Abstract
Staphylococcus aureus uses the SaeRS two-component system to control the expression of many virulence factors such as alpha-hemolysin and coagulase; however, the molecular mechanism of this signaling has not yet been elucidated. Here, using the P1 promoter of the sae operon as a model target DNA, we demonstrated that the unphosphorylated response regulator SaeR does not bind to the P1 promoter DNA, while its C-terminal DNA binding domain alone does. The DNA binding activity of full-length SaeR could be restored by sensor kinase SaeS-induced phosphorylation. Phosphorylated SaeR is more resistant to digestion by trypsin, suggesting conformational changes. DNase I footprinting assays revealed that the SaeR protection region in the P1 promoter contains a direct repeat sequence (GTTAAN6GTTAA [where N is any nucleotide]). This sequence is critical to the binding of phosphorylated SaeR. Mutational changes in the repeat sequence greatly reduced both the in vitro binding of SaeR and the in vivo function of the P1 promoter. From these results, we concluded that SaeR recognizes the direct repeat sequence as a binding site and that binding requires phosphorylation by SaeS.
Staphylococcus aureus is a common gram-positive human pathogen that colonizes skin, the anterior nares, and other mucosal surfaces (2). S. aureus can cause a wide range of diseases from soft tissue infections to life-threatening infections such as toxic shock syndrome, necrotizing pneumonia, and endocarditis (2, 36). The bacterium is so versatile that it can infect almost all human body parts. Its versatility is, in part, due to the variety of virulence factors that it produces (e.g., surface proteins, toxins, and immune modulators). The expression of these virulence factors is coordinated by a network of multiple DNA binding proteins (e.g., SigB, Rot, MgrA, SarA, and SarA homologues) and two-component systems (e.g., agr, srrAB, arlRS, vraSR, and saeRS) (8, 10, 16, 31, 38, 42, 51).
The two-component system is a signal transduction mechanism by which bacteria and lower eukaryotes monitor and respond to environmental stress cues such as nutrient concentrations, ionic strength, and membrane disturbances (27, 47). Typically, the two-component system consists of a sensor histidine kinase (HK) and response regulator (RR) proteins (26). The sensor HK is a membrane protein composed of a signal binding domain and an autokinase domain. The RR is a cytoplasmic protein made up of an N-terminal regulatory domain and the C-terminal effector domain. In a typical RR, the regulatory domain inhibits the function of the effector domain, usually DNA binding activity. Upon sensing its cognate environmental cues, the sensor HK autophosphorylates a histidine residue and then transfers the phosphate group to an aspartate residue in the N-terminal regulator domain of the RR. Commonly, it is the phosphorylated RR that mediates the necessary physiological changes, typically by binding to its target promoters and modulating the level of transcription (26, 27, 49). The phosphorylated RR is dephosphorylated by its intrinsic or the cognate HK-induced phosphorylated RR phosphatase activity, which resets the system to the prestimulus state (27, 47).
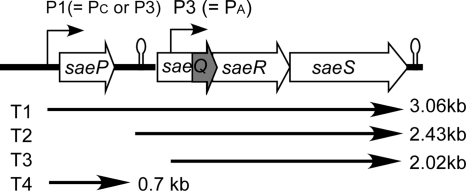
The sae locus is a staphylococcal two-component system critical for the proper expression of exoproteins (18, 19, 21). This locus is composed of four open reading frames (ORFs): saeP (ORF4), saeQ (ORF3), saeR, and saeS (Fig. 1). The two genes saeS and saeR encode the sensor HK and the RR, respectively. SaeS is a 351-amino-acid (aa) polypeptide with two transmembrane segments at the N terminus. The two membrane segments are separated by only nine extracellular amino acid residues (1), which is regarded as too small a sequence to be a signal binding domain. SaeS, therefore, can be classified as an intramembrane sensing HK, which is hypothesized to monitor membrane disturbances (40). The remaining parts of SaeS are in the cytoplasm and contain three subdomains: HAMP (HKs, adenylyl cyclases, methyl binding proteins, and phosphatases; aa 61 to 114), HisKA (His kinase A; aa 122 to 189), and HATPase_c (HK-like ATPases; aa 234 to 348) (SMART analysis [http://smart.embl-heidelberg.de/]). The amino acid residue for autophosphorylation is predicted to be His131 (18). SaeR is a 228-aa polypeptide with an N-terminal regulatory domain and a C-terminal effector domain with potential DNA binding activity. In the regulatory domain, aspartate 51 is predicted to be phosphorylated by SaeS (18). Although phosphorylated SaeR is assumed to be the mediator of signaling, neither DNA binding nor phosphorylation of SaeR has been shown. The functions of SaeP and SaeQ are completely unknown. SaeP is predicted to be a 146-aa membrane protein which is not involved in signal transduction (1). Because the saeQ ORF contains a smaller ORF, SaeQ can be either a 157-aa protein or a 60-aa polypeptide (Fig. 1). SaeQ is presumed to be involved in signal transduction (1); however, no direct biochemical or genetic evidence has shown the mechanism of its involvement.
FIG. 1.
The sae locus of S. aureus. Alternative names of the P1 and P3 promoters are in parentheses. The saeQ ORF contains a smaller ORF (shown in gray). The transcript species are indicated under the ORF map along with their sizes. This map is adapted from a report by Geiger et al. (17).
The sae locus has two promoters, for which various different names have been used in the literature (Fig. 1). The promoter upstream of saeP has been called P1, Pc, or P3, while the downstream promoter can be P3 or Pa (1, 17, 33). Throughout this study, we will use P1 for the upstream promoter and P3 for the downstream promoter only because those names are found more often in the literature. The P1 promoter is positively autoregulated by the sae locus (17, 20, 22), making it a good model system for sae regulation studies. The promoter is also positively regulated by agr and negatively regulated by sigB and rot (17, 33). The activity of the promoter is influenced by several environmental stressors. A low pH or a high NaCl concentration represses promoter activity, while stressors such as subinhibitory concentrations of β-lactam antibiotics, H2O2, or alpha-defensin activate it (17, 30). However, it is not known how transcription regulators or environmental stressors affect P1 promoter activity.
The SaeRS two-component system plays an important role in staphylococcal gene expression and virulence. Most of all, the two-component system is required for the expression of many virulence factors, including cell wall proteins (Spa, FnbA), cell wall-associated proteins (Map/Eap and Emp), and secreted proteins (SspA, Nuc, Coa, Hla, Hlb, Hlc, SCIN, and CHIPS) (19, 21-23, 34, 43). In particular, the saeRS system is an essential positive regulator of alpha-toxin (Hla) production under both in vitro and in vivo conditions (22, 23, 50). A recent study with clinical isolate MW2 showed that the SaeRS system influences the expression of 212 genes (48), whose functions range from virulence to energy metabolism, ion transport, and DNA repair. Lastly, the sae locus is important for the in vivo survival of S. aureus. Knockout mutations of the locus reduced the survival of the bacterium in animal models including device-related infections (22), murine pyelonephritis (34), intraperitoneal injection (44), and sepsis (48). The sae locus also contributes to the apoptosis/death of lung epithelial cells (34), bacterial survival in neutrophils, and killing of neutrophils (48).
Despite its importance in staphylococcal virulence and gene expression, the molecular mechanism of this signaling pathway has not been defined. In this study, we report the identification of the SaeR binding sequence and show that phosphorylation is essential for the DNA binding of SaeR and the signaling process.
MATERIALS AND METHODS
Unless stated otherwise, all of the chemicals used in this study were purchased from Sigma-Aldrich and Fisher and the restriction enzymes were from New England Biolabs.
Bacterial strains, plasmids, and culture conditions.
The bacterial strains used in this study are listed in Table 1. Since the genome sequence of strain USA300-0114 is not known, we sequenced the entire sae operon of the strain and confirmed that the strain has the same sae sequence as strain Newman, except that it does not have the L18P substitution mutation in SaeS. Throughout this study, staphylococci were grown in tryptic soy broth (TSB), except for transduction procedures, for which heart infusion broth supplemented with 5 mM CaCl2 was used. Escherichia coli strains were grown in Luria-Bertani broth (LB). When necessary, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 10 μg/ml; chloramphenicol, 5 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Species and strain | Relevant characteristics | Source |
|---|---|---|
| E. coli | ||
| DH5α | Stratagene | |
| BL21 star(DE3) | Invitrogen | |
| S. aureus | ||
| RN4220 | Restriction deficient, prophage cured | 29 |
| Newman | Clinical isolate, L18P substitution in SaeS | 14 |
| ΦΝΞ-9725 | Strain Newman with Tn917 insertion in saeS | 4 |
| USA300-0114 | Clinical isolate, no L18P substitution in SaeS | NARSAa |
| TB3 | S. aureus Newman with three prophages deleted Δ(ϕNM124) | 3 |
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
Recombinant protein expression and purification. (i) Full-length SaeR.
The 684-bp ORF of saeR was PCR amplified from strain Newman chromosomal DNA with primers SaeRFor_NdeI (5′-GTGTACACATATGACCCACTTACTGATCGTGGATGATGAAC-3′) and SaeRRev_XhoI (5′-GTAGGCACTCGAGTTATCGGCTCCTTTCAAATTTATATCCTAATC-3′). After being digested by NdeI and XhoI, the PCR product was cloned into pET28a (Novagen). The resulting plasmid was transformed first into DH5α and then into BL21 star(DE3). The BL21 star(DE3) strain carrying the plasmid was grown in LB to an optical density at 600 nm (OD600) of 0.6, and then 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. After overnight induction at room temperature, the cells were harvested and frozen at −80°C. The expressed protein was purified from the frozen cells with a HisTrap column (GE Healthcare, Inc.) by following the column manufacturer's recommendations. The purified protein was supplemented with 20% glycerol and stored at −80°C.
(ii) SaeRC (SaeRΔN103).
For expression of the C-terminal effector domain of SaeR (SaeRC), we used the ligation-independent cloning method (13). The coding regions of SaeRC (aa 104 to 228) was PCR amplified from strain Newman chromosomal DNA with primers SaeRΔN103_for (5′-TACTTCCAATCCAATGCCAGTCCAAGGGAACTCGTTTTACGTATTA-3′) and SaeRΔN103_rev (5′-TTATCCACTTCCAATGTTATCGGCTCCTTTCAAATTTATATCCTAATC-3′). The PCR products were treated with T4 DNA polymerase in the presence of dCTP for 30 min at room temperature. Target vector pMCSG19 (13) was digested with SspI, gel purified, and then treated with T4 DNA polymerase in the presence of dGTP for 15 min at 16°C. The T4 DNA polymerase-treated plasmid vector and PCR product were gel purified, mixed, incubated for 5 min at room temperature, and then transformed into E. coli strain DH5α. The resulting plasmid was transformed again into BL21 star(DE3) containing pRK1037(Science Reagents, Inc.), and the transformants were selected on LB agar plates with 150 μg/ml ampicillin and 30 μg/ml kanamycin. The protein was expressed and purified by the same procedures described for full-length SaeR, except that the cells were incubated at 16°C, not at room temperature.
(iii) SaeSC (SaeSΔN92).
Except the primers for PCR amplification of the coding region, the production of the cytoplasmic domain of SaeS (SaeSC aa 93 to 351) was carried out as described for SaeRC above. The primers used were SaeSΔN92 _for (5′-TACTTCCAATCCAATGCCAAAGAAATTTATGAATTAAATCAATC-3′) and SaeSΔN92 _rev (5′-TTATCCACTTCCAATGTTATCGGCTCCTTTCAAATTTATATCCTAATC-3′).
Electrophoretic mobility shift assays (EMSA).
The primers used in EMSA are listed in Table 2. DNA probes were PCR amplified and radiolabeled with T4 polynucleotide kinase (NEB) and [γ-32P]ATP (Perkin-Elmer). The radioactive probe (2 ng) was mixed with various amounts of the test protein in 25 μl of gel shift loading buffer (10 mM Tris-HCl, pH 7.4, 50 mM KCl, 5 mM MgCl2, 10% glycerol, 3 μg/ml sheared salmon sperm DNA). After being incubated at room temperature for 10 min, the samples were analyzed by 8% polyacrylamide gel electrophoresis (100 V for prerun, 85 V for 85 min for sample separation). The gels were dried and subjected to autoradiography on a phosphor screen (BAS-IP; Fuji).
TABLE 2.
Primers used in EMSA
| Primer | Sequencea |
|---|---|
| P1-F | 5′AACGAATTCTTGGTACTTGTATTTAATCGTCTATC |
| P1-R | 5′AAAGGTACCGTTGTGATAACAGCACCAGCTGC3′ |
| hla_I_F | 5′GTGTACAAACGAAAAAGTATCGTATGTATTTTTAATATAG3′ |
| hla_I_R | 5′GTGTACATAAAATAGTTTCATTTTAATCCCCTATC3′ |
| hla_II_F | 5′GTGTACAGATTACAATATAAAAATACAAATATCTTAG3′ |
| hla_II_R | 5′GTACAATCTATTAGATATTTCTATGTAATGGCAAAATTTATTC3′ |
| empF1 | 5′ACAGAATTCAATTATTTATAATGCACC3′ |
| empR1 | 5′CCTAAGCTTTTATATAGACTCAATATTATAAC3′ |
| eapF | 5′TTTGAATTCCACCATCATTATCACTCC3′ |
| eapR | 5′CATCCCGGGAAATTATCTCTCCTTTTTTG3′ |
| vwbF1 | 5′TTCGAATTCAGATAGCGATTCGGACTC3′ |
| vwbR1 | 5′CCTAAGCTTTAATTTTCCCTAATTAAC3′ |
| arlR-F | 5′CAATAGTGAAAAGTCAGTATATGAC3′ |
| arlR-R | 5′AACGGTACCATTTGCGTCATTTGTACACCTC3′ |
Restriction enzyme sites are underlined.
Phosphorylation of SaeR for EMSA.
The purified SaeR protein (20 μM) was mixed with purified SaeSC (1 μM) in phosphorylation buffer (10 mM Tris-HCl, pH 7.4, 50 mM KCl, 5 mM MgCl2, 10% glycerol). One millimolar ATP was then added, and the mixture was incubated at room temperature for 5 min before the addition of 32P-labeled DNA probe. For EMSA or footprinting analyses, the entire reaction mixture was used.
DNase I footprinting assays.
The test proteins were mixed with radiolabeled probe in reaction buffer (10 mM Tris-HCl, pH 7.4, 50 ml KCl, 5 mM MgCl2, 1 mM CaCl2, 1 μg/ml yeast tRNA). After the addition of DNase I (0.1 U; New England Biolabs), the samples were incubated at room temperature for 2 min. The reaction was terminated by the addition of 25 μl of stop solution (50 mM Tris-HCl, pH 8.0, 1% [wt/vol] sodium dodecyl sulfate [SDS], 20 mM EDTA) and extracted first with 50 μl of phenol-chloroform (1/1) and then with 50 μl of chloroform. DNA was precipitated with ethanol, washed with 500 μl of 70% ice-cold ethanol, and suspended in 10 μl of loading buffer (98% deionized formamide, 10 mM EDTA, 0.025% [wt/vol] xylene cyanol FF, 0.025% [wt/vol] bromphenol blue). After being denatured at 95°C for 3 min, samples were separated on an 8% urea-polyacrylamide gel. Sequencing ladders consisting of A+G and T+C for the P1 promoter of the sae operon were made by the standard Maxam-Gilbert method (41).
Phosphorylation of SaeR by SaeSC and cell lysates.
Cell lysates were prepared from strain Newman, a saeS transposon insertion mutant (ΦΝΞ-9725), and strain USA300-0114 (= NRS384), which does not contain the L18P substitution mutation in SaeS. The cells were grown in TSB to mid-log phase (OD600 = 0.6) and collected by centrifugation. The collected cells were suspended in 1 ml of TSM (50 mM Tris-HCl, pH 7.5, 0.5 M sucrose, 10 mM MgCl2) to which was added 10 μl lysostaphin (2 mg/ml) and incubated at 37°C for 30 min. A 0.6-ml volume of 1.3 M KCl was then added to the cell lysates, and the resulting cell lysates were stored at −80°C. The phosphorylation assays were performed in the presence of 10 μM SaeR and 2 μM SaeSC or 2-μl cell lysate volumes in phosphorylation buffer (10 mM Tris-HCl, pH 7.4, 50 mM KCl, 5 mM MgCl2, 10% glycerol). The final reaction mixture volume was 20 μl. To initiate the phosphorylation reaction, 0.5 μl [γ-32P]ATP (80 μCi) was added. The reaction mixtures remained at room temperature for 10 min, and then the reactions were stopped by the addition of 10 μl of 2× SDS loading buffer. Samples were analyzed by 13% SDS-polyacrylamide gel electrophoresis (PAGE).
Assay of phosphotransfer between P-SaeSC and SaeR.
SaeSC (3 μM) was preincubated with 3 μM [γ-32P]ATP in 105 μl of phosphorylation buffer for 1 h at room temperature. As a reference sample, 15 μl was mixed with 15 μl of 2× SDS loading buffer and kept at room temperature. To initiate the phosphotransfer reaction, SaeR (9 μM) was added to phosphorylated SaeSC. At various time points (0 min, 3 min, 30 min, 2 h, 4 h, and 20 h), 15 μl of the sample was mixed with SDS loading buffer and stored at room temperature until electrophoresis. The samples were analyzed by 13% SDS-PAGE and autoradiography.
Limited trypsin digestion assays.
To phosphorylate SaeR, SaeR (50 μM) was mixed with 1 μM SaeSC in the presence of 1 mM ATP and incubated at room temperature for 5 min. Because the phosphorylated SaeR protein sample contains SaeSC, to avoid any artifacts from the presence of SaeSC, an equal amount of SaeSC was added to the unphosphorylated SaeR sample but without ATP. Trypsin digestion was initiated by the addition of 0.2 μg/μl trypsin in phosphorylation buffer containing 0.1 mM EDTA. The samples were incubated at 37°C. Aliquots of 10 μl were removed from the reaction mixture at defined time intervals and mixed with 10 μl of 2× SDS loading buffer. Samples were separated by 14% SDS-PAGE and stained with Coomassie blue R-250. The digestion patterns were quantified by Quantity One (Bio-Rad).
Random mutagenesis of the P1 promoter.
The P1 promoter region was amplified by the error-prone enzymes Taq polymerase (NEB) and Mutazyme (Stratagene) with primers P1-F and P1-R (Table 2). After being purified, the PCR product was digested with BamHI/KpnI and then ligated with pYJ-lacZ, a promoterless lacZ reporter plasmid derived from pYJ335 (28). The ligated DNA was electroporated into E. coli DH5α. Plasmids were purified from the colonies on the transformation plate and electroporated into S. aureus strain RN4220, and then the cells were spread on tryptic soy agar (TSA) containing erythromycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 80 μg/ml) (TSAerm X-Gal). Colonies displaying white or reduced blue color were isolated. In other trials, the RN4220 colonies were pooled and lysed with φ85. The phage lysates were used to transduce the plasmids into strain TB3, a strain derived from strain Newman by the deletion of three prophages. This strain has a wild-type sae phenotype and, because of the absence of three prophages, is useful for complementation studies. The transduced TB3 cells were spread on TSAerm X-Gal. The colonies displaying white or reduced blue color were isolated. To verify that the phenotype was caused by mutations in the P1 promoter, plasmids were purified and the P1 promoter was sequenced. When more than one mutation was found, each mutation was generated by PCR-mediated site-directed mutagenesis; then, the resulting mutation was tested in strain Newman to find the one responsible for the phenotype. During the lacZ assay, however, we found that in strain Newman, the plasmid carrying the wild-type P1 promoter was unstable and the majority of the cells displayed a white color on TSAerm X-Gal. Characterization of the plasmid purified from the cells revealed that a deletion mutation occurred at the N terminus of LacZ, suggesting that the overexpression of β-galactosidase from the high-copy plasmid is toxic to the cells. Therefore, we moved all of the mutations of the P1 promoter to pCL55-lacZ, a lacZ reporter plasmid derived from integration plasmid pCL55 (32). The mutated P1 promoters were PCR amplified by the high-fidelity DNA polymerase Phusion (NEB) with primers P1-F and P1-R (Table 2) and cloned into pCL55-lacZ as described for pYJ-lacZ above. All of the mutant P1 promoter sequences in the pCL-lacZ constructs were verified by DNA sequencing analysis.
LacZ assay.
The test strains were grown in TSB containing 5 μg/ml chloramphenicol (TSBchl-5) and incubated at 37°C for 16 h. After being collected by centrifugation, the cells were suspended in AB buffer (100 mM potassium phosphate, 100 mM NaCl, pH 7.0) and treated with lysostaphin (0.1 μg/ml) at 37°C for 15 min. After the addition of 900 μl of ABT buffer (AB buffer containing 0.1% Triton X-100) to the lysostaphin-treated cells, 50 μl of the cell lysate was mixed with 10 μl of MUG (4-methylumbelliferyl-β-d-galactopyranoside, 4 mg/ml; Sigma) and incubated at room temperature for 1 h. Then, 20 μl of the reaction mixture was mixed with 180 μl ABT buffer in a black 96-well plate and the emission of fluorescence was measured by a plate reader (355-nm excitation, 445-nm emission). The LacZ activity was normalized by cell density at 600 nm, and then the relative activity was calculated by setting the LacZ activity from the wild-type P1 promoter to 100%. The assay was repeated at least twice with similar results.
RESULTS
The N-terminal regulatory domain inhibits the DNA binding activity of the C-terminal effector domain in SaeR.
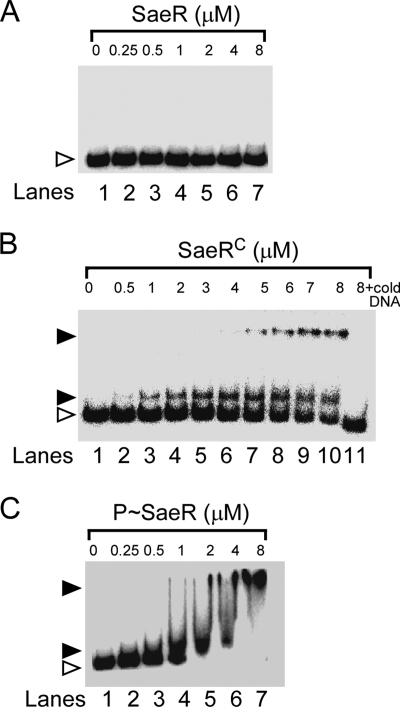
As a first step in identifying the molecular mechanism of sae-mediated signal transduction, we examined the DNA binding activity of the RR SaeR. We expressed the full-length SaeR protein in E. coli with a His6 tag at its N terminus and then purified the protein with a Ni column. As a DNA binding substrate, we used the radiolabeled P1 promoter of the sae operon because the promoter is highly autoregulated by sae (17, 20, 22). Using purified His6-SaeR and radiolabeled P1 promoter DNA, we performed EMSA. As shown in Fig. 2A, full-length SaeR did not bind to the P1 promoter.
FIG. 2.
DNA binding activities of unphosphorylated SaeR (A), the C-terminal effector domain of SaeR (B), and phosphorylated SaeR (C) to the P1 promoter region. In a typical assay, 2 ng of a γ-32P-end-labeled P1 promoter fragment was incubated with the indicated concentration of protein in the presence of 3 μg/ml salmon sperm DNA at room temperature for 15 min. Free DNA is indicated by a white arrowhead, and bound DNA is indicated by a black arrowhead. SaeR, unphosphorylated SaeR; SaeRC, the C-terminal effector domain of SaeR; P-SaeR, phosphorylated SaeR; + cold DNA, addition of a 10-fold excess of unlabeled DNA.
Since the DNA binding activity of the effector domain is commonly inhibited by the N-terminal regulatory domain, we envisioned that if we deleted the N-terminal domain, the C-terminal effector domain alone might be able to bind to its target DNA. To test this hypothesis, we produced the C-terminal effector domain (SaeRC aa 104 to 228) with a His6 tag as described in Materials and Methods and performed EMSA with the radiolabeled P1 promoter DNA again. As shown in Fig. 2B, unlike full-length SaeR, SaeRC bound to the P1 promoter, suggesting that the N-terminal regulatory domain indeed inhibits DNA binding activity. The binding affinity of SaeRC for the P1 promoter was weak. Even at 8 μM SaeRC, approximately 30% of the substrate DNA remained unbound. In a densitometry analysis, the dissociation constant of SaeRC was calculated to be 6 μM. In the EMSA, two shifted bands were observed, suggesting the existence of multiple SaeR binding sites in the P1 promoter. The addition of a 10-fold excess of unlabeled P1 DNA abolished the binding of the radiolabeled DNA, demonstrating the specificity of the protein-DNA interaction (Fig. 2B, lane 11). Based on these results, we concluded that the C-terminal effector domain of SaeR has target DNA binding activity and that in full-length SaeR, this activity is inhibited by the N-terminal regulatory domain.
Phosphorylation is essential for the DNA binding activity of SaeR.
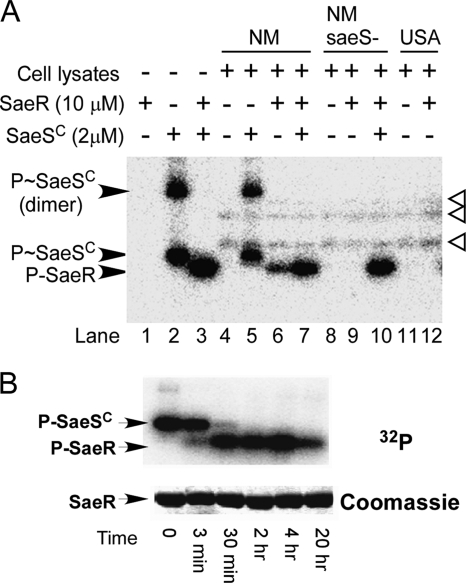
Typically, phosphorylation of the N-terminal regulatory domain abolishes its inhibitory effect on the C-terminal effector domain. To examine whether phosphorylation of SaeR can restore the DNA binding activity of SaeR, we decided to phosphorylate SaeR with the sensor kinase SaeS. Since SaeS has its kinase activity in the cytoplasmic domain, we expressed the cytoplasmic autokinase domain of SaeS (SaeSC aa 93 to 351) with an N-terminal His6 tag. We purified His6-SaeSC and tested the autophosphorylation function of SaeSC by adding [γ-32P]ATP and Mg2+ to the protein. As shown in Fig. 3A, autophosphorylation of SaeSC was clearly observed (lanes 2), proving that the SaeSC protein used in the test does contain autokinase activity. When SaeR was added to SaeSC (SaeR/SaeSC ratio, 5:1), phosphorylation of SaeR was observed (Fig. 3A, lane 3), suggesting that the purified SaeSC protein contains phosphotransferase activity and can be used to phosphorylate SaeR. Interestingly, under these experimental conditions, no phosphorylated SaeSC was detected, implying a very fast phosphotransfer reaction.
FIG. 3.
SaeR phosphorylation by the cytoplasmic domain of SaeS and strain Newman cell lysates. (A) SaeSC (2 μM) or cell lysates from strains Newman (NM), ΦΝΞ-9725 (NMsaeS-), and USA300-0114 (USA) were mixed with SaeR (10 μM) in the presence of [γ-32P]ATP. All of the reactions were performed for 10 min at room temperature. (B) SaeSC (3 μM) was phosphorylated with [γ-32P]ATP, and then SaeR (9 μM) was added. P-SaeSC, phosphorylated SaeSC; P-SaeR, phosphorylated SaeR. The three additional protein bands phosphorylated in the strain Newman cell lysates are indicated by white arrowheads.
In strain Newman, SaeS is constitutively active due to an amino acid substitution mutation (L18P) within the first transmembrane segment (1). Therefore, we anticipated that cell lysates from strain Newman would be able to phosphorylate SaeR. Indeed, when added to SaeR, strain Newman cell lysates clearly phosphorylated the RR within 10 min (Fig. 3A, lane 6). Further addition of purified SaeSC to the reaction mixture did not change the overall outcome (Fig. 3, lane 7). The fact that the cell lysate from a saeS transposon mutant of strain Newman failed to phosphorylate SaeR (Fig. 3, lane 9) strongly suggests that SaeS is responsible for SaeR phosphorylation. Strain USA300-0114 does not contain the L18P substitution mutation in SaeS. When the cell lysate from strain USA300-0114 was used, no significant SaeR phosphorylation was observed, confirming the hyperactivity of SaeS kinase in strain Newman. Noticeably, in the reaction mixtures with cell lysates, three more radiolabeled protein bands appeared (white arrowheads in Fig. 3A), revealing the occurrence of other phosphorylation events in the cell lysate. The identities of these protein bands are unknown.
Although the above result clearly shows that SaeSC can phosphorylate SaeR, it does not show the phosphotransfer reaction directly due to the fast kinetics of this process. In order to detect that phosphotransfer reaction directly, we first phosphorylated SaeSC with [γ-32P]ATP and then added SaeR at a lower ratio (SaeR/SaeSC ratio, 3:1) than in the previous phosphorylation reaction mixture, where the ratio was 5:1. The results are shown in Fig. 3B. Under this condition, a gradual transfer of the phosphate group was observed. After 3 min, only a fraction of SaeR was phosphorylated and the reaction was not complete until 2 h. Interestingly, no significant change in P-SaeR was observed until 4 h after the reaction. Even after 20 h, a significant portion of SaeR remained phosphorylated, indicating that the phosphorylated form of SaeR is stable under the conditions employed.
To examine whether phosphorylation can restore the binding activity of SaeR, we phosphorylated SaeR by adding purified SaeSC and ATP and used phosphorylated SaeR (P-SaeR) in EMSA. As shown in Fig. 2C, P-SaeR indeed bound to P1 promoter DNA. In a densitometry analysis, the dissociation constant of P-SaeR was calculated to be 0.75 μM, suggesting that the P1 promoter binding affinity of P-SaeR is eight times higher than that of SaeRC (KD = 6 μM). To eliminate the possibility that SaeSC bound to the DNA in the EMSA, we performed EMSA with the mixture of SaeSC and SaeR in the absence of ATP; no binding was observed (data not shown), suggesting that it is P-SaeR, not SaeSC, that is responsible for the DNA binding observed. Taken together, these results demonstrate that phosphorylation of SaeR is essential for efficient binding to target DNA.
Phosphorylation elicits conformational changes in SaeR.
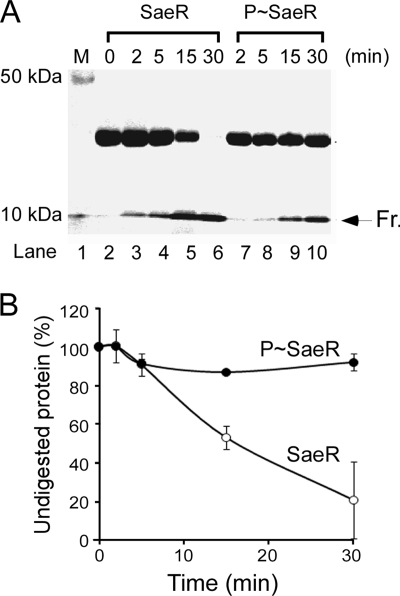
If phosphorylation restores the DNA binding activity of SaeR, we postulated that it would accompany significant conformational changes in SaeR. To examine this postulation, we subjected unphosphorylated SaeR and P-SaeR to a limited trypsin digestion assay where the alteration in protease susceptibility is interpreted as a conformational change in the protein. We digested the two forms of SaeR at a low concentration of trypsin and, by PAGE and Coomassie staining, analyzed the digestion pattern after 2, 5, 15, and 30 min. The digestion results obtained are shown in Fig. 4A. Although almost all of the unphosphorylated SaeR protein was digested by trypsin within 30 min, the majority of the P-SaeR protein remained intact, even after 30 min of digestion (lane 10). This rather dramatic alteration in trypsin susceptibility strongly suggests that phosphorylation elicits significant conformational changes in SaeR which, in turn, lead to restoration of the DNA binding activity of the protein.
FIG. 4.
Limited trypsin digestion analysis of SaeR and P-SaeR. (A) SaeR (50 μM) was phosphorylated by SaeSC (1 μM) in the presence of 1 mM ATP. An equal amount of SaeSC (1 μM) was added to unphosphorylated SaeR (50 μM) but without ATP. The proteins were mixed with 0.2 μg/μl trypsin and incubated at 37°C. Aliquots of 10 μl were removed from the reaction mixtures at different time intervals and quenched by the addition of 10 μl 2× SDS loading buffer, followed by heating at 90°C for 5 min. Samples were analyzed by 14% SDS-PAGE and Coomassie straining. Fr, a 10-kDa SaeR fragment resistant to trypsin digestion. (B) The staining results were quantified by Quantity One (Bio-Rad). The error bars represent the standard deviations calculated from two independent experiments.
DNase I footprinting assays identify the SaeR binding sequence.
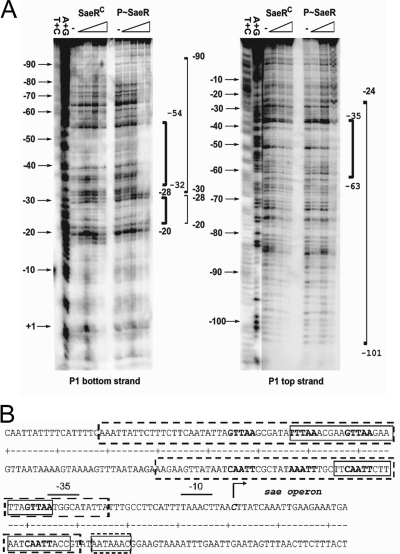
Since both SaeRC and P-SaeR bound to the P1 promoter, to identify the SaeR binding sequence, we decided to perform a DNase I footprinting assay with the proteins. The radiolabeled P1 promoter was incubated with increasing amounts of either SaeRC or P-SaeR, and then DNase I was added. After a 2-min incubation, the reaction was terminated and the product was analyzed by PAGE and autoradiography. As shown in Fig. 5A, SaeRC bound to the P1 promoter in the region from −20 to −54 (bottom strand) or from −35 to −63 (top strand). On the other hand, P-SaeR bound to wider regions of the P1 promoter from −20 to −90 (bottom strand) or from −24 to −101 (top strand).
FIG. 5.
Identification of SaeR binding sequences. (A) DNase I footprinting analysis of the P1 promoter with SaeRC and P-SaeR. Sequencing of the DNA probe was carried out by the Maxam-Gilbert method. The nucleotide positions are indicated to the left of the footprinting image. The regions protected by SaeRC are in bold brackets, and the regions protected by P-SaeR are in plain brackets. Values represent distances from the transcription start site, which was set to +1. (B) P1 promoter sequence with a summary of the DNase I footprinting assay results. The −10 and −35 promoter regions are indicated by solid lines above the sequence. SaeRC-protected regions are in solid boxes, and the P-SaeR-protected regions are in dotted-line boxes. The direct repeat sequences are in boldface. The transcription start site is indicated by a right-angled arrow; and the corresponding nucleotide is in boldface italics.
Because most RRs bind to their target as a dimer, we envisioned that the SaeR binding sequence would be a repeat sequence. Indeed, close inspection of the protected region (−20 to −101 position) revealed a direct repeat sequence (GTTAAN6GTTAA) between positions −35 and −50 and an imperfect repeat sequence (GTTAAN6TTTAA) between positions −56 and −71 (Fig. 5B). To examine whether the repeat sequences are SaeR binding sites, we searched the strain Newman genome, by using Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/), for the genes containing the perfect direct repeat sequence (GTTAAN6GTTAA) in the promoter region. The search results are shown in Table 3. The search identified 21 genes carrying the direct repeat sequence. Of the 21 genes, two pairs of genes (NWMN_0165/0166 and NWMN_1706/1707) shared the same sequence. More importantly, 13 (57%) were genes of the sae regulon (30, 34, 45, 46, 48), raising the possibility that the direct repeat sequence is the SaeR binding site.
TABLE 3.
Genes containing the perfect direct repeat sequence GTTAAN6GTTAA in the promoter region
| Locus IDb | Gene | Matching sequencec | Product |
|---|---|---|---|
| NWMN_0165 | TCTAGTTAATATATAGTTAATGTC | Hypothetical protein | |
| NWMN_0166a | coa | TCTAGTTAATATATAGTTAATGTC | Coagulase precursor |
| NWMN_0362a | TAAAGTTAATCAAGAGTTAAGATG | Hypothetical protein | |
| NWMN_0369 | set2nmg | AATAGTTAAAAACAGGTTAATGT | Enterotoxin-like toxin |
| NWMN_0393 | set6nm | AATAGTTAAAAAGAGGTTAATTCA | Enterotoxin-like toxin |
| NWMN_0394a | set7nm | AATAGTTAAAAAGAGGTTAATTCA | Enterotoxin-like toxin |
| NWMN_0677a | saeP | CGAAGTTAAGAATTAGTTAATGGC | Hypothetical protein |
| NWMN_1066a | TTTAGTTAATAAATAGTTAATGTA | Hypothetical protein | |
| NWMN_1073a | hla | ACTAGTTAATATATAGTTAATTTT | Alpha-hemolysin precursor |
| NWMN_1074 | ACTAGTTAATATATAGTTAATTTT | Hypothetical protein | |
| NWMN_1533 | hisS | AGCCGTTAAACGTACGTTAAACGT | Histidyl-tRNA synthetase |
| NWMN_1706a | splA | AAACGTTAATAAGTGGTTAATTAA | Serine protease |
| NWMN_1707 | AAACGTTAATAAGTGGTTAATTAA | Hypothetical protein | |
| NWMN_1708 | TTTAGTTAATAGATAGTTAATACA | Homologue to Ear | |
| NWMN_1719a | lukE | AATAGTTAATAATCAGTTAATTTT | Leukocidin LukE precursor |
| NWMN_1880a | sak | AAATGTTAAATATTTGTTAATTAT | Staphylokinase precursor |
| NWMN_2317a | sbi | ATTAGTTAATAATTAGTTAATTTC | IgG binding protein Sbi |
| NWMN_2319a | hlgC | AATTGTTAATGAACAGTTAATTAT | Gamma-hemolysin component C |
| NWMN_2397a | fnbB | GCGAGTTAATAAAAAGTTAAGATT | Fibrinogen binding protein B |
| NWMN_2399a | fnbA | GCGAGTTAATGAAAAGTTAAGATT | Fibrinogen binding protein A |
| NWMN_2592 | AACCGTTAACAACACGTTAACGGG | 2-Oxoglutarate/malate translocator |
Known gene of the sae regulon.
Genome of strain Newman.
Four flanking nucleotides are also shown.
Interestingly, Table 3 does not contain some of the well-known members of the sae regulon such as map (also called eap), hlb, and scn. In addition, a recent microarray analysis of strain MW2 showed that the SaeRS system affected the transcription of 212 genes (48), suggesting that the 21 genes identified may not represent the entire sae regulon. Therefore, we repeated the sequence search by allowing one mismatch in the sequence; the results are shown in Table 4. The search identified a total of 155 genes, including map/eap, hlb, and scn. Of these 155 genes, 36 (23%) were reported to be regulated by sae, further supporting the idea that the repeat sequence is the SaeR binding site.
TABLE 4.
Genes containing the direct repeat sequence with no more than one mismatch
| Gene function and IDa | Name | Start | Sequence | End | Product |
|---|---|---|---|---|---|
| Virulence/defense | |||||
| NWMNN_0166b | coa | −103 | GTTAATGCTTTGTTTA | −88 | Coagulase precursor |
| −82 | GTTAATATATAGTTAA | −67 | |||
| NWMNN_0388 | set1nm | −109 | GTTAAATGAGGTTTAA | −94 | Staphylococcal enterotoxin-like toxin |
| NWMNN_0389 | set2nm | −109 | GTTAAAAACAGGTTAA | −94 | Staphylococcal enterotoxin-like toxin |
| NWMNN_0390 | set3nm | −110 | GTTAAAAGGGGTTTAA | −95 | Staphylococcal enterotoxin-like toxin |
| −223 | ATTAAAAAGAAGTTAA | −208 | |||
| NWMNN_0391 | set4nm | −109 | GTTAAACAAGGTTTAA | −94 | Staphylococcal enterotoxin-like toxin |
| NWMNN_0393 | set6nm | −145 | GTTCAAAAATAGTTAA | −130 | Staphylococcal enterotoxin-like toxin |
| NWMNN_0393 | set6nm | −134 | GTTAAAAAGAGGTTAA | −119 | Staphylococcal enterotoxin-like toxin |
| NWMNN_0394 | set7nm | −144 | GTTCAAAAATAGTTAA | −129 | Staphylococcal enterotoxin-like toxin |
| −133 | GTTAAAAAGAGGTTAA | −118 | |||
| NWMNN_0395 | set8nm | −110 | ATTAAACGAGTGTTAA | −95 | Staphylococcal enterotoxin-like toxin |
| −141 | GTTAATGAAGAGCTAA | −126 | |||
| −130 | CTTAAATCATTGTTAA | −115 | |||
| NWMNN_0396 | set9nm | −175 | ATTAAAAATCAGTTAA | −160 | Staphylococcal enterotoxin-like toxin |
| −110 | TTTAAATGAGCGTTAA | −95 | |||
| NWMNN_0397 | set10nm | −110 | TTTAAATCGAGGTTAA | −95 | Staphylococcal enterotoxin-like toxin |
| NWMNN_0400 | set11nm | −231 | GTTAAATAAAGATTAA | −216 | Staphylococcal enterotoxin-like toxin |
| −153 | GTTAACTATTTATTAA | −138 | |||
| −123 | ATTAATTTTTAGTTAA | −108 | |||
| −112 | GTTAAAGTAAGTTTAA | −97 | |||
| −152 | TTTAATAAATAGTTAA | −137 | |||
| NWMNN_2619 | −190 | GTTTAATAGAGGTTAA | −175 | Phenol-soluble modulin alpha 1 | |
| NWMNN_0758 | ssp/emp | −126 | GTTAAGACAACGTTTA | −111 | Extracellular matrix and plasma binding protein |
| −115 | GTTTACTTCAAGTTAA | −100 | |||
| NWMNN_1066 | −99 | ATTAATGTTTAGTTAA | −84 | Similar to fibrinogen binding protein | |
| −88 | GTTAATAAATAGTTAA | −73 | |||
| NWMNN_1069 | efb | −81 | ATTAATAATTAGTTAA | −66 | Similar to fibrinogen binding protein |
| NWMNN_1070 | −76 | TTTAATGACAGGTTAA | −61 | Similar to fibrinogen binding protein | |
| NWMNN_1073 | hla | −405 | GTTAATATATAGTTAA | −390 | Alpha-hemolysin precursor |
| −190 | TTTAAATAAAAGTTAA | −175 | |||
| NWMNN_1084 | −171 | TTTAAAATACAGTTAA | −156 | Phenol-soluble modulin beta 1 | |
| NWMNN_1716 | bsaA2 | −115 | GTTAATTTTTTGTAAA | −100 | Lantibiotic precursor |
| NWMNN_1719 | lukE | −334 | GTTAATAATCAGTTAA | −319 | Leukocidin LukE precursor |
| NWMNN_1600 | −178 | GTTATAATGTAGTTAA | −163 | Universal stress protein family protein | |
| −86 | TTTAATGAACAGTTAA | −71 | |||
| NWMNN_1664 | −357 | GTTAAAATATTTTTAA | −342 | Arsenical resistance operon repressor | |
| −356 | GTTAAAAATATTTTAA | −341 | |||
| NWMNN_1872b | map/eap | −88 | ATTAATATTCAGTTAA | −73 | MHCc class II analog protein |
| NWMNN_1873b | hlb | −307 | ATTAATATTCAGTTAA | −292 | Truncated beta-hemolysin |
| NWMNN_1876 | scn | −77 | GTTAATGAATAATTAA | −62 | Staphylococcal complement inhibitor SCIN |
| NWMNN_1877 | chp | −144 | TTTAATTTTTAGTTAA | −129 | Chemotaxis-inhibiting protein CHIPS |
| −112 | ATTAATTTCAAGTTAA | −97 | |||
| NWMNN_1880 | sak | −175 | GTTAAATATTTGTTAA | −160 | Staphylokinase precursor |
| −164 | GTTAATTATTTTTTAA | −149 | |||
| NWMNN_1928b | lukS | −99 | TTTAATAAATAGTTAA | −84 | Leukocidin/hemolysin toxin family S subunit |
| NWMNN_2061 | fmtB /mrp/sasB | −61 | CTTAATCAACTGTTAA | −46 | Methicillin resistance determinant FmtB protein, |
| NWMNN_2317 | sbi | −92 | GTTAATAATTAGTTAA | −77 | IgG binding protein Sbi |
| NWMNN_2318 | hlgA | −346 | GTTAACGAATAGTTCA | −331 | Gamma-hemolysin component A |
| −85 | TTTAACGAACAGTTAA | −70 | |||
| −84 | ATTAACTGTTCGTTAA | −69 | |||
| NWMNN_2319 | hlgC | −351 | GTTAATAAACAATTAA | −336 | Gamma-hemolysin component C |
| −86 | GTTAATGAACAGTTAA | −71 | |||
| NWMNN_2397 | fnbB | −114 | GTTAATAAAAAGTTAA | −99 | Fibronectin binding protein B precursor |
| NWMNN_2399 | fnbA | −137 | ATTAATTTTATGTTAA | −122 | Fibronectin binding protein A precursor |
| −114 | GTTAATGAAAAGTTAA | −99 | |||
| NWMNN_2586b | drp35 | −219 | GTTATAAGCATGTTAA | −204 | Drp35 |
| Cell envelope | |||||
| NWMNN_0606 | tagA | −141 | GTTAAACTAATTTTAA | −126 | Teichoic acid biosynthesis protein A |
| NWMNN_1882 | −183 | GTTAAATGGTTATTAA | −168 | Phage holin | |
| NWMNN_2051 | −163 | GTTGAAAATTTGTTAA | −148 | Lytic regulatory protein | |
| NWMNN_2395 | gtaB | −55 | GTTAAAATGACGTTGA | −40 | UTP-glucose-1-phosphateuridyl transferase |
| Transcription | |||||
| NWMNN_0038 | −110 | GTTATCATAGAGTTAA | −95 | Similar to LysR family regulatory protein | |
| NWMNN_0636 | −82 | GTTAAATTTACGTTTA | −67 | AraC family regulatory protein | |
| NWMNN_0677 | saeP | −99 | GTTAAGCGATATTTAA | −84 | Conserved hypothetical protein |
| −78 | GTTAAGAATTAGTTAA | −63 | |||
| NWMNN_0925 | −31 | ATTAAATTTTTGTTAA | −16 | Similar to cell envelope-related transcriptional attenuator | |
| NWMNN_1328 | arlR | −169 | GTTAATAAATAATTAA | −154 | Two-component RR |
| Translation | |||||
| NWMNN_0490 | gltX | −321 | GTTAATTTGAAGTTTA | −306 | Glutamyl-tRNA synthetase |
| −139 | TTTAAGGTCATGTTAA | −124 | |||
| NWMNN_0723 | prfB | −336 | GTGAAGAAGTTGTTAA | −321 | Peptide chain release factor 2 |
| NWMNN_1478 | rpsU | −243 | GTTAAAACAACGTTAC | −228 | 30S ribosomal protein S21 |
| NWMNN_1488b | rpsT | −81 | GTTAGACTTTTGTTAA | −66 | 30S ribosomal protein S20 |
| NWMNN_1519 | alaS | −290 | GTTAATGATTTGTTTA | −275 | Alanyl-tRNA synthetase |
| NWMNN_1533 | hisS | −260 | GTTAAACGTACGTTAA | −245 | Histidyl-tRNA synthetase |
| Protein metabolism | |||||
| NWMNN_0824 | ppi | −50 | CTTAAAAGTATGTTAA | −35 | Cyclophilin type peptidyl-prolyl cis-trans isomerase |
| NWMNN_0957 | pdf1 | −59 | GTAAAAAGGTTGTTAA | −44 | Polypeptide deformylase 2 |
| NWMNN_1057 | trxA | −143 | GTTAAAATAATGTAAA | −128 | Thioredoxin |
| NWMNN_1539 | secDF | −157 | GTTAAATTTAAATTAA | −142 | Preprotein translocase component SecDF |
| NWMNN_1706b | splA | −121 | TTTAATAAAACGTTAA | −106 | Serine protease SplA |
| −110 | GTTAATAAGTGGTTAA | −95 | |||
| −99 | GTTAATTAATATTTAA | −84 | |||
| NWMNN_2203 | ssaA | −212 | ATTAATTATCTGTTAA | −197 | Secretory antigen precursor SsaA |
| NWMNN_2354 | −331 | TTTAATGATAAGTTAA | −316 | Glutamyl-aminopeptidase | |
| −276 | GTTAATGCGTTGTTAT | −261 | |||
| Ion transport and metabolism | |||||
| NWMNN_0049 | −73 | GTTAAGATTAGGTAAA | −58 | Similar to Na+ Pi-cotransporter | |
| −62 | GTAAATTTAATGTTAA | −47 | |||
| NWMNN_0071 | −115 | TTTAAGGTATAGTTAA | −100 | Glucose/ribitol dehydrogenase | |
| NWMNN_0123 | −371 | ATTAAGAATTTGTTAA | −356 | Similar to surfactin synthetase | |
| NWMNN_0130 | −172 | GTTAAATCGTTCTTAA | −157 | Branched-chain amino acid transport system II carrier protein | |
| −171 | GTTAAGAACGATTTAA | −156 | |||
| NWMNN_0158 | uhpT | −130 | ATTAATAAATAGTTAA | −115 | Sugar phosphate transport protein |
| NWMNN_0418 | ndhF | −345 | GTTTAATAGAGGTTAA | −330 | NADH dehydrogenase subunit 5 |
| NWMNN_0476 | folP | −17 | ATTAAAGGGTGGTTAA | −2 | Dihydropteroate synthase chain A |
| −16 | GTTAACCACCCTTTAA | −1 | |||
| NWMNN_0577 | adh1 | −370 | ATTAATCTGTAGTTAA | −355 | Alcohol dehydrogenase |
| NWMNN_0623 | −283 | GTTTAAACCTTGTTAA | −268 | Similar to branched-chain amino acid transportsystem II carrier protein | |
| NWMNN_0630 | vraF | −80 | GTTAGTCATATGTTAA | −65 | ABC transporter ATP binding protein VraF |
| NWMNN_0690 | −149 | GGTAATCTCCAGTTAA | −134 | Osmoprotectant ABC transporter, ATP binding protein | |
| NWMNN_0705 | −84 | ATTAAAGAAGGGTTAA | −69 | Ferrichrome ABC transporter lipoprotein | |
| NWMNN_0853 | −110 | GTTAATAAAATTTTAA | −95 | 3-Oxoacyl-(acyl-carrier-protein) synthase III | |
| NWMNN_1040 | isdB | −165 | GTTAAATAAAATTTAA | −150 | Iron-regulated heme-iron binding protein IsdB |
| NWMNN_1060 | sdhC | −65 | GTTAAGCGTACGTTTA | −50 | Succinate dehydrogenase cytochrome b558 subunit |
| NWMNN_1078b | argF | −359 | GCTAAAACTATGTTAA | −344 | Ornithine carbamoyltransferase |
| NWMNN_1300 | pstS | −129 | GTTAAGAAAATGTAAA | −114 | Phosphate ABC transporter, phosphate binding protein PstS |
| NWMNN_1305 | asd | −61 | GTAAAATGATTGTTAA | −46 | Aspartate-semialdehyde dehydrogenase |
| NWMNN_1331 | −80 | GTTTAAAGCATGTTAA | −65 | Acetyltransferase, GNAT family protein | |
| −168 | ATTAATTGATTGTTAA | −153 | |||
| NWMNN_1337 | dfrA | −58 | GTTAAATTAAAGATAA | −43 | Dihydrofolate reductase |
| NWMNN_1349 | ald | −122 | GTTAATAAATATTTAA | −107 | Alanine dehydrogenase 2 |
| NWMNN_1412 | zwf | −194 | GTTATGTCTACGTTAA | −179 | Glucose-6-phosphate 1-dehydrogenase |
| NWMNN_1432 | accB | −190 | TTTAATCAAGAGTTAA | −175 | Acetyl coenzyme A carboxylase, biotin carboxyl carrier protein |
| NWMNN_1453 | −145 | GTTAATTTAAATTTAA | −130 | 5-Formyltetrahydrofolate cyclo-ligase | |
| NWMNN_1500 | −16 | GGTAAAGGTGAGTTAA | −1 | Hydrolase, HAD-superfamily, subfamily IIIA | |
| NWMNN_1593 | pfk | −47 | GGTAATTTAGAGTTAA | −32 | 6-Phosphofructokinase |
| NWMNN_1595 | accD | −188 | TTTAATTAAACGTTAA | −173 | Acetyl coenzyme A carboxylase beta subunit |
| −76 | GTTAAAAACGGGTTTA | −61 | |||
| NWMNN_1871 | −22 | TTTAAAGAGAGGTTAA | −7 | Aspartate transaminase | |
| NWMNN_1929b | −288 | TTTAATAAATAGTTAA | −273 | Similar to succinyl-diaminopimelate desuccinylase | |
| NWMNN_2047 | manA1 | −194 | GTTAAAGTACTGTAAA | −179 | Mannose-6-phosphate isomerase, class I |
| NWMNN_2201 | −122 | GTTAATTTAAGGTTAT | −107 | Dehydrogenase family protein | |
| NWMNN_2206 | −240 | GTTAAATGGATTTTAA | −225 | Monooxygenase family protein | |
| −239 | GTTAAAATCCATTTAA | −224 | |||
| NWMNN_2271 | −82 | GTTAATGTATTTTTAA | −67 | Acetyltransferase, GNAT family protein | |
| NWMNN_2272 | −124 | GTTAATGTATTTTTAA | −109 | Zinc-containing alcohol dehydrogenase | |
| NWMNN_2592 | −220 | GTTAACAACACGTTAA | −205 | 2-Oxoglutarate/malate translocator | |
| −219 | GTTAACGTGTTGTTAA | −204 | |||
| NWMNN_2467 | −107 | CTTAATTAAGGGTTAA | −92 | O-Acetyltransferase OatA | |
| NWMNN_2526 | phoB | −87 | GTTAAAAATATGTAAA | −72 | Alkaline phosphatase III precursor |
| DNA replication/ recombination/repair | |||||
| NWMNN_0090 | −27 | GGTAATAGCCGGTTAA | −12 | Similar to replication initiation protein | |
| NWMNN_0448 | holB | −170 | GTTAATAACTTATTAA | −155 | DNA polymerase III delta subunit |
| NWMNN_1364 | dnaD | −36 | ATTAAGTAATGGTTAA | −21 | DNA replication protein DnaD |
| Nucleotide metabolism | |||||
| NWMNN_0249 | −37 | TTTAAAATACAGTTAA | −22 | 5′ nucleotidase, lipoprotein e(P4) family protein | |
| NWMNN_0699 | nrdI | −310 | GTTAAATTACTTTTAA | −295 | Ribonucleotide reduction-related NrdI protein |
| NWMNN_0809 | −86 | GTTAATGAACAGTTAT | −71 | Similar to pyridine nucleotide-disulfide oxidoreductase | |
| NWMNN_1118 | −345 | GTTAATTATCCGCTAA | −330 | Pseudouridylate synthase | |
| −249 | ATTAAAATTAAGTTAA | −234 | |||
| NWMNN_1119b | pryR | −54 | GTTACAATCATGTTAA | −39 | Pyrimidine operon regulatory protein |
| NWMNN_1120b | gmk | −236 | GTTACAATCATGTTAA | −221 | Guanylate kinase |
| NWMNN_1143 | rnc | −52 | TTTAATTTTACGTTAA | −37 | RNase III |
| NWMNN_1185 | ftsK | −42 | GTTAAAACGACGTTAT | −27 | DNA translocase FtsK/SpoIIIE family protein |
| NWMNN_1411 | −134 | GTTATGTCTACGTTAA | −119 | Similar to RNase Z | |
| NWMNN_2091 | −55 | GTTACTTAAGCGTTAA | −40 | Similar to quinone oxidoreductase | |
| Unknown function | |||||
| NWMNN_0039 | −49 | GTTAAAATAGCATTAA | −34 | Conserved hypothetical protein | |
| NWMNN_0087 | −86 | GTAAATTTTTCGTTAA | −71 | Conserved hypothetical protein | |
| NWMNN_0157 | −22 | ATTAATAAATAGTTAA | −7 | Conserved hypothetical protein | |
| NWMNN_0165b | −123 | GTTAATATATAGTTAA | −108 | Conserved hypothetical protein | |
| −102 | GTTAATGCTTTGTTTA | −87 | |||
| NWMNN_0295 | −55 | GTTACAAAGAGGTTAA | −40 | Similar to ORF041 of Bacteriophage 96 | |
| NWMNN_0362 | −95 | GTTAATCAAGAGTTAA | −80 | Conserved hypothetical protein | |
| −84 | GTTAAGATGAATTTAA | −69 | |||
| NWMNN_0370 | −43 | GTTAAGATGCTGTTAT | −28 | Conserved hypothetical protein | |
| NWMNN_0387 | −378 | GTTAAATGAGGTTTAA | −363 | Conserved hypothetical protein | |
| NWMNN_0402b | −90 | TTTAATAAATAGTTAA | −75 | Conserved hypothetical protein | |
| NWMNN_0403b | lpl1nm | −254 | TTTAATAAATAGTTAA | −239 | Staphylococcal tandem lipoprotein |
| NWMNN_0503 | −103 | GTTAAAACCCCGTTAT | −88 | Similar to small methyltransferase | |
| NWMNN_0546 | −109 | CTTAATAGCTTGTTAA | −94 | Conserved hypothetical protein | |
| NWMNN_0605 | −98 | GTTAAACTAATTTTAA | −83 | Conserved hypothetical protein | |
| NWMNN_0632 | −239 | GTTAAAAATTTGTAAA | −224 | Conserved hypothetical protein | |
| NWMNN_0687 | −250 | GTTAATTGAATGATAA | −235 | Conserved hypothetical protein | |
| −181 | GTTAAATCAAAGTTGA | −166 | |||
| NWMNN_0746 | −147 | GTTAAATATGTGTTAT | −132 | Conserved hypothetical protein | |
| NWMNN_0759 | −126 | GTTAATAGTTTGTTCA | −111 | Conserved hypothetical protein | |
| −115 | GTTCATCGCAAGTTAA | −100 | |||
| NWMNN_0810 | −246 | GTTAATGAACAGTTAT | −231 | Conserved hypothetical protein | |
| NWMNN_0823 | −29 | CTTAAAAGTATGTTAA | −14 | Conserved hypothetical protein | |
| NWMNN_0834 | −164 | GTTAAGAGAAAATTAA | −149 | Conserved hypothetical protein | |
| NWMNN_0956 | −262 | GTTAACAATAAATTAA | −247 | Similar to Zn-dependent hydrolase | |
| −261 | TTTAATTTATTGTTAA | −246 | |||
| NWMNN_0958 | −321 | GTAAAAAGGTTGTTAA | −306 | Conserved hypothetical protein | |
| NWMNN_1059 | −56 | GTTACAATATGGTTAA | −41 | hypothetical protein | |
| NWMNN_1067 | −99 | ATTAATGAAGTGTTAA | −84 | Conserved hypothetical protein | |
| −98 | GTTAACACTTCATTAA | −83 | |||
| NWMNN_1072 | −46 | GTTAAACTTAAGTTGA | −31 | Conserved hypothetical protein | |
| NWMNN_1077b | −85 | GCTAAAACTATGTTAA | −70 | Conserved hypothetical protein | |
| NWMNN_1152b | −221 | GTTAAATCTAAGTTAC | −206 | Conserved hypothetical protein | |
| NWMNN_1153b | −195 | GTTAAATCTAAGTTAC | −180 | Similar to GTP binding protein | |
| NWMNN_1487b | −280 | GTTAGACTTTTGTTAA | −265 | GTP binding protein LepA | |
| NWMNN_1599 | −42 | GTTATAATGTAGTTAA | −27 | CBS domain DNA binding protein | |
| NWMNN_1663 | −182 | GTTAAAAATATTTTAA | −167 | Conserved hypothetical protein | |
| −181 | GTTAAAATATTTTTAA | −166 | |||
| NWMNN_1669 | −31 | TTTAATAAGTTGTTAA | −16 | Conserved hypothetical protein | |
| NWMNN_1707b | −376 | GTTAATTAATATTTAA | −361 | Hypothetical protein | |
| −365 | GTTAATAAGTGGTTAA | −350 | |||
| −354 | TTTAATAAAACGTTAA | −339 | |||
| NWMNN_1708 | −109 | GTTAATAGATAGTTAA | −94 | Conserved hypothetical protein, homologous to ear (MW0758) | |
| NWMNN_1859 | −71 | GATAAATGATTGTTAA | −56 | Conserved hypothetical protein | |
| NWMNN_1860 | −214 | GATAAATGATTGTTAA | −199 | Conserved hypothetical protein | |
| NWMNN_2075 | −219 | GTTTAAAAAAGGTTAA | −204 | Conserved hypothetical protein | |
| NWMNN_2088 | −23 | GTTAAAGTTAATTTAA | −8 | Conserved hypothetical protein | |
| NWMNN_2202 | −141 | GTTAATTTAAGGTTAT | −126 | Conserved hypothetical protein | |
| NWMNN_2228 | −109 | GTTAAGATGATGTAAA | −94 | Conserved hypothetical protein | |
| NWMNN_2408 | −184 | GTTAAGCGGAATTTAA | −169 | Conserved hypothetical protein | |
| NWMNN_2409 | −378 | GTTAAGCGGAATTTAA | −363 | DedA family protein | |
| NWMNN_2415 | −183 | GTTATAATACAGTTAA | −168 | Conserved hypothetical protein | |
| NWMNN_2432 | −74 | GTTAAGTTGATGTAAA | −59 | Conserved hypothetical protein | |
| NWMNN_2436 | −46 | TTTAAAATCATGTTAA | −31 | Conserved hypothetical protein | |
| NWMNN_2508 | −202 | GTTAATGTGTTGTTCA | −187 | Conserved hypothetical protein | |
| NWMNN_2525 | −126 | GTTAAAAATATGTAAA | −111 | Hypothetical protein | |
| NWMNN_2587b | −40 | GTTATAAGCATGTTAA | −25 | Similar to rhodanese family protein |
The start point and end point are the distance from the translation start codon. Conserved repeat sequences are underllined.
This gene shares the same repeat sequence with an adjacent gene.
MHC, major histocompatibility complex.
The direct repeat sequence is necessary for binding to SaeR.
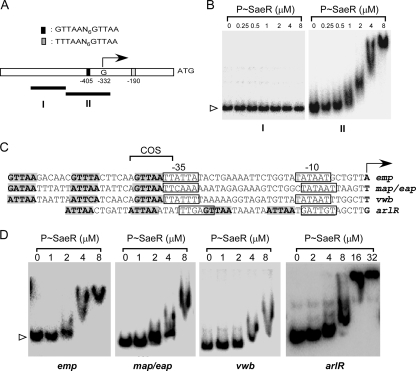
The gene hla encodes alpha-hemolysin and is highly regulated by the sae locus (19, 33, 44, 50). The promoter region of hla contains two direct repeat sequences: an imperfect repeat sequence (TTTAAN6GTTAA, from −190 to −175) and a perfect repeat sequence (GTTAAN6GTTAA, from −405 to −390) (Fig. 6A). It should be noted that the position is the distance from the ATG start codon, not from the transcription start site (11). The transcription start site corresponds to position −332. As the imperfect repeat sequence is located downstream of the transcription start site and is probably not involved in transcriptional regulation, we focused on the perfect repeat sequence at position −405. We PCR amplified two DNA fragments from the hla promoter region: a 180-nucleotide (nt) DNA fragment without the repeat sequence (fragment I) and a 200-nt DNA fragment with the perfect repeat sequence (fragment II). We hypothesized that if the direct repeat sequence is necessary for binding to SaeR, the 180-nt DNA fragment should not bind to P-SaeR while the 200-nt DNA fragment should. After labeling these DNA fragments with [γ-32P]ATP, we mixed each DNA fragment with P-SaeR and performed an EMSA. As shown in Fig. 6B, only DNA fragment II, the DNA with the perfect repeat sequence, bound to P-SaeR, strongly supporting the idea that the direct repeat sequence is the SaeR binding site.
FIG. 6.
DNA binding of P-SaeR to various staphylococcal promoters. (A) Schematic map of the hla promoter. The perfect repeat sequence is represented by a black box, and the imperfect repeat sequence is represented by a gray box. The sequences of the repeats are also shown above the map, where N represents any nucleotide. Values represent distances from the ATG start codon, which is shown to the right. The transcription start site (G) is shown by a right-angled arrow. The two DNA probes used for EMSA are shown as solid lines with the names under them. (B) DNA binding of P-SaeR to two different regions of the hla promoter. The concentration of P-SaeR is indicated at the top. Unbound free DNA probe is indicated by a white arrowhead. I and II represent the hla promoter regions shown in panel A. (C) The direct repeat sequences in the promoters of emp, map/eap, vwb, and arlR. The transcription sites are indicated by a boldfaced letter and a right-angled arrow. The −10 and −35 regions of the promoters are indicated by solid boxes. The direct repeat sequences are in boldface and shaded gray. (D) DNA binding of P-SaeR to the emp, map/eap, vwb, and arlR promoters. Protein concentrations are indicated at the top. For clarity, only unbound free DNA is indicated.
We decided to further examine the role of the direct repeat sequence in SaeR binding with four more genes that contain various combinations of the SaeR binding sequence: emp, map/eap, vwb, and arlR. The genes emp, map/eap, and vwb encode surface proteins important for adhesion and immune evasion (9, 24, 25), while arlR encodes the RR for the arlRS system that regulates the expression of virulence genes such as spa and cap5 (15, 16, 37). The genes emp and map/eap have been reported to be regulated by the sae locus, but vwb and arlR have not. In addition, only emp contains a perfect repeat sequence, while the others have imperfect repeats. It should also be noted that the promoter of arlR has two binding sites but those of the others have 1.5 binding sites (Fig. 6C). After radiolabeling these DNA fragments, we performed an EMSA with P-SaeR, and the results are shown in Fig. 6D. All of the promoters bound to P-SaeR. The binding affinity of emp containing the perfect repeat sequence appeared stronger than those of the others. These results further support the idea that the direct repeat sequence mediates DNA binding to SaeR.
Mutational changes in the direct repeat sequence in the P1 promoter abolish not only SaeR binding but also promoter function.
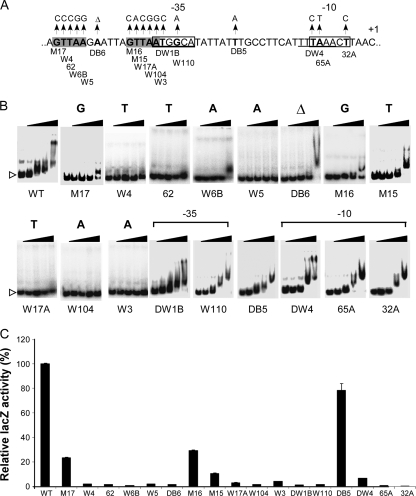
If the direct repeat sequence is the SaeR binding site, mutational changes in the sequence should abolish SaeR binding and promoter activity. To eliminate bias, instead of site-directed mutagenesis, we used a random mutagenesis strategy. We mutated the P1 promoter by amplifying the DNA with error-prone DNA polymerases and then inserted the PCR products into lacZ reporter plasmid pYJ-lacZ, a multicopy plasmid derived from pYJ335 (28). As described in Materials and Methods, the resulting plasmids were inserted into strain RN4220 or TB3, a sae+ strain derived from strain Newman, and the cells were spread on TSA plates containing X-Gal. White colonies were selected and subjected to DNA sequencing analysis. Through this process, we identified eight mutations in the SaeR binding site (W4, 62, W6B, W5, DB6, W17A, W104, and W3) (Fig. 7A), confirming that mutational changes in the binding sequence abolish promoter activity (i.e., white color). Among these mutations, DB6 altered the distance between the repeat sequences from 6 nt to 5 nt, suggesting that not only the nucleotide sequence but also the spacing is important for the binding of SaeR. We also identified five mutations in the promoter sequence in the −35 (DW1B, W110) and −10 (DW4, 65A, 32A) regions. As a control, we included a nucleic acid substitution mutant showing blue color (DB5) on further analysis (Fig. 7A). The mutation is located between the −35 and −10 regions. Since no mutations were obtained for three nucleotides in the SaeR binding site, we mutated these three nucleotides by PCR-mediated site-directed mutagenesis. The resulting mutants are M15, M16, and M17 (Fig. 7A). Strain Newman carrying the mutant plasmids also showed reduced blue color, suggesting that these nucleotides are also important for efficient binding to SaeR.
FIG. 7.
Effects of P1 promoter mutations on binding to P-SaeR and promoter function. (A) Summary of the mutations in the P1 promoter. Each mutation is indicated by an arrow with the name under the sequence. The −10 and −35 regions are in a solid box. The direct repeat sequence is in boldface and shaded gray. +1, transcription start site; Δ, a deletion mutation. (B) Binding of the mutant promoters to P-SaeR. Increasing amounts of P-SaeR (0, 1, 2, 4, and 8 μM) were used for the binding assay. The mutated nucleotides of the direct repeat and mutated promoter regions are shown at the top. Unbound free DNA probe is indicated by a white arrowhead. Δ, a deletion mutation. (C) Activities of the mutated P1 promoters represented by LacZ expression. The values presented are relative LacZ expression, where the LacZ expression from the wild-type promoter was set to 100%. The lacZ assays were repeated two or three times with similar results. Error bars represent standard deviations. WT, wild type.
To examine the correlation between the reduced promoter activity of the mutants and the SaeR binding of the promoters, we performed an EMSA for all of the mutants using P-SaeR. As shown in Fig. 7B, all of the mutations in the SaeR binding site, including spacing mutation DB6, reduced DNA binding to SaeR, confirming that the direct repeat sequence is the SaeR binding site. Some residual SaeR binding was observed with M17, W6B, DB6, M16, and M15, suggesting that the contributions of these positions to SaeR binding are different. As expected, none of the other mutations significantly changed DNA binding to SaeR.
To quantitatively measure the effects of the mutations on the function of the P1 promoter, we measured the LacZ activity of the mutants from 16-h overnight cultures. As shown in Fig. 7C, all of the 11 mutations in the SaeR binding sites greatly reduced LacZ activity, confirming the correlation between SaeR binding and the transcriptional activity of the promoter. Of the mutations, M15, M16, and M17 did not completely abolish promoter activity; compared with the wild-type P1 promoter, the promoters with the mutations still retained 10% to 30% activity. These results confirm that for sae-mediated transcriptional activation, the contribution of each nucleotide position might not be equal. In EMSA, indeed, the promoters with these mutations showed some residual SaeR binding. As expected, all of the mutations in the promoter regions also abolished or greatly reduced LacZ activity while the DB5 mutant showed nearly wild-type promoter activity (80%), showing that not all of the mutations in the P1 promoter region can abolish LacZ activity and the greatly reduced promoter activities in other mutants are due to the loss of specific functions of the DNA sequence (i.e., SaeR binding or RNA polymerase binding). In summary, through a random mutagenesis strategy, we confirmed that (i) the direct repeat sequence GTTAAN6GTTAA is the SaeR binding site and (ii) the binding site is critical for sae-mediated transcriptional activation of the P1 promoter.
DISCUSSION
The SaeRS two-component system is a staphylococcal signaling system that plays a critical role in activating the transcription of many important virulence genes. However, the molecular mechanism of this activation has not yet been elucidated. Here, we report that SaeR binds to the direct repeat sequence GTTAAN6GTTAA and phosphorylation is essential for DNA binding.
Several lines of data support our claim that GTTAAN6GTTAA is the SaeR binding site. First, in DNase footprinting assays, the sequence was found in the P1 promoter regions protected by either SaeRC or P-SaeR (Fig. 5). Second, the sequence was identified in the promoter region of 36 known members of the sae regulon (Table 4). Third, although all of the DNA fragments containing the repeat sequence bound to SaeR, a DNA fragment without the repeat sequence did not (Fig. 6). Lastly, mutations of the repeat sequence severely reduced DNA binding to SaeR (Fig. 7).
Interestingly, all of the genes containing the SaeR binding site in their promoter regions have not been reported to be regulated by the SaeRS system. Of the 155 genes containing the SaeR binding site, only 36 (23%) have been reported to be regulated by SaeRS. One possible explanation for the discrepancy is that some of the genes may be genuinely regulated by the SaeRS system but the regulation has not been detected yet. So far, the sae regulon has been identified mainly by microarray assay or proteomic analysis. These methods might not be sensitive enough to detect all transcriptional changes. Therefore, in the future, more-sensitive techniques such as high-throughput real-time quantitative reverse transcriptase-mediated PCR might identify new genes of the sae regulon. The other possibility is that the binding affinity of some of the SaeR binding sites is too low for them to be genuine targets of sae-mediated regulation under physiologically relevant conditions. In our P1 promoter mutagenesis study, single nucleotide changes in the binding site could abolish binding to SaeR and promoter activity. Therefore, many of the imperfect SaeR binding sites may not bind to SaeR in vivo. For example, the arlR promoter clearly can bind to SaeR at a high concentration of purified P-SaeR in vitro (Fig. 6D); but the arlRS operon is not regulated by the sae two-component system (45). Therefore, the arlR binding shown in Fig. 6D might not be physiologically relevant in vivo. However, some of the well-known sae regulon genes in Table 4, such as efb, hlb, and lukS, have only one imperfect SaeR binding site, suggesting that not only the motif sequence itself but also the context of the motif (e.g., distance from the promoter element, etc.) might play a role in selecting the targets of SaeR binding and sae-mediated regulation.
Conversely, the SaeRS system seems to affect the transcription of the genes that do not contain the SaeR binding site. For example, among the 212 genes of the sae regulon identified by microarray analysis of strain MW2, 176 do not contain the SaeR binding site. How, then, could the SaeRS system have altered the transcription of these genes? One possibility is that the SaeRS system might affect the expression of some of the 176 genes indirectly. For example, as shown in Table 4, four transcriptional regulators (NWMN_0038, NWMN_0636, NWMN_0925, and arlR) have the SaeR binding site in their promoter regions. Although arlR is not involved in downstream regulation by sae, the other three regulators might play a role in it. Since the transcriptional regulatory systems in S. aureus are interconnected and form a very complex network, transcriptional changes in a regulator protein can be transmitted or amplified via the network. In addition, since the sae regulon includes not only virulence genes but also genes involved in various cellular processes such as protein synthesis, ion transport, and energy metabolism (48), we can envision that alterations in those processes might indirectly cause transcriptional changes in other genes. Clearly, more studies are required to identify direct targets and the mechanism of indirect regulation.
In a previous study, Harraghy et al., identified a COS (conserved octanucleotide sequence, AGTTAATT) in 11 genes, of which 6 are genes of the sae regulon such as eap, emp, and efb (24). In emp, a mutational change of three nucleotides (AGTTAATT to TCATAATT, where boldface and underlining indicate the mutated nucleotides and SaeR binding sequences, respectively) decreased emp expression by more than half (24). As shown in Fig. 6C, the COS overlaps the SaeR binding site, and the mutational change in the COS in this study also changed two nucleotides in the SaeR binding site from GTTAA to CATAA. Since we observed in our P1 mutagenesis study that even a single nucleotide change could abolish DNA binding to P-SaeR, we suspect that the two nucleotide changes in this study also abolished DNA binding to SaeR. Therefore, we think the effect of the COS mutation on emp expression is due to the mutated promoter region's inability to bind to SaeR. In fact, in our analysis of the 11 COS-containing genes, 9 genes (eap, emp, vwb, efb, hlgA, hlgC, sbi, lukE, and lukD) have one or more SaeR binding sites (either perfect or imperfect). Therefore, we propose that COS is a part of the SaeR binding site. In their study, the authors reported that neither unphosphorylated SaeR nor the DNA binding domain of SaeR bound to the three promoters (i.e., emp, map/eap, and vwb). In our study, however, although the binding affinity was very low (KD = 6 μM), the DNA binding domain (i.e., SaeRC) clearly bound to its target, the P1 promoter (Fig. 2B). It is possible that, compared with the P1 promoter, the three promoters tested previously have lower binding affinities for SaeRC, which, in turn, made it more difficult to detect protein-DNA binding.
In this study, we demonstrated that phosphorylation is essential for SaeR binding to its target DNA (Fig. 2). Recently, Mainiero et al. also showed that SaeR with a mutation at the phosphorylation site (D51N) could not activate transcription from the hla promoter (39), further confirming the essential role of SaeR phosphorylation in transcriptional regulation. This essentiality of phosphorylation differentiates the SaeRS two-component system from the VraSR (vancomycin resistance-associated sensor/regulator) system, a well-studied staphylococcal two-component system that coordinates cellular responses to antibiotics inhibiting cell wall synthesis (6, 12, 31). VraR, the RR of the VraSR system, recognizes the DNA motif ACT(X)nAGT (where X is any nucleic acid and n = 1 to 3); however, phosphorylation is not required for VraR to bind to the most conserved binding site (7). On the other hand, VraR binding to the less conserved secondary binding site does require phosphorylation. Interestingly, as with the primary SaeR binding site, the secondary binding site of VraR in the vraRS operon overlaps the −35 region and direct interaction between VraR and sigma factor was postulated (7). The different roles of phosphorylation in the SaeRS and VraSR systems suggest that the signaling mechanism of staphylococcal two-component systems might be diverse and cannot be generalized to one model.
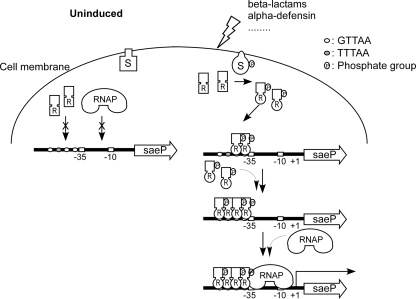
The P1 promoter contains a secondary SaeR binding site with an imperfect repeat sequence (GTTAAN6TTTAA). This secondary site is separated from the primary SaeR binding site (GTTAAN6GTTAA) by 5 nt (Fig. 5B). The EMSA and DNase footprinting analysis results suggest that P-SaeR binds to the secondary binding site as well. In EMSA, two shifted bands were observed, suggesting the existence of two different species of protein-DNA complex (Fig. 2C). In addition, in DNase I footprinting assays, P-SaeR protected not only the primary binding sites but also the secondary binding site (Fig. 5). We envision that P-SaeR binds to the primary binding site first, generating the lower shift band in Fig. 2C, and recruits P-SaeR to the secondary binding site, generating the higher band in EMSA (Fig. 2C). The occupation of the secondary binding site might stabilize P-SaeR binding to the promoter such that P-SaeR can recruit RNA polymerase to the promoter through protein-protein interactions. The close proximity of the primary SaeR binding site and the −35 region resembles the class II promoters in E. coli, where the transcription regulator interacts with sigma factor 70 (5, 35). Therefore, we postulate that the interaction between P-SaeR and the sigma factor recruits RNA polymerase to the P1 promoter. Combining all of the data, we propose, for the first time, a model for the SaeRS signal transduction mechanism in the P1 promoter (Fig. 8). Under uninduced conditions, SaeS is not active and does not phosphorylate SaeR. However, when external environmental stressors (e.g., β-lactam antibiotics or alpha-defensin) are present, SaeS experiences conformational changes and autophosphorylation and then transfers the phosphate group to D51 of SaeR. Phosphorylated SaeR binds to the primary SaeR binding domains, forming a dimer, and recruits more P-SaeR to the secondary binding site. As a result, the SaeR-P1 promoter complex becomes stable and recruits RNA polymerase to the promoter by interactions between SaeR and sigma factor σA, resulting in transcription initiation at the promoter.
FIG. 8.
Model of P1 promoter activation by the SaeRS system. RNAP, RNA polymerase.
Acknowledgments
This study was supported by scientist development grant 0835158N from the American Heart Association (T.B.), AI077564 from the National Institute of Allergy and Infectious Diseases (T.B), AI074658 (C.H.), and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (C.H.).
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Adhikari, R. P., and R. P. Novick. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 3.Bae, T., T. Baba, K. Hiramatsu, and O. Schneewind. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 62:1035-1047. [DOI] [PubMed] [Google Scholar]
- 4.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U. S. A. 101:12312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 6.Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. [DOI] [PubMed] [Google Scholar]
- 7.Belcheva, A., V. Verma, and D. Golemi-Kotra. 2009. DNA-binding activity of the vancomycin resistance associated regulator protein VraR and the role of phosphorylation in transcriptional regulation of the vraSR operon. Biochemistry 48:5592-5601. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjerketorp, J., M. Nilsson, A. Ljungh, J. I. Flock, K. Jacobsson, and L. Frykberg. 2002. A novel von Willebrand factor binding protein expressed by Staphylococcus aureus. Microbiology 148:2037-2044. [DOI] [PubMed] [Google Scholar]
- 10.Cheung, A. L., K. A. Nishina, M. P. Trotonda, and S. Tamber. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 12.Cui, L., H. M. Neoh, M. Shoji, and K. Hiramatsu. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly, M. I., M. Zhou, C. S. Millard, S. Clancy, L. Stols, W. H. Eschenfeldt, F. R. Collart, and A. Joachimiak. 2006. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 47:446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 15.Fournier, B., and A. Klier. 2004. Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology 150:3807-3819. [DOI] [PubMed] [Google Scholar]
- 16.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 17.Geiger, T., C. Goerke, M. Mainiero, D. Kraus, and C. Wolz. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15-22. [DOI] [PubMed] [Google Scholar]
- 19.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 20.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46:246-250. [DOI] [PubMed] [Google Scholar]
- 21.Giraudo, A. T., C. G. Raspanti, A. Calzolari, and R. Nagel. 1994. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40:677-681. [DOI] [PubMed] [Google Scholar]
- 22.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect. Immun. 73:3415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 24.Harraghy, N., D. Homerova, M. Herrmann, and J. Kormanec. 2008. Mapping the transcription start points of the Staphylococcus aureus eap, emp, and vwb promoters reveals a conserved octanucleotide sequence that is essential for expression of these genes. J. Bacteriol. 190:447-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harraghy, N., J. Kormanec, C. Wolz, D. Homerova, C. Goerke, K. Ohlsen, S. Qazi, P. Hill, and M. Herrmann. 2005. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 151:1789-1800. [DOI] [PubMed] [Google Scholar]
- 26.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 27.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. ASM Press, Washington, DC.
- 28.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda, H., M. Kuroda, L. Cui, and K. Hiramatsu. 2007. Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268:98-105. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 32.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 33.Li, D., and A. Cheung. 2008. The repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect. Immun. 76:1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, X., C. Yu, J. Sun, H. Liu, C. Landwehr, D. Holmes, and Y. Ji. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74:4655-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd, G. S., W. Niu, J. Tebbutt, R. H. Ebright, and S. J. Busby. 2002. Requirement for two copies of RNA polymerase alpha subunit C-terminal domain for synergistic transcription activation at complex bacterial promoters. Genes Dev. 16:2557-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 37.Luong, T. T., and C. Y. Lee. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123-3131. [DOI] [PubMed] [Google Scholar]
- 38.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mainiero, M., C. Goerke, T. Geiger, C. Gonser, S. Herbert, and C. Wolz. 2010. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 192:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 41.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 43.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 44.Rampone, H., G. L. Martinez, A. T. Giraudo, A. Calzolari, and R. Nagel. 1996. In vivo expression of exoprotein synthesis with a Sae mutant of Staphylococcus aureus. Can. J. Vet. Res. 60:237-240. [PMC free article] [PubMed] [Google Scholar]
- 45.Rogasch, K., V. Ruhmling, J. Pane-Farre, D. Hoper, C. Weinberg, S. Fuchs, M. Schmudde, B. M. Broker, C. Wolz, M. Hecker, and S. Engelmann. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schäfer, D., T. T. Lam, T. Geiger, M. Mainiero, S. Engelmann, M. Hussain, A. Bosserhoff, M. Frosch, M. Bischoff, C. Wolz, J. Reidl, and B. Sinha. 2009. A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters response to biocide exposure. J. Bacteriol. 191:7306-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 48.Voyich, J. M., C. Vuong, M. Dewald, T. K. Nygaard, S. Kocianova, S. Griffith, J. Jones, C. Iverson, D. E. Sturdevant, K. R. Braughton, A. R. Whitney, M. Otto, and F. R. Deleo. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 199:1698-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 50.Xiong, Y. Q., J. Willard, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2006. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J. Infect. Dis. 194:1267-1275. [DOI] [PubMed] [Google Scholar]
- 51.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]