Abstract
We have defined a sensitivity profile for 22 antibiotics by extending previous work testing the entire KEIO collection of close to 4,000 single-gene knockouts in Escherichia coli for increased susceptibility to 1 of 14 different antibiotics (ciprofloxacin, rifampin [rifampicin], vancomycin, ampicillin, sulfamethoxazole, gentamicin, metronidazole, streptomycin, fusidic acid, tetracycline, chloramphenicol, nitrofurantoin, erythromycin, and triclosan). We screened one or more subinhibitory concentrations of each antibiotic, generating more than 80,000 data points and allowing a reduction of the entire collection to a set of 283 strains that display significantly increased sensitivity to at least one of the antibiotics. We used this reduced set of strains to determine a profile for eight additional antibiotics (spectinomycin, cephradine, aztreonem, colistin, neomycin, enoxacin, tobramycin, and cefoxitin). The profiles for the 22 antibiotics represent a growing catalog of sensitivity fingerprints that can be separated into two components, multidrug-resistant mutants and those mutants that confer relatively specific sensitivity to the antibiotic or type of antibiotic tested. The latter group can be represented by a set of 20 to 60 strains that can be used for the rapid typing of antibiotics by generating a virtual bar code readout of the specific sensitivities. Taken together, these data reveal the complexity of intrinsic resistance and provide additional targets for the design of codrugs (or combinations of drugs) that potentiate existing antibiotics.
The proliferation of antibiotic-resistant bacteria has reached the point where close to 100,000 infections occur in the United States per year that are caused by methicillin-resistant Staphylococcus aureus (MRSA), leading to 18,650 deaths in 2005 (44), a total that is greater than the 17,011 deaths the same year from HIV/AIDS according to the CDC (http://www.cdc.gov/hiv/topics/surveillance/resources/reports/2005report/table7.htm). The spread of resistance (2, 21, 51) has long stimulated efforts to find new antibiotics by a variety of methods, such as altering existing antibiotics, screening chemical (18) or peptide (32, 43) libraries for specific inhibitors (18), or targeting new proteins or processes (11, 32, 35, 47, 58). Recent efforts also have involved detecting new targets through genomics (66, 67), such as bioinformatics screening for novel producers of peptide antimicrobials (8) and finding new sources of antibiotics via metagenomics (68). Some authors have suggested that discovering new molecular scaffolds for antibiotics should be a priority (29). Another approach involves potentiating existing antibiotics by identifying targets for increasing susceptibility to specific antimicrobials (for a review, see reference 19), such as the AmgRS-mediated stress response to the action of aminoglycosides in Pseudomonas aeruginosa (49), or bacterial nitric oxide synthases (33). This has been achieved clinically in the case of inhibitors of β-lactamase with β-lactam antibiotics (see the review by Buynak [15]). A related example is the imipenem-cilastatin combination (36, 38). Cilastatin is an inhibitor of DHP-1, a human renal dehydropeptidase-1 that hydrolyzes the β-lactam imipenem (36). In the laboratory, inhibitors of efflux pumps have been used together with tetracycline in Escherichia coli (63) and levofloxacin in Pseudomonas aeruginosa (56), but these have not been used successfully in a clinical setting. Tools now are available for more extensive systematic searches for targets for such potentiating codrugs. A transposon library has been used to detect genes responsible for increased sensitivity to one of a set of antibiotics in Acinetobacter baylyi (31), comprehensive transposon mutant libraries of Pseudomonas aeruginosa (41, 54) have been used to screen for sensitivity to tobramycin (49) and ciprofloxacin (13), and a somewhat less-defined transposon library has been used to detect sensitivity to one of a set of six antibiotics (28). A comprehensive transposon library also has been constructed for Francisella novicida (30). In yeast, a deletion library has been screened against a set of four DNA-damaging agents (74) and a set of more than 400 small molecules (37). In the work reported here, we have expanded our previous work (73) and used the high-throughput screening of an Escherichia coli knockout collection of close to 4,000 strains, each with a different gene inactivated (6), to look for mutants that are more susceptible to 1 of 14 different antibiotics (the antibiotics and their abbreviations are listed in Table 1). We then used a reduced set of 283 identified strains to examine an additional eight antibiotics with the intent of finding a core set of tester strains. Many mutants are more susceptible to a wide range of antibiotics and represent the basic intrinsic resistance framework, but others are more specific and allow us to define so-called bar codes for the rapid typing of antibiotics with different target activities. These results also help to define new combinational drug targets, as most of the mutants do not result in growth inhibition in the absence of subinhibitory or sublethal concentrations of an antibiotic. Moreover, combining mutations with increased sensitivities would allow the design of more precise tests for the persistence of antibiotics in foods, milk, and various wastewaters.
TABLE 1.
List of antibiotics
| Antibiotic | Category | Primary target | Process affected |
|---|---|---|---|
| Ciprofloxacin (CIP) | Fluoroquinolone | DNA gyrase | DNA replication |
| Enoxacin (ENX) | |||
| Nitrofurantoin (NIT) | Nitrofuran | DNA | DNA |
| Metronidazole (MTR) | Nitroimidazole | DNA | DNA structure |
| Sulfamethoxazole (SFX) | Sulfonamide | Dihydropteroate synthetase | Folate synthesis |
| Rifampin (RIF) | Rifamycin | RNA polymerase | Transcription |
| Gentamicin (GEN) | Aminoglycoside | 30S subunit | Protein synthesis |
| Tobramycin (TOB) | |||
| Neomycin (NEO) | |||
| Streptomycin (STR) | |||
| Spectinomycin (SPT) | Aminocyclitol | 30S subunit | Protein synthesis |
| Tetracycline (TET) | Polyketide | 30S subunit | Protein synthesis |
| Vancomycin (VAN) | Glycopeptide | NAM/NAG peptides | Cell wall synthesis |
| Ampicillin (AMP) | Penicillin | Transpeptidase | Cell wall synthesis |
| Cephradine (RAD) | Cephalosporin | Transpeptidase | Cell wall synthesis |
| Cefoxitin (FOX) | |||
| Azetreonam (ATM) | Monobactam | Transpeptidase | Cell wall synthesis |
| Colistin (CST) | Polymyxin | Cytoplasmic membrane | Cell wall permeability |
| Chloramphenicol (CHL) | Phenicol | 50S subunit | Protein synthesis |
| Erythromycin (ERY) | Macrolide | 50S subunit | Protein synthesis |
| Fusidic acid (FUS) | Fusidic acid | Elongation factor G | Protein synthesis |
| Triclosan (TRI) | Chlorophenol | Enoyl-acyl carrier protein reductase | Fatty acid synthesis |
MATERIALS AND METHODS
E. coli strains.
The KEIO collection was described in Baba et al. (6) and made from the starting strain BW25113 (20). This strain (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) is the starting strain used in the experiments reported here unless otherwise stated.
E. coli genetic methods.
Unless otherwise stated, all genetic methods are as described by Miller (62).
Use of the Deutz cryoreplicator.
The Deutz cryoreplicator (23) contains 96 prongs on individual springs, allowing its frequent application to frozen glycerol cultures. The KEIO collection (4) is maintained on 45 96-well microtiter plates and stored at −80°C in glycerol. Material from frozen microtiter plates was transferred to microtiter wells with 0.5 ml of LB and were incubated overnight, and then the replicator was used to transfer a microdrop to microtiter wells with fresh LB medium containing 50 μg/ml kanamycin to prevent the growth of contaminants. (All of the strains in the KEIO collection are Kanr). After 3 to 4 h of growth, these plates were printed onto LB plates with different concentrations of different antibiotics. For the initial screening, kanamycin was present in the plates, but for retests and MIC determinations kanamycin was not present.
Determination of MICs.
MICs (5) were determined by printing, with the Deutz replicator, approximately 2 × 105 to 4 × 105 CFU to an LB plate with the appropriate concentration of the desired antibiotic and then examining the spots after overnight incubation at 37°C. (This represents a minor deviation from the standard method in that LB medium was used and that somewhat more cells were applied to each spot.) All determinations were carried out on strain sets that were prepared from purified colonies. The tests were repeated up to six times in some cases and at various concentrations (see the supplemental material). The antibiotics used in the Tamae et al. study (73) were retested along with the new antibiotics, using additional concentrations and the somewhat different conditions, to standardize scoring. The concentrations reported for compounds such as the aminoglycosides might be different in the LB medium used here from those in other media, although we did not find reproducible differences when NaCl was left out of the LB medium in the two cases tested (tobramycin and gentamicin). Cross-resistance between some of the aminoglycosides, particularly neomycin and the kanamycin resistance marker in the strains used, might alter the concentrations that are effective, but we found this to be the case only for neomycin. We found prominent patterns for neomycin at the concentrations reported.
Validation of the data set.
A collection of 4,000 strains will contain some errors and some impure strains. The latter problem can be minimized by repurifying and retesting, as was done here. Also, in a number of cases, strains pick up suppressors that will attenuate the effects of the deletion knockouts. We have reisolated several strains to eliminate this effect. Thus, the copies of strains with oxyR and recA are somewhat different from those reported previously (73). Mori and coworkers (75) have subjected the KEIO collection to an intensive analysis aimed at uncovering errors in the collection that might arise from duplications of the target gene. They have generated a list of 14 mutants that are incorrect and another 9 that might be incorrect. We therefore have removed coaE, glmM, parC, prfB, polA, rpoD, and rpsU from our data sets. Ultimately, the most prudent use of such a large collection is to verify any mutants that are particularly important to the final results by PCR analysis and/or sequencing. We examined in detail some of the more important assignments. We took an expanded set of 12 of the mutants from the bar codes that are used to identify specific antibiotics (Table 2) (recF, argO, degP, dacA, glpD, trxB, xseA, sapC, rplI, tufA, ycdZ, and dinB), as well as recA, oxyR, flgB, ppiD, and rpsF. Using two primer pairs for each mutant, we verified in two PCR experiments for each that each carried a kan insert in place of the correct gene and that there was no presence of the original gene sequence elsewhere (via a duplication). In the case of rpsF, we carried out both PCR and sequencing. Moreover, an additional two from Table 2 can be identified by their phenotypes (dam, mutator; lon, mucoid formation). This constitutes a verification of 19 mutants, most of which are key indicators for specific antibiotics. We had reason to suspect the recF deletion mutant, and a similar analysis revealed that the recF deletion strain in the first copy of the KEIO strains is incorrect, although the recF strain in the second is correct, and this has been used in the work reported here. Regarding further validation, we have detected all four of the genes found through biochemical experiments that, when mutated, render the cell more sensitive to CPR (25) and all six of the genes for NIT (64).
TABLE 2.
Diagnostic gene knockout strains for specific antibiotics
| Antibiotic | Strain |
|---|---|
| CIP-ENX | xseA |
| NIT | recO |
| MTR | gshA (recO) |
| SFX | degP |
| RIF | trxB |
| Aminoglycosides | sapC ompF |
| TET | ycdZ |
| VAN | tufA |
| β-lactams | dacA |
| CST | vacJ |
| CAM | argO |
| FUS | rplI |
| TRI | glpD dnaJ |
Chemicals.
Kanamycin, tetracycline, chloramphenicol, rifampin, vancomycin, sulfamethoxazole, gentamicin, ampicillin, spectinomycin, cephradine, colistin, streptomycin, aztreonam, erythromycin, fusidic acid, metronidazole, nitrofurantoin, neomycin, enoxacin, tobramycin, triclosan, and cefoxitin were purchased from Sigma (St. Louis, MO). Ciprofloxacin was purchased from ICN Biomedicals, Inc., Aurora, OH.
RESULTS AND DISCUSSION
Screening for antibiotic hypersensitivity.
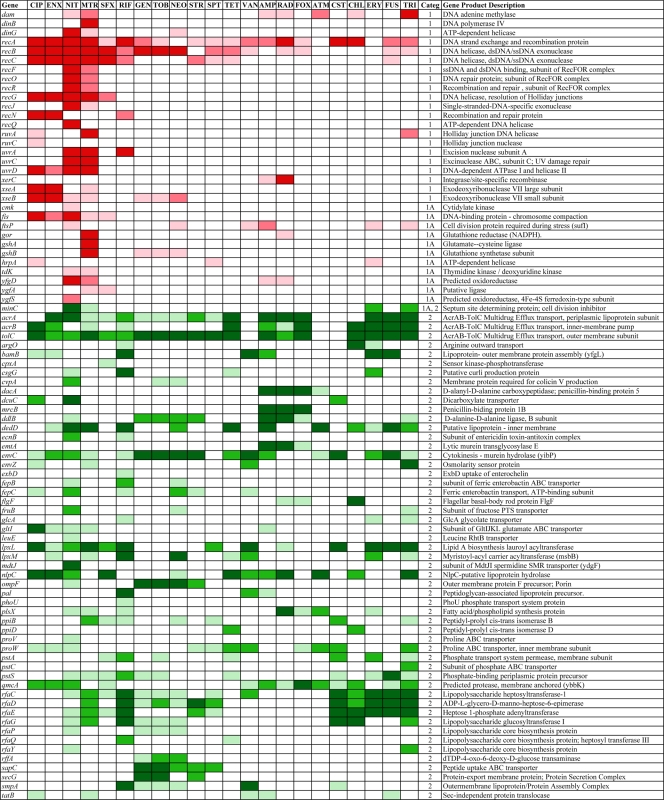
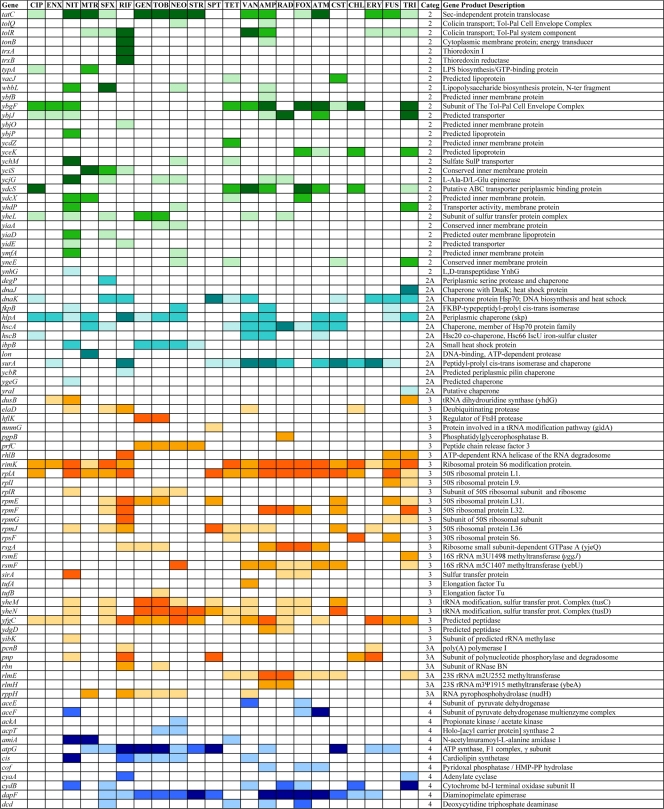
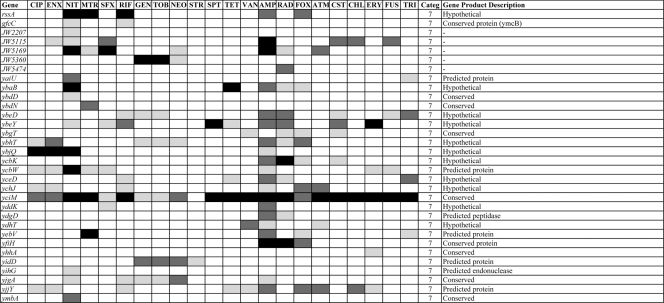
The entire KEIO collection of close to 4,000 strains (6) was screened for mutants that are more sensitive than the wild type to one or more of a set of 14 different antibiotics (Table 1). We first examined CIP, VAN, RIF, AMP, SFX, GEN, and MTR at several different subinhibitory concentrations (73), and we have expanded this to include an additional set of seven compounds (streptomycin, STR; fusidic acid, FUS; tetracycline, TET; chloramphenicol, CHL; nitrofurantoin, NIT; triclosan, TRI; and erythromycin, ERY; see Materials and Methods for details). This generated an initial set of close to 80,000 data points. Mutants showing increased susceptibility then were purified and retested, resulting in a set of 283 strains that displayed significantly increased susceptibility to at least 1 of the 14 antibiotics tested. We then used this set of 283 strains to test an additional eight antibiotics (spectinomycin, SPT; cephradine, RAD; aztreonem, ATM; colistin, CST; neomycin, NEO; enoxacin, ENX; tobramycin, TOB; and cefoxitin, FOX) (Table 1), as well as additional concentrations of the original 14 antibiotics, to show that a sensitivity profile can be determined from this smaller set of strains. Figures 1 and 2 give a broad overview of the data (for quantitative details, see the supplemental material) in which the strongest sensitivities are indicated in darker colors and sensitivities that are less strong are indicated in lighter colors, with different colors and numbers indicating different functions (Fig. 1). The functions were categorized and assigned numbers as follows: 1, DNA recombination, replication, and repair; 1A, DNA related; 2, permeation, membranes, and transport; 2A, chaperoning; 3, protein synthesis; 3A, RNA processing; 4, general metabolic reactions; 5, transcriptional control; 6, prophage encoded; and 7, unassigned functions. The intensity is shown in three levels. The darkest level for each group represents the strongest sensitivities. Many different types of functions are involved, including those concerned with DNA replication, recombination, and repair that are prevalent among the sensitivity profiles for agents that interact with DNA, as well as functions involved in the cell wall and cell membrane, chaperoning, protein synthesis, and general metabolism. Also, close to 30 genes encoding transcriptional regulators appear in Fig. 1. Figure 2 groups all of the detected susceptible mutants deleted for as-yet unassigned genes. In addition to providing a wealth of information on different functions involved in intrinsic resistance, this set of 283 strains can be used to generate a susceptibility fingerprint for antibiotics. For example, note how clearly the two fluoroquinolones CIP and ENX give very similar profiles. However, as detailed below, one set of mutants in Fig. 1 and 2 is defective in functions involved in multidrug resistance, as the resulting phenotype includes increased sensitivity to a range of different antibiotics, while another set of hypersensitive mutants are partially or completely specific for each antibiotic or class of antibiotic. From the data shown in Fig. 1, it is possible to predict whether an antibiotic primarily affects DNA-related functions or not, but it is necessary to separate the different components of the strain set to enable the typing of antibiotics with a simpler set of mutants displaying more specific patterns.
FIG. 1.
Strains with sensitivity to one or more of the 22 antibiotics with three levels of intensities: stronger susceptibilities are in darker shades, medium susceptibilities are in lighter shades, and weak susceptibilities are in the lightest shade. The categories (categ.) are the following: 1, DNA replication, recombination, and repair; 1A, functions indirectly affecting category 1; 2, transport, efflux, cell wall, and cell membrane synthesis; 2A, chaperones and functions related to category 2; 3, protein synthesis; 3A, RNA processing; 4, central metabolic reactions; 5, regulation; and 6, prophage-carried genes and cell adhesion.
FIG. 2.
Hypothetical strains with sensitivity to one or more of the 22 antibiotics. All belong to category 7 for unassigned gene products.
Multidrug-resistant functions.
Many of the 283 mutants shown in Fig. 1 and 2 display hypersensitivity to more than one antibiotic. We have selected those that show increased sensitivity to at least 8 of the 22 antibiotics tested (36%), as shown in Fig. 3: a total of 61 mutants, including genes involved in recombination and recombinational repair of double-strand breaks (recABC), the main efflux pump in E. coli (acrA, acrB, tolC), those involved in cell wall and cell membrane synthesis and integrity, and transporters and chaperones. Note that yciM mutants (Fig. 2) are more sensitive to 21 of the 22 antibiotics tested and tolC mutants to 19 (Fig. 1). Several genes encoding ribosomal proteins (rplA, rpmE, rpmJ) or their modification (rimK) confer intrinsic resistance to a varied set of antibiotics, as their deletion renders the cell more sensitive than the wild type. Figure 3 includes genes encoding partially (ybgF, yfgC) or completely (yciM, ybeY, ycbW, yjjY) uncharacterized functions. Interestingly, some genes encode proteins that are involved in the regulation of gene expression (dksA, fur, hfq, hns, mfd, nusB, rseA, xapR, yciT). In some (and perhaps all) cases, these regulators affect functions that already appear in Fig. 1, such as the DksA transcription factor that regulates genes involved in double-strand break repair (61), in addition to multiple target genes associate with many processes (65). The regulatory genes operate in different ways, such as Hfq, which binds many small noncoding RNAs and is involved in posttranscriptional gene regulation (50), or HNS, a DNA-binding protein involved in DNA topology and compaction, which plays a role in the regulation of many unrelated genes (70 and references therein), or RseA, an anti-sigma factor that inhibits the transcriptional activity of σE (1, 72). Additionally, rfaH mutants lacking an antitermination factor (7, 9) are more sensitive to six antibiotics (Fig. 1).
FIG. 3.
Sixty-one strains with sensitivity to eight or more of the 22 antibiotics.
Specific sensitivities.
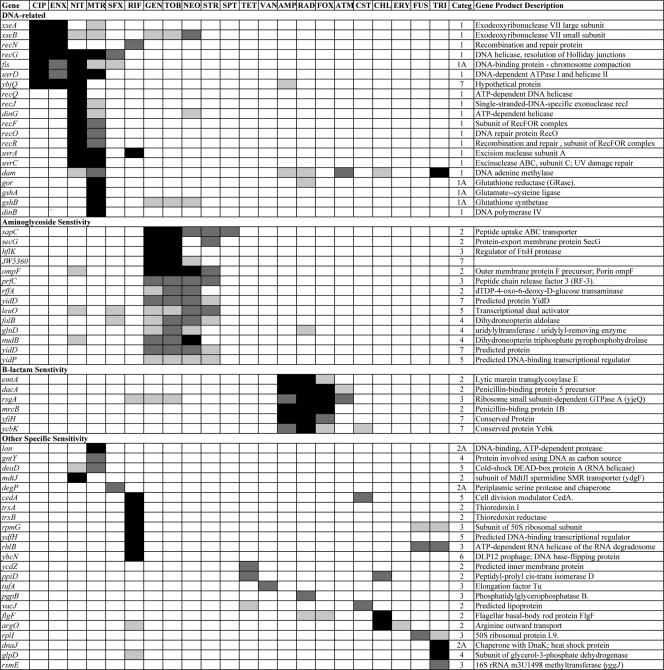
While the data in Fig. 1 and 2 portray a sensitivity profile that is diagnostic for each antibiotic, the charts are complex. However, we can simplify this by selecting only those mutants that display more specific sensitivities to a particular antibiotic or class of antibiotic. Figure 4 shows the results from the latter set of mutants and displays a more useful set of mutants for typing antibiotics (also see the following sections). This selected set or subsets can be used to generate a bar code signature for each antibiotic; as more antimicrobial compounds are tested, they can be added. From Fig. 3 and 4, we can see both general and specific sensitivities within each functional category. For example, with regard to the sensitivities of knockouts of genes related to recombination and repair, the recABC genes appear general (Fig. 3), but many genes in the RecF pathway, as well as those involved in other DNA-related functions, are more specific. One should note that the RecBCD pathway carries out the repair of double-strand DNA breaks, while the RecF pathway is involved principally in the repair of single-strand breaks and gaps (4, 17, 54, 55, 71). The RecF pathway mutants recF, recO, recR, recQ, and recJ, along with dinG and uvrC mutants, are specifically sensitive to NIT and, usually to a lesser extent, MTR, the only two agents that are converted to forms that directly damage DNA. Among these, recQ mutants are absolutely specific for NIT. Also, xseA gene knockouts, lacking an exonuclease, exo VII large subunit (16), and, to a somewhat lesser extent, xseB knockouts, are particularly sensitive to fluoroquinolones. Interestingly, mutants lacking the glutathione reductase system are fairly specific in their hypersensitivity to MTR. However, mutants lacking the thioredoxin/thioredoxin reductase system (trxA, trxB) are sensitive specifically to RIF.
FIG. 4.
Strains with sensitivities unique to a particular class of antibiotics and strains with specific sensitivities.
Antibiotic identification.
We can designate a minimal set of 13 to 15 strains that allowed the identification of almost all of the antibiotics used here, as depicted in Table 2. In some cases, further testing with other mutants from Fig. 4 can distinguish between members of general groups (fluoroquinolones, aminoglycosides, and β-lactams). Of course, other combinations of strains are possible for a list of this type. The one exception is ERY, for which no single strain is specifically sensitive. However, one can use strains such as pnp or rep, which are hypersensitive to ERY, and then identify the other possibilities, such as RIF, with strains diagnostic for RIF.
Targets for codrugs or potentiators.
Considering the studies reported here and elsewhere (13, 28, 31, 37, 73), can we use this type of information to design codrugs that potentiate existing antibiotics? The data in Fig. 1 to 4 offer an expanded set of targets for finding compounds that act by inactivating proteins that provide intrinsic resistance and, thus, could be used in combinational therapy with existing antibiotics. There is an emerging emphasis on considering combinational strategies (19), of which there are several types. One category includes combinations of antibiotics that are either in the same pathway (e.g., cotrimoxazole [14]) or in different pathways (e.g., isoniazid plus rifampin, pyrazinamide, and streptomycin for the treatment of tuberculosis; http://www.cdc.gov; also see reference 19) to yield a more effective treatment and often to overcome resistance to a single drug. Recently, Kishony and coworkers have described the functional classification of drugs according to their pairwise interactions (76). An additional class is represented by synercid (quinupriostin-dalfopristin), a combination of two bacteriostatic drugs that together produce a bactericidal effect (3). A third category comprises drugs used with a codrug that inhibits a resistance pathway, as has been used clinically (15, 36, 38) and in the laboratory (56, 63). Additional targets for codrugs (either small molecules or engineered bacteriophages [57]) involve different pathways of intrinsic resistance or persister cell formation or maintenance (52, 53). Of particular interest in the work reported here are targets for potentiators that involve specific responses to a particular antibiotic or class of antibiotic; focusing on these might yield a useful set of compounds to be used with specific antibiotics. These include the following.
(i) Rifampin.
Finding inhibitors of the trxA- or trxB-encoded enzymes (thioredoxin 1 and thioredoxin reductase, respectively) (10, 22), the rhlB-encoded RNA helicase, or the coaE-encoded dephospho-coenzyme A kinase would sensitize cells to lower levels of RIF or increase the potency of RIF at higher levels.
(ii) Triclosan.
TRI works by specifically inhibiting the fabI gene product that catalyzes a key step in fatty acid biosynthesis (60). Mutants with deletions of the dam gene lack adenine methylase and are almost uniquely sensitive to TRI. Mashhoon, Reich, and coworkers have identified a number of compounds that specifically inhibit bacterial methylases, including the Dam methylase (58, 59). Using these inhibitors in the presence of TRI on the wild-type strain would be equivalent to using a dam mutant strain with TRI and, thus, could be an effective drug combination. Because glpD mutants are specifically inhibited by TRI, the inhibitor of the GlpD enzyme, glycerol-3-phosphate dehydrogenase, also would be an effective codrug for TRI. Interestingly, Lewis has shown that GlpD is a key protein involved in the generation of persister cells in the population (52, 53).
(iii) Nitrofurantoin.
An inhibitor of the RecQ helicase, a homolog of the human helicase lacking in Bloom's Syndrome patients (27), would be an effective and specific codrug for NIT. Yet another codrug candidate would be an inhibitor of the MdtJI spermidine SMR transporter.
(iv) Aminoglycosides.
There are numerous functions that are specifically involved in protecting the cell from aminoglycosides (Fig. 4), and perhaps SapC and SecG are the best targets. (SecG has been suggested as a target for potentiators of aminoglycosides in experiments by others [46]).
(v) β-Lactams.
Inhibitors of some of the penicillin binding proteins pinpointed in Fig. 4 (e.g., the dacA- and mrcB-encoded functions) would potentiate β-lactams.
Some of the functions identified in Fig. 1 and 2 represent pathways that are not directly connected to the presumed primary target of some of the antibiotics and reflect the complexity of antibiotic action (34). Studying some of these pathways in depth should provide additional insights regarding antibiotic action. For instance, Collins and coworkers have argued that the generation of hydroxyl radicals leading to double-strand breaks is a major contributor to cell death for bactericidal antibiotics (26, 45, 46). From Fig. 1 it is evident that a number of strains lacking some aspect of the RecBCD system involved in the recombinational repair of double-strand breaks (recA, recB, or recC) are more sensitive to many antibiotics, and mildly increased sensitivity to some of the four aminoglycosides tested is exhibited by mutants lacking the ability to respond to DNA damage or damaging agents (e.g., dinG, xseB, gshB). We can use the specific sensitivity profiles defined here to type new antibiotics with significantly less effort than that for microarray studies, particularly if we employ the reduced set of strains shown in Fig. 4 or in Table 2. In principle, these data complement the data from microarrays (12, 40, 42, 69), since each method has a different, although not totally independent, basis for scoring. Microarrays measure the change in gene expression in response to a subinhibitory concentration of an antibiotic, whereas the profiles such as those in Fig. 1 to 4 measure specific phenotypes. Moreover, some of the strains shown in Fig. 4 can be used in the initial screening for specific antibiotics. Previous workers have used engineered strains to aid in the detection of antibiotics from soil isolates (24, 39).
As noted above, the data in this work further define the intrinsic resistome, as has work with other microorganisms (13, 28, 31, 37); as we dismantle the cell's intrinsic protection, the cell becomes more sensitive. We have shown that certain double mutants are even more sensitive than their parents alone (73). We can apply the knowledge of the property of double and triple mutants to construct improved strains that are useful for detecting the presence of antibiotics in the environment, such as in milk or hospital wastewater, as is done with the Delvotest SP-NT and the Copan milk test (48), and have already developed some preliminary tests.
Supplementary Material
Acknowledgments
We thank Julian Davies and Lynn Silver for helpful discussions and comments on the manuscript.
E.B. was supported by the Whitcome Fellowship. This work was supported by a grant from the National Institutes of Health (ES0110875).
Footnotes
Published ahead of print on 11 January 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross.2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy.2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037-1050. [DOI] [PubMed] [Google Scholar]
- 3.Allington, D. R., and M. P. Rivey.2001. Quinupristin/dalfopristin: a therapeutic review. Clin. Ther. 23:24-44. [DOI] [PubMed] [Google Scholar]
- 4.Amundsen, S. K., and G. R. Smith.2003. Interchangeable parts of the Escherichia coli recombinational machinery. Cell 112:741-744. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, J. M.2001. Determination of minimum inhibitory concentrations. Antimicrob. Agents Chemother. 48:5-16. [DOI] [PubMed] [Google Scholar]
- 6.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori.2006. Construction of Escherichia coli K-12 in-frame single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey, M. J. A., C. Hughes, and V. Koronakis.1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 8.Begley, M., P. D. Cotter, C. Hill, and R. P. Ross.2009. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 75:5451-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beloin, C., K. Michaelis, K. Lindner, P. Landini, J. Hacker, J.-M. Ghigo, and U. Dobrindt.2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjur, E., S. Eriksson-Ygberg, F. Aslund, and M. Rhen.2006. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 74:5140-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowser, T. E., V. J. Bartlett, M. C. Grier, A. K. Verma, T. Warchol, S. B. Levy, and M. N. Alekshun.2007. Novel anti-infection agents: small-molecule inhibitors of bacterial transcription factors. Bioorg. Med. Chem. Lett. 17:5652-5655. [DOI] [PubMed] [Google Scholar]
- 12.Brazas, M. D., and R. E. W. Hancock.2005. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 10:1245-1252. [DOI] [PubMed] [Google Scholar]
- 13.Breidenstein, E. B., B. K. Khaira, I. Wiegand, J. Overhage, and R. E. Hancock.2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brumfitt, W., and J. M. Hamilton-Miller.1994. Limitations of and indications for the use of co-trimoxazole. J. Chemother. 6:3-11. [DOI] [PubMed] [Google Scholar]
- 15.Buynak, J. D.2006. Understanding the longevity of the β-lactam antibiotics and of antibiotic/β-lactamase inhibitor combinations. Biochem. Pharmacol. 71:930-940. [DOI] [PubMed] [Google Scholar]
- 16.Chase, J. W., B. A. Rabin, J. B. Murphy, K. L. Stone, and K. R. Williams.1986. Escherichia coli exonuclease VII. Cloning and sequencing of the gene encoding the large subunit (xseA). J. Biol. Chem. 261:14929-14935. [PubMed] [Google Scholar]
- 17.Clark, A. J., and K. B. Low.1988. Pathways and systems of homologous recombination in Escherichia coli, p. 155-215. In K. B. Low (ed.), The recombination of genetic material. Academic Press, San Diego, CA.
- 18.Costi, M. P., A. Gelanin, D. Barlocco, S. Ghelli, F. Soragni, F. Reniero, T. Rossi, A. Ruberto, C. Guillou, A. Cavazzuti, C. Casolari, and S. Ferrari.2006. Antibacterial agent discovery using thymidylate synthase biolibrary screening. J. Med. Chem. 49:5958-5968. [DOI] [PubMed] [Google Scholar]
- 19.Cottarel, G., and J. Wierzbowski.2007. Combination drugs, an emerging option for antibacterial therapy. Trends Biotech. 25:547-555. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner.2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies, J.2007. Microbes have the last word. EMBO Rep. 8:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debarbieux, L., and J. Beckwith.1998. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc. Natl. Acad. Sci. USA 95:10751-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deutz, W. A., L. Rüedi, R. Hermann, K. O'Connor, J. Büchs, and B. Witholt.2000. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 66:2641-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeVito, J. A., J. A. Mills, V. G. Liu, A. Agarwal, C. F. Sizemore, Z. Yao, D. M. Stoughton, M. G. Cappiello. M. D. Barbosa, L. A. Foster, and D. L. Pompliano.2002. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 20:478-483. [DOI] [PubMed] [Google Scholar]
- 25.Drlica, K., and X. Zhao.1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwyer, D. J., M. A. Kohanski, and J. J. Collins.2009. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 12:482-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German.1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 28.Fajardo, A., N. Martinez-Martin, M. Mercadillo, J. C. Galan, B. Ghysels, S. Matthijs, P. Cornelis, L. Wiehlmann, B. Tuemmler, F. Baquero, and J. L. Martinez.2008. The neglected intrinsic resistome of bacterial pathogens. PloS One 3:e1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischbach, M. A., and C. T. Walsh.2009. Antibiotics for emerging pathogens. Science 325:1089-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher, L. A., E. Ramage, M. A. Jacobs, R. Kaul, M. Brittnacher, and C. Manoil.2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. USA 104:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez, M. J., and A. A. Neyfakh.2006. Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob. Agents Chemother. 50:3562-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunderson, C. W., and A. M. Segall.2006. DNA repair, a novel antibacterial target: Holliday junction-trapping peptides induce DNA damage and chromosome segregation defects. Mol. Microbiol. 59:1129-1148. [DOI] [PubMed] [Google Scholar]
- 33.Gusarov, I., K. Shatalin, M. Starodubtseva, and E. Nudler.2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hancock, R. E. W.2007. The complexities of antibiotic action. Mol. Syst. Biol. 3:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins, D. A., M. E. Pomianek, C. M. Cramel, R. K. Taylor, M. F. Semmelhack, and B. L. Bassler.2007. The major Vibrio cholerae autoindicer and its role in virulence factor production. Nature 450:883-886. [DOI] [PubMed] [Google Scholar]
- 36.Hikida, M., K. Kawashima, M. Yoshida, and S. Mitsuhashi.1992. Inactivation of new carbapenem antibiotics by dehydropeptiase-1 from porcine and human renal cortex. J. Antimicrob. Chemother. 30:129-134. [DOI] [PubMed] [Google Scholar]
- 37.Hillenmeyer, M. E., E. Fung, J. Wildenhain, S. E. Pierce, S. Hoon, W. Lee, M. Proctor, R. P. St. Onge, M. Tyers, D. Koller, R. B. Altman, R. W. Davis, C. Nislow, and G. Giaever.2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman, J., J. Trimble, and G. M. Brophy.2009. Safety of imipenem/cilastatin in neurocritical care patients. Neurocrit. Care 10:403-407. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh, P., S. A. Siegel, B. Rogers, D. Davis, and K. Lewis.1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. USA 95:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer.2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, R. C. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil.2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaldalu, N., R. Mei, and K. Lewis.2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kepple, K. V., J. L. Boldt, and A. M. Segall.2005. Holliday junction-binding peptides inhibit distinct junction-processing enzymes. Proc. Natl. Acad. Sci. USA 102:6867-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin.2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 45.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins.2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 46.Kohanski, M. A., D. J. Dwyer, J. Wierzbowski, G. Cottarel, and J. J. Collins.2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuijl, C., N. D. L. Savage, M. Marsman, A. W. Tuin, L. Janssen, D. A. Egan, M. Ketema, R. van den Nieuwendijk, S. J. F. van den Eeden, A. Geluk, A. Poot, G. van der Marel, R. L. Beijersbergen, H. Overkleeft, T. H. M. Ottenhoff, and J. Neefjes.2007. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 450:725-730. [DOI] [PubMed] [Google Scholar]
- 48.Le Breton, M.-H., M.-C. Savoy-Perroud, and J.-M. Diserens.2007. Validation and comparison of the Copan milk test and Delvotest SP-NT for the detection of antimicrobials in milk. Anal. Chim. Acta 586:280-283. [DOI] [PubMed] [Google Scholar]
- 49.Lee, S., A. Hinz, E. Bauerle, A. Angermeyer, K. Juhasova, Y. Kaneko, P. K. Singh, and C. Manoil.2009. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. USA 106:14570-14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, T., and A. L. Feig.2008. The RNA binding protein Hfq interacts specifically with tRNAs. RNA 14:514-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levy, S. B., and B. Marshall.2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 52.Lewis, K.2007. Persister cells, dormancy, and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 53.Lewis, K.2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107-131. [DOI] [PubMed] [Google Scholar]
- 54.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel.2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lloyd, R. G., and K. B. Low.1996. Homologous recombination, p. 2236-2255. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 56.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, H. Ishida, and V. J. Lee.2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu, T. K., and J. J. Collins.2009. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. USA 106:4629-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mashhoon, N., M. Carroll, C. Pruss, J. Eberhard, S. Ishikawa, R. A. Estabrook, and N. Reich.2004. Functional characterization of Escherichia coli DNA adenine methyltransferase, a novel target for antibiotics. J. Biol. Chem. 279:52075-52081. [DOI] [PubMed] [Google Scholar]
- 59.Mashhoon, N., C. Pruss, M. Carroll, P. H. Johnson, and N. O. Reich.2006. Selective inhibitors of bacterial DNA adenine methyltransferase. J. Biomol. Screen. 11:497-510. [DOI] [PubMed] [Google Scholar]
- 60.McMurry, L. M., M. Oethinger, and S. B. Levy.1998. Triclosan targets lipid synthesis. Nature 394:531-532. [DOI] [PubMed] [Google Scholar]
- 61.Meddows, T. R., A. P. Savory, J. I. Grove, T. Moore, and R. G. Lloyd.2005. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol. Microbiol. 57:97-110. [DOI] [PubMed] [Google Scholar]
- 62.Miller, J. H.1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, p. 194-195. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 63.Nelson, M. L., and S. B. Levy.1999. Reversal of tetracycline resistance by different bacterial tetracycline resistance determinants by an inhibitor of the Tet(B) antiport protein. Antimicrob. Agents Chemother. 43:1719-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ona, K. R., C. T. Courcelle, and J. Courcelle.2009. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J. Bacteriol. 191:4959-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paul, B. J., M. B. Berkmen, and R. L. Gourse.2005. DksA potentiates direct activation of amino acid promoters by ppGpp. J. Bacteriol. 102:7823-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Payne, D. J., M. N. Gwynn, D. J. Holmes, and M. Rosenberg.2004. Genomic approaches to antibacterial discovery. Methods Mol. Biol. 266:231-259. [DOI] [PubMed] [Google Scholar]
- 67.Pucci, M. J.2007. Novel genetic techniques and approaches in the microbial genomics era: identification and/or validation of targets for the discovery of new antibacterial agents. Drugs R D 8:2010-2212. [DOI] [PubMed] [Google Scholar]
- 68.Schloss, P. D., and J. Handelsman.2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 69.Shaw, K. J., N. Miller, X. Liu, D. Lerner, J. Wan, A. Bittner, and B. J. Morrow.2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5:105-122. [DOI] [PubMed] [Google Scholar]
- 70.Skoko, D., D. Yoo, H. Bai, B. Schnurr, J. Yan, S. M. McLeod, J. F. Marko, and R. C. Johnson.2006. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 364:777-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spies, M., and S. C. Kowalczykowski.2005. Homologous recombination by RecBCD and RecF pathways, p. 389-403. In N. P. Higgins (ed.), The bacterial chromosome. ACM Press, Washington, DC.
- 72.Tam, C., B. Collinet, G. Lau, S. Raina, and D. Missiakia.2002. Interaction of the conserved region 4.2 of σE with the RseA anti-sigma factor. J. Biol. Chem. 277:27282-27287. [DOI] [PubMed] [Google Scholar]
- 73.Tamae, C., A. Liu, K. Kim, D. Sitz, J. Hong, E. Becket, A. Bui, P. Solaimani, K. P. Tran, H. Yang, and J. H. Miller.2008. Determination of antibiotic hypersensitivity among 4,000 single gene knockout mutants of Escherichia coli. J. Bacteriol. 190:5981-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Workman, C. T., H. C. Mak, S. McCuine, J. Tagne, M. Agarwal, O. Ozier, T. J. Begley, L. D. Samson, and T. Ideker.2006. A systems approach to mapping DNA damage response pathways. Science 312:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamamoto, N., K. Nakahigashi, T. Nakamichi, M. Yoshino, Y. Takai, Y. Touda, A. Furubayashi, S. Kinjyo, H. Dose, M. Hasegawa, K. A. Datsenko, T. Nakayashiki, M. Tomita, B. L. Wanner, and H. Mori.2009. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5:335-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeh, P., A. I. Tschumi, and R. Kishony.2006. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 28:489-494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.