Abstract
Despite data suggesting that the adenovirus E1A protein of 243 amino acids creates an S-phase environment in quiescent cells by overcoming the nucleosomal repression of E2F-regulated genes, the precise mechanisms underlying E1A's ability in this process have not yet been defined at the biochemical level. In this study, we show by kinetic analysis that E1A, as opposed to an E1A mutant failing to bind p130, can temporally eliminate corepressor complexes consisting of p130-E2F4 and HDAC1/2-mSin3B from the promoters of E2F-regulated genes in quiescent cells. Once the complexes are removed, the di-methylation of H3K9 at these promoters becomes dramatically diminished, and this in turn allows for the acetylation of H3K9/14 and the recruitment of activating E2F family members, which is then followed by the transcriptional activity of the E2F-regulated genes. Remarkably, although an E1A mutant that can no longer bind to a histone acetyltransferase (PCAF) is as capable as wild-type E1A in eliminating corepressor complexes and methyl groups from the promoters of these genes, it cannot mediate the acetylation of H3K9/14 or induce their transcription. These findings suggest that corepressors as well as coactivators are acted upon by E1A to derepress E2F-regulated genes in quiescent cells. Thus, our results highlight for the first time a functional relationship between E1A and two transcriptional pathways of differing functions for transitioning cells out of quiescence and into S phase.
Human adenoviruses normally infect quiescent or terminally differentiated cells. Central to this infectivity is the action of the small-size adenovirus E1A protein of 243 amino acid (aa) residues (243R), which creates a condition favorable for viral replication (1). As such, this protein is principally responsible for transitioning cells out of quiescence and into S phase or for reactivating DNA synthesis in terminally differentiated muscle cells (3, 17, 25). Earlier studies have shown that the activities of E1A in this context are largely dependent upon its ability to physically associate with members of the retinoblastoma family of proteins, e.g., pRb and p130 (6, 10, 17). Both of these proteins are widely known for their ability to regulate the E2F family of transcription factors (E2F1 to E2F5), which play pivotal roles in regulating the expression of genes involved in cell cycle reentry and DNA synthesis (2). In general, the functions of the E2Fs serve broad roles, with E2F1 to -3 acting as transcriptional activators and E2F4 to -5 as transcriptional repressors. The remaining E2Fs (E2F6 to -8) can also act as transcriptional repressors, but in an Rb-independent manner (29). In cycling cells, pRb is believed to inhibit the activating function of E2F1 by recruiting chromatin-modifying complexes with histone deacetylase (HDAC) or histone methyltransferase activity to E2F-regulated genes (8). However, such recruitment may be important only for repressing key E2F promoters under specific conditions (e.g., Ras-induced senescence) since chromatin immunoprecipitation (ChIP) assays have yet to detect pRb at the promoters of known E2F-dependent genes in both quiescent and proliferating cells (10, 22, 27).
ChIP experiments, however, have revealed the occupancy of p130 as well as E2F4 at the promoters of several E2F-regulated genes in cells restricted to quiescence or in early G1 (10, 22, 27). This approach showed that the corepressor complex HDAC1-mSin3B was bound to these promoters as well (22). A role for this complex has been proposed in silencing E2F-regulated genes in quiescent cells by continually deacetylating the histones in association with their promoters (10, 22, 27). The recruitment of HDAC1 to the E2F promoters in quiescent cells appears to be mediated by p130 (22), and circumstantial evidence suggests that p130 may also be involved in recruiting the histone methylase SUV39H1(10), which is largely responsible for catalyzing the methylation of histone H3 on lysine 9 (H3K9) (23).
Our laboratory has previously examined the function of E1A after its delivery into quiescent cells by a “Tet-on” inducible expression system (10). With this approach, we were able to show that E1A could surprisingly reorganize chromatin structure at the promoters of selected E2F-dependent genes in these cells and, as a result, induce their transcription (10). More specifically, our experiments revealed that E1A could transiently occupy these promoters after its expression in quiescent cells and consequently dissociate a residing p130-E2F4 complex. Following this elimination, the balance of histone H3K9 methylation at the E2F-dependent promoters shifted to an acetylated state. Despite these advances in our understanding of E1A's activities in quiescent cells, a complete characterization of the mechanisms behind E1A's ability to induce transcription from E2F-dependent promoters and thereby lead these cells back into the cell cycle has not been achieved. Here we show that E1A can interact with at least two transcriptional pathways which have opposing functions for regulating E2F-dependent genes in quiescent cells and that this provides a plausible explanation for how E1A drives these cells into a proliferative state.
MATERIALS AND METHODS
Inducible cell lines, cell culture, and synchronization.
Tet-on inducible BALB/c-3T3 cell lines containing an empty vector (minus E1A) or a gene expressing either a wild-type (WT) E1A or a mutant E1A have previously been described (9). Blasticidin/zeocin-resistant clones were selected based on having no E1A expression in the absence of doxycycline (Dox) but a modest amount of E1A expression after the addition of 200 ng/ml of Dox. Each of the E1A-inducible cell lines were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with antibiotics, l-glutamine, blasticidin/zeocin, and 10% fetal bovine serum (Tet system approved; Clontech). The respective cell lines, plated on either 60-mm or 100-mm plates, were made quiescent by shifting them to medium containing 0.05% fetal bovine serum (FBS) for a period of 60 h, as previously described (9).
Antibodies, immunoprecipitation, and Western blotting.
The preparation of whole-cell and nuclear extracts and the conditions for immunoprecipitation and Western blotting have previously been described by our laboratory (10, 17). Anti-pRb (C-15, M-153, and IF8), anti-p130 (C-20), anti-MyoD (M-318), anti-E2F1 (C-20), anti-E2F2 (C-20), anti-E2F3a/b (C-18), anti-E2F4 (C-20), anti-p300 (C-20 and N-15), anti-HDAC1 (H-11 and H-51), HDAC2 (C-8 and H-54), and normal rabbit or mouse IgG antibodies were purchased from Santa Cruz Biotechnology. Antibodies against acetylated histone H3 (Lys-9), di-methyl histone H3 (Lys-9), and anti-p300 (05-257) were obtained from Upstate Biotechnology, Inc. An antibody specific for PCAF (C1469) or an acetylated histone H3 (Lys-9/Lys-14) was purchased from Cell Signaling. Anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was obtained from BioDesign, and the M73 monoclonal antibody specific for E1A was kindly provided by E. Harlow (11).
Immunofluorescence and monitoring of DNA synthesis.
The E1A-inducible cell lines were grown on glass coverslips to a confluence of ∼60% and then brought to a state of quiescence as described above. The cultures were then treated with Dox (0.2 μg/ml) and after 12 h labeled with 25 μM 5-bromo-deoxyuridine (BrdU) up to the time they were harvested, which was 1 h later. Afterward, the cells were fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100, as previously described by our laboratory (10, 17). Primary antibodies used for immunofluorescence were the monoclonal antibody to E1A (M73) and the anti-BrdU monoclonal antibody conjugated to fluorescein isothiocyanate (FITC) (Boehringer Mannheim). The secondary antibody for E1A staining was Texas Red-conjugated rabbit anti-mouse IgG (Jackson Laboratory). Specimens for immunofluorescence were examined using a Nikon Optiphot-2 fluorescence microscope and then digitally captured (Oncor Video Imaging System).
RT-PCR analysis.
Total RNA was prepared from quiescent wild-type and mutant E1A-inducible cell lines using Trizol reagent (Invitrogen) and according to the manufacturer's instructions. One hundred nanograms of RNA was amplified by reverse transcription (RT)-PCR using the OneStep RT-PCR kit (Invitrogen) with cDNA-specific primer sequences. Thirty (cyclin A, Cdc6, and E2F1) or 20 (GAPDH) cycles of PCR were used, and each product was resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
ChIP and ChIP reimmunoprecipitation.
The preparation of chromatin and the immunoprecipitation of such for ChIP analysis were performed as previously described by our laboratory (10, 18). Briefly, E1A-inducible cell lines that had been treated with or without Dox were cross-linked with 1% formaldehyde for 10 min at room temperature. The isolated nuclei from these cells were then sonicated under conditions yielding an average length of 400 to 600 bp. After the sonicated chromatin was purified by a CsCl step gradient, it was dialyzed against a defined buffer and then precleared, accordingly. Ten percent of the total amount of chromatin (∼100 μg) was then put aside for control, and the remainder was immunoprecipitated using either 2 μg or 10 μl of the antibody of interest. After the immunoselected DNA was reverse cross-linked as described previously (10, 18), the chromatin was resuspended in DNase-free water until further use.
For serial ChIPs, ∼200 μg of sonicated chromatin was first immunoprecipitated, in parallel, with an antibody specific for E1A (M73), p130, or PCAF. The immune-selective DNA fragments were then released from the primary immunoprecipitations as previously described (10, 18) and afterward reimmunoprecipitated with normal rabbit IgG (NR IgG) or an antibody specific for E1A, p130, or PCAF.
Semiquantitative radioactive PCR was performed in a reaction volume of 25 μl consisting of 5 μl of immunoselected DNA, 10 pmol of primers specific for the gene of interest, 0.25 units of Taq DNA polymerase (Roche), and 0.2 μCi of [α-32P]dCTP in PCR buffer. All PCRs were performed using an MJ Research thermal cycler, and the linear range of PCR products (134 to 222 bp) for all primers used was determined empirically using different amounts of fragmented chromatin DNA. PCR products were fractionated on 8% native polyacrylamide gels and visualized by autoradiography. For input control, 10 μg of cross-linked chromatin was reverse cross-linked, accordingly (10), and after processing as described above, the DNA was dissolved in DNase-free water and 0.05% of the total was assayed by PCR using the GAPDH, cyclin A, Cdc6, or E2F1 primer set. Primer sets for PCR include Cdc6 (forward), 5′-TGATGAGTGACAACTAATCGA-3′; Cdc6 (reverse), 5′-GAGCTTTGCACTCTTGAGG-3′; cyclin A (forward), 5′-ATCCTATGAGGCTACAGATC-3′; cyclin A (reverse), 5′-TGGTCTTGTAGTTCAAGTAG-3′; E2F1 (forward), 5′-CGGAGCCTCCTGCGTCACAGCCGC-3′; E2F1 (reverse), 5′-CGGCCGCCGCTGCCTGCAAAGTCC-3′; GAPDH (forward), 5′-AGGCCAAACTAGCAGCTAGG-3′; and GAPDH (reverse), 5′-GGGCTAGTCTATCATTGCAG-3′.
RESULTS
E1A requires PCAF activity to induce cellular DNA synthesis.
We have previously described the creation of two Tet-on E1A-inducible BALB/c 3T3 cell lines (10), one of which is capable of conditionally expressing the E1A 243R protein (wild-type [WT] E1A), while the other expresses an E1A mutant (E1A.928) that is no longer able to bind pRb and p130 (Fig. 1A) (10). By using both of these cell lines, we were able to reveal for the first time that E1A could alter chromatin structure at the promoters of E2F-regulated genes in quiescent cells and that one aspect of this change entailed the conversion of methylation to acetylation on lysine 9 (K9) of nucleosomal histone H3 (10). To better understand E1A's role in mediating the acetylation of histone H3 at the promoters of these genes during quiescence, we constructed two more E1A-inducible cell lines that conditionally express E1A mutants that can no longer bind to a protein or to a protein complex having histone acetyltransferase (HAT) activity. One of these E1A-inducible cell lines harbors the E1A Δ26-35 mutant, which is defective in the binding of a large protein called TRRAP, while retaining its ability to bind pRb, p300, and CBP (4, 9, 15, 24). TRRAP is commonly shared between several HAT complexes, including that of PCAF and GCN5, and both of these multisubunit complexes are primarily involved in histone H3K9/14 modifications (19-20). Since our new data indicate that E1A can promote the acetylation of both H3K9 and H3K14 at the promoters of E2F-regulated genes in quiescent cells (see below) (10), we determined whether this E1A-mediated event involved the PCAF complex by focusing our efforts on the E1A Δ26-35 mutant-inducible cell line.
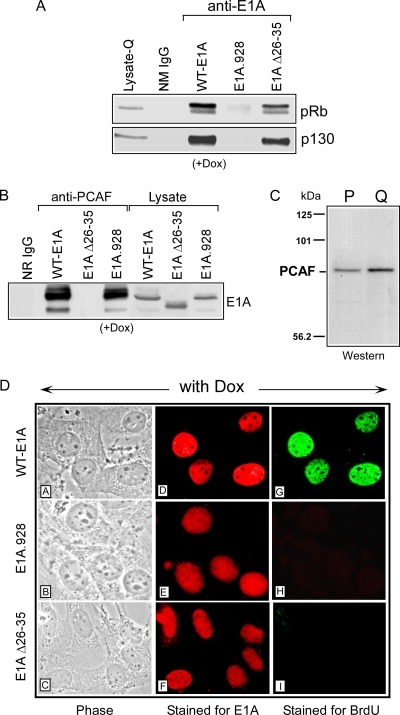
FIG. 1.
Induction of DNA synthesis in quiescent cells by E1A correlates with pRb/p130 or PCAF interaction. (A and B) Whole-cell extracts were prepared from quiescent wild-type E1A-, E1A.928-, and E1A Δ26-35 mutant-inducible BALB/c 3T3 cell lines after treatment with Dox for 6 h. Extracts from these cells were then immunoprecipitated in parallel using a control antibody (NR IgG), the E1A-specific antibody M73, and an antibody specific for the PCAF protein. Immune complexes were resolved on SDS polyacrylamide gels alongside straight extract from each of the E1A-inducible cell lines with Dox, for control. Western blot analysis was performed using the M73 antibody or antibodies specific for p130 and pRB. (C) Nuclear extracts were prepared from proliferating or quiescent wild-type E1A-inducible BALB/c 3T3 cells and then subjected to Western blot analysis, using an antibody specific for PCAF. (D) Each of the E1A-inducible cell lines was grown and rendered quiescent by serum starvation on glass coverslips and then treated with Dox for 12 h. Afterward, 25 μM BrdU was added to the media, and after 1 h, the cells were fixed and sequentially stained for E1A and BrdU incorporation (see Materials and Methods). Panels DA to DC represent phase-contrast photomicroscopy of fixed wild-type and mutant E1A cells. Wild-type E1A cells in panels DD and DG, which correspond to the same field shown in panel DA, display E1A staining (red) and BrdU incorporation (green). E1A.928 (panels DE and DH) and the E1A Δ26-35 mutant (panels DF and DI) (and panels DB and DC [phase]) show E1A staining but are negative for BrdU.
We first monitored the expression of the inducible E1A Δ26-35 mutant by adding the tetracycline analog Dox to cultured cells that had been brought to quiescence by serum starvation. We found that this mutant was maximally expressed by 4 h and at levels similar to that of wild-type E1A or the mutant E1A.928 protein after they were induced by Dox under the same conditions (Fig. 1B and data not shown). To confirm the integrity of the E1A Δ26-35 mutant after being induced in the quiescent cell line, we performed coimmunoprecipitations to determine whether it retained its ability to bind at least pRb, p130, and p300/CPB (9). In this experiment, we found that this mutant was almost as efficient as wild-type E1A in associating with these proteins (Fig. 1A and data not shown). Perhaps more important is that the Δ26-35 E1A mutant was always incapable of interacting with PCAF or a PCAF-containing complex compared to wild-type E1A and the E1A.928 mutant (Fig. 1B). Notably, the antibody that was used in this experiment was also able to recognize a protein matching the size of PCAF in extracts of proliferating or quiescent E1A-inducible cells, as determined by Western blotting (Fig. 1C).
Finally, after treating quiescent mouse cells with Dox for 13 h, we found that the E1A Δ26-35 mutant was phenotypically similar to the E1A.928 mutant, in that it could not stimulate these cells to enter S phase compared to wild-type E1A, as judged by BrdU incorporation (Fig. 1C) (10). With wild-type E1A, the percentage of labeled nuclei within this time frame was ∼60% and comparable to that which had been previously observed after a purified E1A 243R protein was microinjected into quiescent BALB/c 3T3 cells (5). Because those studies and the studies herein show that E1A alone can induce and shorten the G0-to-S phase interval in quiescent cells much earlier than when functioning in the company of other viral proteins, this could be one reason why the expression of the inducible Δ26-35 E1A mutant does not result in any measurable induction of DNA synthesis in our quiescent system. This is consistent with the fact that an adenovirus mutant (dl1102) with a deletion of aa 26 to 35 has been shown in quiescent baby rat kidney (BRK) cells to be moderately defective in inducing DNA synthesis when measured against wild-type E1A over a 72-h period (12). Taken together, these data lend support to the idea that E1A may be required to interact with at least two different regulatory pathways to bring quiescent cells into a proliferative state.
E1A eliminates HDAC1/2-mSin3B from the promoters in quiescent cells.
Evidence indicates that the regulation of E2F-dependent genes in quiescent cells involves the recruitment of HDAC1/2 and the transcriptional repressor mSin3B (22). In order to further explore the role of E1A in reorganizing the chromatin environment at the promoters of cyclin A and Cdc6 in quiescent cells (10), we determined whether HDAC1/2 and/or mSin3B were in fact being recruited to these genes after switching each of our E1A-inducible cell lines into a state of quiescence. After performing ChIP experiments with primers flanking potential E2F-binding sites (Fig. 2A), we found that the promoters of cyclin A and Cdc6 in the absence of Dox were indeed being occupied by HDAC1/2 and mSin3B (Fig. 2B and data not shown). Consistent with our previous studies and with the results of others (10, 22), we found that E2F4 and p130 were occupying these promoters as well (Fig. 2B). As expected, there was no enrichment of cyclin A and Cdc6 after an irrelevant antibody was used in the ChIP experiments (Fig. 2B).
FIG. 2.
Detection of corepressors associated with the promoters of cyclin A and Cdc6 in quiescent E1A-inducible cell lines. (A) Diagram of promoters used in this study. Inverted arrows above each schematic indicate the positions of the PCR primers used to detect promoter fragments, whereas the right angle arrows represent the transcriptional start sites (+1) determined in earlier studies. Solid boxes identify the E2F binding sites within the respective promoters. (B) All of the E1A-inducible cell lines were rendered quiescent by serum deprivation and then subjected to ChIP analysis. Input lane represents 0.05% of total chromatin with primer sets for cyclin A and Cdc6. Control reactions with the use of an irrelevant antibody (Ab) specific for the transcription factor MyoD are shown. Proximal promoter sequences for each gene were amplified by PCR using primers illustrated in the schematic and then detected by autoradiography as described in Materials and Methods.
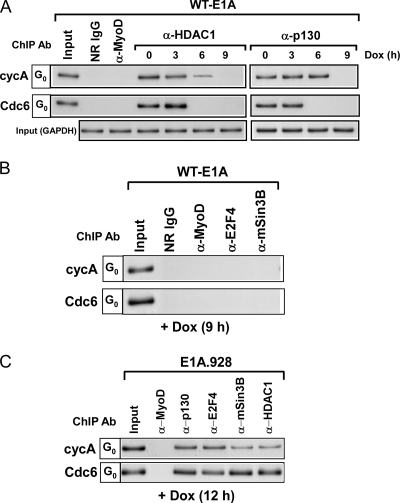
That HDAC1/2-mSin3B may require p130 in order to occupy E2F-dependent promoters is supported by the fact that HDAC1/2 and to a variable degree mSin3B cannot be detected at these promoters in quiescent p107−− and p130−− mouse embryonic fibroblasts (MEFs) (22). Moreover, other studies indicate that p130 can physically interact with HDAC1 (26). Given the evidence for a role of p130 in recruiting HDAC1/2 to E2F-dependent promoters during quiescence and the fact that E1A can eliminate p130 from these promoters (10), we examined the occupancy of p130 and HDAC1 at the promoters of Cdc6 and cyclin A following E1A induction in quiescent cells. Surprisingly, we found by ChIP analysis that the chromatin-associated HDAC1 and p130 were dissociating almost identically over time from both of these promoters after E1A was expressed in these cells (Fig. 3A). Furthermore, both E2F4 and the mSin3B repressor were likewise eliminated from Cdc6 and cyclin A promoters after the expression of E1A (Fig. 3B). In contrast, these promoters did not undergo a loss in p130-E2F4 and HDAC1/2-mSin3B when the E1A.928 mutant incapable of p130 interaction (Fig. 1A) was expressed in the quiescent cells (Fig. 3C). These results strongly suggest that the silencing of E2F-responsive genes in quiescent cells depends in part on p130's ability to recruit corepressor complexes, such as HDAC1/mSin3B, to their promoters. This idea is strengthened by the fact that once E1A causes p130 to dissociate from the promoters of cyclin A and Cdc6, these genes become actively transcribed, as illustrated in Fig. 4A by RT-PCR analysis and in our previous study (10).
FIG. 3.
Wild-type E1A affects the removal of corepressor complexes from the promoters of cyclin A and Cdc6, whereas the E1A.928 mutant cannot. (A and B) Quiescent wild-type E1A-inducible cells were treated with or without Dox for the indicated times, and afterward ChIP analyses were performed using antibodies specific for p130, E2F4, HDAC1, and mSin3B. Control reactions were performed with NR IgG and anti-MyoD. Bottom row in panel A represents PCR analysis with GAPDH primers on input chromatin (0.05% of total) and confirms the use of equivalent amounts of chromatin in each of the ChIP reactions. (C) Quiescent mutant E1A.928-inducible cells were treated with Dox for 12 h, and afterward ChIP assays were performed using anti-p130, anti-E2F4, anti-HDAC1, and anti-mSin3B antibodies. A control reaction was performed in parallel with anti-MyoD antibody.
FIG. 4.
Promoter occupancy by E1A Δ26-35 mutant causes the removal of corepressor complexes from cyclin A and Cdc6 but does not induce their transcription. (A) Total RNA (top sections) was prepared from quiescent wild-type E1A- or E1A Δ26-35 mutant-inducible cells treated with or without Dox for the indicated times and subjected to RT-PCR analysis using cDNA-specific primers described in Materials and Methods. GAPDH expression was used as a loading control, and the “−” lane indicates RT-PCRs in the absence of reverse transcription. The “P” lane represents proliferating wild-type E1A- or E1A Δ26-35 mutant-inducible cells without Dox. Western blot analysis (bottom section) was performed on nuclear extracts prepared from proliferating or quiescent E1A Δ26-35 mutant-inducible cells treated with or without Dox for 12 h. Blots were probed with cyclin A and Cdc6 antibodies. The membrane was also probed with anti-GAPDH to monitor equal loading. (B) Cross-linked chromatin from quiescent E1A Δ26-35 mutant-inducible cells treated with or without Dox for the indicated times were immunoprecipitated, in parallel, with control antibodies (NR IgG and anti-MyoD) and antibodies specific for p130, HDAC1, E2F4, and mSin3B. Precipitated DNA fragments after processing were then analyzed by semiquantitative PCR using primers spanning the E2F sites within the promoters of cyclin A and Cdc6 (Fig. 2A). Bottom rows represent PCR analysis with GAPDH primers as described in the legend for Fig. 3. (C) Chromatin from quiescent E1A Δ26-35 mutant-inducible cells treated with Dox for 6 or 8 h was initially immunoprecipitated in parallel with anti-E1A (M73) or anti-p130 antibody. E1A-DNA or p130-DNA fragments were eluted from the immunoprecipitates and afterward reimmunoprecipitated with either NR IgG or antibodies against p130 or E1A, respectively. The resulting DNA fragments were then subjected to PCR analysis as described in the legend for Fig. 2.
The E1A Δ26-35 mutant occupies and eliminates corepressor complexes from promoters in quiescent cells.
We have previously reported that the E1A.928 mutant cannot activate cyclin A and Cdc6 in quiescent cells (10), and this is most likely due to its inability to bind p130 (Fig. 1A) and therefore displace transcriptional repressors from their promoters (Fig. 3C). Interestingly, although the E1A Δ26-35 mutant can still bind to p130 (Fig. 1A), it cannot induce transcription from the promoters of cyclin A and Cdc6 or therefore the expression of the cyclin A or Cdc6 proteins in quiescent cells as can wild-type E1A (Fig. 4A) (10). This result prompted us to investigate whether the E1A Δ26-35 mutant was capable of causing the p130-E2F4 and HDAC1/2-mSin3B complexes to dissociate from the cyclin A and Cdc6 promoters in quiescent cells. As shown in Fig. 4B, we found by kinetic studies coupled with ChIP assays that this mutant was in fact capable of eliminating these corepressors from the promoters by at least 12 h. Interestingly, this analysis also showed that unlike wild-type E1A, the E1A Δ26-35 mutant could not effect a difference in the time periods in which the corepressors dissociated from the promoters of Cdc6 and cyclin A (Fig. 3A and 4B). The reason for this disparity remains unclear.
We next determined by serial ChIP assays whether the removal of p130-E2F4 and HDAC1/2-mSin3B from cyclin A and Cdc6 was a function of the mutant's ability to occupy the promoters of these genes and thereby affect the activities of p130. In this experiment, equivalent amounts of cross-linked chromatin, in parallel, were immunoprecipitated with antibodies specific for the mutant E1A (M73) or p130. Afterward, the immune-selective DNA fragments were released from each of the antibodies and then reimmunoprecipitated with NR IgG for control or antibodies directed against p130 or mutant E1A. Similar to what was previously observed with wild-type E1A (10), we found that the E1A Δ26-35 mutant was also capable of occupying the same E2F-dependent promoters as p130 in quiescent cells (Fig. 4C).
E1A Δ26-35 mutant can erase the di-methylation of H3K9 but cannot induce its acetylation.
The failure of the E1A Δ26-35 mutant to activate cyclin A and Cdc6 despite its ability to eliminate p130-E2F4 and HDAC1/2-mSin3B from their promoters prompted us to determine what effect, if any, this mutant had on the histones that surrounded these genes. Given that the di-methylation of H3K9 is known to be highly indicative of transcriptional silencing (13) and that it becomes erased at the promoters of cyclin A and Cdc6 after wild-type E1A is expressed in quiescent cells (10), we investigated whether the E1A Δ26-35 mutant was equally capable of inducing the de-methylation of H3K9 as well. For comparison, we first examined the effect of wild-type E1A on this established marking. In agreement with our earlier studies (10), we found by ChIP analysis that the methylation of H3K9 at cyclin A or Cdc6 dramatically diminished and was subsequently replaced by acetylation after wild-type E1A was expressed in quiescent cells (Fig. 5A). In addition, this experiment also enabled us to detect for the first time an E1A-mediated histone H3 acetylation at Lys-14 (Fig. 5A).
FIG. 5.
The E1A Δ26-35 mutant causes the demethylation but not the acetylation of H3 or the binding of E2Fs at the promoters of cyclin A and Cdc6 in quiescent cells. (A) Cross-linked chromatin (left) was prepared from quiescent wild-type E1A-inducible cells without Dox and then immunoprecipitated in parallel with NR IgG or anti-H3-methyl-Lys-9 antibodies. Precipitated DNA fragments were analyzed by PCR as described in the legend for Fig. 3. Cross-linked chromatin (right) prepared from quiescent wild-type E1A-inducible cells treated with Dox for 12 h was immunoprecipitated in parallel with NR IgG, anti-H3-methyl-Lys-9, anti-H3-acetyl-Lys-9, or anti-H3-acetyl-Lys-14 antibodies. Precipitated DNA fragments were analyzed by PCR as described in the legend for Fig. 3. (B) E1A Δ26-35 mutant-inducible cells were rendered quiescent and then treated with or without Dox for 3-h intervals, ranging from 0 to 12 h. ChIP assays were then performed with the use of anti-H3-methyl-Lys-9, anti-H3-acetyl-Lys-9, anti-MyoD, or NR IgG antibodies. (C) Quiescent E1A.928-inducible cells were treated with Dox for 12 h and then subjected to ChIP analysis using NR IgG and antibodies specific for H3K9 di-methylation, H3K9 acetylation, or H3K14 acetylation. Precipitated DNA fragments were analyzed by PCR as described in the legend for Fig. 2.
Analysis of the E1A Δ26-35 mutant in the same experiment yielded results that were significantly different. Although kinetic studies coupled with ChIP assays indicated that this mutant was similar to wild-type E1A in diminishing the methylation of H3K9 at cyclin A and Cdc6 (Fig. 5B) (10), the mutant was still unable to induce the acetylation of K9 and K14 on the same histone (Fig. 5B; data not shown). Since p130 is seen to dissociate from the promoters of cyclin A and Cdc6 with kinetics similar to the demethylation of H3K9 after the E1A Δ26-35 mutant is expressed in quiescent cells (Fig. 4B and 5B), we hypothesized that p130 might have a role in the removal of the methyl groups from this histone. This speculation is supported in part by the inactivity of the E1A.928 mutant, which can neither dissociate p130 nor coincidentally induce the demethylation of H3K9 when expressed in quiescent cells (Fig. 2B and 5C). Furthermore, there is evidence to suggest that p130 can physically associate with the histone H3K9-specific methyltransferase SUV39H1 (10, 21), indicating that p130 may be involved in recruiting this enzyme to the promoters of E2F-responsive genes in quiescent cells.
The E1A Δ26-35 mutant does not effect the recruitment of activating E2Fs to promoters in quiescent cells.
It is well established that the transcriptional factors E2F1, E2F2, and E2F3a play a role in stimulating E2F-reponsive promoters as cells transition out of quiescence and progress into the cell cycle (29). It is not surprising then that after wild-type E1A is expressed in quiescent cells, all three of these factors temporally bind to the promoters of cyclin A and Cdc6 and contribute to the timely activation of these genes, as shown in our previous work (10). We therefore performed ChIP assays to determine whether any of the E2F factors replaced E2F4 at these promoters after the E1A Δ26-35 mutant was expressed in quiescent cells. As expected, the ChIP data show that E2F4 is recruited to these promoters during quiescence but that it becomes noticeably absent after the expression of the E1A Δ26-35 mutant (Fig. 6A). We then monitored the potential recruitment of E2F1, E2F2, or E2F3a to the promoters under study, and in striking contrast to what we have previously shown with wild-type E1A (10), we could not detect the occupancy of any of these factors after the expression of the E1A Δ26-35 mutant (Fig. 6A). We concluded that there were at least two reasons that could account for this result: (i) as previously shown (16), a relaxed chromatin structure provided by histone acetylation, which the E1A Δ26-35 mutant fails to induce (Fig. 5B), may be required for E2F1-to-3a access, or (ii) each of the E2F genes remains transcriptionally inactive during the expression of the E1A Δ26-35 mutant. Because E2F1 is responsible in part for activating E2F2 and E2F3 transcription as cells reenter the cell cycle (16), we focused on the second reason by determining whether the promoter of this gene was in fact being induced in quiescent cells after the expression of either wild-type E1A or the E1A Δ26-35 mutant. To begin, ChIP experiments, with the use of primers flanking potential E2F-binding sites within the E2F1 promoter (Fig. 2A), showed the occupancy of the repressor complexes p130-E2F4 and HDAC1/2-mSin3B during quiescence (Fig. 6B). Moreover, our ChIP analysis also indicated that the promoter was associated with a di-methylated H3K9 and a histone H3K9/14 without a detectable level of acetylation (Fig. 6C). These results were consistent with the fact that E2F1 was transcriptionally inactive in quiescent cells, as determined by RT-PCR analysis (Fig. 6D, lane 2). As expected, after E1A was induced in these cells by the addition of Dox, we found that this gene was now being transcriptionally expressed (Fig. 6D, lanes 3 to 6). However, E2F1 transcripts were not detected by this analysis after the E1A Δ26-35 mutant or the E1A.928 mutant was expressed in the quiescent cells (Fig. 6D and data not shown).
FIG. 6.
Analysis of the E2F1 promoter before and after the expression of wild-type E1A in quiescent cells. (A) Quiescent E1A Δ26-35 mutant-inducible cells were treated with or without Dox for the indicated times, and afterward ChIP assays, as described in the legend for Fig. 3, were performed in parallel using antibodies specific for E2F1, E2F2, E2F3, and E2F4. Control antibodies include NR IgG and anti-MyoD antibody. (B and C) Cross-linked chromatin was prepared from quiescent cells without the addition of Dox and then subjected to ChIP assays as described in the legend for Fig. 2 using antibodies specific for p130, E2F4, mSin3B, HDAC1, H3K9 di-methylation, H3K9 acetylation, or H3K14 acetylation. Control antibodies include anti-MyoD. Precipitated products were then analyzed by semiquantitative PCR using primers surrounding the E2F binding sites within the E2F1 promoter (Fig. 2A). (D) Total RNA was prepared from quiescent wild-type E1A-inducible or E1A Δ26-35 mutant-inducible cells treated with or without Dox for the indicated times. RT-PCR analysis was then performed in parallel on this RNA as described in the legend for Fig. 4A. The “P” lane represents proliferating wild-type E1A- or E1A Δ26-35 mutant-inducible cells. (E) Quiescent wild-type E1A-inducible cells were treated with or without Dox for the indicated times and then subjected to ChIP analysis as described in the legend for Fig. 3 using antibodies specific for p130, E2F4, mSin3B, HDAC1, H3K9 di-methylation, H3K9 acetylation, or H3K14 acetylation. Control antibodies include anti-MyoD and NR IgG. (F) Quiescent E1A Δ26-35 mutant- or E1A.928-inducible cells treated with Dox for 12 h were subject to ChIP analysis as described above using antibodies specific for p130, E2F4, H3K9 di-methylation, or H3K9 acetylation. The control antibody includes NR IgG.
We also found by ChIP analysis that E1A's ability to activate E2F1 in quiescent cells is mediated in part by the dissociation of p130-E2F4 and HDAC1/2-mSin3B, the loss of H3K9 di-methylation, and the acetylation of H3K9/14 at its promoter (Fig. 6E and data not shown). Except for the acetylation of H3K9, these alterations at the E2F1 promoter were also induced by the E1A Δ26-35 mutant (Fig. 6F), and this essentially matched its effect on the cyclin A and Cdc6 promoters (Fig. 4 and 5). Not surprisingly, none of the aforementioned changes could be observed at the E2F1 promoter after the expression of the E1A.928 mutant (Fig. 6F). Taken together, these data suggest that the E1A Δ26-35 mutant cannot mediate the activation of E2F1 in quiescent cells, and because this factor is required for activating E2F2 and E2F3a (16), we speculate that this may be the reason why this mutant is unable to effect the recruitment of E2Fs to the promoters of cyclin A and Cdc6.
The recruitment of a PCAF-containing complex to E2F-dependent promoters requires E1A.
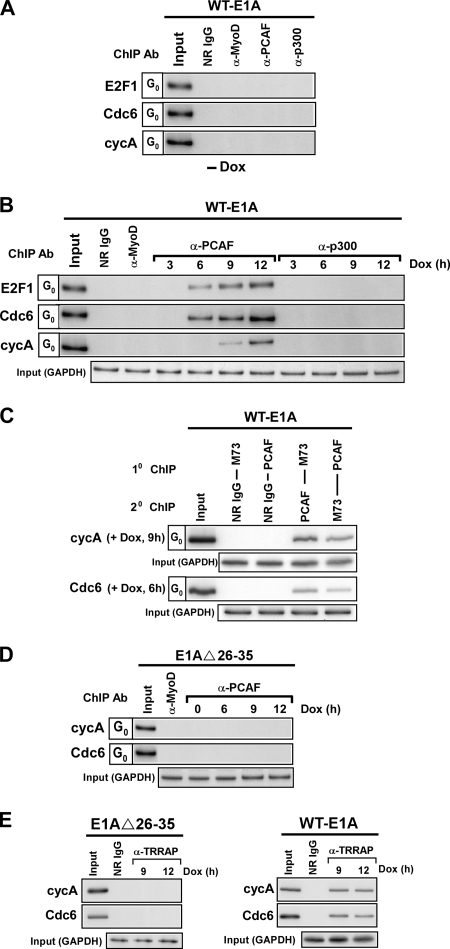
Our data indicate that unlike wild-type E1A, the E1A Δ26-35 mutant cannot mediate the activation of cyclin A, Cdc6, and E2F1 or induce cellular DNA synthesis in quiescent cells (Fig. 1A and 4A). Remarkably, the only apparent difference between these two proteins in effecting a change at these E2F-dependent genes is that the E1A Δ26-35 mutant cannot induce the subsequent acetylation of H3K9/14 in association with their promoters (Fig. 5B and 6F and data not shown). Thus, the recruitment of a HAT activity to the promoters of cyclin A, Cdc6, and E2F1 in order to help induce their transcription in quiescent cells might be facilitated by wild-type E1A but is deficient in the E1A Δ26-35 mutant. To investigate this possibility and to test for the recruitment of HATs, such as PCAF and p300, to the promoters of the cyclin A, Cdc6, and E2F1 genes, we first performed kinetic studies combined with ChIP assays on quiescent cells with and without the expression of wild-type E1A. Despite the presence of both these proteins in quiescent cells (Fig. 1C and data not shown), neither PCAF nor p300 is being recruited to the promoters of cyclin A, Cdc6, and E2F1 while these cells are in this state (Fig. 7A). Quite strikingly, however, after wild-type E1A is expressed in these cells, PCAF appears at the promoters of these genes and, except for cyclin A, with similar kinetics (Fig. 7B). Moreover, this association occurs after the p130-E2F4 and HDAC1/2-mSin3B complexes have been eliminated from each of the promoters and coincidently with the acetylation of H3K9 (Fig. 3A and 6E) (10). The binding of p300, on the other hand, was virtually undetectable at cyclin A, Cdc6, and E2F1, whether wild-type E1A was expressed or not (Fig. 7A and B). For purposes of efficacy, this experiment was repeated with several anti-p300 antibodies with similar results (data not shown). Finally, our failure to detect p300 at the E2F target genes in quiescent cells with or without the expression of wild-type E1A is consistent with previous experiments that showed the absence of p300 near the transcriptional start site (TSS) of a number of E2F-responsive genes in both quiescent and serum-stimulated T98G cells (28). Interestingly, despite the inability of p300 to bind near the TSS, ChIPs combined with microarrays have shown the presence of p300 within 2 kb of the TSS of E2F-dependent genes after contact-inhibited human IMR90 cells were infected with the adenovirus mutant dl1500, which expresses only the 243R E1A protein, but not after the cells were infected with a mutant adenovirus (RG2) encoding an E1A 243R protein defective for the binding of p300/CBP (7). To substantiate further that E1A may be involved in recruiting PCAF activity to E2F-regulated genes during quiescence, we performed a serial ChIP experiment to determine whether these two proteins could be found together on the same E2F-dependent promoter in quiescent cells. As shown in Fig. 7C, direct evidence of physical binding of both E1A and PCAF to the same promoters of Cdc6 and cyclin A at 6 and 9 h, respectively, was evident by the ChIP reimmunoprecipitation assays. Because E1A and a PCAF-containing complex can associate with one another in nuclear extracts of quiescent cells (Fig. 1B), we interpreted these results to suggest that E1A is participating in the recruitment of a PCAF-containing complex to the promoters of E2F-dependent genes and that this step is important for ultimately acetylating H3 histones in association with these promoters. This possibility is supported by the lack of detectable PCAF and TRRAP at the promoters of cyclin A or Cdc6 in quiescent cells following the expression of the E1A Δ26-35 mutant (Fig. 7D and E) and by the failure of this mutant to stimulate the acetylation of H3K9 at these promoters, despite the fact that it can induce the departure of repressor complexes, which in turn leads to the diminishing of H3K9 di-methylation (Fig. 4B and 5B). Taken together, these results suggest a new paradigm for the role of E1A in transitioning cells out of quiescence and inducing their entry into S phase.
FIG. 7.
E1A is responsible for recruiting a PCAF-containing complex to the promoters of E2F-dependent genes in quiescent cells. (A) ChIP assays were performed in parallel on quiescent wild-type E1A-inducible cells without the addition of Dox, as described in the legend for Fig. 3. The antibodies used in this analysis included anti-PCAF, anti-p300, or NR IgG and anti-MyoD for control. (B) ChIP analysis, as described in the legend for Fig. 3, was performed on quiescent wild-type E1A-inducible cells after being treated with or without Dox for the indicated times. The antibodies used in this analysis included anti-PCAF, anti-p300, or NR IgG and anti-MyoD for control. (C) Serial ChIP assays were performed on quiescent wild-type E1A-inducible cells after treatment with Dox for the indicated times using initially antibodies specific for E1A (M73) or PCAF. E1A-DNA, or PCAF-DNA fragments were reimmunoprecipitated with either NR IgG or antibodies against PCAF or E1A (M73), respectively. The resulting DNA fragments were then subjected to PCR analysis as described in the legend for Fig. 3. (D) Cross-linked chromatin from quiescent E1A Δ26-35 mutant-inducible cells treated with or without Dox for the indicated times was subjected to ChIP analysis using anti-PCAF or anti-MyoD antibodies, for control. The illustration (bottom rows) represents PCR analysis with GAPDH primers on input chromatin (0.05% of total) and confirms the use of equivalent amounts of chromatin in each of the ChIP reactions. (E) Cross-linked chromatin from wild-type or E1A Δ26-35 mutant-inducible cells treated with or without Dox for the indicated times was subjected to ChIP analysis using anti-TRRAP antibody or NR IgG, for control. Bottom rows in the illustration represent PCR analysis with GAPDH primers on input chromatin (0.05%).
DISCUSSION
In summary, our data demonstrate that the adenovirus E1A 243R protein is involved in at least two key steps in activating E2F-responsive phase genes. First, it clears their promoters of corepressor complexes, which in turn diminishes the methylation of nucleosomal H3 histones at Lys-9, and second, it stimulates the recruitment of activator elements to the promoters.
The use of wild-type and mutant E1A-inducible cell lines have allowed us to uncover novel mechanisms by which the E1A protein induces quiescent cells to reenter the cell cycle. In our previous work, this experimental approach enabled us to establish, for the first time, an important role for E1A in altering the epigenetic program controlling the silencing of E2F-regulated genes in quiescent cells (10). The work we present here extends this finding and suggests that E1A directly interacts with at least two opposing transcriptional pathways to accomplish this activity. In the end, this leads to the activation of a specific class of genes involved in cell cycle reentry. One of the pathways includes the corepressor complexes p130-E2F4 and HDAC1/2-mSin3B, which are found in association with E2F-responsive genes in quiescent cells. Our results indicate that E1A can cause these complexes to separate from these genes after it has occupied their promoters in quiescent cells. Given that E1A can physically bind to p130 (8), we speculate that this interaction facilitates the dissociation of p130-E2F4 from the E2F-dependent promoters, and since the HDAC1/2-mSin3B complex is likely tethered to the same promoter through its association with p130-E2F4 (Fig. 3) (22), this may be the reason why it too is eliminated. We have also shown that once the transcriptional repressors are lost from the E2F-dependent promoters, the methylation of histone H3K9 that lies in close proximity to the transcriptional start site becomes rapidly diminished. As of yet, the mechanisms responsible for this alteration are unclear, since we have been unable to find a lysine-specific demethylase that is affected by E1A (data not shown).
The other pathway that E1A alters during quiescence is also involved in transcriptional regulation and includes a PCAF-containing complex. This histone H3-specific HAT has been reported to regulate global acetylation and/or the expression of specific genes (20). Our experiments suggest that E1A recruits this complex to E2F-dependent genes once the corepressor complexes have been dissociated from their promoters. This recruitment then allows for the acetylation of nucleosomal H3K9/K14, which in turn contributes to the activation of genes such as E2F1, cyclin A, and Cdc6, all of which are critical to transitioning cells out of quiescence and into S phase. Although H3K9/K14 are considered major targets for the PCAF complex (20), there is evidence to suggest that p300/CBP can also acetylate H3K14 in vitro (14). However, we have been unable to detect the recruitment of this HAT to any of the E2F-dependent promoters that were analyzed in this study. Nevertheless, future experiments must address whether p300/CBP is recruited to these promoters at a later time or as cells progress through G1.
Our observation of a role for E1A in activating E2F1 in quiescent cells is likely important for activating E2F2 and E2F3a. Others have shown that the activation of these genes depends in part on the E2F1 protein and that their expression is necessary for cells to reenter the proliferative process (16). Indeed, we have previously demonstrated that E2F1, E2F2, and E2F3a temporally bind to the promoters of cyclin A and Cdc6 after E1A has induced the loss of promoter occupancy for p130-E2F4 (10). Moreover, the fact that E2F1 mRNA is not detected and that none of the E2Fs assemble at the target gene promoters in quiescent cells expressing the E1A Δ26-35 mutant is consistent with the idea that the E2F genes are activated only after the E2F1 protein has accumulated in cells reentering the cell cycle. Although we have observed a clear role for PCAF activity in the activation of E2F1 in our studies, we cannot exclude the possibility that once the other E2Fs are activated, they begin to participate in the acetylation of E2F promoter-associated nucleosomal histones, as well (27). There is evidence to suggest that the acetylation of H3 and H4 at E2F-dependent genes, particularly in late G1, is associated with the recruitment of E2F1 to -3 to their promoters and that Tip60 HAT could account for the E2F-mediated acetylation (28).
Finally, E1A's ability to target two transcriptional pathways involved in the activation or repression of genes allows this viral protein to serve as a potent mediator of remodeling chromatin to activate a select group of E2F-regulated genes essential to the reentry of quiescent cells into G1 and ultimately into S phase. It will be essential to understand the full range of E1A's ability in this endeavor in order to ensure the production of adenovirus after the infectivity of quiescent cells.
Acknowledgments
We thank David Samols for careful reading of the manuscript.
The project described was supported in part by grants from the NIH NIGMS to M.L.H.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673-7685. [DOI] [PubMed] [Google Scholar]
- 2.Blais, A., and B. D. Dynlacht. 2007. E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 19:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crescenzi, M., S. Soddu, and F. Tato. 1995. Mitotic cycle reactivation in terminally differentiated cells by adenovirus infection. J. Cell. Physiol. 162:26-35. [DOI] [PubMed] [Google Scholar]
- 4.Deleu, L., S. Shellard, K. Alevizopoulos, B. Amati, and H. Land. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270-8275. [DOI] [PubMed] [Google Scholar]
- 5.Dobrowolski, S. F., D. W. Stacey, M. L. Harter, J. T. Stine, and S. W. Hiebert. 1994. An E2F dominant negative mutant blocks E1A induced cell cycle progression. Oncogene 9:2605-2612. [PubMed] [Google Scholar]
- 6.Felsani, A., A. M. Mileo, and M. G. Paggi. 2006. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25:5277-5285. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari, R., M. Pellegrini, G. A. Horwitz, W. Xie, A. J. Berk, and S. K. Kurdistani. 2008. Epigenetic reprogramming by adenovirus e1a. Science 321:1086-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolov, M. V., and N. J. Dyson. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 117:2173-2181. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, M. K., and M. L. Harter. 2003. A viral mechanism for remodeling chromatin structure in G0 cells. Mol. Cell 12:255-260. [DOI] [PubMed] [Google Scholar]
- 11.Harlow, E., B. R. Franza, Jr., and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howe, J. A., J. S. Mymryk, C. Egan, P. E. Branton, and S. T. Bayley. 1990. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 87:5883-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hublitz, P., M. Albert, and A. H. Peters. 2009. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 53:335-354. [DOI] [PubMed] [Google Scholar]
- 14.Kalkhoven, E. 2004. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 68:1145-1155. [DOI] [PubMed] [Google Scholar]
- 15.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836-2841. [DOI] [PubMed] [Google Scholar]
- 16.Leung, J. Y., G. L. Ehmann, P. H. Giangrande, and J. R. Nevins. 2008. A role for Myc in facilitating transcription activation by E2F1. Oncogene 27:4172-4179. [DOI] [PubMed] [Google Scholar]
- 17.Mal, A., D. Chattopadhyay, M. K. Ghosh, R. Y. C. Poon, T. Hunter, and M. L. Harter. 2000. p21 and retinoblastoma protein control the absence of DNA replication in terminally differentiated muscle cells. J. Cell Biol. 149:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mal, A., and M. L. Harter. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murr, R., T. Vaissiere, C. Sawan, V. Shukla, and Z. Herceg. 2007. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene 26:5358-5372. [DOI] [PubMed] [Google Scholar]
- 20.Nagy, Z., and L. Tora. 2007. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26:5341-5357. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas, E., C. Roumillac, and D. Trouche. 2003. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol. Cell. Biol. 23:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F meditates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 24.Samuelson, A. V., M. Narita, H. M. Chan, J. Jin, E. de Stanchina, M. E. McCurrach, M. Fuchs, D. M. Livingston, and S. W. Lowe. 2005. p400 is required for E1A to promote apoptosis. J. Biol. Chem. 280:21915-21923. [DOI] [PubMed] [Google Scholar]
- 25.Spindler, K. R., C. Y. Eng., and A. J. Berk. 1985. An adenovirus early region 1A protein is required for maximal viral DNA replication in growth-arrested human cells. J. Virol. 53:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiegler, P., A. De Luca, L. Bagella, and A. Giordano. 1998. The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res. 58:5049-5052. [PubMed] [Google Scholar]
- 27.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 28.Taubert, S., C. Gorrini, S. R. Frank, T. Parisi, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2004. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 24:4546-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Heuvel, S., and N. J. Dyson. 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9:713-724. [DOI] [PubMed] [Google Scholar]