Abstract
Previous studies have shown that the Helicobacter pylori ArsRS two-component signal transduction system contributes to acid-responsive gene expression. To identify additional members of the ArsRS regulon and further investigate the regulatory role of the ArsRS system, we analyzed protein expression in wild-type and arsS null mutant strains. Numerous proteins were differentially expressed in an arsS mutant strain compared to a wild-type strain when the bacteria were cultured at pH 5.0 and also when they were cultured at pH 7.0. Genes encoding 14 of these proteins were directly regulated by the ArsRS system, based on observed binding of ArsR to the relevant promoter regions. The ArsRS-regulated proteins identified in this study contribute to acid resistance (urease and amidase), acetone metabolism (acetone carboxylase), resistance to oxidative stress (thioredoxin reductase), quorum sensing (Pfs), and several other functions. These results provide further definition of the ArsRS regulon and underscore the importance of the ArsRS system in regulating expression of H. pylori proteins during bacterial growth at both neutral pH and acidic pH.
Persistent colonization of the human stomach with Helicobacter pylori results in gastric inflammation and contributes to the pathogenesis of peptic ulceration, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma (4, 43). H. pylori is highly adapted for colonization of the human stomach, and multiple mechanisms allow the bacteria to proliferate in this highly acidic environment (3, 11). One mechanism by which H. pylori tolerates gastric acidity involves the enzymatic generation of ammonia from various substrates. Enzymes involved in ammonia production include urease (a nickel-containing enzyme composed of subunits UreA and UreB) (36) and amidases (38).
In comparison to many other bacteria, H. pylori possesses relatively few transcriptional regulatory systems (2, 46). The ArsRS two-component signal transduction system has an important role in allowing H. pylori to sense and regulate target genes in response to changes in pH and is required for colonization of the stomach (25, 29, 31-33, 50, 51). This two-component system is comprised of a sensor kinase (ArsS, corresponding to HP0165 in H. pylori 26695) and a response regulator (ArsR, corresponding to HP0166) (20, 28; reviewed in references 21 and 34). The ArsRS two-component system regulates multiple genes involved in acid resistance, including genes encoding members of the urease complex (33), amidases (32), and carbonic anhydrase (50). The accepted model for pH-dependent regulation by this two-component system involves detection of a change in the environmental pH, which leads to phosphorylation of the ArsS sensor kinase protein (34). The phosphate from the activated ArsS protein is then transferred to the cognate response regulator ArsR, which in turn binds to and modulates the expression of target genes. In support of this model, several studies have detected direct binding of ArsR to the promoter regions of various acid-responsive target genes (12, 32, 33, 50, 51). In addition to the regulation of H. pylori gene expression by the phosphorylated form of ArsR, ArsR in its unphosphorylated state also likely plays a key role in the regulation of H. pylori genes. This view is supported by the observation that strains encoding a form of ArsR with a mutated phosphorylation site are viable, whereas arsR null mutant strains are nonviable (12).
In previous studies, members of the ArsRS regulon have been identified by isolating DNA sequences that bind to ArsR (12) and by analyzing gene expression in arsS mutant strains compared to wild-type strains (15, 19, 25, 32, 51). Several strain-specific variations in ArsRS regulons have been identified (15). Comparative transcriptional profiling experiments have been performed using wild-type and arsS mutant strains cultured at either pH 5.0 or pH 7.0 (15, 25, 32, 51). Validation of the array results has been carried out for only a few of the genes that are differentially expressed in wild-type and arsS mutant strains (19, 32, 33, 50, 51), and thus the ArsRS regulon has not yet been completely characterized. In this study, we set out to characterize further the regulon controlled by the ArsRS two-component system and to investigate further the regulatory role of the ArsRS system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori reference strain J99 (2) was used in this study. Bacterial cultures were routinely passaged every 2 days on Trypticase soy agar plates containing 5% sheep blood at 37°C in ambient air containing 5% CO2. For proteomic experiments, wild-type and mutant H. pylori strains were grown overnight in modified brucella broth supplemented with 5% fetal bovine serum (BB-FBS) (pH 7.0) to an optical density at 600 nm (OD600) of 0.5 to 0.6. The cultures were then subcultured into fresh BB-FBS broth (adjusted to either pH 5.0 or 7.0) for either 1.5 h or 3.5 h (25). Bacteria were harvested and stored at −80°C prior to proteomic analysis. Escherichia coli strains were grown in Luria-Bertani (LB) medium supplemented with ampicillin (50 μg/ml) or kanamycin (25 μg/ml) as necessary.
Generation of H. pylori arsS sensor kinase mutants.
To generate arsS mutant strains, two independent single-colony isolates of wild-type H. pylori strain J99 (designated J99A and J99B), each of which was catalase, urease, and cagA positive, were transformed with the nonreplicating plasmid p165Km1 as previously described (25). Transformants were selected on brucella agar plates supplemented with kanamycin (10 μg/ml). Insertion of a kanamycin resistance cassette into the chromosomal arsS gene was confirmed by PCR analysis. The arsS mutant strains were designated J99A (arsS::kan) and J99B (arsS::kan).
Difference gel electrophoresis (DIGE) and mass spectrometry (MS).
Wild-type and isogenic arsS mutant strains [J99A and J99A (arsS::kan) or J99B and J99B (arsS::kan)] were each cultured in quadruplicate at pH 5.0 and pH 7.0 as described above. Bacteria were lysed for 30 min with lysis buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)} as previously described (10). Bacterial lysates (0.25 mg protein per sample) were precipitated separately with methanol and chloroform (54) and resuspended in 30 μl of labeling buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, 5 mM magnesium acetate). The N-hydroxysuccinimide (NHS)-ester dyes Cy2, Cy3, and Cy5 were used for minimal labeling, using the mixed-internal-standard methodology of Alban et al. (1). Two-thirds (167 μg) of each of the 16 experimental samples (quadruplicate preparations of wild-type strain J99 and the arsS mutant grown either at pH 5.0 or pH 7.0) was individually labeled with 200 pmol of either Cy3 or Cy5. In similar fashion, the remaining portions of each of the 16 experimental samples (83 μg) were combined and labeled with 800 pmol of Cy2 to generate the mixed internal standard. Labeling was performed for 30 min on ice in the dark, after which the reaction was quenched by treatment with 10 mM lysine for 10 min, followed by the addition of an equal volume of 2× rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 4 mg/ml dithiothreitol [DTT]). Pairs of Cy3/Cy5-labeled samples were mixed with an equal aliquot of Cy2-labeled mixed internal standard, and 500 μg of total protein was resolved on each gel. A sample loading matrix was created to randomize the two independent samples coresolved on each gel, and a dye swap labeling method was used to ensure that independent samples within an experimental group were never all labeled with the same Cy dye.
Proteins were resolved using eight coordinated DIGE gels for quantification essentially as described previously (1, 16, 17; reviewed in reference 24). All two-dimensional gel- and DIGE-associated instrumentation was manufactured by GE Healthcare (Piscataway, NJ). First-dimension separations were performed on a manifold-equipped IPGphor II first-dimension isoelectric focusing (IEF) unit, and second-dimension 12% SDS-PAGE was performed using hand-cast gels for which one plate was presilanized using an Ettan DALT 12 unit. Tripartite samples were passively rehydrated into 24-cm immobilized pH gradient (IPG) strips (pH 4 to 7 range) for 24 h and subjected to simultaneous isoelectric focusing per the manufacturer's recommendations. Focused proteins were equilibrated into the second-dimension loading buffer by incubating the IEF strips in equilibration buffer (30% glycerol, 2% SDS, 6 M urea, 50 mM Tris [pH 8.8], trace bromophenol blue) supplemented with 1% DTT for 20 min at room temperature, followed by 2.5% iodoacetamide in fresh equilibration buffer for an additional 20 min at room temperature. IPG strips were then seated on top of 12% homogenous polyacrylamide gels, and second-dimension SDS-PAGE was performed on all eight gels simultaneously at <1 W/gel overnight followed by 20 W/gel until the dye front had migrated off the gel. Cy2/3/5-specific 16-bit data files were acquired at 100-μm resolution separately by dye-specific excitation and emission wavelengths using a Typhoon 9400 variable-mode imager, and gels were poststained for total protein content with SyproRuby (Molecular Probes/Invitrogen) per the manufacturer's instructions.
The DeCyder v6.5 suite of software tools (GE Healthcare) was used for DIGE analysis. The normalized volume ratio of each individual protein spot feature from a Cy3- or Cy5-labeled sample was directly quantified relative to the Cy2 signal from the pooled-sample internal standard corresponding to the same spot feature. This was performed for all resolved features in a single gel, where no gel-to-gel variation exists between the three coresolved signals. The individual signals from the Cy2 standard were then used to normalize and compare Cy3/Cy2 and Cy5/Cy2 abundance ratios across the eight-gel set, enabling statistical confidence to be associated with each change in abundance or charge-altering posttranslational modification using Student's t test and analysis of variance (ANOVA). Unsupervised principal component analysis (PCA) (17, 18) was performed using the DeCyder Extended Data Analysis (EDA) module.
Proteins of interest were robotically excised and digested into peptides in-gel with modified porcine trypsin protease (Trypsin Gold; Promega), and the resulting peptides were applied to a stainless steel target using an integrated Spot Handling Workstation (GE Healthcare) per the manufacturer's recommendations. Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) and data-dependent TOF/TOF tandem MS/MS were performed on a Voyager 4700 (Applied Biosystems). The resulting peptide mass maps and the associated fragmentation spectra were collectively used to interrogate H. pylori sequences (Swiss-Prot and NCBI nonredundant databases) to generate statistically significant candidate identifications using GPS Explorer software (Applied Biosystems) running the MASCOT search algorithm (http://www.matrixscience.com). Database searches allowing for complete carbamidomethylation of cysteine, partial oxidation of methionine residues, and one missed cleavage were performed. Molecular weight search (MOWSE) scores, number of matched ions, number of matching ions with independent MS/MS matches, percent protein sequence coverage, and correlation of gel region with predicted molecular weight (MW) and pI were collectively considered for each protein identification.
EMSAs.
Full-length (26-kDa) ArsR and the DNA-binding domain of ArsR (13 kDa) were expressed in E. coli and purified as described previously (20). The primers listed in Table S1 in the supplemental material were used to PCR amplify 100- to 150-bp regions located upstream of the translation initiation sites of the selected target genes. For each primer set, the forward primer was 5′ biotinylated. To phosphorylate the full-length ArsR protein in vitro, the purified recombinant protein (3 μM) was incubated for 30 min at 25°C in phosphorylation buffer (50 mM Tris [pH 7.5], 5 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol) containing 50 mM acetyl phosphate (12). The recombinant ArsR proteins were incubated with biotinylated DNA probes (100 pM) for 20 min in a binding buffer [10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 50 ng/μl poly(dI-dC), 0.05% NP-40, 2.5% glycerol, 5 mM MgCl2]. For competition assays, a 20-fold excess of nonbiotinylated probe (compared to biotinylated probe) was included in the binding mixture. Loading buffer was then added, and the samples were subjected to electrophoresis in a 6% polyacrylamide native gel in 0.5× TBE (50 mM Tris [pH 8.3], 45 mM boric acid, 5 mM EDTA). Samples were electrophoretically transferred to a nylon membrane (Bio-Rad), and the transferred DNA was cross-linked to the membrane using a UV Stratalinker 1800 (Stratagene). Biotin-labeled DNA was detected using the Light Shift chemiluminescence electrophoretic mobility shift assay (EMSA) kit (Pierce) and visualized on X-ray film.
Analysis of AI-2 production.
H. pylori strains were cultured overnight in brucella broth supplemented with 5% FBS and inoculated into fresh culture medium at an OD600 of 0.2. At various time points, aliquots of the culture were removed and cell-free conditioned media (CM) prepared as previously described (23, 26). The conditioned media then were added (10% [vol/vol] final concentration) to cultures of the Vibrio harveyi reporter strain BB170 (6, 44). Following 5 h of incubation, the luminescence of V. harveyi cultures was quantified using a luminometer (OpticompII; MCM Instruments). Luminescence induction values were calculated as the ratio of luminescence observed following the addition of H. pylori conditioned medium to that observed following the addition of sterile brucella broth.
RESULTS
Proteomic analysis of wild-type and arsS mutant strains.
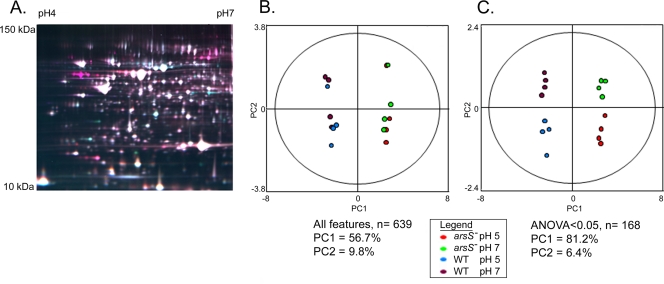
As a first step in investigating the role of the sensor kinase ArsS in H. pylori protein expression, we analyzed a pair of wild-type and isogenic arsS mutant strains designated strains J99A and J99A (arsS::kan). These strains were each cultured for 3.5 h in medium adjusted to starting pH values of 5.0 and 7.0. The pH of the pH 5.0 cultures increased slightly during the course of the experiment; at the time of harvest, the pH of wild-type cultures was 5.8 ± 0.3 and the pH of arsS mutant cultures was 5.5 ± 0.2. Bacterial cell lysates were prepared, and proteins were labeled with Cy3 or Cy5. All 16 extracts (quadruplicate samples of wild-type and arsS mutant strains, each cultured at pH 5.0 or 7.0) were then simultaneously coresolved across eight two-dimensional (2D) DIGE gels that included a Cy2-labeled 16-mix pooled-sample internal standard, as described in Materials and Methods (Fig. 1A). A total of 639 resolved protein spot features were matched across all eight gels, and the signals for each feature were normalized as described in the Materials and Methods. Principal component analysis (17, 18) was then used to assess the variation in expression patterns among these 639 features. The first principal component (PC1) comprised 56.7% of the variation, and this variation clearly distinguished between the wild-type and arsS mutant strains analyzed in this experiment (Fig. 1B). Neither the second principal component, comprising the second greatest source of variation (PC2 = 9.8%), nor any of the other principal components distinguished between the two different growth states (pH 5.0 and pH 7.0). This analysis, at the level of all matched features, demonstrated that most of the variation in protein expression reflected differences between the wild-type and mutant strains rather than differences arising from technical errors in sample handling or unanticipated variation in growth conditions.
FIG. 1.
Proteomic analysis of wild-type and arsS mutant [J99A and J99A (arsS::kan)] strains, cultured at pH 5.0 and pH 7.0. (A) Representative 2D DIGE gel. A false-color overlay is shown for demonstrative purposes (Cy2 [blue], internal standard; Cy3 [green], strain J99 cultured at pH 5.0; Cy5 [red], arsS mutant cultured at pH 5.0). (B) Unsupervised principal component analysis was used to assess variation in protein expression patterns among the 16 samples. A total of 639 resolved protein spot features were analyzed. The first principal component (PC1) comprised 56.7% of the variation and clearly distinguished between the wild-type and arsS mutant strains. The second principal component, comprising the second greatest source of variation, did not distinguish between the two different growth states (pH 5.0 versus pH 7.0). (C) Principal component analyses of 168 protein features that were altered in expression in any one of the four groups of samples relative to the others (based on ANOVA P values of <0.05). PC1 comprises over 81% of the variation among the 168 features in this subset, and the variation represented in PC2 (6.4%) distinguishes between bacteria grown at pH 5.0 versus pH 7.0.
We next narrowed the principal component analysis to focus on a subset of protein features that were significantly altered in expression in any one of the four groups of samples (wild-type and arsS mutant strains, each cultured at pH 5.0 and pH 7.0) relative to the others (based on ANOVA P values of <0.05). PC1 now comprised over 81% of the variation among the 168 features in this subset (Fig. 1C), and this variation again distinguished between the wild-type and arsS mutant strains. PC2 comprised 6.4% of the variation, and this variation distinguished between bacteria grown at pH 5.0 versus pH 7.0 (Fig. 1C). Thus, principal component analysis demonstrated distinct protein expression patterns in the four groups of samples.
As ArsS is known to be a regulator of acid-responsive gene expression (25, 32, 33, 50, 51), we sought to identify proteins that were differentially expressed in wild-type strain H. pylori J99A compared to the isogenic arsS mutant strain [J99A (arsS::kan)], each grown at pH 5.0. Forty-one spot features, corresponding to 25 differentially expressed proteins, were identified after in-gel digestion of proteins and mass spectrometry analysis of the resulting peptides (see Table S2 in the supplemental material). Of these, 13 were expressed at higher levels in the arsS mutant than in the wild-type strain, and 12 were expressed at lower levels in the mutant strain than in the wild-type strain (Table 1, experiment 1). Several of the differentially expressed proteins were present in a series across the first dimension of IEF, and mass spectrometry analysis of these spot features provided matches to the same intact protein, likely indicating the presence of charge-altering modifications.
TABLE 1.
Comparison of protein expression patterns in wild-type H. pylori strain J99 and isogenic arsS mutants
| Function and proteinb | Locus tagsc | Expt 1a |
Expt 2 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| pH 5 |
pH 7 |
pH 5 |
pH 7 |
||||||
| arsS/J99 ratiod | P valuee | arsS/J99 ratio | P value | arsS/J99 ratio | P value | arsS/J99 ratio | P value | ||
| Acid acclimation | |||||||||
| AmiE | jhp0279, HP0294 | 0.27 | <0.001 | 0.35 | 0.003 | 0.16 | <0.001 | 0.13 | <0.001 |
| HypB | jhp0837, HP0900 | 0.63 | 0.010 | 0.66 | 0.006 | 0.52 | <0.001 | 0.52 | <0.001 |
| RocF | jhp1427, HP1399 | 1.89 | 0.002 | 1.62 | <0.001 | ||||
| UreB | jhp0067, HP0072 | 0.25 | <0.001 | 0.26 | <0.001 | 0.16 | <0.001 | 0.15 | <0.001 |
| UreG | jhp0063, HP0068 | 0.28 | <0.001 | 0.31 | <0.001 | 0.26 | <0.001 | 0.26 | <0.001 |
| UreH | jhp0062, HP0067 | 0.27 | <0.001 | 0.29 | <0.001 | ||||
| Acetone metabolism | |||||||||
| AcxA | jhp0633, HP0695 | 2.36 | 0.010 | 1.89 | 0.038 | 2.82 | <0.001 | 2.69 | 0.001 |
| AcxB | jhp0632, HP0696 | 2.18 | 0.011 | 1.81 | 0.026 | 2.74 | <0.001 | 2.68 | <0.001 |
| AcxC | jhp0631, HP0697 | 2.32 | 0.037 | 2.31 | 0.011 | 2.95 | <0.001 | 2.73 | <0.001 |
| Motility | |||||||||
| FlaA | jhp0548, HP0601 | 56.48 | <0.001 | 87.69 | <0.001 | 1.42 | <0.001 | 1.27 | 0.27 |
| FlaB | jhp0107, HP0115 | 34.49 | <0.001 | 24.43 | <0.001 | ||||
| FlgE | jhp0804, HP0870 | 15.2 | <0.001 | 14.22 | <0.001 | 1.42 | 0.004 | 1.6 | 0.019 |
| FlgK | jhp1047, HP1119 | 8.96 | <0.001 | 9.84 | <0.001 | 1.63 | 0.005 | 1.58 | 0.039 |
| FlgL | jhp0280, HP0295 | 7.65 | 0.001 | 9.64 | <0.001 | ||||
| FliD | jhp0689, HP0752 | 6.76 | <0.001 | 5.69 | <0.001 | ||||
| Oxidative stress resistance | |||||||||
| TrxR1 | jhp0764, HP0825 | 0.65 | 0.002 | 0.58 | <0.001 | 0.6 | 0.008 | 0.59 | 0.002 |
| TrxR2 | jhp1091, HP1164 | 0.65 | <0.001 | 0.64 | 0.003 | ||||
| Other | |||||||||
| AspB | jhp0568, HP0624 | 0.71 | 0.009 | 0.69 | 0.008 | ||||
| FabD | jhp0083, HP0090 | 1.51 | 0.000 | 1.45 | <0.001 | 1.5 | 0.012 | 1.51 | 0.001 |
| FutB | jhp0596, HP0651 | 1.61 | 0.001 | 1.59 | 0.014 | ||||
| HomD | jhp1346, HP1453 | 2.43 | 0.001 | 2.66 | 0.002 | 2.09 | 0.006 | 1.99 | 0.004 |
| Hypothetical protein | jhp0149, HP0162 | 0.19 | 0.000 | 0.19 | <0.001 | 0.22 | <0.001 | 0.2 | <0.001 |
| MetB | jhp0098, HP0106 | 0.41 | 0.000 | 0.43 | <0.001 | ||||
| Pfs | jhp0082, HP0089 | 1.37 | 0.014 | 1.25 | 0.024 | 1.51 | 0.005 | 1.47 | 0.006 |
| ProS | jhp0223, HP0238 | 0.78 | 0.030 | 0.66 | 0.003 | ||||
| TatD | jhp1481, HP1573 | 0.55 | 0.000 | 0.57 | 0.023 | ||||
Experiment 1 tested strains J99A and J99A (arsS::kan) (acid exposure for 3.5 h), and experiment 2 tested strains J99B and J99B (arsS::kan) (acid exposure for 1.5 h).
Proteins expressed differentially in wild-type and arsS mutant strains are listed.
Corresponding locus tags for the respective proteins in H. pylori strain J99 (jhp prefix) and strain 26695 (HP prefix).
Average volume ratios of the indicated protein features (arsS mutant/wild type). A ratio of >1 indicates that the protein was expressed at higher levels in the arsS mutant strain than in the wild-type strain.
P values when comparing protein expression in wild-type and arsS mutant strains (Student's t test).
In addition to analyzing protein expression in wild-type and arsS mutant strains grown at pH 5.0, we analyzed protein expression in these two strains grown at pH 7.0. The same 25 proteins that were differentially expressed in the wild-type and mutant strains cultured at pH 5.0 were also differentially expressed when the two strains were cultured at pH 7.0 (Table 1, experiment 1). Relatively few pH-responsive changes in protein expression were detected in either the wild-type strain or the arsS mutant strain. The most striking of these pH-responsive changes were acid-induced upregulation of AccB (biotin carboxyl carrier protein) (2) and ComH (competence protein) (41) (data not shown). These changes were detected in both the wild-type and arsS mutant strains, which indicated that pH-dependent alteration in expression of these two proteins is ArsS independent.
Analysis of a second pair of wild-type and arsS mutant strains.
An unexpected result of the initial proteomic experiment was the relative paucity of acid-induced changes in protein expression. Therefore, we performed another proteomic experiment in which conditions for acid exposure were modified to more closely mimic conditions that were used for previous transcriptional studies (25, 27). For this next experiment, we selected an independent clone of wild-type strain J99 and generated a new arsS mutant strain. These strains [designated J99B and J99B (arsS::kan)] were cultured in medium adjusted to pH 5.0 and pH 7.0, as described for the previous proteomic experiment, but the bacteria were harvested after growth for 90 min (compared to growth for 3.5 h in the first experiment). At the time of harvest, there was only a slight increase in the pH of the pH 5.0 cultures; the final pH of wild-type strain J99 cultures was 5.3 ± 0.1, and the pH of arsS mutant cultures was 5.1 ± 0.2. Lysates were then processed and analyzed by 2D DIGE. The 2D DIGE data from this experiment were analyzed in an independent manner, without reference to the 2D DIGE data collected in the first proteomic experiment.
Overall, the results of this second proteomic analysis (designated experiment 2), were similar to the results of the first proteomic analysis (experiment 1). Specifically, the first principal component (PC1) for all matched features (n = 861) comprised 34.9% of the variation, and this variation clearly distinguished between the wild-type and mutant strains (data not shown). When focusing on protein features (n = 174) where one of the four groups of samples (wild-type and arsS mutant, each cultured at pH 5.0 or pH 7.0) was significantly different from the others (ANOVA P values of <0.05), the first principal component (PC1 = 76%) indicated that the largest source of variation represented differences between the wild-type and arsS mutant strains (data not shown). Consistent with the results of experiment 1, very few pH-responsive changes were observed in the second proteomic experiment.
We identified 15 proteins that were differentially expressed in wild-type and arsS mutant strains in both of the proteomic experiments, and several other differentially expressed proteins were identified in only one experiment (Table 1). Identification of a differentially expressed protein in only one of the two proteomic experiments could occur if the resolution of a protein was more satisfactory in one experiment than the other. For example, multiple previous studies have reported that rocF is a member of the ArsRS regulon (15, 32, 51), but differential expression of RocF was detected in only one of the two proteomic analyses. Identification of a differentially expressed protein in only one of the two proteomic experiments could also occur if an unrecognized secondary mutation was present in one of the analyzed pairs of wild-type and mutant strains. When comparing the results of the two proteomic experiments, the most striking difference involved the expression of proteins involved in motility. In the first experiment, six proteins involved in motility were expressed at markedly higher levels in the arsS mutant strain than in the wild-type strain (∼7- to 56-fold differences in an analysis at pH 5.0). In the second experiment, we detected slightly increased expression of three of these proteins in the arsS mutant strain compared to the wild-type strain, but the magnitude of the difference in protein expression (∼1.5-fold difference) was much smaller than what was observed in the first experiment.
As shown in Table 1, multiple proteins contributing to acid resistance (including UreB, UreG, UreH, and an amidase [AmiE] [36-39]) were reduced in expression in the arsS mutant strains compared to the wild-type strains. Differential expression of the urease subunit UreA was not detected in the 2D DIGE experiments, probably because its isoelectric point (predicted pI = 8.68) was outside the range selected for analysis (pH 4 to 7). However, immunoblotting of H. pylori cell extracts with an anti-UreA antibody (Santa Cruz Biotechnology) indicated that UreA expression was reduced in the arsS mutant compared to the wild-type strain (data not shown).
In contrast to the proteins described above that contribute to acid resistance, several other proteins were expressed at higher levels in the arsS mutant strains than in the wild-type strains (Table 1). These included three subunits of acetone carboxylase (AcxA, AcxB, and AcxC) that are predicted to be cotranscribed in an operon (9), as well as two other proteins (FabD [malonyl coenzyme A-acyl carrier protein transacylase] and Pfs [methyladenosine nucleosidase]) that are also predicted to be cotranscribed in an operon.
Binding of ArsR to DNA promoter probes corresponding to differentially expressed proteins.
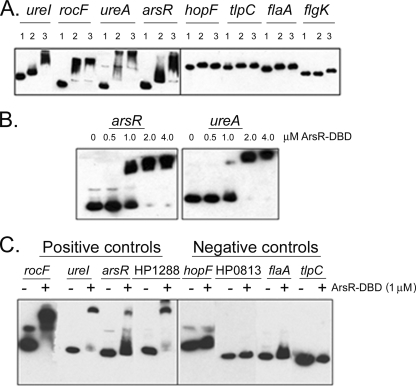
To investigate whether the expression of proteins listed in Table 1 was directly regulated by the ArsRS system, we next performed a series of experiments to investigate whether ArsR could bind to the promoter regions of genes of interest. The full-length (26-kDa) ArsR protein was expressed and purified as described in Materials and Methods, and binding of this protein to biotinylated DNA fragments (probes) was then assessed in EMSAs. In initial experiments, we validated the ability of the full-length recombinant ArsR protein to bind to promoter regions of several genes that were previously reported to be members of the ArsRS regulon (ureI, rocF, ureA, and arsR) (12, 33, 51). ArsR was tested at a concentration of 3 μM, as described previously (8). Consistent with previous results (8, 32, 33), ArsR retarded the migration of these positive-control promoter probes (Fig. 2A). We also examined the ability of ArsR to bind to promoter regions of several genes (hopF, tlpC, flaA, and flgK) previously reported to be transcribed in an ArsRS-independent manner (51). In agreement with the results of previous studies (51), the migration of these negative-control promoter probes was not retarded by ArsR (Fig. 2A).
FIG. 2.
Binding of ArsR and ArsR-DBD to DNA promoter probes. (A) Full-length ArsR binding to promoter regions of target genes previously reported to be regulated by ArsR. Fragments of 150 bp upstream of the translational initiation sites of the target genes were PCR amplified, using biotinylated primers. DNA fragments were incubated with buffer alone, unphosphorylated ArsR (3 μM final concentration), or ArsR that was phosphorylated by incubation with acetyl phosphate (see Materials and Methods). Fragments tested included positive-control promoter probes (ureI, rocF, ureA, and arsR) known to be bound by ArsR and negative-control promoter probes (hopF, tlpC, flaA, and flgK). Samples were subjected to nondenaturing PAGE and transferred to nylon membranes, and biotinylated DNA was visualized using chemiluminescent techniques. Lane 1, no protein; lane 2, unphosphorylated ArsR protein; lane 3, phosphorylated ArsR. (B) ArsR-DBD binding to arsR and ureA promoter probes. Increasing concentrations of the ArsR-DBD protein were added to DNA fragments, and the samples were then subjected to nondenaturing PAGE. Separation and visualization of biotinylated DNA were performed as described for panel A. (C) ArsR-DBD binding to promoter regions of target genes previously reported to be regulated by ArsR. Promoter probes were incubated with 1.0 μM ArsR-DBD. Fragments tested included positive-control promoter probes (rocF, ureI, arsR, and HP1288) known to be bound by ArsR and negative-control promoter probes (hopF, HP0813, flaA, and tlpC) (32, 51).
ArsR caused a detectable shift in the migration of each of the positive-control promoter probes in the absence of prior treatment with acetyl phosphate, which is consistent with previous observations (51). In the absence of phosphorylation, response regulators typically exist in a dynamic equilibrium between activated and inactive conformations (13, 42); therefore, at the relatively high protein concentrations used in this experiment (3 μM), detectable binding of nonphosphorylated ArsR to cognate binding sites is consistent with expectations. The migration of several positive-control promoter probes was retarded to a greater extent if the full-length ArsR protein was preincubated with acetyl phosphate than if it was not preincubated with acetyl phosphate. This is likely attributable to an increased binding affinity of phosphorylated ArsR to cognate binding sites.
In further experiments, we examined the binding of the DNA-binding domain of ArsR (i.e., ArsR-DBD) to the same positive-control and negative-control promoter regions described above. Similar to full-length (26-kDa) ArsR, the ArsR-DBD protein (13 kDa) bound to ureA and arsR promoter probes, and binding occurred in a concentration-dependent fashion (Fig. 2B). The minimum concentration of ArsR-DBD that caused detectable retardation in migration of these promoter probes was 1.0 μM, and therefore, binding of ArsR-DBD to all other promoter probes was subsequently carried out at this concentration. As shown in Fig. 2C, ArsR-DBD retarded the migration of promoter probes (rocF, ureI, arsR, and HP1288) that were previously reported to be bound by the full-length (26-kDa) ArsR protein (i.e., positive controls) (51). Moreover, ArsR-DBD did not bind to the promoter probes that were tested as negative controls (hopF, HP0813, flaA, and tlpC) (51). Thus, the recombinant ArsR-DBD demonstrated a DNA-binding specificity similar to that of the full-length ArsR protein. As the full-length ArsR protein was less soluble and less stable in solution than ArsR-DBD, we performed all subsequent EMSAs with the ArsR-DBD protein.
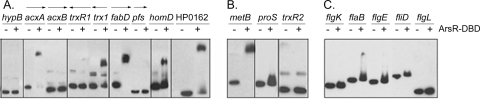
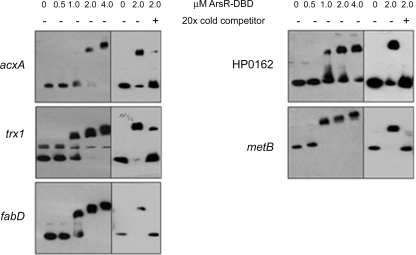
Many of the differentially expressed proteins listed in Table 1 are encoded by genes that previously had not been considered members of the ArsRS regulon or that had been identified as candidate members of the ArsRS regulon based on array studies but were not validated by gel shift assays or related approaches. To evaluate whether the corresponding genes were directly regulated by the ArsRS system, we tested the ability of ArsR-DBD to bind to promoter regions of these genes (Table 2). As shown in Fig. 3, ArsR-DBD bound to biotinylated promoter probes for acxA, fabD, homD, HP0162, and metB. Binding of ArsR-DBD to these biotinylated promoter probes occurred in a concentration-dependent fashion, and binding was competitively inhibited by the addition of excess unlabeled probes (Fig. 4). No binding of ArsR-DBD to DNA sequences upstream from acxB or pfs was detected. This is probably attributable to the localization of these genes immediately downstream of acxA and fabD; we hypothesize that acxA/acxB/acxC and fabD/pfs are cotranscribed (Fig. 3). Similarly, ArsR-DBD did not bind to DNA sequences upstream from trxR1, but it did bind to a promoter probe for trx1, a gene located immediately upstream of trxR1 (Fig. 3 and 4). We hypothesize that trx1 and trxR1 are cotranscribed. In summary, these EMSA results indicate that ArsR binds directly to the promoter regions located upstream from acxA, acxB, acxC, trxR1, fabD, pfs, homD, HP0162, and metB and provide evidence that the ArsRS system directly regulates expression of these genes.
TABLE 2.
Members of the ArsRS regulon identified by proteomic analysis and confirmed by ArsR-DNA binding assays
| Protein | Proteomic analysisa | ArsR bindingb |
|
|---|---|---|---|
| Current studyc | Previous studiesd | ||
| AmiE | ++ | ND | + |
| AcxA | ++ | + | ND |
| AcxB | ++ | + | ND |
| AcxC | ++ | + | ND |
| FabD | ++ | + | ND |
| HP0162 | ++ | + | ND |
| HomD | ++ | + | ND |
| MetB | + | + | ND |
| Pfs | ++ | + | ND |
| RocF | + | + | + |
| TrxR1 | ++ | + | − |
| UreB | ++ | + | + |
| UreG | ++ | + | + |
| UreH | + | + | + |
++, proteins differentially expressed in arsS mutant strains compared to wild-type strains in both proteomic experiments. +, proteins differentially expressed in only one of the two proteomic experiments.
ArsR binding to promoter regions of target genes (or upstream genes within operons), as determined by EMSAs or DNA footprinting analysis. +, ArsR binding; −, absence of binding; ND, binding of ArsR was not determined.
FIG. 3.
Binding of ArsR-DBD to DNA promoter probes corresponding to proteins that were differentially expressed in wild-type and arsS mutant strains. EMSAs were performed using 1.0 μM ArsR-DBD and promoter regions of genes whose protein products were differentially expressed in the arsS mutant and wild-type strains. EMSAs were performed as described in Materials and Methods. The predicted relationships of cotranscribed genes are indicated by directional arrows. (A) Analysis of genes encoding proteins whose expression was altered in both proteomic experiments 1 and 2; (B) analysis of genes encoding proteins whose expression was altered in only one of the proteomic experiments; (C) analysis of genes encoding flagellar proteins that were differentially expressed in experiment 1.
FIG. 4.
Specificity in the binding of ArsR-DBD to target DNA sequences. Various concentrations (0 to 4.0 μM) of the DNA-binding domain of ArsR (ArsR-DBD) were added to promoter probes for genes encoding differentially expressed proteins, and the ability of ArsR-DBD to bind these DNA sequences was examined using nondenaturing gel electrophoresis. For competition assays, reaction mixtures contained 2.0 μM biotinylated ArsR-DBD with or without a 20-fold excess of nonbiotinylated probe.
ArsR-DBD did not bind to promoter regions located upstream from several of the genes listed in Table 1. For example, ArsR-DBD did not bind to the promoter regions located upstream of the flagellar genes flaA, flgK, flgE, fliD, and flgL (Fig. 2 and 3C) and did not bind to promoter regions for hypB or proS (Fig. 3A and B). This suggests that the observed differential expression of the corresponding proteins in 2D DIGE experiments was not mediated directly by the ArsRS system.
Comparison of AI-2 production in wild-type and arsS mutant strains.
One of the differentially expressed proteins identified in both proteomic analyses was Pfs (5′-methyladenosine nucleosidase). Pfs is an enzyme that regulates the production of S-ribosylhomocysteine, the substrate that LuxS converts to the autoinducer molecule AI-2 (7, 35). We hypothesized that increased expression of Pfs in the arsS mutant strain compared to the wild-type strain might result in increased production of AI-2. To test this hypothesis, we compared AI-2 production in the wild-type strain and the arsS mutant strain. As shown in Fig. 5, the addition of conditioned medium from an arsS mutant strain [J99B (arsS::kan)] to a V. harveyi reporter strain resulted in significantly greater luminescence than did conditioned medium from the wild-type strain (P value of <0.05 by the paired Student t test). These results provide evidence that increased expression of Pfs in the arsS mutant strain is associated with increased production of AI-2.
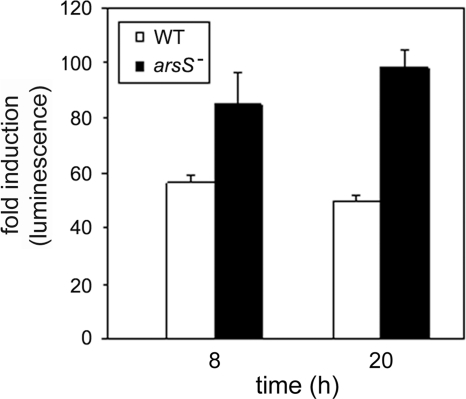
FIG. 5.
AI-2 production in H. pylori wild-type and arsS mutant strains. Wild-type (WT) or arsS mutant H. pylori strains were cultured in broth for 8 h or 20 h, resulting in similar optical densities of the wild-type and mutant cultures. Conditioned media from these cultures were assayed in a bioluminescence assay using V. harveyi BB170 as an AI-2 reporter strain (see Materials and Methods). The figure indicates luminescence induced by conditioned media from H. pylori cultures relative to luminescence induced by sterile brucella broth. Conditioned medium from the arsS mutant strain induced significantly greater luminescence than did conditioned medium from the wild-type strain (P < 0.05 by Student's t test). The data represent results (means ± standard deviations) for triplicate samples from a single experiment. Similar results were obtained in three independent experiments.
DISCUSSION
In this study, we investigated the regulatory role of the ArsRS two-component signaling system by comparing the proteomes of wild-type and isogenic arsS mutant strains. Several previous studies have shown that the ArsRS two-component system is involved in the regulation of H. pylori gene expression in response to pH changes in the environment (31, 32, 50, 51), and therefore, we analyzed protein expression during bacterial growth at both pH 5.0 and pH 7.0. Since transcriptional alterations are detectable when H. pylori is cultured under pH 5.0 conditions compared to pH 7.0 conditions (27, 53) and differences in the transcription of wild-type and arsS mutant strains are detectable when these strains are cultured at pH 5.0 (31, 32, 50, 51), we expected to detect specific pH-responsive changes in protein expression in the wild-type strain but not in the arsS mutant strain. In fact, we detected very few pH-responsive changes in either strain. A failure to detect acid-induced alterations in protein expression could potentially indicate that the low-pH conditions used in this study were not optimal to allow acid-induced alterations in protein translation or, alternatively, that posttranscriptional regulatory processes may have blunted the expected acid-induced alterations in protein expression (5). We did not perform experiments in which the bacteria were cultured at a pH of <5.0, because H. pylori growth in vitro is inhibited under such conditions (40). Despite a limited ability to detect pH-responsive changes in protein expression, we were nevertheless able to detect numerous proteins that were differentially expressed in arsS mutant strains compared to wild-type strains. These proteins were differentially expressed in arsS mutant strains compared to wild-type strains not only when the bacteria were cultured at pH 5.0 but also when they were cultured at pH 7.0 (Table 1). These data indicate that the ArsRS two-component system has an important regulatory role even in the absence of a low-pH stimulus, and they suggest that the ArsRS system might be responsive to other environmental factors in addition to low pH.
By using mass spectrometry, we identified 15 electrophoretically resolved proteins that were differentially expressed in experiments involving two different pairs of wild-type and arsS mutant strains. Multiple additional proteins were differentially expressed in only one of the two proteomic experiments. To further evaluate candidate ArsRS-regulated proteins identified in this study, we performed gel shift experiments. Among the 26 proteins identified as candidate members of the ArsRS regulon, direct binding of ArsR to promoter regions of five of these proteins (UreB, UreH, UreG, AmiE, and RocF) had been demonstrated previously (12, 32, 33, 51). We examined ArsR binding to promoter regions of 16 additional genes (including all 10 genes whose protein products were differentially expressed in both proteomic experiments and 6 genes whose protein products were differentially expressed in only one of the two proteomic experiments), and we detected binding of ArsR to 9 of these genes (Table 2). Thus, we conclude that 14 of the differentially expressed proteins identified by 2D DIGE analysis are directly regulated by the ArsRS system (Table 2).
We were unable to detect a direct role of the ArsRS system in regulating several of the differentially expressed proteins identified in this study. In particular, we did not detect binding of ArsR to promoter regions of five flagellar genes tested. In experiment 1, we observed dramatic differences in the expression of multiple flagellar components when comparing the wild-type and arsS mutant strains, but these dramatic differences were not observed in the second experiment. We did not perform additional experiments (such as complementation analysis) to investigate the basis for the differences in flagellar protein expression observed in the first proteomic experiment, and therefore it is unclear whether these alterations resulted from an unrecognized secondary mutation or other factors. However, the absence of detectable ArsR binding to the promoter regions of the corresponding flagellar genes provides evidence that these motility-associated genes are not members of the ArsRS regulon.
When performing gel shift experiments in this study, we assessed binding of ArsR to target promoters by using either recombinant full-length ArsR or the DNA-binding domain of ArsR. In general, we did not detect any differences in the specificity of target DNA binding by full-length ArsR compared to ArsR-DBD. The ability of the DNA-binding domain of a response regulator to bind to or activate target genes has also been demonstrated in studies of E. coli two-component systems (14, 45), and in the case of the response regulator PhoB, the C-terminal DNA-binding domain binds to a target pho box sequence with an affinity seven times higher than that of the unphosphorylated full-length protein (14).
Previous studies have attempted to identify members of the ArsRS regulon, using a variety of approaches. The results of these previous studies have not been entirely uniform, perhaps due to differences in methodology among the studies and the use of several different H. pylori strains (12, 15, 25, 32, 51). In a previous study (25), we utilized DNA macroarrays to evaluate H. pylori gene transcription in wild-type and arsS mutant strains, each grown at pH 5.0 or pH 7.0. We analyzed the data from these previous transcriptional profiling experiments to allow a comparison with the proteomic results described in the current study. Among the 15 proteins that were differentially expressed in wild-type and arsS mutant strains in two separate proteomic experiments in the current study (Table 1), 8 of the corresponding genes (acxAB, fabD, flaA, hypB, pfs, trxR1, and ureB) were differentially expressed at the transcriptional level in wild-type and arsS mutant strains during growth at pH 5.0 (data not shown). For two of the seven remaining proteins identified in both proteomic experiments, the corresponding genes (ureG and amiE) had previously been shown to be directly regulated by ArsR (12, 32, 33, 51). The differences between previous transcriptional profiling results and the current proteomic results may be attributable to limitations of the previously employed array technologies and also might be attributable to the occurrence of posttranscriptional regulatory phenomena. When comparing the results of the current study with previous transcriptional profiling results, another relevant factor is that the current proteomic analysis focused on proteins with isoelectric points of between 4 and 7, and therefore ArsRS-regulated proteins with higher isoelectric points, such as UreA, were not detected using the current proteomic approach.
One of the ArsRS-regulated proteins identified in the current study is Pfs, a methyladenosine nucleosidase that plays a key role in the activated methyl cycle of bacteria (reviewed in reference 49) by converting the toxic S-adenosyl-l-homocysteine to S-ribosyl-l-homocysteine. The latter product can then be converted by the LuxS protein to the quorum signal AI-2 (49). Consistent with the observed increased expression of Pfs in the arsS mutant strain compared to the wild-type strain, AI-2 levels were higher in the arsS mutant than in the parental wild-type strain. Previous studies have shown that the expression of flaA can be modulated by AI-2 (26), and therefore, increased AI-2 levels in the arsS mutant compared to the wild-type strain could potentially account for the detectable differences in FlaA protein expression that were observed in the second proteomic experiment.
The list of newly recognized ArsRS regulon members identified in this study includes three subunits of acetone carboxylase (AcxA, AcxB, and AcxC), which are predicted to be cotranscribed in an operon. H. pylori is able to use acetone as an alternative carbon source, and mutagenesis of the acx operon reduced the ability of H. pylori to colonize mice (9). A previous study showed that the acx operon is regulated by an orphan response regulator known as HP1021 (30). The fabD-pfs operon also appears to be regulated by both the ArsRS system and the orphan response regulator HP1021 (this study and reference 30). Thus, there seems to be dual regulation of these target genes by the ArsRS two-component system and the HP1021 response regulator. There are multiple other examples of H. pylori genes that seem to be regulated by both the ArsRS system and other transcriptional regulatory systems. For instance, in a previous study, we demonstrated overlap in genes regulated by the CrdRS two-component system and members of the ArsRS regulon (25). There is also overlap among acid-responsive genes that are regulated by both the ArsRS and FlgRS two-component systems (52). Similarly, the urease operon is regulated by both ArsRS and the nickel regulator NikR, while AmiE is regulated by both ArsRS and the iron utilization regulator Fur (47, 48). The regulation of H. pylori genes by multiple two-component systems potentially allows finely regulated gene expression in response to changing environmental stimuli (22).
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Service of the Department of Veterans Affairs and the National Institutes of Health (grants AI39657, AI068009, and CA116087). Proteomic work was supported by grant P30 DK058404, the Vanderbilt Ingram Cancer Center, and the Vanderbilt Academic Venture Capital Fund.
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3:36-44. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Amieva, M. R., and E. M. El-Omar. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134:306-323. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 1:63-96. [DOI] [PubMed] [Google Scholar]
- 5.Barnard, F. M., M. F. Loughlin, H. P. Fainberg, M. P. Messenger, D. W. Ussery, P. Williams, and P. J. Jenks. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 51:15-32. [DOI] [PubMed] [Google Scholar]
- 6.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brahmachary, P., G. Wang, S. L. Benoit, M. V. Weinberg, R. J. Maier, and T. R. Hoover. 2008. The human gastric pathogen Helicobacter pylori has a potential acetone carboxylase that enhances its ability to colonize mice. BMC Microbiol. 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busler, V. J., V. J. Torres, M. S. McClain, O. Tirado, D. B. Friedman, and T. L. Cover. 2006. Protein-protein interactions among Helicobacter pylori cag proteins. J. Bacteriol. 188:4787-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cover, T. L., and M. J. Blaser. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyer, C. M., and F. W. Dahlquist. 2006. Switched or not? The structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 188:7354-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellison, D. W., and W. R. McCleary. 2000. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J. Bacteriol. 182:6592-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 184:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman, D. B., S. Hill, J. W. Keller, N. B. Merchant, S. E. Levy, R. J. Coffey, and R. M. Caprioli. 2004. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 4:793-811. [DOI] [PubMed] [Google Scholar]
- 17.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman, D. B., S. E. Wang, C. W. Whitwell, R. M. Caprioli, and C. L. Arteaga. 2007. Multivariable difference gel electrophoresis and mass spectrometry: a case study on transforming growth factor-beta and ERBB2 signaling. Mol. Cell Proteomics 6:150-169. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin, A. C., D. M. Weinberger, C. B. Ford, J. C. Nelson, J. D. Snider, J. D. Hall, C. I. Paules, R. M. Peek, Jr., and M. H. Forsyth. 2008. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology 154:2231-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, S. S., B. N. Borin, T. L. Cover, and A. M. Krezel. 2009. Structural analysis of the DNA-binding domain of the Helicobacter pylori response regulator ArsR. J. Biol. Chem. 284:6536-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joseph, B., and D. Beier. 2007. Global analysis of two-component gene regulation in Helicobacter pylori by mutation analysis and transcriptional profiling. Methods Enzymol. 423:514-530. [DOI] [PubMed] [Google Scholar]
- 22.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, W. K., K. Ogura, J. T. Loh, T. L. Cover, and D. E. Berg. 2006. Quantitative effect of luxS gene inactivation on the fitness of Helicobacter pylori. Appl. Environ. Microbiol. 72:6615-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lilley, K. S., and D. B. Friedman. 2004. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev. Proteomics 1:401-409. [DOI] [PubMed] [Google Scholar]
- 25.Loh, J. T., and T. L. Cover. 2006. Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infect. Immun. 74:3052-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh, J. T., M. H. Forsyth, and T. L. Cover. 2004. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect. Immun. 72:5506-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, S., M. Gotz, and D. Beier. 2009. Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS One 4:e6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panthel, K., P. Dietz, R. Haas, and D. Beier. 2003. Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect. Immun. 71:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pflock, M., M. Bathon, J. Schar, S. Muller, H. Mollenkopf, T. F. Meyer, and D. Beier. 2007. The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J. Bacteriol. 189:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pflock, M., P. Dietz, J. Schar, and D. Beier. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol. Lett. 234:51-61. [DOI] [PubMed] [Google Scholar]
- 32.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pflock, M., S. Kennard, I. Delany, V. Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pflock, M., S. Kennard, N. Finsterer, and D. Beier. 2006. Acid-responsive gene regulation in the human pathogen Helicobacter pylori. J. Biotechnol. 126:52-60. [DOI] [PubMed] [Google Scholar]
- 35.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 36.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 37.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 38.Skouloubris, S., A. Labigne, and H. De Reuse. 2001. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol. Microbiol. 40:596-609. [DOI] [PubMed] [Google Scholar]
- 39.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25:989-998. [DOI] [PubMed] [Google Scholar]
- 40.Slonczewski, J. L., D. J. McGee, J. Phillips, C. Kirkpatrick, and H. L. Mobley. 2000. pH-dependent protein profiles of Helicobacter pylori analyzed by two-dimensional gels. Helicobacter 5:240-247. [DOI] [PubMed] [Google Scholar]
- 41.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock, A. M., and J. Guhaniyogi. 2006. A new perspective on response regulator activation. J. Bacteriol. 188:7328-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 44.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapparel, C., A. Monod, and W. L. Kelley. 2006. The DNA-binding domain of the Escherichia coli CpxR two-component response regulator is constitutively active and cannot be fully attenuated by fused adjacent heterologous regulatory domains. Microbiology 152:431-441. [DOI] [PubMed] [Google Scholar]
- 46.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 47.van Vliet, A. H., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Vliet, A. H., J. Stoof, S. W. Poppelaars, S. Bereswill, G. Homuth, M. Kist, E. J. Kuipers, and J. G. Kusters. 2003. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori fur repressor. J. Biol. Chem. 278:9052-9057. [DOI] [PubMed] [Google Scholar]
- 49.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 50.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2007. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J. Bacteriol. 189:2426-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2006. Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori. J. Bacteriol. 188:1750-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2009. The pH-responsive regulon of HP0244 (FlgS), the cytoplasmic histidine kinase of Helicobacter pylori. J. Bacteriol. 191:449-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.