a ligand for corneal cell binding and ingestion of Pseudomonas aeruginosa. Investig. Ophthalmol. Vis. Sci. 37:976-986. [PubMed] [Google Scholar]