Abstract
Molecular tools are valuable for determining evolutionary history and the prevalence of drug-resistant malaria parasites. These tools have helped to predict decreased sensitivity to antimalarials and fixation of multidrug resistance genotypes in some regions. In order to assess how historical drug policies impacted Plasmodium falciparum in Venezuela, we examined molecular changes in genes associated with drug resistance. We examined pfmdr1 and pfcrt in samples from Sifontes, Venezuela, and integrated our findings with earlier work describing dhfr and dhps in these samples. We characterized pfmdr1 genotypes and copy number variation, pfcrt genotypes, and proximal microsatellites in 93 samples originating from surveillance from 2003 to 2004. Multicopy pfmdr1 was found in 12% of the samples. Two pfmdr1 alleles, Y184F/N1042D/D1246Y (37%) and Y184F/S1034C/N1042D/D1246Y (63%), were found. These alleles share ancestry, and no evidence of strong selective pressure on mutations was found. pfcrt chloroquine resistance alleles are fixed with two alleles: StctVMNT (91%) and SagtVMNT (9%). These alleles are associated with strong selection. There was also an association between pfcrt, pfmdr1, dhfr, and dhps genotypes/haplotypes. Duplication of pfmdr1 suggests a potential shift in mefloquine sensitivity in this region, which warrants further study. A bottleneck occurred in P. falciparum in Sifontes, Venezuela, and multidrug resistance genotypes are present. This population could be targeted for malaria elimination programs to prevent the possible spread of multidrug-resistant parasites.
Amplification of the Plasmodium falciparum multidrug resistance gene (pfmdr1) has been implicated in mefloquine (MQ) resistance in Thailand and Cambodia (1, 17, 27, 28, 34, 41), but not elsewhere. It is not known if amplification has occurred in Venezuela, where MQ monotherapy was used between 2001 and 2004 and the combination of artesunate (AS) and MQ thereafter. pfmdr1 duplication is also implicated in resistance to lumefantrine, halofantrine, quinine, and AS (39) and may decrease resistance to chloroquine (CQ) (43). Also, single-nucleotide mutations in pfmdr1, such as N86Y, Y184F, S1034C, N1042D, and D1246Y (the mutated amino acid is shown in boldface type), are postulated to modulate drug response. While these mutations may or may not contribute to CQ resistance (40), mutations at codons 1034, 1042, and 1246 make parasites more sensitive to MQ (40). Studies suggest at least two lineages of mutant pfmdr1 genotypes have evolved in South America (4, 21).
In South America, CQ and sulfadoxine-pyrimethamine (SP) were used to treat P. falciparum prior to the use of artemisinin-based combination therapy (ACT). Resistance to CQ and SP evolved independently in South America (18, 23). Point mutations in the P. falciparum chloroquine resistance transporter (pfcrt) gene are correlated with CQ resistance (10). The pfcrt point mutation K76T is critical, but C72S, M74I, N75E, and N75K are also associated with resistance (48). There are at least four different origins of CQ resistance pfcrt alleles: one in Papua New Guinea (SVMNT), where the genotype represents amino acids at codons 72 to 76, one in Southeast Asia (CVIET) that spread to Africa, and two in South America (SVMNT/CVMNT in Brazil/Peru and CVMET/CVMNT in Ecuador/Colombia) (49).
Molecular surveillance showed that, after drug removal, CQ resistance genotypes, in Malawi and China (16, 46), and SP resistance genotypes, in the Peruvian Amazon (52), declined. Therefore, the reduction in the frequency of resistant parasites likely occurred because resistant parasite populations are at a fitness disadvantage in the absence of drug pressure. In Bolivar State, Venezuela, mutant pfcrt alleles remained after the removal of CQ in 1986 (6) and mutant dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) genes remained fixed after SP removal (19). Whether the recent use of MQ and AS-MQ led to the evolution of pfmdr1 genotypes associated with AS and MQ resistance is unknown.
This study in the state of Bolivar, Venezuela assessed the following: (i) whether pfmdr1 duplication has occurred, (ii) the frequency of pfmdr1 and pfcrt mutations, (iii) whether MQ and CQ drug pressure has affected variation surrounding these genes, and (iv) linkage disequilibrium between dhfr, dhps, pfcrt, and pfmdr1 alleles.
(Part of this research [some data pertaining to pfmdr1 and pfcrt genotypes, microsatellites, and copy number] was presented at the 57th Annual Meeting of the American Society of Tropical Medicine and Hygiene, New Orleans, LA, 2008, and the 58th Annual Meeting of the American Society of Tropical Medicine and Hygiene, Washington, DC, 2009.)
MATERIALS AND METHODS
Study site and subjects.
The municipality of Sifontes, located in the state of Bolivar, Venezuela, is an epicenter of multidrug-resistant P. falciparum with 35 to 40% of malaria cases in Venezuela in 1999 (5, 44). We tested 93 blood samples taken from a Sifontes surveillance study during 2003 to 2004. Patients were adults with confirmed P. falciparum parasitemia and generally uncomplicated malaria. Previously characterization of these samples for dhfr and dhps, microsatellites around these genes, and additional neutral microsatellites did not reveal any multiple infections; each sample possessed a single genotype at all loci (19). Informed consent was obtained from patients, and the study protocol was approved by the bioethics commission of the Instituto de Altos Estudios in Venezuela.
DNA isolation, amplification, and genotyping methods.
DNA was isolated from whole blood using the QIAamp DNA minikit (Qiagen, Valencia, CA). Genomic DNA was used for sequencing and real-time PCR. Amplified DNA (REPLI-g whole genome amplification kit; Qiagen, Valencia, CA) was used for microsatellite characterization.
pfmdr1 copy number was determined by TaqMan real-time PCR (Stratagene MX3005P; Agilent Technologies, La Jolla, CA) with published primers and probes (34) labeled with 3′ black hole quencher (BHQ) and 5′ FAM (6-carboxyfluorescein) (pfmdr1) or 5′ HEX (hexachlorofluorescein) (Table 1). Amplification reactions were multiplexed. Samples were run in triplicate, with clone 3D7 as a normalizer. Two reference DNAs were included, Dd2 has 3 or 4 copies of pfmdr1 (47), and W2-mef has 2 copies (15). Assays were repeated if threshold cycle (CT) values were >32 or if the 95% confidence interval around the estimation was >0.4 (34). Copy number was calculated with the comparative ΔΔCT method (34). Copy number estimates were rounded to the nearest integer, and parasites with greater than 1.5 copies were considered multicopy (34). Following the convention established by Price et al. (34), the copy numbers reported are based on mean values after rounding, even if the final 95% confidence intervals calculated contained more than one integer after rounding. Two-tailed 95% confidence intervals were calculated from the individual replicate ΔΔCT calculations (50).
TABLE 1.
Summary of methods used to study pfcrt and pfmdr1
| Gene [method(s)] | Primary PCR |
Nested PCR |
Sequencing primers | ||
|---|---|---|---|---|---|
| Primer sequences (5′-3′)a | PCR cycling conditions | Primer sequences (5′-3′)a | PCR cycling conditions | ||
| pfmdr1 copy no. (TaqMan assay)b | 10 min at 95°C; 50 cycles of 20 s at 95°C and 1 min at 60°C | NAc | NA | NA | |
| pfmdr1 codons 86 and 184 (real-time PCR) | 10 min at 96°C; 50 cycles of 15 s at 96°C and 1 min at 59°C (codon 86) or 1 min at 61°C (codon 184) | NA | NA | NA | |
| pfmdr1 codons 1034, 1042, and 1246 (PCR and sequencing) | GCATTTAGTTCAGATGATGAAATG (F) and CCATATGGTCCAACATTTGTATC (R) | Initial denaturation of 3 min at 95°C; 40 cycles of 30 s at 93°C, 40 s at 58°C, and 45 s at 72°C; final extension step of 10 min at 72°C | TATGCATACTGTTATTAATTATGG (F) and TTCGATAAATTCATCTATAGCAG (R) | Initial denaturation of 10 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 59°C, and 1 min at 72°C; final extension at 72°C for 10 min | Used both nested primers for sequencing |
| pfcrt codons 72 to 76 (PCR and sequencing) | TTTTTCCCTTGTCGACCTTAAC (F) and AGGAATAAACAATAAAGAACATAATCATAC (R) | Initial denaturation of 10 min at 94°C; 38 cycles of 30 s at 94°C, 30 s at 56°C, and 45 s at 72°C; final extension step of 20 min at 72°C | NA | NA | Used reverse primer for sequencing |
We examined pfmdr1 for mutations in codons 86, 184, 1034, 1042, and 1246. Mutations in codons 86 and 184 of pfmdr1 were detected using a Stratagene MX3005P real-time PCR system (35) (Table 1). Both probes were labeled with a minor groove binder-nonfluorescent quencher. Wild-type probes were labeled with FAM, and mutant probes were labeled with VIC (Applied Biosystems, Foster City, CA). Direct sequencing was used to analyze polymorphisms in codons 1034, 1042, and 1246 (Table 1).
A 264-bp region of pfcrt containing codons 72 to 76 was amplified in 89 samples (Table 1). Residual dye terminators were removed by ethanol precipitation followed by a 70% ethanol wash. Pellets were resuspended in 10 μl HiDi formamide (Applied Biosystems, Foster City, CA) and sequenced using an ABI PRISM 3130xl genetic analyzer (Applied Biosystems, Foster City, CA).
Microsatellite analysis.
Samples were assayed for 12 microsatellite loci spanning 499.5 kb around pfcrt on chromosome 7 and 15 microsatellite loci, spanning 544.8 kb around pfmdr1 on chromosome 5, previously identified (24, 26, 49). Upstream distances were calculated from the gene's start codon and downstream distances were calculated from the gene's stop codon. PCR primer sequences are provided in Table S1 in the supplemental material following the cycling conditions detailed in references 25 and 37. PCR products were separated on an Applied Biosystems 3130xl sequencer and scored using GeneMapper software v.3.7 (Applied Biosystems, Foster City, CA). Multiple alleles were not detected (see Table S2 in the supplemental material), supporting earlier results (19) that suggested the samples were all single infections. Two samples were removed due to contamination. Haplotypes were classified as different if they contained ≥2 different alleles across all loci. eBurst (9) was used to examine the microsatellite haplotypes of both pfcrt (−4.8 kb upstream to 3.9 kb downstream) and pfmdr1 (−4.2 to 3.7 kb). Missing data were reported but not considered when defining haplotypes. Previously published data for microsatellite loci and dhfr and dhps genotypes were also incorporated (19) in our data analyses.
Statistical analysis.
The expected heterozygosity (He) was calculated for each locus as [n/(n − 1)][1 − Σpi2], where n is the number of isolates sampled and pi is the frequency of the ith allele (26). The sampling variance for He was calculated as [2(n − 1)/n3][2(n − 2)][Σpi3 − (Σpi2)2] (26). The Excel Microsatellite tool kit calculated the number of alleles per locus and allele frequencies (29). An α of 0.05 was our threshold of statistical significance. Significant associations between microsatellite pairs were determined using an exact test of linkage disequilibrium (36) with 10,000 Monte Carlo steps in Arlequin version 3.1 (8). We also noted whether there was any linkage between pfcrt, pfmdr1, dhfr, or dhps genotypes and between pfcrt and pfmdr1 genes versus dhfr and dhps genes using the same conditions. In a panmictic population, the null hypothesis is linkage equilibrium between loci located on different chromosomes. P values for microsatellites were examined after a Bonferroni-Holms correction (38).
RESULTS
pfmdr1 copy number variation and genotypes.
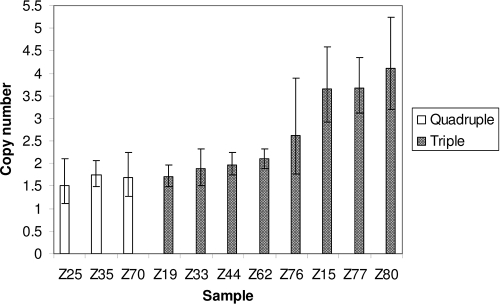
pfmdr1 copy number analysis was successful for 90 samples: 79 samples (88%) had 1 copy, 7 samples (8%) had 2 copies, 1 sample (1%) had 3 copies, and 3 samples (3%) had 4 copies (Fig. 1). Complete pfmdr1 genotyping was successful for 78 samples. We found only two pfmdr1 mutant alleles, Y184F/N1042D/D1246Y (triple mutant) and Y184F/S1034C/N1042D/D1246Y (quadruple mutant) at frequencies of 37% (n = 29) and 63% (n = 50), respectively (the mutated amino acid is shown in boldface type). Codon 86 was wild type in all 83 samples.
FIG. 1.
Estimates of pfmdr1 copy number with 95% confidence intervals (error bars) from samples with pfmdr1 duplication. Open bars represent quadruple mutant pfmdr1 parasites, while shaded bars represent those with triple mutant pfmdr1 parasites. A total of 90 samples were successfully tested.
pfcrt genotypes.
No wild-type CVMNK pfcrt genotypes were present. We found two alleles StctVMNT and SagtVMNT at a frequency of 91% (n = 81) and 9% (n = 8), respectively, where tct and agt refer to the bases coding for S.
Microsatellite characterization.
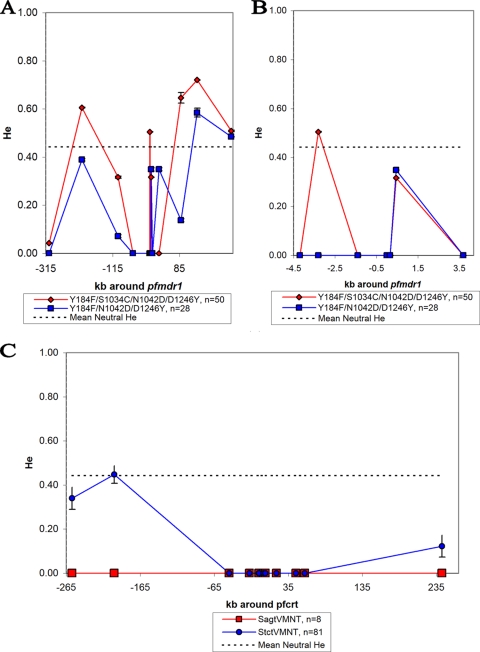
Table S2 in the supplemental material details the microsatellite haplotypes around pfcrt and pfmdr1. Table 2 shows the overall He at each microsatellite marker, and Fig. 2 shows He around each gene after separating samples by gene allele. The mean He of loci surrounding pfmdr1 was 0.25. He was reduced in the microsatellite loci closest to pfmdr1 (Fig. 2). Quadruple pfmdr1 mutant parasites carrying the pfcrt allele SagtVMNT had only one haplotype. Duplication of pfmdr1 was found on both triple and quadruple mutant pfmdr1 lineages. There was no marked difference in the He curves of single and multicopy pfmdr1 parasites (data not shown).
TABLE 2.
Number of alleles and expected heterozygosity per microsatellite locus for pfmdr1 and pfcrt
| Locusa | Ab | Hec (mean ± SD) |
|---|---|---|
| Loci around pfmdr1 on chromosome 5 | ||
| −305 kb | 2 | 0.0227 ± 0.0006 |
| −207 kb | 3 | 0.5424 ± 0.0014 |
| −99 kb | 2 | 0.2360 ± 0.0029 |
| −54 kb | 1 | 0.0000 ± 0.0000 |
| −4.2 kb | 1 | 0.0000 ± 0.0006 |
| −3.4 kb | 2 | 0.4086 ± 0.0007 |
| −1.4 kb | 1 | 0.0000 ± 0.0000 |
| Within gene | 1 | 0.0000 ± 0.0000 |
| 0.16 kb | 1 | 0.0000 ± 0.0000 |
| 0.45 kb | 2 | 0.3524 ± 0.0025 |
| 3.7 kb | 1 | 0.0000 ± 0.0000 |
| 23 kb | 2 | 0.3905 ± 0.0020 |
| 89 kb | 4 | 0.5405 ± 0.0029 |
| 137 kb | 4 | 0.7247 ± 0.0049 |
| 239.8 kb | 2 | 0.5058 ± 0.0007 |
| Mean | 1.93 | 0.2487 |
| Loci around pfcrt on chromosome 7 | ||
| −257 kb | 2 | 0.3209 ± 0.0026 |
| −200 kb | 2 | 0.4796 ± 0.0004 |
| −45 kb | 1 | 0.0000 ± 0.0000 |
| −17.7 kb | 1 | 0.0000 ± 0.0000 |
| −4.8 kb | 2 | 0.1655 ± 0.0024 |
| −4.5 kb | 2 | 0.0000 ± 0.0000 |
| 1.5 kb | 1 | 0.0000 ± 0.0000 |
| 3.9 kb | 1 | 0.0000 ± 0.0000 |
| 18.8 kb | 1 | 0.0000 ± 0.0000 |
| 45.3 kb | 1 | 0.0000 ± 0.0000 |
| 57.1 kb | 1 | 0.0000 ± 0.0000 |
| 242.5 kb | 3 | 0.2630 ± 0.0058 |
| Mean | 1.18 | 0.0728 |
The locus positions of upstream loci are measured with respect to the start codon of the gene, and the downstream loci are measured with respect to the stop codon.
Number of alleles (A) per microsatellite locus for pfmdr1 and pfcrt.
Expected heterozygosity (He) per microsatellite locus for pfmdr1 and pfcrt.
FIG. 2.
Graphical displays of He ± 1 standard deviation around pfmdr1 and pfcrt. The dashed line in each graph is the mean neutral He calculated from loci on chromosomes 2 and 3 (19). On the x axis, negative numbers are positions 5′ to the start codon of the gene and positive numbers are positions 3′ to stop codon of the gene. (A) The entire region surrounding pfmdr1 characterized by microsatellite markers on chromosome 5. (B) Close-up of the pfmdr1 region with low He. (C) The entire region surrounding pfcrt characterized by microsatellite markers on chromosome 7. For SagtVMNT, the error bars for the microsatellite markers are all 0 due to the lack of variation.
The mean He of loci surrounding pfcrt was 0.07 (Table 2). There are only 1 or 2 alleles at each of the microsatellite loci around pfcrt, with the exception of 242.5 kb (Table 2). Little variation was found immediately surrounding pfcrt with the exception of markers at −4.8 kb (Table 2). The majority of the variation around pfcrt was attributable to the different haplotypes of StctVMNT or SagtVMNT lineages. For StctVMNT, an increase in variation is seen 200 kb 5′, but no increase is seen within 57.1 kb 3′ of pfcrt (Fig. 2). The lack of variation in the loci around SagtVMNT is striking; however, the small number of parasites with this genotype warrants caution.
The results of visual inspection (see Table S2 in the supplemental material) and eBurst analysis (data not shown) suggest that the pfcrt genotypes StctVMNT and SagtVMNT are closely related, with the only evidence for differentiation or mutation found at the −4.8 kb marker and 242.5 kb. The triple and quadruple pfmdr1 genotypes are related and clustered around a single haplotype (203, 126, 196, 206, 221, 191, and 168; Table S2).
Linkage disequilibrium between genotypes and haplotypes.
Previously, dhfr and dhps were shown to be in linkage disequilibrium (LD) in this population (19). LD existed between each pair of genes: pfcrt versus pfmdr1 (P = 0.02), dhfr versus dhps, (P = 0.00), pfcrt versus dhfr (P = 0.00), pfcrt versus dhps (P = 0.00), pfmdr1 versus dhfr (P = 0.03), and pfmdr1 versus dhps (P = 0.02). LD was also significant for a comparison of combined pfcrt and pfmdr1 genotypes versus combined dhfr and dhps genotypes (P = 0.00). Each gene occurs on a separate chromosome and, here, had two alleles. A maximum of 16 possible combinations of dhfr, dhps, pfcrt, and pfmdr1 alleles would be expected, assuming independent assortment. We saw only three combinations: (i) SagtVMNT pfcrt/quadruple mutant pfmdr1/double mutant dhfr/double mutant dhps, (ii) StctVMNT pfcrt/triple mutant pfmdr1/triple mutant dhfr/triple mutant dhps, and (iii) StctVMNT pfcrt/quadruple mutant pfmdr1/triple mutant dhfr/triple mutant dhps. In addition, only the two StctVMNT “types” had multiple copies of pfmdr1.
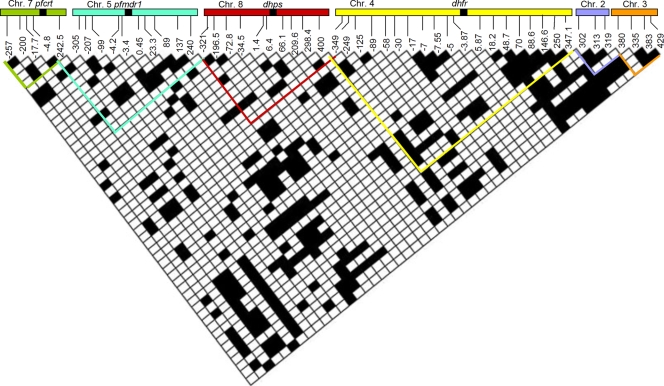
There was extensive linkage disequilibrium among microsatellites around all four genes and neutral markers (Fig. 3). We compared 1,275 pairs of loci on chromosomes 2, 3, 4, 5, 7, and 8. We expected 63.75 pairs (0.05 × 1,275 pairs) to be statistically significant in a panmictic population; here, 325 pairs, or 26%, showed significant disequilibrium.
FIG. 3.
Pairwise linkage disequilibrium between microsatellite loci on different chromosomes. Each box represents one comparison between polymorphic pairs of loci; nonpolymorphic pairwise comparisons are not included. Bonferroni's correction for multiple comparisons was conducted for each comparison. Black cells represent statistical significance at P = 0.01, and white cells were not statistically significant. The location of each microsatellite locus is given on the x axis (loci are named according to their position relative to pfcrt, pfmdr1, dhfr, dhps, or position along chromosome 2 or 3 according to the 3D7 genome sequence available from PlasmoDB v6.3). In addition, the values given for dhfr and dhps have been updated from those used previously (19). Previously reported values are indicated first and new, corrected values are indicated second: for dhfr, −350 = −349, −250 = −249, −10 = −7, −5.3 = −5, 20 = 18.2, 50 = 48.7, 90 = 90.5, 150 = 146.6, and 350 = 347.1; for dhps, −397 = 400, −297 = 298.4, −211 = 209.6, −66.6 = 66.1, −7.4 = 6.4, −2.5 = 1.4, 33.1 = −34.5, 71.6 = −72.8, 198 = −196.5, and 301 = −321. Chr., chromosome.
The SagtVMNT allele had only a single microsatellite haplotype and was found with one quadruple mutant pfmdr1 haplotype and none of the parasites with multicopy pfmdr1. This pfcrt/pfmdr1 haplotype appeared with only a single double mutant dhps (minor variation at 301 kb) and one dhfr (minor variation noted at −16.7, 5.8, and 347.1 kb) microsatellite haplotype. This variation continued with the neutral markers, where 7/8 of the SagtVMNT samples shared a haplotype (the eighth differed at 2/7 markers), suggesting clonal expansion. Conversely, the StctVMNT pfcrt genotype occurs with both pfmdr1 genotypes, multiple related haplotypes of the triple dhfr and triple dhps alleles (n = 82), and with multiple neutral marker profiles. StctVMNT had a single haplotype, with the exception of variation at −257, −200 kb, and 242.5 kb.
DISCUSSION
Our findings raise concerns about the potential development of de novo MQ resistance in South America. Twelve percent of the samples carried multiple copies of pfmdr1. This had previously been reported only in Southeast Asia, where it was linked to MQ failure and decreased ACT efficacy (1, 17, 27, 28, 34, 41). The retrospective nature of our study prevented testing whether pfmdr1 duplication was induced by MQ monotherapy or its implications for MQ treatment. However, a few studies have shown reduced MQ sensitivity in vitro or prophylaxis failure in South America (22). Our data highlight the importance of testing more recently collected samples for shifts in pfmdr1 copy number prevalence and potential MQ resistance. In contrast to Venezuela, there is no evidence of multicopy pfmdr1 in isolates from the Peruvian Amazon, where AS-MQ therapy has been the first-line treatment since 2001 (4).
We found only two alleles for pfmdr1, Y184F/N1042D/D1246Y and Y184F/S1034C/N1042D/D1246Y, and pfmdr1 duplication occurred with both alleles. Parasites with the Y184F mutation and higher copy number are reported to have higher 50% inhibition concentrations (IC50) in vitro to MQ and other drugs (31). Previously reported multicopy pfmdr1 occasionally carried a mutation at codon 86, but not mutations at codon 1034, 1042, or 1246 (31, 33, 34). At least two explanations have been hypothesized: (i) the mutation at codon 1042 imposes a severe fitness cost on parasites with multiple copies of pfmdr1 (31) or (ii) there is underreporting of mutations due to the limited number of studies (3). Our results support the latter hypothesis because all parasites carrying multicopy pfmdr1 had the mutation at codon 1042 and two or three other mutations. However, if a fitness cost is associated with the mutation at codon 1042, then the additional pfmdr1 mutations seen in this population may be compensatory.
In contrast to the multiple origins of pfmdr1 amplification, point mutations associated with pfmdr1 resistance have a common founder lineage in our study. There is a shared haplotype for both the triple mutant and quadruple mutants between −4.2 and 3.7 kb and an additional quadruple mutant haplotype. The latter, while differing at both −3.4 and 0.45 kb, appears to be due to slippage (see Table S2 in the supplemental material). These data suggest that the triple mutant is ancestral to the quadruple mutant or vice versa. While the overall He around pfmdr1 is lower than in Southeast Asia, we see a similar relative reduction in variation close to pfmdr1 (24). There is a smaller region of reduced He around pfmdr1 than pfcrt (Fig. 2). This suggests that pfmdr1 may have (i) experienced little to no selection or (ii) the selective event(s) for pfmdr1 occurred earlier than for pfcrt, allowing recombination to break down any linkage. The latter possibility appears less likely, given the recent history of antimalarial policy. Additionally, point mutations in pfmdr1 may be under selection by multiple drugs, which could complicate the signal of selection (40). The two most recent influences on pfmdr1 in Sifontes, Venezuela, are MQ and CQ, which may have differing directions of selection for mutations at codons 1042, 1034, and 1246. Our data could be interpreted as evidence of selection for multiple alleles or soft selective sweeps, as shown at the Thailand-Myanmar border (24).
Recent drug policy in Sifontes, Venezuela, may have influenced preexisting pfmdr1 alleles, since nothing in our data indicates that mutations occurred locally. For example, South American isolates collected in 1984 carried the same quadruple mutant pfmdr1 genotype found in our samples, though we could not compare microsatellite haplotypes (11). The quadruple mutant pfmdr1 allele has been seen in Peru, Guyana, and Brazil (21, 51), and the triple mutant allele has been seen in Peru (4) and Colombia (21). Whether all of these alleles share microsatellite haplotypes is unknown. However, pfmdr1 haplotypes in Guyana and Brazil are more closely related to each other than those found in Colombia (21). Our data indicate that Venezuelan pfmdr1 haplotypes are closely related to one of the two major haplotypes (MDR-A1 and MDR-A8) found in the Peruvian Amazon (4). If we assume that these pfmdr1 alleles existed prior to the gene duplication event(s), then pfmdr1 duplication evolved multiple times in South America, as seen in Southeast Asia (24).
To clarify whether reduced He around pfcrt is due to a sweep or bottleneck, we looked for a U-shaped depression in He surrounding the gene. For StctVMNT, a selective sweep is suggested by the reduced He in a long surrounding region and the observation that distant markers are approaching the mean heterozygosity of neutral markers (Fig. 2). The lack of variation surrounding SagtVMNT may be due to a smaller sample size or a bottleneck followed by clonal expansion. The second possibility appears more likely given the lack of variation associated with dhfr, dhps, and pfmdr1 genotypes/haplotypes. Additional data are required to test whether a selective sweep influenced He around SagtVMNT. Nonetheless, the depressed He around the SagtVMNT allele, compared to that around StctVMNT, suggests that it is a recent introduction with a smaller number of founders.
The evolutionary relationship between SagtVMNT and StctVMNT in South America is unclear in the literature. Proximal microsatellite alleles are shared between the two genotypes, suggesting that the two alleles are closely related. Some of the remaining variation in StctVMNT haplotypes could be explained by recombination with the SagtVMNT haplotype. While the limited variation around pfcrt does not define which allele arose first, our results suggest they originated from the same lineage (21, 45). Our haplotype data also suggest that SagtVMNT was introduced to Sifontes, Venezuela, along with a related StctVMNT haplotype (see Table 2 in the supplemental material). It had been hypothesized that SagtVMNT originated in Mato Grosso, Brazil (45), but SagtVMNT is also found in our study and in Guyana, Peru, Suriname, and Venezuela (6, 7, 21, 30), which makes its point of origin obscure.
There are at least three possible explanations for the fixation of CQ resistance pfcrt SVMNT alleles in Sifontes, Venezuela. First, the at-risk population may continue to expose P. falciparum indirectly due to CQ-based Plasmodium vivax treatment. Second, SVMNT may have little or no fitness disadvantage in the absence of drug pressure. In Africa, CQ-resistant parasites with CVIET declined after CQ was withdrawn, but CVIET is more likely to revert to CQ sensitivity in the presence of verapamil than SVMNT is, suggesting that the alleles differ in biological fitness (20). Third, there are no wild-type parasites present to replace the less-fit CQ resistance genotype. This is supported by our results and earlier work, which found only SagtVMNT and StctVMNT in Sifontes and Gran Sabana in Venezuela in 1998 to 2000 (6).
Fixation of CQ resistance in Sifontes, Venezuela, is likely to continue because of its isolation and the fixation of CQ resistance in neighboring populations. According to one study, fixation of the K76T mutation has occurred across Bolivar (6). Sifontes, Venezuela, is isolated from the Orinoco river basin flow, which influences travel through the state of Bolivar (5, 44), and to the west, it is separated from Bolivar by a region of higher elevation and a large reservoir. To the south, Sifontes is separated from most of Gran Sabana and Brazil by a mountain range, though a road does connect them. Even if migration occurs from Brazil, the K76T mutation was fixed in Manaus in 2000 to 2002 (n = 38) (45). To the east, there are few geographic barriers with Guyana, where SagtVMNT occurs at high frequency and two studies indicate that the K76T mutation is fixed or nearly fixed (21, 32).
The association between alleles of pfcrt, pfmdr1, dhfr, and dhps among our samples indicates inbreeding, a bottleneck, and/or that each subsequent resistant gene was established from a population already fixed for other resistant genes. Our results for the SagtVMNT pfcrt lineage support clonal propagation. These eight samples carried a single quadruple mutant pfmdr1 haplotype and always exhibited the double dhfr mutation (N51I/S108N) and double dhps mutation (A437G/A581G), as well as an exclusive neutral marker haplotype. Our results for StctVMNT also support this hypothesis, albeit with a larger starting population. Only a small portion of the pfcrt alleles found in another study of Bolivar (CVIET, CVMET, CVMNT, and CVMNK) (6) were seen in Sifontes, Venezuela. This lack of allelic diversity, in comparison to the rest of the state, extends to dhfr and dhps genotypes (6, 19).
Clonal propagation is argued to play a significant role in the population structure of P. falciparum in Venezuela (42). Low transmission leads to high rates of self fertilization, and thus de facto clonal propagation. For example, with 1% recombination, markers 5 cM apart could maintain linkage disequilibrium for longer than 400 years (2). Our results suggest that the level of transmission, genetic diversity, and migration should be considered when predicting whether drug resistance alleles will decline after new drugs are introduced.
Demographic history may also explain the strong linkage disequilibrium across multiple chromosomes. P. falciparum populations in Sifontes, Venezuela, likely originated from a recent population expansion after a bottleneck. In 1970, the state of Bolivar had a malarious zone to the west and another in the middle of the state, yet in Sifontes, Venezuela, malaria had been eradicated (12). By 1983, P. falciparum reemerged in El Dorado, the capital of Sifontes, Venezuela, and presumptively acted as a founding population (44). Since CQ and SP resistance were already present in the 1970s in Venezuela (13, 14), it is unlikely that the drug resistance alleles originated in Sifontes, Venezuela; resistance was noted elsewhere before and during the time Sifontes was malaria free. It has been postulated that the SP resistance alleles in Bolivar came from Brazil (6). Therefore, the limited diversity and linkage we see across all markers and genes in this population may be due to rapid expansion from a small parasite population over 20 years, resulting in a semiclonal population of multidrug-resistant parasites.
Our results suggest how multidrug-resistant P. falciparum can develop in isolated populations with low genetic diversity. If resistance to an antimalarial (CQ) reaches fixation, then a mutant allele is at no fitness disadvantage until a fitter allele with fewer mutations appears through back mutation or migration. Successful back mutation is unlikely due to the low probability of facilitatory mutations and genetic drift. Successful migration is unlikely given this region's isolation and the lack of nearby wild-type source populations in South America. Given these restrictions, if resistance is fixed for a drug (CQ) and a second drug (SP) is introduced, then resistance to the second will occur on a background of prior resistance. Such multidrug-resistant strains will remain stable and increase in the population as inbreeding renders chromosomal reassortment ineffective. The generation of MQ resistance multicopy pfmdr1 in CQ- and SP-resistant parasites may give resistance to additional drugs, like halofantrin, quinine, and AS (31, 33, 34, 40), and challenge the effectiveness of ACT.
Whatever the mechanism, potentially MQ-resistant P. falciparum are evolving on a background of CQ and SP resistance in Sifontes, Venezuela. Therefore, this is a region of special concern for malaria treatment and elimination, because migrants could spread multidrug resistance to other countries. It remains to be seen whether pfmdr1 duplication in Sifontes, Venezuela, has resulted in increased levels of MQ resistance and less AS-MQ sensitivity. Future molecular surveillance will be critical for determining whether the prevalence of pfmdr1 duplication has increased since the time of our study and whether it is associated with ACT resistance.
Supplementary Material
Acknowledgments
This work was financially supported in part by the Antimicrobial Drug Resistance Working Group, Centers for Disease Control and Prevention, Atlanta Research and Education Foundation, Atlanta VA Medical Center, and the U.S. Agency for International Development-supported Amazon Malaria Initiative. S. Griffing was supported by a National Science Foundation Graduate Research Fellowship. Luke Syphard and Sankar Sridaran were supported by Emerging Infectious Diseases fellowships from the Association of Public Health Laboratories. Andrea M. McCollum, Tonya Mixson-Hayden, and Sumiti Vinayak were supported by the Atlanta Research and Education Foundation. A. A. Escalante is supported by grant R01GM084320 from the National Institutes of Health.
Footnotes
Published ahead of print on 9 February 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Alker, A. P., P. Lim, R. Sem, N. K. Shah, P. Yi, D. M. Bouth, R. Tsuyuoka, J. D. Maguire, T. Fandeur, and F. Ariey.2007. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am. J. Trop. Med. Hyg. 76:641-647. [PubMed] [Google Scholar]
- 2.Anderson, T. J., B. Haubold, J. T. Williams, J. G. Estrada-Francos, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, and N. French.2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, T. J., and C. Roper.2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 94:269-280. [DOI] [PubMed] [Google Scholar]
- 4.Bacon, D. J., A. M. McCollum, S. M. Griffing, C. Salas, V. Soberon, M. Santolalla, R. Haley, P. Tsukayama, C. Lucas, A. A. Escalante, and V. Udhayakumar.2009. Dynamics of malaria drug resistance patterns in the Amazon basin region following changes in Peruvian national treatment policy for uncomplicated malaria. Antimicrob. Agents Chemother. 53:2042-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caraballo, A., and A. Rodriguez-Acosta.1999. Chemotherapy of malaria and resistance to antimalarial drugs in Guayana area, Venezuela. Am. J. Trop. Med. Hyg. 61:120-124. [DOI] [PubMed] [Google Scholar]
- 6.Contreras, C. E., J. F. Cortese, A. Carballo, and C. V. Plowe.2002. Genetics of drug-resistant Plasmodium falciparum malaria in the Venezuelan state of Bolivar. Am. J. Trop. Med. Hyg. 67:400-405. [DOI] [PubMed] [Google Scholar]
- 7.Cortese, J. F., A. Caraballo, C. E. Contreras, and C. V. Plowe.2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186:999-1006. [DOI] [PubMed] [Google Scholar]
- 8.Excoffier, L., G. Laval, and S. Schneider.2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinformatics 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt.2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. B. Ursos, A. B. Sidhu, B. Naudé, and K. W. Deitsch.2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foote, S. J., D. E. Kyle, R. K. Martin, A. M. J. Oduola, K. Forsyth, D. J. Kemp, and A. F. Cowman.1990. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255-258. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Martin, G.1972. Status of malaria eradication in the Americas. Am. J. Trop. Med. Hyg. 21:617-623. [DOI] [PubMed] [Google Scholar]
- 13.Godoy, G. A., G. Volcan, O. Marenco Reales, R. Guevara, and A. Texeira.1975. Demonstration of chloroquine diphosphate resistance in Plasmodium falciparum strains naturally infecting man in an area of the Bolivar State, Venezuela. Rev. Inst. Med. Trop. Sao Paulo 17:38-48. [PubMed] [Google Scholar]
- 14.Godoy, G. A., G. S. Volcan, R. Guevara, C. Medrano, J. Castro, and A. Texeira.1977. Venezuelan strains of Plasmodium falciparum resistant to sulfa and pyrimethamine as demonstrated by in vitro test. Rev. Latinoam. Microbiol. 19:229-231. [PubMed] [Google Scholar]
- 15.Hodel, E. M., J. Marfurt, D. Müller, A. Rippert, S. Borrmann, I. Müller, J. C. Reeder, P. Siba, B. Genton, and H. P. Beck.2008. Lack of multiple copies of pfmdr1 gene in Papua New Guinea. Trans. R. Soc. Trop. Med. Hyg. 102:1151-1153. [DOI] [PubMed] [Google Scholar]
- 16.Kublin, J. G., J. F. Cortese, E. M. Njunju, R. A. G. Mukadam, J. J. Wirima, P. N. Kazembe, A. A. Djimdé, B. Kouriba, T. E. Taylor, and C. V. Plowe.2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 187:1870-1875. [DOI] [PubMed] [Google Scholar]
- 17.Lim, P., A. P. Alker, N. Khim, N. K. Shah, S. Incardona, S. Doung, P. Yi, D. M. Bouth, C. Bouchier, and O. M. Puijalon.2009. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar. J. 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maberti, S.1960. Desarrollo de resistencia a la pyrimetamine, apresentación de 15 casos estudiados en Trujillo, Venezuela. Arch. Venez. Med. Trop. Parasitol. Med. 3:239-259. [PubMed] [Google Scholar]
- 19.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante.2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehlotra, R. K., H. Fujioka, P. D. Roepe, O. Janneh, L. Ursos, V. Jacobs-Lorena, D. T. McNamara, M. J. Bockarie, J. W. Kazura, and D. E. Kyle.2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc. Natl. Acad. Sci. U. S. A. 98:12689-12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehlotra, R. K., G. Mattera, M. J. Bockarie, J. D. Maguire, J. K. Baird, Y. D. Sharma, M. Alifrangis, G. Dorsey, P. J. Rosenthal, and D. J. Fryauff.2008. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 52:2212-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mockenhaupt, F. P.1995. Mefloquine resistance in Plasmodium falciparum. Parasitol. Today 11:248-253. [DOI] [PubMed] [Google Scholar]
- 23.Moore, D. V., and J. E. Lanier.1961. Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am. J. Trop. Med. Hyg. 10:5-9. [DOI] [PubMed] [Google Scholar]
- 24.Nair, S., D. Nash, D. Sudimack, A. Jaidee, M. Barends, A. Uhlemann, S. Krishna, F. Nosten, and T. J. Anderson.2007. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol. Biol. Evol. 24:562. [DOI] [PubMed] [Google Scholar]
- 25.Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay, P. N. Newton, J. P. Guthmann, F. M. Smithuis, T. T. Hien, N. J. White, F. Nosten, and T. J. Anderson.2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20:1526-1536. [DOI] [PubMed] [Google Scholar]
- 26.Nash, D., S. Nair, M. Mayxay, P. N. Newton, J. P. Guthmann, F. Nosten, and T. J. Anderson.2005. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc. R. Soc. Lond. B. Biol. Sci. 272:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, A. L., A. Purfield, P. McDaniel, N. Uthaimongkol, N. Buathong, S. Sriwichai, R. S. Miller, C. Wongsrichanalai, and S. R. Meshnick.2005. pfmdr1 genotyping and in vivo mefloquine resistance on the Thai-Myanmar border. Am. J. Trop. Med. Hyg. 72:586-592. [PubMed] [Google Scholar]
- 28.Noedl, H., Y. Se, K. Schaecher, B. L. Smith, D. Socheat, and M. M. Fukuda.2008. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359:2619-2620. [DOI] [PubMed] [Google Scholar]
- 29.Park, S. D. E.2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. dissertation. University of Dublin, Dublin, Ireland.
- 30.Peek, R., T. O. M. Van Gool, D. Panchoe, S. Greve, E. Bus, and L. Resida.2005. Drug resistance and genetic diversity of Plasmodium falciparum parasites from Suriname. Am. J. Trop. Med. Hyg. 73:833-838. [PubMed] [Google Scholar]
- 31.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. S. Miller, and S. R. Meshnick.2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plummer, W. B., L. M. P. Pereira, and C. V. F. Carrington.2004. pfcrt and pfmdr1 alleles associated with chloroquine resistance in Plasmodium falciparum from Guyana, South America. Mem. Inst. Oswaldo Cruz 99:389-392. [DOI] [PubMed] [Google Scholar]
- 33.Price, R. N., C. Cassar, A. Brockman, M. Duraisingh, M. Van Vugt, N. J. White, F. Nosten, and S. Krishna.1999. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 43:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, and N. J. White.2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purfield, A., A. Nelson, A. Laoboonchai, K. Congpuong, P. McDaniel, R. S. Miller, K. Welch, C. Wongsrichanalai, and S. R. Meshnick.2004. A new method for detection of pfmdr1 mutations in Plasmodium falciparum DNA using real-time PCR. Malar. J. 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond, M., and F. Rousset.1995. An exact test for population differentiation. Evolution 49:1280-1283. [DOI] [PubMed] [Google Scholar]
- 37.Roper, C., R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp.2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174-1181. [DOI] [PubMed] [Google Scholar]
- 38.Sankoh, A. J., M. F. Huque, and S. D. Dubey.1997. Some comments on frequently used multiple endpoint adjustment methods in clinical trials. Stat. Med. 16:2529-2542. [DOI] [PubMed] [Google Scholar]
- 39.Sidhu, A. B., A. Uhlemann, S. G. Valderramos, J. Valderramos, S. Krishna, and D. A. Fidock.2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidhu, A. B., S. G. Valderramos, and D. A. Fidock.2005. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913-926. [DOI] [PubMed] [Google Scholar]
- 41.Uhlemann, A. C., R. McGready, E. A. Ashley, A. Brockman, P. Singhasivanon, S. Krishna, N. J. White, F. Nosten, and R. N. Price.2007. Intrahost selection of Plasmodium falciparum pfmdr1 alleles after antimalarial treatment on the northwestern border of Thailand. J. Infect. Dis. 195:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urdaneta, L., A. Lal, C. Barnabe, B. Oury, I. Goldman, F. J. Ayala, and M. Tibayrenc.2001. Evidence for clonal propagation in natural isolates of Plasmodium falciparum from Venezuela. Proc. Natl. Acad. Sci. U. S. A. 98:6725-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valderramos, S. G., and D. A. Fidock.2006. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 27:594-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velásquez, A.1996. Fundamentos teoricos y epidimiologicos de un sistema de farmacovigilancia de la malaria en el estado Bolivar, Venezuela. Rev. Peru. Epidemol. 9:5-7. [Google Scholar]
- 45.Vieira, P. P., M. U. Ferreira, M. G. Alecrim, W. D. Alecrim, L. H. P. Silva, M. M. Sihuincha, D. A. Joy, J. B. Mu, X. Z. Su, and M. G. Zalis.2004. pfcrt polymorphism and the spread of chloroquine resistance in Plasmodium falciparum populations across the Amazon Basin. J. Infect. Dis. 190:417-424. [DOI] [PubMed] [Google Scholar]
- 46.Wang, X., J. Mu, G. Li, P. Chen, X. Guo, L. Fu, L. I. N. Chen, X. Su, and T. E. Wellems.2005. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am. J. Trop. Med. Hyg. 72:410-414. [PubMed] [Google Scholar]
- 47.Wellems, T. E., L. J. Panton, I. Y. Gluzman, V. E. Do Rosario, R. W. Gwadz, A. Walker-Jonah, and D. J. Krogstad.1990. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature 345:253-255. [DOI] [PubMed] [Google Scholar]
- 48.Wellems, T. E., and C. V. Plowe.2001. Chloroquine-resistant malaria. J. Infect. Dis. 184:770-776. [DOI] [PubMed] [Google Scholar]
- 49.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Su.2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320-323. [DOI] [PubMed] [Google Scholar]
- 50.Yuan, J., A. Reed, F. Chen, and C. Stewart.2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zalis, M. G.1998. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am. J. Trop. Med. Hyg. 58:630-637. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, Z., S. M. Griffing, A. M. de Oliveira, A. M. McCollum, W. M. Quezada, N. Arrospide, A. A. Escalante, and V. Udhayakumar.2008. Decline in sulfadoxine-pyrimethamine-resistant alleles after change in drug policy in the Amazon region of Peru. Antimicrob. Agents Chemother. 52:739-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.