Abstract
The influence of antibiotic dosages and bacterial mutator phenotypes on the emergence of linezolid-resistant mutants was evaluated in an in vitro pharmacokinetic-pharmacodynamic model. A twice-daily 0.5-h infusion of a 200-, 600-, or 800-mg dose for 48 h was simulated against four strains (MIC, 2 μg/ml): Staphylococcus aureus RN4220 and its mutator derivative MutS2, Enterococcus faecalis ATCC 29212, and a mutator clinical strain of E. faecalis, Ef1497. The peak concentrations (4.38 to 4.79, 13.4 to 14.6, and 19.2 to 19.5 μg/ml) and half-lives at β-phase (5.01 to 6.72 h) fit human plasma linezolid pharmacokinetics. Due to its bacteriostatic property, the cumulative percentages of the dosing interval during which the drug concentration exceeded the MIC (T > MIC), 66.6 and 69.1% of the dosing interval, were not significant, except for Ef1497, with an 800-mg dose and a T > MIC of 80.9%. At the standard 600-mg dosage, resistant mutants (2- to 8-fold MIC increases) were selected only with Ef1497. A lower, 200-mg dosage did not select resistant mutants of E. faecalis ATCC 29212, but a higher, 800-mg dosage against Ef1497 did not prevent their emergence. For the most resistant mutant (MIC, 16 μg/ml), characterization of 23S rRNA genes revealed the substitution A2453G in two of the four operons, which was previously described only in in vitro mutants of archaebacteria. Nevertheless, this mutant did not yield further mutants under 600- or 200-mg treatment. In conclusion, linezolid was consistently efficient against S. aureus strains. The emergence of resistant E. faecalis mutants was probably favored by the rapid decline of linezolid concentrations against a strong mutator, a phenotype less exceptional in E. faecalis than in S. aureus.
Linezolid, the first of the new oxazolidinone class of antibacterial drugs, acts by binding the 50S ribosomal subunit, blocking the formation of the functional 70S initiation complex, and inhibiting protein synthesis (9). Due to this unique mechanism of action, linezolid has become an important therapeutic option against multidrug-resistant Gram-positive organisms (38). Selection for linezolid mutational resistance has been predicted to be difficult, based on preliminary in vitro static tests (38). Accordingly, most of the bacterial species harbor multiple copies of the ribosomal operon (e.g., five or six in Staphylococcus aureus and four in Enterococcus faecalis), and significant resistance requires mutations in more than one copy of the 23S rRNA gene (7, 38). Nevertheless, treatment failures due to the emergence of linezolid-resistant mutants have occurred, rarely with staphylococci (43, 50) but less infrequently with enterococci (21, 31, 42). These mutants exhibit changes in the peptidyltransferase region (domain V) of their 23S rRNA genes, particularly the G2576T substitution (38). They account for most of the rare clinical resistant strains, although other resistance mechanisms have been described (33, 52).
Linezolid clinical failures are often associated with prolonged monotherapy and could have both pharmacological and microbiological causes. Low tissue penetration cannot be implicated, since the low protein binding (31%) (15) reflects a large free, and therefore diffusible, fraction. Moreover, the volume of distribution at steady state in healthy adults is 0.5 to 0.6 liters/kg of body weight, which approximates that of body water (30), and linezolid concentrations in interstitial fluid are very close to those of plasma (20). Thus, therapeutic failures could be explained by achieving inefficient concentrations at the infection site at the usual doses due to interindividual variability of drug pharmacokinetics (13, 35, 48). This might be particularly critical for strains, called mutators, that exhibit unusually high spontaneous mutation rates (53) due to altered systems of correction and prevention of errors during DNA replication (11). In clinical strains, the mutator phenotype is often related to the inactivation of the main postreplicative DNA repair pathway, the mismatch repair (MMR) system (39, 40, 51). While there are some differences among bacteria, the presence of two essential components, MutS and MutL, is required. MutS recognizes the DNA mismatch and recruits MutL, targeting the mutation for an excision. Then, a new strand is synthesized and the mismatch is corrected (11). Mutators can be detected by the disk diffusion method by the presence of “squatter” colonies within the growth inhibition zone of antibiotics with a high rate of resistance, such as rifampin (14). Further confirmation is obtained by the determination of the frequency of resistant mutants (up to 100-fold higher than normal) (14, 26). They occur at variable frequencies according to the species and the type of infection. Thus, their rate is around 1% in natural populations of Escherichia coli and Salmonella (26) but less than 0.2% in S. aureus (36), except in particular situations (14.6% in cystic fibrosis) (40). No data are available for enterococci.
Previous investigations using in vitro pharmacokinetic-pharmacodynamic (PK-PD) models have evaluated the efficiency of linezolid (1, 6, 22, 24, 28, 49). However, most of these studies used a one-compartment model, which does not avoid dilution of the bacterial population, and none has investigated the impact of the mutator phenotype on the selection of resistant mutants. The purposes of this study were to use a two-compartment in vitro PK-PD model previously developed in our laboratory (2) to assess the potential effectiveness of linezolid at concentrations within the interindividual fluctuation window and against S. aureus and E. faecalis strains presenting either a wild-type or a mutator phenotype, and to characterize the resistant mutants that might be selected.
MATERIALS AND METHODS
The PK-PD model, simulated dose regimens, and reagents.
The two-compartment PK-PD model (2) consisted of a central compartment (CCp) represented by a thermostable flask with a magnetic stirrer and a dialysis cartridge (F40S; Fresenius, Fresnes, France) containing porous hollow polysulfone fibers that allowed bidirectional passage of broth and antibiotic but retained bacteria and heavy molecules. The extracapillary space of the cartridge represented the peripheral compartment (PCp), where the inoculum was injected through a port septum. Antibiotic administration into the CCp was performed with a computer-controlled syringe pump. Antibiotic-free broth was pumped from a reservoir to the CCp at the same flow as antibiotic-containing broth was pumped from the CCp to the elimination reservoir. A human total (free plus bound)-plasma pharmacokinetic profile of linezolid after a twice-daily 0.5-h infusion of 200, 600, or 800 mg for 48 h was simulated. The CCp dilution and elimination flow rates were adjusted according to reference pharmacokinetic parameters obtained after a 0.5-h infusion of 600 mg in humans, i.e., a maximum concentration of drug in plasma (Cmax) of 14.1 μg/ml and a half-life at β-phase (t1/2β) of 5.1 h (13). For simulation of 200- and 800-mg dose regimens, the reference Cmax was estimated by the proportionality rule at 4.7 μg/ml and 18.8 μg/ml, respectively. All experiments were performed in cation-adjusted and protein-free Mueller-Hinton (MH) broth (AES Chemunex, Bruz, France) at 37°C. Linezolid was obtained as a commercial infusion solution (Zyvoxid; 600 mg/300 ml; Pfizer).
Bacterial strains, antibiotic susceptibility testing, and determination of mutant prevention concentration (MPC).
The activity of linezolid was first evaluated against four strains: two S. aureus strains, RN4220 (derived from the NCTC 8325-4 strain) and its mutS knockout mutant, RN4220MutS2, with a mutator phenotype (a gift from R. Leclercq, University of Caen, Caen, France) (39), and two E. faecalis strains, the reference strain, ATCC 29212, and the clinical mutator strain, E. faecalis Ef1497. The last strain was one of the four mutators detected among 233 consecutive and nonredundant enterococcal isolates collected in a 2001 extrahospital survey (22a). The mutators produced “squatter” colonies within the growth inhibition zones of rifampin, fosfomycin, and streptomycin by the disk diffusion method. Ef1497 exhibited a frequency of rifampin-resistant mutants of 7.3 × 10−6, i.e., 100-fold higher than the other tested strains, including E. faecalis ATCC 29212. All four investigated staphylococcal and enterococcal strains gave a linezolid MIC of 2 μg/ml by the agar dilution method (http://www.sfm.asso.fr). Linezolid-resistant mutants arose during assays with Ef1497. Cross-resistances were investigated for 23 of these clones by determination of chloramphenicol, erythromycin, lincomycin, ciprofloxacin (Sigma-Aldrich, Saint Quentin Fallavier, France), and pristinamycin (kindly provided by Sanofi-Aventis) MICs. The most resistant mutant, Ef1497MutM3 (MIC, 16 μg/ml), also underwent linezolid treatment to investigate the possible occurrence of supplementary step mutations. E. coli DH5α was used for cloning experiments.
The linezolid MPC was determined by the technique of Zhao and Drlica (54). In brief, an 18-h bacterial culture was concentrated by centrifugation and spread on MH agar containing 128 to 0.125 μg/ml of linezolid. Diluted cultures of 105 to 109 CFU/ml were also plated on MH agar without the addition of linezolid. The MPC was defined as the concentration at which no mutant appeared in the presence of a high inoculum of 1010 CFU/ml after incubation at 37°C for 24 h.
Operating procedure and antimicrobial assays.
An overnight culture of the strain under investigation was transferred into MH broth. High inocula were used in order to increase the probability of emergence of resistant mutants. Thus, after an 18-h incubation at 37°C under agitation, the bacterial suspension was concentrated 50 times by centrifugation and inoculated into the PCp of the model 3 h before the first sampling to obtain an exponentially growing culture of 108 (E. faecalis) or 109 (S. aureus) CFU/ml. A total of 24 (at 0 [3 h after inoculation], 0.3, 0.4, 0.5, 0.75, 1, 1.50, 2, 3, 4, 6, 12, 12.3, 12.4, 12.5, 12.75, 13, 13.5, 14, 15, 16, 18, 24, and 48 h) samples of 300 μl each were drawn from the PCp manually with sterile Vacutainer tubes (VWR, Strasbourg, France). A 150-μl aliquot of each sample was used for the determination of antibiotic concentrations by a validated high-pressure liquid chromatography (HPLC) method (3). A 100-μl volume from 16 PCp samples (at 0, 0.5, 1, 2, 3, 4, 6, 12, 12.5, 13, 14, 15, 16, 18, 24, and 48 h) was devoted to the bacterial growth quantification in the presence or absence of antibiotic by dilution plating and enumeration. The emergence of resistant mutants was monitored by plating the same diluted samples (at 0, 0.5, 12, 12.5, 24, and 48 h) on MH agar supplemented with linezolid at 0.5, 1, and 4× MIC (1, 2, and 8 μg/ml for S. aureus RN4220 and RN4220MutS2 and E. faecalis ATCC 29212 and Ef1497, and 8, 16, and 32 μg/ml for the resistant mutant, Ef1497MutM3). In order to avoid a carryover effect, the first 10-fold-diluted sample was not plated, giving a lower detection of 2 to 3 log10 CFU/ml. All experiments were performed in duplicate.
Pharmacokinetic and pharmacodynamic data analysis. (i) Pharmacokinetic analysis.
Compartmental analysis of experimental total (free plus bound) drug concentration data was performed with the software Pharmacokin (22b). The goodness of fit for each concentration-time curve was evaluated by the correlation coefficient between experimental and software-calculated data. The Cmax, the time to reach the maximal concentration (Tmax), and the residual concentration at the end of the administration interval (Cres) were taken directly from concentration-time profiles, whereas the t1/2β, the area under the concentration-time curve (AUC) within the first two dosing intervals, the mean residence time (MRT), the total clearance (Cltot), the apparent volume of distribution (V), and the volume of distribution at steady state (Vss) were calculated.
(ii) Quantification of bacterial growth and evaluation of the antibacterial effect.
The MIC-related pharmacokinetic parameters—the inhibitory quotient (Cmax/MIC) (16), the AUC divided by the MIC (AUC/MIC) (29, 34), the cumulative percentage of the dosing interval period during which the drug concentration exceeded the MIC (T > MIC) (34), the indices of bacterial killing in the presence of antibiotic (the bacterial killing and regrowth curve from 0 to 24 h [AUBC0-24], the area between the control growth curve from 0 to 24 h [AUGC0-24], the bacterial killing and regrowth curve from the zero point to 24 h [ABBC0-24][17], and the difference between the bacterial counts at the beginning of the treatment and at a defined time [Δlog CFU/milliliter] [17, 29])—were calculated.
Molecular analysis of linezolid-resistant mutants derived from Ef1497.
Genomic DNA was extracted from overnight cultures of E. faecalis Ef1497 and derived linezolid-resistant mutants (44). Then, the domain V region was amplified with Ampli Taq Gold (Applied Biosystems Division, Perkin-Elmer [PE], Courtaboeuf, France) using primers A and B (31). To detect the prevalent G2576T mutation, the resulting 389-bp amplicons were digested by the enzyme BfaI (an isoschizomer of MaeI; New England BioLabs Inc., Saint-Quentin, France), which generates two digestion fragments in case of mutation (31). A PCR product containing a BfaI site was added as a control for endonuclease functionality.
For the most resistant mutant, Ef1497MutM3, the 389-bp amplicon was ligated to the pGEM-T easy vector (Promega, Charbonnières-les-Bains, France) and used to transform electrocompetent strains of E. coli DH5α. Then, the cloned PCR products were sequenced by an automated fluorescence method based on dye terminator chemistry (AmpliTaq DNA Polymerase FS Dye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems Division, PE) and the ABI-3130xl sequencer (Applied Biosystems Division, PE). In addition, on the basis of the published E. faecalis genome V583 (GenBank accession number AE016830) (37), two of the four copies of the E. faecalis 23S rRNA gene were separated by amplification, taking advantage of an additional 102-bp region in the intergenic region of the rrnB and rrnC operons located upstream of the 23S rRNA gene. Thus, the upper primers A_DF (5′-GGTCTACTCTCAAAACATTC-3′) and B_CF (5′-TCCATTGATAGCTTTTGCTATCAG-3′) for the rrnA and rrnD and the rrnB and rrnC operons, respectively, were designed to amplify the 23S rRNA gene, together with a common lower primer 1-4R (5′-CTGGGTGTTGTTTCTTATTGAG-3′). The amplicons were reamplified using the primer pair A and B and sequenced as indicated above.
RESULTS
Antibiotic assays and pharmacokinetic data.
The HPLC method with UV detection allowed a quantification limit of 0.39 μg/ml with a 50-μl sample size for linezolid in MH broth, and good selectivity. The standard curve was linear between 0.39 and 25 μg/ml, with a mean correlation coefficient of 0.9999 (n = 6). The online extraction of linezolid from a C8 precolumn led to recoveries of 93.9% ± 1.47%, 93.5% ± 1.03%, and 93.3% ± 1.12% for quality control samples of 1, 10, and 20 μg/ml. The intraday and interday coefficients of variation within the linearity range varied from 0.70 to 6.77% and from 0.83 to 7.35%, respectively. The intraday and interday accuracies ranged from 2.00 to 9.83% and from 2.11 to 11.8%, respectively. The mean concentration-time curves obtained during simulation of 0.5-h infusions of 200, 600, and 800 mg linezolid (Fig. 1 to 4, A1 and B1) allowed us to determine pharmacokinetic parameters (Table 1), and the coefficient of correlation between experimental data and the calculated pharmacokinetic profile was always greater than 0.98.
FIG. 1.
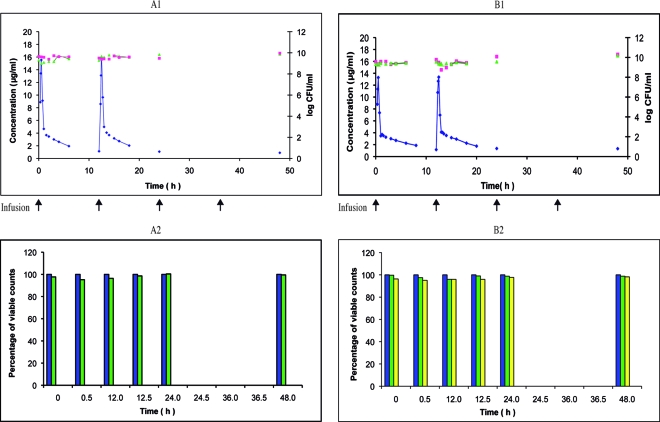
Linezolid twice-daily 0.5-h infusion of 600 mg over 48 h against S. aureus RN4220 and RN4220MutS2. Shown are the pharmacokinetic and pharmacodynamic effects of linezolid on S. aureus RN4220 (A1) and its mutant S. aureus RN4220MutS2 (B1) during the simulation of twice-daily 0.5-h infusion of 600 mg over 48 h in the in vitro PK-PD model (mean; n = 2). ⧫, PCp concentration-time curve; ▴, control growth curve; ▪, killing and regrowth curve. (A2 and B2) Emergence of linezolid-resistant mutants of S. aureus RN4220 (A2) and RN4220MutS2 (B2) during the corresponding simulations (mean percentage of viable counts; n = 2). Blue, control; green, resistant to 1.0 μg/ml; yellow, resistant to 2.0 μg/ml.
FIG. 4.
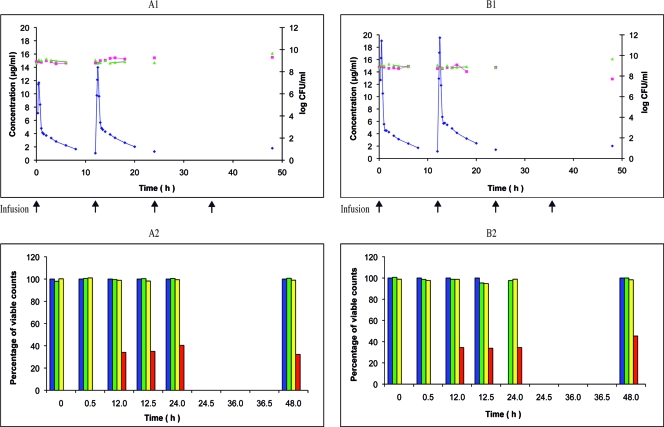
Linezolid twice-daily 0.5-h infusion of 200 or 600 mg over 48 h against E. faecalis Ef1497MutM3. Shown are the pharmacokinetic and pharmacodynamic effects of linezolid on E. faecalis Ef1497MutM3 during the simulation of twice-daily 0.5-h infusion of 200 mg (A1) and 600 mg (B1) over 48 h in the in vitro PK-PD model (mean; n = 2). ⧫, PCp concentration-time curve; ▴, control growth curve; ▪, killing and regrowth curve. (A2 and B2) Emergence of linezolid-resistant mutants of Ef1497MutM3 during the corresponding simulations (mean percentage of viable counts; n = 2). Blue, control; red, resistant to 8.0 μg/ml; orange, resistant to 16 μg/ml.
TABLE 1.
Mean pharmacokinetic parameters for the PCp after simulation with twice-daily 0.5-h infusion of 200, 600, or 800 mg linezolid, and the corresponding human reference data
| Parameter | Experimental data (mean value) for a dose (mg)/12 h of: |
Human dataa (n = 10) for a dose of 600 mg/12 h | |||||
|---|---|---|---|---|---|---|---|
| 200 (n = 4) |
600 (n = 10) |
800 (n = 2) |
|||||
| 0-12b | 12-24 | 0-12 | 12-24 | 0-12 | 12-24 | ||
| Cmax (μg/ml) | 4.38 | 4.79 | 13.4 | 14.6 | 19.2 | 19.5 | 14.1 ± 2.8 |
| Tmax (h) | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | |
| Cres (μg/ml) | 0.39 | 0.31 | 1.17 | 1.26 | 1.15 | 1.42 | |
| t1/2β (h) | 6.72 | 6.27 | 6.54 | 6.18 | 5.01 | 5.2 | 5.1 ± 2.6 |
| MRT (h) | 8.65 | 7.95 | 8.54 | 8.10 | 6.55 | 6.81 | |
| AUC (h × μg/ml) | 10.1 | 10.1 | 32.4 | 35.4 | 38.2 | 47.7 | 88.1 ± 34.0c |
| CLtot (liters/h) | 14.9 | 15.1 | 13.9 | 12.9 | 17.2 | 13.7 | |
| V (liter)s | 140.7 | 136.7 | 129.7 | 114.9 | 124.5 | 102.2 | |
| Vss (liters) | 119.7 | 116.9 | 112.1 | 100.2 | 103.9 | 89.3 | |
| Correlation coefficient | 0.987 | 0.983 | 0.986 | 0.989 | 0.995 | 0.994 | |
Determination of the MPC.
The MPC could not be determined for either S. aureus or E. faecalis ATCC 29212. Indeed, no resistant mutants were obtained with an inoculum of 1010 CFU/ml in the presence of concentrations greater than the MIC of linezolid in three independent assays. In contrast, from E. faecalis Ef1497, in two independent experiments, three mutants emerged (C-1 and C-2 were obtained in the first experiment and C-3 in the second) at a concentration of 4 μg/ml. Therefore, the MPC for this strain was estimated at 8 μg/ml.
Pharmacodynamic data. (i) Growth control curves.
The initial inocula (3 h after inoculation of the PCp) obtained with the E. faecalis strains were slightly lower than those of the S. aureus strains, 5.21 × 108 to 5.90 × 108 CFU/ml versus 1.98 × 109 to 2.02 × 109 CFU/ml, due to a difference in the growth rates. After a 48-h experiment, the same magnitude of difference persisted: 2.00 × 109 to 2.85 × 109 CFU/ml versus 7.23 × 109 to 1.50 × 1010 CFU/ml. For all growth control curves, the increase in the bacterial concentration was always less than 1 log10 CFU/ml.
(ii) Killing curves.
The initial inocula were 4.47 × 109 and 3.60 × 109 CFU/ml for S. aureus RN4220 and RN4220MutS2, respectively. For the E. faecalis strains, they ranged from 3.50 × 108 to 8.10 × 108 CFU/ml. Due to the inherent bacteriostatic property of linezolid (18), no appreciable reductions in the number of viable organisms were noted for all strains under investigation. The AUGC0-24 ranged from 214.1 to 228.2 h × log CFU/ml, whereas the AUBC0-24 ranged from 210.5 to 229.4 h × log CFU/ml, leading to low ABBC0-24 values (−2.66 to 5.11 h × log CFU/ml) (Table 2). The values of Δlog CFU/ml were related to the time-dependent activity and the small bactericidal effect of linezolid in the absence of natural defenses. Maximal values of Cmax/MIC and AUC0-24/MIC were obtained with an 800-mg dose against E. faecalis Ef1497 (MIC, 2 μg/ml) and were 9.76 and 42.9 h, respectively.
TABLE 2.
MIC-related pharmacokinetic parameters and antibacterial effect indices for simulation of 0.5-h infusion of 200, 600, or 800 mg linezolid per 12 h over 48 h against S. aureus and E. faecalis strains
| Parameter | Value |
|||||||
|---|---|---|---|---|---|---|---|---|
|
S. aureus RN4220 (MIC = 2 μg/ml) |
S. aureus RN4220MutS2 (MIC = 2 μg/ml) |
E. faecalis ATCC 29212 (MIC = 2 μg/ml) |
E. faecalis Ef1497 (MIC = 2 μg/ml) |
E. faecalis Ef1497MutM3 (MIC = 16 μg/ml) |
||||
| 600 (4.47 × 109)a | 600 (3.60 × 109) | 200 (3.57 × 108) | 600 (7.47 × 108) | 600 (8.10 × 108) | 800 (7.60 × 108) | 200 (3.50 × 108) | 600 (7.27 × 108) | |
| Δlog CFU/ml | ||||||||
| t0.5 | −0.04 | −0.21 | −0.08 | 0.04 | −0.01 | 0.08 | −0.009 | −0.01 |
| t1 | −0.07 | 0.02 | 0.14 | −0.12 | −0.05 | −0.02 | 0.02 | −0.01 |
| t2 | −0.26 | 0.02 | 0.09 | −0.08 | 0.08 | −0.13 | 0.06 | −0.05 |
| t12 | −0.15 | 0.21 | 0.14 | 0.15 | −0.12 | −0.16 | 0.29 | 0.19 |
| t12.5 | −0.26 | −0.06 | 0.26 | 0.10 | −0.14 | −0.03 | 0.23 | 0.22 |
| t13 | −0.22 | −0.81 | 0.28 | 0.16 | 0.09 | −0.16 | 0.23 | 0.25 |
| t14 | −0.24 | −0.58 | 0.55 | 0.18 | 0.13 | −0.05 | 0.41 | 0.28 |
| t24 | −0.17 | 0.52 | 0.83 | 0.32 | 0.35 | −0.04 | 0.52 | 0.44 |
| t48 | 0.32 | 0.74 | 1.23 | 0.53 | 0.39 | −1.16 | 0.79 | 0.45 |
| Cmax /MIC | ||||||||
| 0.5 h | 7.77 | 6.64 | 2.01 | 7.17 | 5.83 | 9.51 | 0.30 | 0.88 |
| 12.5 h | 7.95 | 6.64 | 2.10 | 7.70 | 6.99 | 9.76 | 0.34 | 0.90 |
| AUC0-24/MIC (h) | 32.9 | 32.9 | 10.2 | 37.1 | 35.4 | 42.9 | 1.24 | 3.90 |
| T > MIC (% dosing interval) | 50.0 | 66.6 | 8.08 | 69.1 | 69.0 | 80.9 | 0.0 | 0.0 |
| AUBC0-24(h × log CFU/ml) | 229.4 | 228.8 | 214.0 | 217.8 | 215.6 | 210.5 | 212.2 | 217.1 |
| ABBC0-24(h × log CFU/ml) | −1.21 | −2.27 | 5.11 | 1.32 | −1.49 | 3.60 | 2.32 | −2.66 |
Dose (mg/12 h) (inoculum [CFU/ml]).
(iii) Detection of linezolid-resistant mutants.
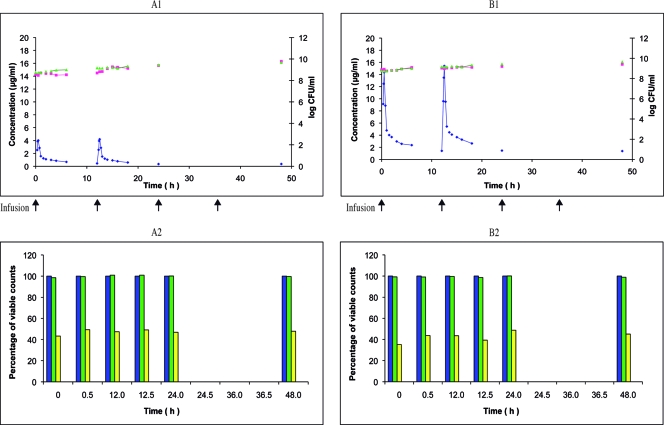
At the nominal dose of 600 mg, survivors of the wild-type strain S. aureus RN4220 taken during the whole study period and plated on MH agar supplemented with linezolid did not grow at the MIC and at the concentration above the MIC (Fig. 1A2). With the mutator strain S. aureus RN4220MutS2, even before any contact with linezolid (i.e., at time zero) and during the whole study, around 100% of the population was able to grow at the linezolid MIC but not at 4× MIC (Fig. 1B2). However, further subcultures of the colonies grown at the MIC failed. The wild-type strain E. faecalis ATCC 29212 exhibited similar behavior: from T0 to T48, around 40% of the population, initially and under the simulated linezolid treatment, grew at the MIC, although these clones could not be subcultured (Fig. 2B2). With the mutator strain E. faecalis Ef1497, after T0, around 100% of the population developed at 2 μg/ml, and after T12, ca. 30% to 40% developed at up to 8 μg/ml (Fig. 3A2). These clones were easily subcultured and showed stable linezolid resistance, reflecting the emergence of resistant mutants (Fig. 4A2).
FIG. 2.
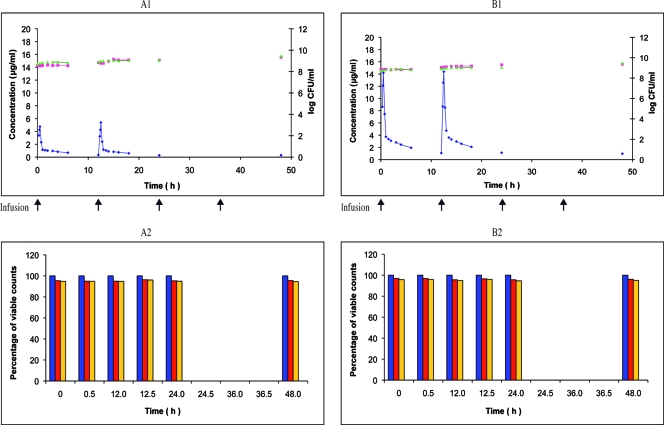
Linezolid twice-daily 0.5-h infusion of 200 or 600 mg over 48 h against E. faecalis ATCC 29212. Shown are the pharmacokinetic and pharmacodynamic effects of linezolid on E. faecalis ATCC 29212 during the simulation of twice-daily 0.5-h infusion of 200 mg (A1) and 600 mg (B1) over 48 h in the in vitro PK-PD model (mean; n = 2). ⧫, PCp concentration-time curve; ▴, control growth curve; ▪, killing and regrowth curve. (A2 and B2) Emergence of linezolid-resistant mutants of E. faecalis ATCC 29212 during the corresponding simulations (mean percentage of viable counts; n = 2). Blue, control; green, resistant to 1.0 μg/ml; yellow, resistant to 2.0 μg/ml.
FIG. 3.
Linezolid twice-daily 0.5-h infusion of 600 or 800 mg over 48 h against E. faecalis Ef1497. Shown are the pharmacokinetic and pharmacodynamic effects of linezolid on E. faecalis Ef1497 during the simulation of twice-daily 0.5-h infusion of 600 mg (A1) and 800 mg (B1) over 48 h in the in vitro PK-PD model (mean; n = 2). ⧫, PCp concentration-time curve; ▴, control growth curve; ▪, killing and regrowth curve. (A2 and B2) Emergence of linezolid-resistant mutants of Ef1497 during the corresponding simulations (mean percentage of viable counts; n = 2). Blue, control; green, resistant to 1.0 μg/ml; yellow, resistant to 2.0 μg/ml; red, resistant to 8.0 μg/ml.
Thus, in our investigations the impacts of the linezolid doses and the mutator phenotype on the selection of resistant mutants focused on E. faecalis. At first, a regimen of 200 mg twice a day was tested on the wild-type strain ATCC 29212. The response was similar to that obtained at the reference dosage of 600 mg twice a day: 40% of the survivors developed at the MIC after T0 but could not be subcultured, and no resistant mutants were selected (Fig. 2A2). Then, a regimen of 800 mg twice a day was tested on the mutator strain Ef1497. Again, the response was similar to that obtained at the reference dosage of 600 mg twice a day: 100% of the survivors developed at the MIC after T0, and 30 to 50% at 4× MIC were resistant mutants (Fig. 3B2).
Subsequently, the influence of linezolid treatment on the previously selected mutator and linezolid-resistant mutant Ef1497MutM3 was studied. Using the standard 600-mg regimen, almost 100% of the population tolerated the linezolid MIC, but these clones were not viable, and no further resistant mutant was isolated (Fig. 4B2). With a lower regimen of 200 mg, no change was observed (Fig. 4A2).
(iv) MIC determination in linezolid-resistant mutants derived from Ef1497.
The linezolid MICs were determined for 20 clones of E. faecalis Ef1497 selected at random during simulation of 600-mg (n = 13) and 800-mg (n = 7) doses and grown on plates supplemented with 8 μg/ml, together with the 3 mutants obtained during the MPC determination. The linezolid MICs were 4 μg/liter (2-fold superior to the MIC for the wild-type strain), except for Ef1497MutM3, which showed a MIC reaching 16 μg/ml. In addition, the possible coresistance phenotypes for all 23 mutants were investigated. No increases in the MICs of chloramphenicol (4 to 8 μg/ml), erythromycin (2 to 4 μg/ml), lincomycin (64 to 128 μg/ml), pristinamycin (2 to 4 μg/ml), and ciprofloxacin (2 to 4 μg/ml) compared to Ef1497 were detected.
Molecular characterization of linezolid-resistant mutants derived from Ef1497.
Since the main linezolid resistance mechanism is target modification (32), PCR amplifications of the domain V region of the 23S rRNA gene were performed with the primer pair A/B for the 23 linezolid-resistant mutants derived from Ef1497. Restriction of the amplicons using BfaI revealed that the linezolid resistances of the mutants, including Ef1497MutM3, were not due to the expected mutation, G2576T (data not shown and references 8, 31, and 42). Further characterization was carried out for the most resistant mutant, Ef1497MutM3. The amplicons obtained with the primer pair A/B were separated by cloning and sequenced. The results showed that Ef1497MutM3 had the same sequence of the domain V region as did E. faecalis V583, except for position 2453, which was a guanine instead of an adenine (as in Ef1497, used as a control) for 7 of the 12 inserts analyzed. In addition, after amplifications with the primer pairs A_DF/1-4R and B_CF/1-4R and subsequent nested PCR using the A and B primers, sequence analysis of the domain V region of Ef1497MutM3 revealed the presence at position 2453 of an adenine for the rrnB and rrnC operons but a guanine for the rrnA and rrnD operons.
DISCUSSION
In this study, using an in vitro PK-PD model, we attempted to elucidate the reasons for linezolid treatment failures due to the selection of resistant mutants and the means to prevent them. Since linezolid binding by serum proteins is low and not concentration dependent (30), and since concentrations in interstitial fluid are close to those of plasma (13), non-protein-supplemented broth was used to simulate human total (free plus bound) concentrations of the drug. Pharmacokinetic reference parameters (Cmax and t1/2β) provided by the model after simulation of a 0.5-h infusion of 600 mg (Table 1) were similar to those from human data (13), showing the reliability of the model and the method of determination (3). However, interindividual variations in linezolid pharmacokinetics have been observed (47, 48). Cmax data obtained with 200 and 800 mg matched human data fluctuation well, but the AUC0-24 values were lower, essentially due to different modes of calculation (13). At the end of the infusion of a 600-mg dose, the concentration of “total” linezolid was seven times the MIC (2 μg/ml) of the strains under investigation, and for an 800-mg dose, the Cmax reached nine times the MIC and was even higher than the MIC for E. faecalis Ef1497MutM3 (16 μg/ml). After a 200-mg dose, the Cmax was approximately two times a MIC of 2 μg/ml. Then, the concentrations decreased rapidly, and the corresponding trough concentrations at t12 were below 2 μg/ml for 600 and 800 mg and below 1 μg/ml for 200 mg over a 24-h administration. Nevertheless, linezolid accumulation was observed in healthy volunteers after multiple administrations (10).
The pharmacology of an antibiotic takes into account not only its pharmacokinetics, but also its activity. Several parameters have been described as predictive of the effectiveness of antibiotics with concentration-dependent time-kill activity, i.e., Cmax/MIC and AUC0-24/MIC (19). The major pharmacodynamic parameter for a better prediction of antibiotics with time-dependent activity has been shown to be T > MIC (4). However, as found in this study but in contrast with previous experiments in in vitro PK-PD models (6, 49), linezolid is essentially bacteriostatic (18), and the treatment behavior was simulated without natural host defenses. Therefore, T > MIC values of 66.6 and 69.1% obtained with a simulated dose of 600 mg had no significant effect on the curves of S. aureus RN4220 and RN4220MutS2 or E. faecalis ATCC 29212 survivors. A T > MIC of 80.9% obtained with a simulated dose of 800 mg led to a slight bactericidal effect on E. faecalis Ef1497, with an ABBC0-24 of 3.60 h × log CFU/ml. This effect occurred after some latency. Indeed, at t0.5, the time of peak concentration, Δlog CFU/ml had a positive value (0.08 CFU/ml). At t1, the decrease of the bacterial population was −0.02 CFU/ml, and it fell to −0.13 CFU/ml at t2. This phenomenon, more or less pronounced at other dosing intervals, is in accordance with the time-dependent effect of this antibiotic. During the whole study period, S. aureus RN4220MutS2 (about 100% of the population) and E. faecalis ATCC 29212 (40%) and Ef1497 (100%), present in the model at 5 × 108 to 5 × 109 CFU/ml, were reproducibly able to grow on plates containing the linezolid MIC determined with a standard inoculum of ca. 105 CFU/spot, although the clones obtained could not be subcultured. The same phenomenon was observed when MICs were determined with the same high inocula as those used in the PK-PD model (data not shown). Thus, except for the nonmutator S. aureus, similar to a previous report (25), we observed a slight inoculum effect that was more marked in mutator than in nonmutator strains. This effect cannot be easily explained; it could reflect the induction of a transitory resistance mechanism(s) or a kind of tolerant state facilitating further acquisition of mutations.
No emergence of resistant mutants was observed with either S. aureus strain under our experimental conditions. In preliminary assays, linezolid-resistant mutants of S. aureus were isolated at an extremely low frequency, i.e., <10−11 (56). RN4220MutS2 is only a moderate mutator, exhibiting a 24-fold increase in mutation frequency with rifampin, in comparison to RN4220 (39). Moreover, conflicting results have been reported for the correlation between S. aureus hypermutability and the emergence of antibiotic resistance (12, 40, 41, 45). These data might explain why the MPC could not be determined and why linezolid-resistant mutants were not selected under the standard dosage. A stronger mutator like RN4220MutL (36, 39) might have given different results. The paucity of linezolid failures during treatment of staphylococcal infections might be related to the usually low prevalence of highly hypermutable S. aureus strains (36).
Linezolid-resistant mutants could also not be selected from the nonmutator strain of E. faecalis, either during MPC determination or in the PK-PD model, even using a lower regimen of 200 mg. In contrast, linezolid-resistant mutants appeared with the mutator strain Ef1497, more easily in the model than in static tests (MPC determination), and even at a higher dosage of 800 mg. Such different behavior strongly suggests that the mutator phenotype plays a key role in the emergence of linezolid-resistant mutants of E. faecalis under therapy. These mutants arose at the 12th hour and persisted despite repeated administration of linezolid and peaks above the MPC (8 μg/ml), in accordance with previous data (1). Such emergence might be explained by the rapid decline of the concentrations below efficient levels, i.e., the MPC. The persistence of the mutants is likely related to the bacteriostatic property of the drug: resistant cells are inhibited only by concentrations higher than the MIC or the MPC, and they can regrow when the antibiotic levels decline. Thus, increases in linezolid dosages do not lower the risk of emergence of resistant mutants. The most resistant mutant, Ef1497MutM3, exhibited a linezolid MIC of 16 μg/ml, i.e., on the same order of magnitude as those observed for clinical linezolid-resistant mutants (4 to 64 μg/ml). Supplementary step mutants were not selected from Ef1497MutM3 at the usual (600-mg) or lower (200-mg) dosages, maybe because such mutations would be lethal. Also, prolonged simulated treatment should have contributed to their emergence in this case as for other strains and regimens. The more frequent occurrence of linezolid failures with enterococci, compared to staphylococci, might be related to a higher prevalence of the mutator phenotype. Indeed, this prevalence, evaluated here for the first time in E. faecalis, reached 1.7%, i.e., about 20 times more than in S. aureus (36).
The mechanism of linezolid resistance in mutants selected from the E. faecalis mutator strain Ef1497 in the PK-PD model has been studied. Detection of the expected mutation, G2576T, common in E. faecalis strains (7, 31, 42), failed. In agreement with this, no coresistances were evidenced with chloramphenicol and quinupristin-dalfopristin, which have sites overlapping with those of linezolid, as in G2576T S. aureus mutants (5). Other mutations in 23S rRNA genes (e.g., G2505A, G2447T, and T2500A) (7, 31, 42) or other mechanisms, such as deletions in the L4 protein described in Streptococcus pneumoniae (52) or the transferable 23S rRNA methyltransferase Cfr in staphylococcal isolates (33), might be involved, but these mechanisms also confer multidrug resistance phenotypes. In the same way, no coresistances were observed with ciprofloxacin, which is a substrate, like oxazolidinones, for most multidrug efflux pumps of Gram-negative organisms (46).
The mechanism of linezolid resistance was elucidated for the most resistant mutant, Ef1497MutM3. The characterization of the 23S rRNA gene domain V revealed the presence of the substitution A2453G. So far, this mutation has been described only in in vitro mutants of Halobacterium halobium and is located in the single 23S rRNA gene of that archaebacterium, allowing linezolid resistance to be multiplied by 44 (23); coresistances in this mutant were not mentioned. The types of E. faecalis mutants (G2576T and G2505A) obtained in an in vivo animal model have been reported to be dependent on the linezolid regimen (7). Perhaps the unusual A2453G mutation found in Ef1497MutM3 was generated by the specific conditions of our assay. In addition, we demonstrated that this A2453G mutation was present in two of the four copies of the 23S rRNA genes of Ef1497MutM3. The level of linezolid resistance is known to increase with the number of mutated copies of the E. faecalis 23S rRNA gene (7, 31). The presence of the mutator phenotype in Ef1497 could enhance not only the occurrence of a mutation in one 23S rRNA gene, but also the recombination process between wild-type and mutated copies (53). Accordingly, the in vitro linezolid resistance was selected at a higher frequency in E. faecalis JH2-2 than in a recA recombination-deficient mutant (27).
In conclusion, in our PK-PD model, the emergence of linezolid-resistant mutants was primarily influenced by the mutational capabilities of the species and the strain; increases in dosages of this bacteriostatic and time-dependent antibiotic with a rapid decline of the concentrations did not influence the emergence and survival of the resistant mutants. Thus, when linezolid must be prescribed, hypermutable strains should be detected by the disk diffusion method (14), and the antibiotic should be administered in combination (22, 55), particularly for prolonged treatment. Finally, mutators should be included in the preliminary tests of any new antibiotic.
Acknowledgments
The sequencing experiments were performed at the Genotyping and Sequencing Facility of Bordeaux under grants from the Conseil Régional d'Aquitaine (no. 20030304002FA and 20040305003FA) and from the European Union (FEDER no. 2003227). This work was supported in part by a grant from Pfizer France Laboratories.
We thank Catherine André and Laure Coulange for technical assistance and Isabelle Maachi-Guillot (Haut Levêque Hospital Medical Device Service) and Sylvie Darmagnac (Fresenius Medical Care) for providing disposable devices.
Footnotes
Published ahead of print on 25 January 2010.
REFERENCES
- 1.Allen, G. P., and B. C. Bierman.2009. In vitro analysis of resistance selection by linezolid in vancomycin-susceptible and -resistant Enterococcus faecalis and Enterococcus faecium. Int. J. Antimicrob. Agents 34:21-24. [DOI] [PubMed] [Google Scholar]
- 2.Ba, B. B., A. Bernard, A. Iliadis, C. Quentin, D. Ducint, R. Etienne, M. Fourtillan, I. Maachi-Guillot, and M. C. Saux.2001. New approach for accurate simulation of human pharmacokinetics in an in vitro pharmacodynamic model: application to ciprofloxacin. J. Antimicrob. Chemother. 47:223-227. [DOI] [PubMed] [Google Scholar]
- 3.Ba, B. B., B. Bikie Bi Nso, C. Quentin, and M. C. Saux.2007. Determination of linezolid in growth media by high-performance liquid chromatography with on-line extraction. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 854:104-108. [DOI] [PubMed] [Google Scholar]
- 4.Barger, A., C. Fuhst, and B. Wiedemann.2003. Pharmacological indices in antibiotic therapy. J. Antimicrob. Chemother. 52:893-898. [DOI] [PubMed] [Google Scholar]
- 5.Besier, S., A. Ludwig, J. Zander, V. Brade, and T. A. Wichelhaus.2008. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob. Agents Chemother. 52:1570-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boak, L. M., J. Li, C. R. Rayner, and R. L. Nation.2007. Pharmacokinetic/pharmacodynamic factors influencing emergence of resistance to linezolid in an in vitro model. Antimicrob. Agents Chemother. 51:1287-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgeois-Nicolaos, N., L. Massias, B. Couson, M. J. Butel, A. Andremont, and F. Doucet-Populaire.2007. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J. Infect. Dis. 195:1480-1488. [DOI] [PubMed] [Google Scholar]
- 8.Bozdogan, B., and P. C. Appelbaum.2004. Oxazolidinones: activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 23:113-119. [DOI] [PubMed] [Google Scholar]
- 9.Bozdogan, B., D. Esel, C. Whitener, F. A. Browne, and P. C. Appelbaum.2003. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J. Antimicrob. Chemother. 52:864-868. [DOI] [PubMed] [Google Scholar]
- 10.Burkhardt, O., K. Borner, N. von der Höh, P. Köppe, M. W. Pletz, C. E. Nord, and H. Lode.2002. Single- and multiple-dose pharmacokinetics of linezolid and co-amoxiclav in healthy human volunteers. J. Antimicrob. Chemother. 50:707-712. [DOI] [PubMed] [Google Scholar]
- 11.Chopra, I., A. J. O'Neill, and K. Miller.2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Updat. 6:137-145. [DOI] [PubMed] [Google Scholar]
- 12.Daurel, C., A. L. Prunier, F. Chau, L. Garry, R. Leclercq, and B. Fantin.2007. Role of hypermutability on bacterial fitness and emergence of resistance in experimental osteomyelitis due to Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 51:344-349. [DOI] [PubMed] [Google Scholar]
- 13.Dehghanyar, P., C. Bürger, M. Zeitlinger, F. Islinger, F. Kovar, M. Müller, C. Kloft, and C. Joukhadar.2005. Penetration of linezolid into soft tissues of healthy volunteers after single and multiple doses. Antimicrob. Agents Chemother. 49:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denamur, E., S. Bonacorsi, A. Giraud, P. Duriez, F. Hilali, C. Amorin, E. Bingen, A. Andremont, B. Picard, F. Taddei, and I. Matic.2002. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J. Bacteriol. 184:605-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diekema, D. J., and R. N. Jones.2001. Oxazolidinone antibiotics. Lancet 358:1975-1982. [DOI] [PubMed] [Google Scholar]
- 16.Ellner, P. D., and H. C. Neu.1981. The inhibitory quotient. A method for interpreting minimum inhibitory concentration data. JAMA 246:1575-1578. [DOI] [PubMed] [Google Scholar]
- 17.Firsov, A. A., S. N. Vostrov, A. A. Shevchenko, and G. Cornaglia.1997. Parameters of bacterial killing and regrowth kinetics and antimicrobial effect examined in terms of area under the concentration-time curve relationships: action of ciprofloxacin against Escherichia coli in an in vitro dynamic model. Antimicrob. Agents Chemother. 41:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French, G.2001. Linezolid. Int. J. Clin. Pract. 55:59-63. [PubMed] [Google Scholar]
- 19.Frimodt-Møller, N.2002. How predictive is PK/PD for antibacterial agents? Int. J. Antimicrob. Agents 19:333-339. [DOI] [PubMed] [Google Scholar]
- 20.Gee, T., R. Ellis, G. Marshall, J. Andrews, J. Ashby, and R. Wise.2001. Pharmacokinetics and tissue penetration of linezolid following multiple oral doses. Antimicrob. Agents Chemother. 45:1843-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzales, R. D., P. C. Schreckenberger, M. B. Graham, S. Kelkar, K. DenBesten, and J. P. Quinn.2001. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 357:1179. [DOI] [PubMed] [Google Scholar]
- 22.Huang, V., and M. J. Rybak.2005. Pharmacodynamics of cefepime alone and in combination with various antimicrobials against methicillin-resistant Staphylococcus aureus in an in vitro pharmacodynamic infection model. Antimicrob. Agents Chemother. 49:302-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Jullin, J., et al.2001. Prog. Abstr. 21st Interdisciplinary Meet. Anti-infect. Chemother., abstr. 127/P1, p. 131.
- 22b.Kister, G., et al.1998. Prog. Abstr. 2nd Sci. Meet. Assoc. Pharm. Faculties Pharmacologists, abstr. 13, p. 14.
- 23.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin.1999. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 24.LaPlante, K. L., S. N. Leonard, D. R. Andes, W. A. Craig, and M. J. Rybak.2008. Activities of clindamycin, daptomycin, doxycycline, linezolid, trimethoprim-sulfamethoxazole, and vancomycin against community-associated methicillin-resistant Staphylococcus aureus with inducible clindamycin resistance in murine thigh infection and in vitro pharmacodynamic models. Antimicrob. Agents Chemother. 52:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaPlante, K. L., and M. J. Rybak.2004. Impact of high-inoculum Staphylococcus aureus on the activities of nafcillin, vancomycin, linezolid, and daptomycin, alone and in combination with gentamicin, in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 48:4665-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula.1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 27.Lobritz, M., R. Hutton-Thomas, S. Marshall, and L. B. Rice.2003. Recombination proficiency influences frequency and locus of mutational resistance to linezolid in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3318-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louie, A., H. S. Heine, K. Kim, D. L. Brown, B. VanScoy, W. Liu, M. Kinzig-Schippers, F. Sorgel, and G. L. Drusano.2008. Use of an in vitro pharmacodynamic model to derive a linezolid regimen that optimizes bacterial kill and prevents emergence of resistance in Bacillus anthracis. Antimicrob. Agents Chemother. 52:2486-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacGowan, A., C. Rogers, H. A. Holt, M. Wootton, and K. Bowker.2000. Assessment of different antibacterial effect measures used in in vitro models of infection and subsequent use in pharmacodynamic correlations for moxifloxacin. J. Antimicrob. Chemother. 46:73-78. [DOI] [PubMed] [Google Scholar]
- 30.MacGowan, A. P.2003. Pharmacokinetic and pharmacodynamic profile of linezolid in healthy volunteers and patients with Gram-positive infections. J. Antimicrob. Chemother. 51(Suppl. 2):ii, 17-25. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice.2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meka, V. G., S. K. Pillai, G. Sakoulas, C. Wennersten, L. Venkataraman, P. C. DeGirolami, G. M. Eliopoulos, R. C. Moellering, Jr., and H. S. Gold.2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J. Infect. Dis. 190:311-317. [DOI] [PubMed] [Google Scholar]
- 33.Mendes, R. E., L. M. Deshpande, M. Castanheira, J. DiPersio, M. A. Saubolle, and R. N. Jones.2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouton, J. W., M. N. Dudley, O. Cars, H. Derendorf, and G. L. Drusano.2005. Standardization of pharmacokinetic/pharmacodynamic (PK/PD) terminology for anti-infective drugs: an update. J. Antimicrob. Chemother. 55:601-607. [DOI] [PubMed] [Google Scholar]
- 35.Muller, M., A. dela Pena, and H. Derendorf.2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: distribution in tissue. Antimicrob. Agents Chemother. 48:1441-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Neill, A. J., and I. Chopra.2002. Insertional inactivation of mutS in Staphylococcus aureus reveals potential for elevated mutation frequencies, although the prevalence of mutators in clinical isolates is low. J. Antimicrob. Chemother. 50:161-169. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser.2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 38.Pillai, S. K., G. Sakoulas, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., M. J. Ferraro, and H. S. Gold.2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J. Infect. Dis. 186:1603-1607. [DOI] [PubMed] [Google Scholar]
- 39.Prunier, A. L., and R. Leclercq.2005. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J. Bacteriol. 187:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclercq.2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187:1709-1716. [DOI] [PubMed] [Google Scholar]
- 41.Prunier, A. L., B. Malbruny, D. Tandé, B. Picard, and R. Leclercq.2002. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob. Agents Chemother. 46:3054-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin.2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, S. M., A. F. Freeman, S. M. Harrington, S. M. Holland, P. R. Murray, and A. M. Zelazny.2006. Linezolid-resistant Staphylococcus aureus in two pediatric patients receiving low-dose linezolid therapy. Pediatr. Infect. Dis. J. 25:562-564. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., F. Fritsch, and T. Maniatis (ed.).1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Schaaff, F., A. Reipert, and G. Bierbaum.2002. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher, A., R. Trittler, J. A. Bohnert, K. Kümmerer, J. M. Pagès, and W. V. Kern.2007. Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. J. Antimicrob. Chemother. 59:1261-1264. [DOI] [PubMed] [Google Scholar]
- 47.Slatter, J. G., D. J. Stalker, K. L. Feenstra, I. R. Welshman, J. B. Bruss, J. P. Sams, M. G. Johnson, P. E. Sanders, M. J. Hauer, P. E. Fagerness, R. P. Stryd, G. W. Peng, and E. M. Shobe.2001. Pharmacokinetics, metabolism, and excretion of linezolid following an oral dose of [(14)C]linezolid to healthy human subjects. Drug Metab. Dispos. 29:1136-1145. [PubMed] [Google Scholar]
- 48.Stein, G. E., S. Schooley, C. A. Peloquin, A. Missavage, and D. H. Havlichek.2007. Linezolid tissue penetration and serum activity against strains of methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility in diabetic patients with foot infections. J. Antimicrob. Chemother. 60:819-823. [DOI] [PubMed] [Google Scholar]
- 49.Strukova, E. N., M. V. Smirnova, S. N. Vostrov, I. Y. Lubenko, A. A. Firsov, S. H. Zinner, and Y. A. Portnoy.2009. Linezolid pharmacodynamics with Staphylococcus aureus in an in vitro dynamic model. Int. J. Antimicrob. Agents 33:251-254. [DOI] [PubMed] [Google Scholar]
- 50.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, Jr., and M. J. Ferraro.2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 51.Willems, R. J., J. Top., D. J. Smith, D. I. Roper, S. E. North, and N. Woodford.2003. Mutations in the DNA mismatch repair proteins MutS and MutL of oxazolidinone-resistant or -susceptible Enterococcus faecium. Antimicrob. Agents Chemother. 47:3061-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolter, N., A. M. Smith, D. J. Farrell, W. Schaffner, M. Moore, C. G. Whitney, J. H. Jorgensen, and K. P. Klugman.2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49:3554-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worth, L., Jr., T. Bader, J. Yang, and S. Clark.1998. Role of MutS ATPase activity in MutS, L-dependent block of in vitro strand transfer. J. Biol. Chem. 273:23176-23182. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, X., and K. Drlica.2001. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin. Infect. Dis. 33(Suppl. 3):S147-S156. [DOI] [PubMed] [Google Scholar]
- 55.Zinner, S. H., D. Gilbert, I. Y. Lubenko, K. Greer, and A. A. Firsov.2008. Selection of linezolid-resistant Enterococcus faecium in an in vitro dynamic model: protective effect of doxycycline. J. Antimicrob. Chemother. 61:629-635. [DOI] [PubMed] [Google Scholar]
- 56.Zurenko, G. E., B. H. Yagi, R. D. Schaadt, J. W. Allison, J. O. Kilburn, S. E. Glickman, D. K. Hutchinson, M. R. Barbachyn, and S. J. Brickner.1996. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob. Agents Chemother. 40:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]