Abstract
Fifty-eight fusaria isolated from 50 Italian patients between 2004 and 2007 were subject to multilocus DNA sequence typing to characterize the spectrum of species and circulating sequence types (STs) associated with dermatological infections, especially onychomycoses and paronychia, and other fusarioses in northern and central Italy. Sequence typing revealed that the isolates were nearly evenly divided among the Fusarium solani species complex (FSSC; n = 18), the F. oxysporum species complex (FOSC; n = 20), and the Gibberella (Fusarium) fujikuroi species complex (GFSC; n = 20). The three-locus typing scheme used for members of the FSSC identified 18 novel STs distributed among six phylogenetically distinct species, yielding an index of discrimination of 1.0. Phylogenetic analysis of the FOSC two-locus data set identified nine STs, including four which were novel, and nine isolates of ST 33, the previously described widespread clonal lineage. With the inclusion of eight epidemiologically unrelated ST 33 isolates, the FOSC typing scheme scored a discrimination index of 0.787. The two-locus GFSC typing scheme, which was primarily designed to identify species, received the lowest discrimination index, with a score of 0.492. The GFSC scheme, however, was used to successfully identify 17 isolates as F. verticillioides, 2 as F. sacchari, and 1 as F. guttiforme. This is the first report that F. guttiforme causes a human mycotic infection, which was supported by detailed morphological analysis. In addition, the results of a pathogenicity experiment revealed that the human isolate of F. guttiforme was able to induce fusariosis of pineapple, heretofore its only known host.
Fusarium is a large genus of hyaline filamentous molds best known as the most important group of mycotoxigenic plant pathogens (15, 21, 24). Fusaria have also emerged over the past 3 decades as opportunistic pathogens of immunocompetent and immunocompromised hosts (27). Infections in healthy individuals typically remain localized and include keratitis, especially in association with ocular trauma; onychomycosis of the toenails or fingernails; allergic sinusitis; paronychia; and dermatomycoses (11, 13, 18, 28, 34). By way of contrast, fusarioses in artificially immunosuppressed or immunocompromised patients can become life-threatening systemic infections if they become disseminated within the bloodstream, especially if the individual is persistently and severely neutropenic (50). Deeply invasive fusarial infections pose a significant challenge to infectious disease specialists because, with the exception of limited success with liposomal amphotericin B, most species show high levels of resistance to all antifungals currently available (1, 4, 5, 36, 39).
Several species-level molecular phylogenetic analyses of medically important fusaria over the past decade have revealed that the most commonly reported species (i.e., Fusarium solani, F. moniliforme, F. oxysporum, F. incarnatum, F. chlamydosporum, and F. dimerum) actually represent species complexes that collectively contain approximately 70 medically relevant species (31, 32, 35, 36, 37, 52). Typing schemes have been developed for these six species complexes and have focused on the identification of species limits on the basis of multilocus genealogical concordance (46), in contrast to schemes for the high-resolution typing of the strains within a species (6, 29). The fusarial studies have revealed that the majority of clinically important fusaria cannot be identified by the use of morphology alone (36, 37), and therefore, accurate reporting of the etiological agent typically requires a DNA sequence-based identification (7). These studies have also revealed the necessity of developing species and sequence type (ST) nomenclatures within the F. solani species complex (FSSC), the F. oxysporum species complex (FOSC), the F. incarnatum/F. equiseti species complex (FIESC), and the F. chlamydosporum species complex (FCSC), in which close to two-thirds of the species (i.e., ∼40) lack Latin binomials. Fortunately, all of the clinically relevant species within the Gibberella (Fusarium) fujikuroi species complex (GFSC; until recently, generally reported as F. moniliforme) and most of those within the F. dimerum species complex (FDSC) have been formally described (31, 42). Sequence-based typing has revealed that members of the FSSC, FOSC, and GFSC are collectively the cause of approximately 80% of all fusarial infections of humans and other animals, with members of the other three complexes accounting for most of the remainder.
Due in part to the increased awareness of clinicians, coupled with their possible insurgence, onychomycoses and acute and chronic cutaneous infections caused by Fusarium spp. are increasingly being reported in immunocompetent patients (10, 16, 18, 22, 40, 43). Paronychial and other dermatomycotic fusarial infections, including dermatitis, represent a challenge for the clinician, since an incorrect diagnosis may lead to topical steroidal therapy and local and/or systemic antibiotic therapy, which can delay recovery and increase morbidity (23). Moreover, onychomycotic infections require careful monitoring within the neutropenic patient population because they can progress into life-threatening disseminated infections (50).
The present molecular phylogenetic study was conducted to (i) identify the etiological agents of 46 dermatological fusaria and 12 other fusarial pathogens associated with diverse mycotic infections occurring in the Lombardy and Piedmont regions in northern Italy and in the Marche region in central Italy during the period from 2004 through 2007; (ii) assess the prediction, on the basis of the available typing data, that most of the clinically relevant fusaria are nested within the FSSC, FOSC, or GFSC; (iii) determine the index of discrimination of the DNA typing schemes employed (19); and (iv) assess whether the novel human pathogen Fusarium guttiforme, which was recovered from a human paronychial infection, is pathogenic for its only known host, Ananas comosus (L.) Merr. (pineapple).
MATERIALS AND METHODS
Fungal strains.
The 58 human pathogenic fusaria analyzed in the present study were obtained from four different hospitals in Milan (designated hospitals M-1, M-2, M-3, and M-4) and from one hospital each in Varese, Novara, Torino, and Ancona (Table 1). Close to three-quarters of the isolates (n = 46) were obtained from the Dermatology Unit of Sesto San Giovanni Hospital in Milan (designated hospital M-1 in Table 1). Part of this collection was provided by the Medical Mycology Committee (CoSM) of the Associazione Microbiologi Clinici Italiani, Milan, Italy. All strains are stored cryogenically in the Agricultural Research Service (NRRL) Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, and in the collection of the Centro per la Conservazione e Valorizzazione della Biodiversità Vegetale (CBV), University of Sassari, Sassari, Italy, for future reference. The reference strains used in this study, including F. guttiforme (NRRL 25295), F. virguliforme O'Donnell and T. Aoki (NRRL 22825), F. tucumaniae T. Aoki, O'Donnell, Yosh, Homma, and Lattanzi (NRRL 31096), F. sacchari (E. J. Butler and Hafiz Khan) W. Gams (NRRL 13999), and F. foetens Schroers, O'Donnell, Baayen, and Hooftman (NRRL 38302, NRRL 31852), are also available from the ARS Culture Collection (NRRL).
TABLE 1.
Isolates subjected to DNA MLST
| MLSTa | NRRL no. | VB no.b | Diagnosis | Isolate source | Date isolated (mo/day/yr or yr) | Origin | Hospitalc | Age (yr)d | Sexd |
|---|---|---|---|---|---|---|---|---|---|
| F. guttiforme | 53131 | VN | Paronychiae | Finger | 12/14/2007 | Milan | M-1 | 65 | F |
| F. sacchari | 44901 | NC 2 | Paronychia | Finger | 7/28/2005 | Milan | M-1 | 51 | F |
| F. sacchari | 44905 | NC 1 | Paronychia | Finger | 7/12/2005 | Milan | M-1 | 51 | F |
| F. verticillioides | 44891 | MTP 2 | Paronychia | Finger | 1/18/2006 | Milan | M-1 | 67 | F |
| F. verticillioides | 44894 | MTP 1 | Paronychia | Finger | 1/5/2006 | Milan | M-1 | 67 | F |
| F. verticillioides | 44895 | SB dx | Onychomycosis | Toes | 9/19/2006 | Milan | M-1 | 45 | F |
| F. verticillioides | 44897 | GJ | Paronychiaf | Toes | 12/1/2005 | Milan | M-1 | 48 | F |
| F. verticillioides | 44898 | RL 2 | Onychomycosis | Toe | 10/12/2005 | Milan | M-1 | 55 | F |
| F. verticillioides | 46442 | AL | Onychomycosis | Toe | 3/16/2005 | Milan | M-1 | 64 | F |
| F. verticillioides | 46444 | CA | Onychomycosis | Toes | 3/3/2005 | Milan | M-1 | 54 | F |
| F. verticillioides | 46599 | PM | Onychomycosis | Toe | 6/5/2007 | Milan | M-1 | 40 | F |
| F. verticillioides | 53119 | ANC 6 | Sepsis in leukemia | Blood | 2007 | Ancona | A | ||
| F. verticillioides | 53122 | NIG 3 | Sepsis in leukemia | Blood | 2007 | Milan | M-3 | ||
| F. verticillioides | 53123 | NOV | Sepsis in leukemia | Blood | 2007 | Novara | N | ||
| F. verticillioides | 53125 | NOV | Sepsis in kidney transpant | Blood | 2007 | Novara | N | ||
| F. verticillioides | 53126 | NOV | Chronic sinusitis | Maxillary sinus pus | 2007 | Novara | N | ||
| F. verticillioides | 53127 | NOV | Acute lymphoblastic leukemia | Nasal swab and bal | 2007 | Novara | N | ||
| F. verticillioides | 53129 | VAR 8 | Pneumonia | Bronchoalveolar lavage | 2007 | Varese | V | ||
| F. verticillioides | 53130 | VIM 1 | Keratitis | Ocular swab | 2007 | Milan | M-4 | ||
| F. verticillioides | 53133 | ANC 5 | Keratitis | Ocular swab | 2007 | Ancona | A | ||
| FOSC ST 33 | 44893 | GS | Paronychia | Finger | 9/29/2005 | Milan | M-1 | 38 | M |
| FOSC ST 33 | 44899 | AVE | Onychomycosis | Toe | 11/14/2006 | Milan | M-1 | 51 | F |
| FOSC ST 33 | 46436 | ML | Onychomycosis | Toes | 5/19/2007 | Milan | M-1 | 75 | F |
| FOSC ST 33 | 46439 | GR | Onychomycosis | Toe | 1/17/2007 | Milan | M-1 | 80 | M |
| FOSC ST 33 | 46595 | ML 2 | Onychomycosis | Toes | 6/5/2007 | Milan | M-1 | 75 | F |
| FOSC ST 33 | 46600 | CCJ | Dermatomycosis | Foot | 7/14/2007 | Milan | M-1 | 27 | M |
| FOSC ST 33 | 46601 | GC | Onychomycosis | Toe | 7/14/2007 | Milan | M-1 | 45 | F |
| FOSC ST 33 | 46603 | DP | Onychomycosis | Toe | 10/15/2004 | Milan | M-1 | 61 | M |
| FOSC ST 33 | 46606 | FC | Onychomycosis | Toe | 7/26/2005 | Milan | M-1 | 37 | F |
| FOSC ST 43 | 46441 | NR | Onychomycosis | Toe | 1/17/2007 | Milan | M-1 | 70 | F |
| FOSC ST 43 | 46607 | AM | Onychomycosis | Toe | 6/5/2007 | Milan | M-1 | 70 | F |
| FOSC ST 52 | 53121 | MIL 8 | Soft tissue Kaposi's sarcoma | Pus | 2007 | Milan | M-2 | ||
| FOSC ST 90 | 46605 | SC | Onychomycosis | Toe | 10/16/2004 | Milan | M-1 | 59 | F |
| FOSC ST 93 | 52681 | MV 1 | Onychomycosis | Toe | 7/10/2007 | Milan | M-1 | 61 | F |
| FOSC ST 93 | 52682 | MV 2 | Onychomycosis | Toe | 7/24/2007 | Milan | M-1 | 61 | F |
| FOSC ST 93 | 52683 | MV 3 | Onychomycosis | Toe | 8/4/2007 | Milan | M-1 | 61 | F |
| FOSC ST 288 | 44900 | ZA | Onychomycosis | Toe | 9/23/2006 | Milan | M-1 | 43 | M |
| FOSC ST 289 | 44902 | RL 1 | Onychomycosis | Toe | 9/21/2005 | Milan | M-1 | 55 | F |
| FOSC ST 290 | 46597 | RM | Onychomycosis | Toe | 6/5/2007 | Milan | M-1 | 67 | F |
| FOSC ST 291 | 46602 | ACG | Onychomycosis | Toes | 3/10/2007 | Milan | M-1 | 59 | F |
| FSSC 1-f | 46604 | TC | Onychomycosis | Toe | 3/10/2007 | Milan | M-1 | 50 | M |
| FSSC 1-g | 46440 | VP | Onychomycosis | Finger | 3/22/2005 | Milan | M-1 | 50 | F |
| FSSC 2-aa | 46438 | GL 1 | Onychomycosis | Toe | 5/19/2007 | Milan | M-1 | 40 | F |
| FSSC 2-bb | 46443 | SS | Dermatomycosis | Foot | 12/15/2004 | Milan | M-1 | 31 | M |
| FSSC 2-cc | 53132 | BA | Onychomycosis | Toe | 12/14/2007 | Milan | M-1 | 66 | F |
| FSSC 2-y | 44890 | GSK | Dermatomycosis | Foot | 12/1/2005 | Milan | M-1 | 37 | M |
| FSSC 2-z | 46437 | GL | Onychomycosis | Toe | 4/11/2007 | Milan | M-1 | 40 | F |
| FSSC 3 + 4-lll | 44906 | SB sx | Onychomycosis | Toes | 6/19/2006 | Milan | M-1 | 45 | F |
| FSSC 3 + 4-mmm | 52680 | CFR 2 | Onychomycosis | Toe | 7/28/2007 | Milan | M-1 | 36 | F |
| FSSC 3 + 4-nnn | 52832 | SA | Onychomycosis | Toe | 11/24/2007 | Milan | M-1 | 31 | F |
| FSSC 3 + 4-ooo | 53120 | ANC 8 | Sepsis | Blood | 2007 | Ancona | A | ||
| FSSC 3 + 4-ppp | 53128 | TOM 1 | Meningitis | Cerebrospinal fluid | 2007 | Torino | T | ||
| FSSC 5-aa | 44903 | GF | Onychomycosis | Toe | 6/9/2006 | Milan | M-1 | 21 | M |
| FSSC 5-bb | 46598 | CA | Onychomycosis | Toe | 6/5/2007 | Milan | M-1 | 35 | F |
| FSSC 5-z | 44896 | MC | Onychomycosis | Toe | 6/9/2006 | Milan | M-1 | 35 | F |
| FSSC 6-k | 44892 | TF | Dermatitis | Face | 6/9/2006 | Milan | M-1 | 75 | M |
| FSSC 6-l | 44904 | EN | Onychomycosis | Toe | 6/9/2006 | Milan | M-1 | 35 | M |
| FSSC 35-b | 46596 | ME | Onychomycosis | Toe | 6/5/2007 | Milan | M-1 | 40 | M |
A two-locus typing scheme was used to identify 20 isolates within the Gibberella (Fusarium) fujikuroi species complex (i.e., F. guttiforme, F. sacchari, and F. verticillioides); the STs of 20 FOSC isolates were identified by using a two-locus typing scheme (32); a three-locus typing scheme (36) was used to characterize 18 members of FSSC. Arabic numerals are used to designate FSSC species, and lowercase roman letters are used to identify unique haplotypes within each species.
VB no., Virgilio Balmas' patient code. Molecular typing of two isolates obtained successively from three separate patients (highlighted in boldface) revealed mixed fusarial infections: Fusarium verticillioides NRRL 44895 (SB dx) and FSSC 3 + 4-lll NRRL 44906 (SB sx), F. verticillioides NRRL 44898 (RL 2) and FOSC ST 289 NRRL 44902 (RL 1), and FSSC 2-z NRRL 46437 (GL) and FSSC 2-aa NRRL 46438 (GL 1). By way of contrast, sequential isolates obtained from the four patients highlighted in italics (patient codes ML, MTP, MV, and NC) appeared to be clonally propagated, in that isolates from each individual were genetically identical.
Locations of eight Italian hospitals sampled listed by city: A, Ancona (Marche region, central Italy); M-1, Sesto San Giovanni Hospital, Milan; M-2, hospital 2, Milan; M-3, hospital 3, Milan; M-4, hospital 4, Milan; V, Varese (Lombardy region, northern Italy); N, Novara; T, Torino (Piedmont region, northern Italy).
Age and gender are reported only for the 38 dermatology patients, 26 of whom (70.7%) were women. Fusaria were sequentially cultured from seven female patients. F, female; M, male.
Paronychia in the absence of onychomycosis.
Isolation yielded a mixed culture of F. verticillioides and an Acremonium sp.
Genotypic and phenotypic identification.
The 58 Italian isolates received as Fusarium spp. were initially identified to the species complex or species level by using a partial translation elongation factor (EF-1α) gene sequence as a BLAST query against the Fusarium database (http://isolate.fusariumdb.org/), as described previously (14). To obtain total genomic DNA, mycelium was grown in yeast-malt broth, freeze-dried, and then extracted with hexadecyltrimethylammonium bromide (CTAB; Sigma, St. Louis, MO) by a published protocol (31). The conditions used for PCR amplification of the partial EF-1α gene, the internal transcribed spacer (ITS) region plus domains D1 and D2 of the nuclear large-subunit (LSU) rDNA, and a 1.8-kb portion of the second-largest subunit of RNA polymerase (RPB2) are described by O'Donnell and colleagues (31, 33, 36); and amplification of the nuclear ribosomal intergenic spacer (IGS) rDNA has been described by O'Donnell et al. (32). The three-locus FSSC, two-locus FOSC, and two-locus GFSC data sets were analyzed by maximum parsimony (MP) with the PAUP* (version 4.0b10) program (44). The shortest MP trees were identified by using searches that employed 1,000 random sequence addition replicates and reconnection (tree bisection-reconnection [TBR]) branch swapping. Clade support was assessed by nonparametric MP bootstrapping by employing 10 random addition sequences per replicate, 1,000 pseudoreplicates of the data, and TBR branch swapping. Previously published sequences of Fusarium virguliforme and F. tucumaniae, which were used to root the FSSC phylogeny, and those of F. foetens, which were used to root the FOSC phylogram, are available from GenBank (http://www.ncbi.nlm.nih.gov/).
The phenotypic methods used followed those reported by Aoki et al. (3) for the morphological characterization of the single isolate of Fusarium guttiforme NRRL 53131 cultured from a human paronychial infection. Isolate NRRL 53131 was grown on potato dextrose agar (PDA; Difco, Detroit, MI) and synthetic low-nutrient agar (SNA) (25) in 9-cm plastic petri dishes. The cultures were incubated at 20°C in complete darkness, under continuous fluorescent light (FL40S-W; Mitsubishi), or under daylight, prior to examination of the cultural and morphological characteristics. Cultures were grown on PDA to obtain data on colony color and odor. The color scheme of Kornerup and Wanscher (20) was used for the phenotypic analysis. Fifty randomly selected conidia were measured to obtain the average and standard deviation (SD) of their size.
Pathogenicity test with detached pineapple leaves.
Pathogenicity of the novel human pathogenic Fusarium guttiforme isolate NRRL 53131 for pineapple, its only known host, was tested by using a detached-leaf assay. Ex-holotype isolate F. guttiforme NRRL 25295 was included as a positive control, and F. culmorum isolate ISPaVe-MCf 21 (INRA 117 [9]) and a water blank were employed as separate negative controls. Leaf segments (1 by 1 cm) were excised from the basal portion of a pineapple plant and were then inoculated by pipetting 10 μl of a spore suspension (1 × 106 ml−1) of one of the fungal strains or 10 μl of sterile distilled water as a negative control onto an artificial lesion on each detached leaf induced with a sterile scalpel. Inoculated leaf segments were incubated in 9-cm plastic petri plates at 25°C under white light (12-h photoperiod) for 14 days. Isolates were considered pathogenic if they conidiated on the wounded leaf surface and within the leaf tissues.
Nucleotide sequence accession numbers.
The DNA sequences obtained in the present study have been deposited in GenBank under accession numbers GU170539 to GU170656.
RESULTS AND DISCUSSION
Multilocus DNA sequence-based genotyping.
The primary objective of the present molecular phylogenetic study was to characterize the circulating species and STs associated with dermatological fusarioses (n = 46) and those isolated from 12 other localized or systemic fusarial infections from 50 patients in northern and central Italy between 2004 and 2007. All of the dermatological isolates were obtained from Sesto San Giovanni Hospital in Milan, and all isolates except for those recovered sequentially from five patients appeared to be unrelated epidemiologically (Table 1). Similar to the findings of López et al. (22), the 26 female patients diagnosed with dermatological infections in the present study ranged in age from 31 to 75 years (mean, 52.9 years) and comprised the source of close to three-quarters of the isolates (n = 34); the 12 male patients ranged in age from 21 to 80 years (mean, 44.8 years) (Table 1). Interestingly, women accounted for all seven dermatology patients from whom fusaria were sequentially isolated (Table 1). The potential risk factors identified by López et al. (22) to explain the gender bias included higher frequencies of manicure, pedicure, and barefooted walking by women. The results of the partial EF-1α gene BLAST queries of the Fusarium database (14) revealed that the 58 isolates were nearly evenly divided among the three species complexes predicted to be the most prevalent: 18 FSSC isolates, 20 FOSC isolates, and 20 GFSC isolates. The distribution of unique haplotypes was 18 FSSC isolates, 9 FOSC isolates, and 5 GFSC isolates. Once the isolates were identified to the species complex level, they were subjected to one of the following three complex-specific typing schemes. Members of the FSSC were subjected to a three-locus typing scheme that used partial sequences of the EF-1α gene (756-bp alignment), the RPB2 gene (1,831-bp alignment), and the ITS rDNA region plus domains D1 and D2 of the LSU rDNA (1,095-bp alignment) to identify the species and STs, as described previously (36). The 20 FOSC isolates were subjected to a two-locus typing scheme that used the partial EF-1α gene (688 bp) and most of the IGS rDNA region (2,325 bp) to identify STs, as described by O'Donnell et al. (32). Lastly, the 20 isolates within the GFSC were identified to the species level by conducting phylogenetic analyses of partial EF-1α gene (683 bp) and RPB2 gene (1,635 bp) sequences after they were added to comprehensive data sets for members of this complex (31, 34; K. O'Donnell, unpublished data).
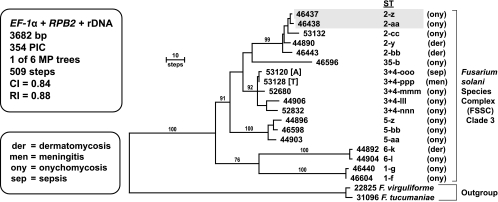
On the basis of the results of more inclusive analyses (30, 36), the sequences of two FSSC clade 2 species (Fusarium virguliforme and F. tucumaniae) were used to root the phylogram (Fig. 1). All of the clinically relevant isolates were nested within FSSC clade 3, as reported previously (36, 52). Maximum-parsimony analysis of the three-locus FSSC data set resolved 18 novel STs distributed among six human pathogenic species (FSSC 1, 2, 3 + 4, 5, 6, and 35), yielding an index of discrimination of 1.0 (19). Interestingly, typing of FSSC isolates from soil in Sardinia by the same three-locus scheme revealed similarly high levels of novel FSSC genetic diversity, as all nine STs within FSSC 5 and both STs within FSSC 9 in the aforementioned molecular ecology study were novel (8). Sixteen of the 18 FSSC isolates were recovered from dermatological infections from 15 patients treated at Sesto San Giovanni Hospital in Milan and consisted of isolates that caused onychomycoses (n = 13) and dermatomycoses (n = 3). Although the only two FSSC isolates from disseminated infections were represented by closely related but epidemiologically unrelated STs within the same species, NRRL 53120 (FSSC 3 + 4-ooo) in Ancona and NRRL 53128 (FSSC 3 + 4-ppp) in Torino, the ability to disseminate hematogenously is widespread among the FSSC clade 3 species (36, 52). An interesting case is represented by the identification of two closely related haplotypes, NRRL 46443 (FSSC 2-bb), isolated in December 2004, and NRRL 44890 (FSSC 2-y), isolated in December 2005, which were obtained from separate dermatomycotic infections of the feet of two brothers living in the same house.
FIG. 1.
One of six most-parsimonious trees inferred from MP analysis of the combined three-locus data set for 18 NRRL isolates within the FSSC. Sequences of two FSSC clade 2 species, F. virguliforme and F. tucumaniae, were used to root the phylogram. The in-group species and their multilocus STs are identified by Arabic numbers and lowercase roman letters, respectively. All in-group isolates except for NRRL 53120 and NRRL 53128 were from Sesto San Giovanni Hospital in Milan; NRRL 53120 was from Ancona (A), and NRRL 53128 was from Torino (T). Gray shading is used to identify two isolates (46437 and 46438) representing different STs recovered from the toe of a patient with onychomycosis in April and May 2007. The numbers above the nodes represent MP bootstrap support of ≥70% from 1,000 pseudoreplicates of the data. PIC, parsimony informative characters; CI, consistency index; RI, retention index.
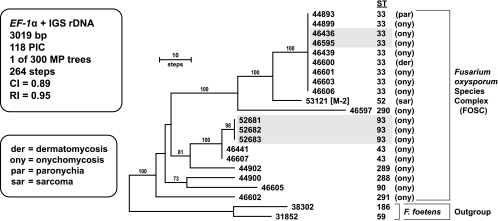
Sequences of Fusarium foetens, the sister group of the FOSC (41), were used to root the FOSC phylogeny of this complex (Fig. 2). The typing scheme consisted of partial sequences of the EF-1α gene (688-bp alignment) and nearly complete sequences of the IGS rDNA (2,325-bp alignment). Analysis of the two-locus FOSC data set identified nine STs among 19 isolates recovered from 16 patients examined at Sesto San Giovanni Hospital in Milan. Two isolates of ST 33 (formerly FOSC 3-a [34]) and three isolates of ST 93 were successively isolated from separate patients with onychomycosis in 2007. The other isolates appear to be epidemiologically unrelated, in that they were isolated from different patients examined at different times and included seven additional isolates of the widespread clonal lineage ST 33, the most prevalent pathogen sampled within the FOSC. Members of this clonal lineage were responsible for over 70% of the cases of fusariosis analyzed in a previous study, including those associated with a pseudoepidemic at a hospital in San Antonio, TX (35). Although ST 33 has been found only a few times in Europe (35), probably due to limited sampling, results of the present study indicate that it might be relatively common, especially in the vicinity of Milan, in comparison to the prevalence of other two-locus STs within this complex. NRRL 44893 (ST 33) was the sole FOSC isolate responsible for a paronychial infection. One environmental reservoir of ST 33 appears to be water systems, given that it has been recovered from hospital sinks and drains (2, 35) and coupled with the fact that it was the second most common ST recovered from corneal infections and contact lenses and lens cases during the 2005 and 2006 contact lens-associated keratitis outbreak within the United States (11, 34). NRRL 53121 (ST 52), which represented the only nondermatological FOSC isolate, was recovered from the soft tissue of a Kaposi's sarcoma patient at a second hospital in Milan (designated hospital M-2; Table 1). The latter ST appears to be geographically widespread (32), having been isolated from a human foot in Louisiana (NRRL 26370 = FRC O-1732) and machinery oil in New Zealand (NRRL 38540 = ICMP 10432). The clone-corrected index of discrimination for the FOSC two-locus typing scheme was 0.787 (19).
FIG. 2.
One of 300 most-parsimonious phylograms inferred from MP analysis of the combined two-locus data set for 20 isolates within the FOSC. Sequences of the sister taxon of the FOSC, F. foetens, were used to root the phylogeny by the outgroup method. With the exception of NRRL 53121 from Milan hospital 2 (designated hospital M-2), all of the remaining 19 in-group isolates were obtained from Sesto San Giovanni Hospital in Milan. The two-locus ST is indicated to the right of the five-digit NRRL number. Two isolates of ST 33 (46436 and 46595) and three isolates of ST 93 (52681, 52682, and 52683), shaded in gray, were isolated, from separate patients with onychomycosis, respectively, on separate occasions during 2007. MP bootstrap support of ≥70% from 1,000 pseudoreplicates of the data is indicated above the nodes.
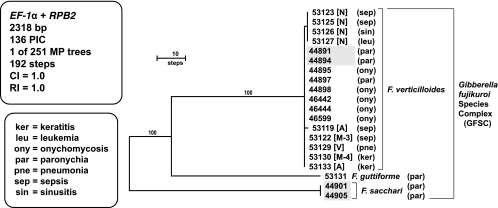
The GFSC phylogeny was rooted with sequences of Fusarium sacchari on the basis of the results of more inclusive analyses (31) (Fig. 3). Molecular typing of the GFSC employed partial sequences of the EF-1α gene (683-bp alignment) and a portion of the RBP2 gene (1,635-bp alignment). In contrast to the multilocus sequence typing (MLST) schemes for FSSC and FOSC, the GFSC two-locus typing scheme was designed to identify species rather than STs within a species. F. verticillioides was the predominant human pathogenic species sampled in the current study (n = 17), consistent with the findings described in a previous report from northern Italy (48). Of the three two-locus haplotypes resolved, one was restricted to a hospital in Novara and another one was restricted to one in Ancona, whereas the predominant ST (n = 12) was geographically widespread, with such isolates being detected in Milan, Ancona, and Varese. Other members of the GFSC included two isolates of F. sacchari from the same patient and one isolate of a novel human pathogen, F. guttiforme. The 11 dermatological isolates were recovered from nine patients seen at the Sesto San Giovanni Hospital in Milan. These included five onychomycotic infections, all of which were caused by F. verticillioides, and four paronychial infections, two of which were caused by F. verticillioides and the other two of which were caused by F. sacchari and F. guttiforme. The remaining nine isolates of F. verticillioides were obtained from nine patients with diverse illnesses (i.e., keratitis, leukemia, pneumonia, sepsis, and sinusitis; Table 1) from five different northern Italian hospitals (Table 1, hospitals M-3 and M-4 in Milan and hospitals in Novara, Ancona, and Varese). Given the study's focus on identifying GFSC species, it is not surprising that the clone-corrected F. verticillioides typing scheme preformed the most poorly of the three employed, having a discrimination index of 0.492 (19).
FIG. 3.
One of 251 phylograms inferred from MP analysis of the combined two-locus data set for 20 isolates within the GFSC by using sequences of F. sacchari to root the tree. MP bootstrap clade support of ≥70% from 1,000 pseudoreplicates of the data is indicated above nodes. The 11 dermatological isolates from onychomycotic and paronychial infections were from Sesto San Giovanni Hospital in Milan. The remaining nine isolates were from hospitals 3 and 4 in Milan (designated hospitals M-3 and M-4, respectively), Novara (N), Ancona (A), and Varese (V). The two shaded isolates of F. sacchari (44901 and 44905) and F. verticillioides (44891 and 44894) were isolated successively during 2005 and 2006, respectively, from separate patients with onychomycosis.
Molecular typing also allowed us to assess whether different species or different STs within a species were responsible for mixed fusarial infections within a patient. Genetically distinct FSSC STs (NRRL 46437 = FSSC 2-z and NRRL 46438 = FSSC 2-aa) were isolated from the right and left toes of the same patient with onychomycosis in April and May 2007. To our knowledge, this represents the first report of different STs of the same species being responsible for a mixed fusarial infection. It is worth noting, however, that Guarro et al. (17) first documented a mixed infection involving two Fusarium species (a member of the FSSC and F. verticillioides) in an immunocompromised patient. Similar to the findings of Guarro et al. (17), NRRL 44898 (F. verticillioides) and NRRL 44902 (FOSC ST 289) were isolated from the same patient with onychomycosis in September and October 2005, as were NRRL 44895 (F. verticillioides) and NRRL 44906 (FSSC 3 + 4-lll) from the right and left toes of a separate patient in June and September 2006.
Case report and morphological characterization of the novel human pathogen Fusarium guttiforme.
A 65-year-old native Italian female in good health presented with a 12-month history of very painful paronychia of the 4th finger of the right hand, from which the novel human pathogen Fusarium guttiforme was subsequently isolated and identified morphologically and molecularly. Slight proximal nail fold retraction was discernible, but there was no discoloration of the nail plate. No spontaneous discharge of pus was observed, even when pressure was exerted on the nail fold. However, the medial part of the nail plate was thin and deteriorated. Despite prior treatment with systemic antibiotics (amoxicillin-clavulanic acid, 2 g per day for 10 days) and topical gentamicin, the symptoms deteriorated. Scrapings from beneath the proximal nail fold examined microscopically revealed the presence of hyphae. Tissue scrapings from beneath the nail fold cultured on Sabouraud's agar amended with chloramphenicol and gentamicin (100 μg ml−1), with and without cycloheximide (100 μg ml−1), for 3 weeks at 25°C in complete darkness yielded a mixed culture of Fusarium and Candida parapsilosis. Identical results were obtained from a second mycological examination conducted 3 weeks later. After treatment with systemic terbinafine (250 mg per day for 2 months), a follow-up visit with the patient revealed that the infection had completely resolved. However, 6 months later there was a relapse of the paronychia, but this time there was a slight swelling of the nail fold, while the features of the nail plate were normal. The patient acknowledged moderate inconvenience. Microbiological assessment yielded a pure culture of C. parapsilosis. Treatment with itraconazole for 7 days gave a complete cure. In summary, the lack of pigmentation of the nail plate, which is characteristic of Candida paronychia, and the prompt recovery with oral terbinafine therapy, which is known to be very active against Fusarium but not Candida, strongly indicate that F. guttiforme played an active pathogenic role in this infection.
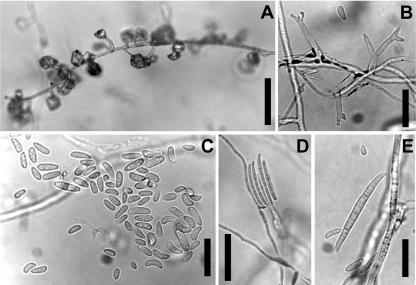
Morphological observations of monosporic cultures of Fusarium guttiforme NRRL 53131 (Fig. 4A to E) yielded fast-growing colonies on PDA at 20°C which lacked a perceptible odor. The aerial mycelium was white with a grayish violet tinge, and the colony surface was floccose. Sporulation in the aerial mycelium on SNA consisted of conidia formed in false heads (i.e., clumped together on a conidiophore loosely resembling a vesicle in Aspergillus; Fig. 4A). Aerial conidiophores were erect or prostrate (i.e., along the agar surface) and were branched or unbranched. Conidiophores terminated in mono- and polyphialides (Fig. 4B) which proliferated sympodially. Aerial conidia (Fig. 4C) were oblong, short clavate, obovoid, fusoid to ellipsoid, straight or slightly curved, 0(−1)-septate, 0-septate: 5 to 17.5 by 2 to 4 μm in total range and 8.3 ± 2.5 by 2.8 ± 0.32 μm (average ± SD). Although true sporodochia were not formed on SNA, multiseptate conidia were formed on phialides produced laterally on the substrate mycelium (Fig. 4D and E). Three (−5)-septate conidia were straight or slightly falcate with a slightly hooked apical cell and a foot-like basal cell, 3-septate: 30.5 to 56.5 by 3 to 5 μm in total range and 42.1 ± 5.4 by 4.1 ± 0.49 μm (average ± SD). Chlamydospores were not observed. In the original description of F. guttiforme (26), multiseptate conidia were not observed. However, in isolate NRRL 53131, multiseptate (mostly 3-septate), falcate conidia with a foot-like cell were formed abundantly from phialides as lateral branches of the vegetative hyphae. Except for this character, the morphological characteristics of isolate NRRL 53131 matched those given in the original species description (26). It is worth mentioning that Leslie and Summerell (21) provided photographs of multiseptate conidia of this species which were very similar to those produced by F. guttiforme NRRL 53131.
FIG. 4.
Fusarium guttiforme NRRL 53131 isolated from paronychia of the hand. (A) Prostrate conidiophore formed in culture on SNA, forming aerial conidia in false heads on the tips of phialides. Bar, 50 μm. (B) Polyphilalides formed on aerial conidiophores. Bar, 20 μm. (C) Short clavate to elliptical, 0-septate aerial conidia. Bar, 20 μm. (D) Conidiogenesis of falcate multiseptate conidia on agar. Bar, 50 μm. (E) Enlargement of a multiseptate conidium. Bar, 20 μm.
Because the only known host of Fusarium guttiforme is pineapple (Ananas comosus), we were interested in determining whether the patient had come into contact with fresh pineapple fruit which harbored this pathogen, as well as further characterizing this isolate to see if it was pathogenic for pineapple. A follow-up interview with the patient revealed that she frequently consumed fresh pineapple after peeling it by hand. Results of the pathogenicity experiment revealed that human pathogenic isolate F. guttiforme NRRL 53131 was able to induce tissue necrosis and sporulate profusely after 14 days of incubation at 25°C when it was inoculated on artificially wounded pineapple leaves. The same symptoms were induced by the ex-holotype-positive control strain of F. guttiforme (NRRL 25295). By way of contrast, separate negative controls consisting of the wheat pathogen F. culmorum and a distilled water blank induced only browning of the wounded surface. The latter symptom is a common reaction of detached plant tissues exposed to air and is unrelated to pathogenicity. The importation of F. guttiforme-containing pineapple into nonindigenous areas where this crop is produced could have destructive results, given that pineapple fusariosis has become a major constraint to the production of this crop in certain regions of South America.
Conclusions.
On the basis of our current knowledge of the phylogenetic diversity of human pathogenic fusaria, results of the present study fit the prediction that members of the FSSC, FOSC, and GFSC would predominate. In contrast to MLST schemes developed for human pathogenic species such as Aspergillus fumigatus (6) and Candida albicans (29), the three-locus FSSC typing scheme was developed primarily to identify species within FSSC clade 3, within which 21 of 35 species appear to be clinically relevant (36). Nevertheless, all 18 FSSC isolates typed in the present study were identified to be novel STs, indicating that the three-locus typing scheme is reasonably robust and that the spectrum of medically important FSSC in northern and central Italy is phylogenetically diverse. The present study adds to our growing knowledge of human pathogenic FOSC in the discovery of four novel singleton STs and the predominance of ST 33 (FOSC 3-a), the previously characterized widespread clonal lineage (35), in Milan. Isolates of ST 33 have also been recovered from clinical cases in Germany, Belgium, and Canada (32), in addition to the water systems of geographically distant hospitals in Seattle, WA, Houston, TX, and Baltimore, MD, in the United States; a soft-drink production plant in New York state (NRRL 38360 = FRC O-2347 [32]); a greenhouse irrigation system in Finland; and confirmed keratitis outbreak infections in California, Pennsylvania, and Vermont (11). Although 30 genetically diverse STs within FOSC have been implicated in infections of humans and other animals (32), the available data suggest that ST 33 is the most important clinically relevant FOSC haplotype. The species and ST/haplotype nomenclature developed in prior studies provide the most efficient and accurate means of reporting on the genetic diversity of these fusaria within the medical, phytopathological, and mycotoxological research communities. The species/ST nomenclature is especially critical, considering that Latin binomials can be applied to only 3 of the 21 clinically important FSSC species and because species limits are unresolved within the genetically diverse FOSC (32). It is fortunate that the recent availability of the whole genome sequences of F. verticillioides, F. oxysporum, and F. graminearum online from the Broad Institute of the Massachusetts Institute of Technology and Harvard University (12; http://www.broad.mit.edu/annotation/fungi/fgi/) and the accessibility of the F. solani f. sp. pisi (FSSC 11 [36]) genome online from the Joint Genome Institute (http://www.jgi.doe.gov) should greatly facilitate the identification of additional phylogenetically informative loci and population-level markers (47, 51), should the necessity arise. In addition, 10 diverse members of the FOSC, including ST 33, are currently being sequenced at the Broad Institute (H. C. Kistler and L.-J. Ma, personal communication). Profiling of phylogenetically informative orthologs (49) from the FOSC core genomes should provide a robust phylogenetic framework for understanding the evolution of human pathogenicity and virulence within this complex (38) and help elucidate whether the reproductive mode is strictly clonal (45). In the absence of morphological apomorphies, the MLST typing schemes for FSSC, FOSC, and other medically important species complexes within Fusarium provide the only means for the accurate assessment of their genetic diversity, thereby facilitating comparative studies at the local, regional, and global scales.
Acknowledgments
Thanks are due to Stacy Sink and Angela Marcello for excellent technical assistance, Nathane Orwig for running the DNA sequences in the NCAUR DNA core facility, Don Fraser for preparation of the tree figures, and the Medical Mycology Committee (CoSM) of the Associazione Microbiologi Clinici Italiani for providing some of the isolates used in this study.
The Ministry of University and Research provided funds (to Q.M.) for a portion of this research (PRIN 2007, Transposon tagging and RNA silencing in the wheat pathogen Fusarium culmorum).
The mention of trade products or firm names does not imply that they are recommended by the U.S. Department of Agriculture over similar products or other firms not mentioned.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Alastruey-Izquierdo, A., M. Cuenca-Estrella, A. Monzón, E. Mellado, and J. L. Rodríguez-Tudela. 2008. Antifungal susceptibility profile of clinical Fusarium spp. isolates identified by molecular methods. J. Antimicrob. Chemother. 61:805-809. [DOI] [PubMed] [Google Scholar]
- 2.Anaissie, E. J., R. T. Kuchar, J. H. Rex, A. Francesconi, M. Kasai, F.-M. Müller, M. Lozano-Chiu, R. C. Summerbell, M. C. Dignani, S. J. Chanock, and T. J. Walsh. 2001. Fusariosis associated with pathogenic Fusarium species colonization of a hospital water system: a new paradigm for the epidemiology of opportunistic mold infections. Clin. Infect. Dis. 33:1871-1878. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, T., K. O'Donnell, Y. Homma, and A. R. Lattanzi. 2003. Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex—Fusarium virguliforme in North America and F. tucumaniae in South America. Mycologia 95:660-684. [PubMed] [Google Scholar]
- 4.Azor, M., J. Gené, J. Cano, and J. Guarro. 2007. Universal in vitro antifungal resistance of genetic clades of the Fusarium solani species complex. Antimicrob. Agents Chemother. 51:1500-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azor, M., J. Gené, J. Cano, P. Manikandan, N. Venkatapathy, and J. Guarro. 2009. Less-frequent Fusarium species of clinical interest: correlation between morphological and molecular identification and antifungal susceptibility. J. Clin. Microbiol. 47:1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bain, J. M., A. Tavanti, A. D. Davidson, M. D. Jacobsen, D. Shaw, N. A. R. Gow, and F. C. Odds. 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J. Clin. Microbiol. 45:1469-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balajee, S. A., A. M. Borman, M. E. Brandt, J. Cano, M. Cuenca-Estrella, E. Dannaoui, J. Guarro, G. Haase, C. C. Kibbler, W. Meyer, K. O'Donnell, C. A. Petti, J. L. Rodriguez-Tudela, D. Sutton, A. Velegraki, and B. L. Wickes. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: where are we and where should we go from here? J. Clin. Microbiol. 47:877-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balmas, V., Q. Migheli, B. Scherm, P. Garau, K. O'Donnell, G. Ceccherelli, S. Kang, and D. M. Geiser. Multilocus molecular phylogenetics indicates high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia, in press. [DOI] [PubMed]
- 9.Boutigny, A.-L., C. Barreau, V. Atanasova-Penichon, M.-N. Verdal-Bonnin, L. Pinson-Gadais, and F. Richard-Forget. 2009. Ferulic acid, an efficient inhibitor of type B trichothecene biosynthesis and Tri gene expression in Fusarium liquid cultures. Mycol. Res. 113:746-753. [DOI] [PubMed] [Google Scholar]
- 10.Calado, N. B., F. Sousa, N. O. Gomes, F. R. Cardoso, L. C. Zaror, and E. P. Milan. 2006. Fusarium nail and skin infections: a report of eight cases from Natal, Brazil. Mycopathologia 161:27-31. [DOI] [PubMed] [Google Scholar]
- 11.Chang, D. C., G. B. Grant, K. O'Donnell, K. A. Wannemuehler, J. Noble-Wang, C. Y. Rao, L. M. Jacobson, C. S. Crowell, R. S. Sneed, F. M. T. Lewis, J. K. Schaffzin, M. A. Kainer, C. A. Genese, E. C. Alfonso, D. B. Jones, A. Srinivasan, S. K. Fridkin, and B. J. Park. 2006. A multistate outbreak of Fusarium keratitis associated with use of a new contact lens solution. JAMA 296:953-963. [DOI] [PubMed] [Google Scholar]
- 12.Cuomo, C. A., U. Güldener, J.-R. Xu, F. Trail, B. G. Turgeon, A. Di Pietro, J. D. Walton, L.-J. Ma, S. E. Baker, M. Rep., G. Adam, J. Antoniw, T. Baldwin, S. Calvo, Y.-L. Chang, D. DeCaprio, L. R. Gale, S. Gnerre, R. S. Goswami, K. Hammond-Kosack, L. J. Harris, K. Hilburn, J. C. Kennell, S. Kroken, J. K. Magnuson, G. Mannhaupt, E. Mauceli, H.-W. Mewes, R. Mitterbauer, G. Muehlbauer, M. Münsterkötter, D. Nelson, K. O'Donnell, T. Ouellet, W. Qi, H. Quesneville, M. I. G. Roncero, K.-Y. Seong, I. V. Tetko, M. Urban, C. Waalwijk, T. J. Ward, J. Yao, B. W. Birren, and H. C. Kistler. 2007. The Fusarium graminearum genome reveals localized polymorphism and pathogen specialization. Science 317:1400-1402. [DOI] [PubMed] [Google Scholar]
- 13.Dignani, M. C., and E. J. Anaissie. 2004. Human fusariosis. Clin. Microbiol. Infect. 10(Suppl. 1):67-75. [DOI] [PubMed] [Google Scholar]
- 14.Geiser, D. M., M. D. M. Jiménez-Gasco, S. Kang, I. Makalowska, N. Veeraraghavan, T. J. Ward, N. Zhang, G. A. Kuldau, and K. O'Donnell. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110:473-479. [Google Scholar]
- 15.Gerlach, W., and H. Nirenberg. 1982. The genus Fusarium—a pictorial atlas. Mitt. Biol. Bundesanst. Land-Forstwirtsch. 209:1-406. [Google Scholar]
- 16.Gianni, C., A. Cerri, and C. Crosti. 1997. Unusual clinical features of fingernail infection by Fusarium oxysporum. Mycoses 40:455-459. [DOI] [PubMed] [Google Scholar]
- 17.Guarro, J., M. Nucci, T. Akiti, and J. Gené. 2000. Mixed infection caused by two species of Fusarium in a human immunodeficiency virus-positive patient. J. Clin. Microbiol. 38:3460-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilhermetti, E., G. Takahachi, C. S. Shinobu, and T. I. E. Svidzinski. 2007. Fusarium spp. as agents of onychomycosis in immunocompetent hosts. Int. J. Dermatol. 46:822-826. [DOI] [PubMed] [Google Scholar]
- 19.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of discrimination. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornerup, A., and J. H. Wanscher. 1978. Methuen handbook of colour, 3rd ed. Methuen, London, United Kingdom.
- 21.Leslie, J. F., and B. A. Summerell. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames, IA.
- 22.López, N. C., C. Casas, L. Sopo, A. Rojas, P. D. Portillo, M. C. C. de García, and S. Restrepo. 2008. Fusarium species detected in onychomycosis in Columbia. Mycoses 52:350-356. [DOI] [PubMed] [Google Scholar]
- 23.Mays, S. R., and P. R. Cohen. 2006. Emerging dermatologic issues in the oncology patient. Semin. Cutaneous Med. Surg. 25:179-189. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, P. E., T. A. Toussoun, and W. F. O. Marasas. 1983. Fusarium species: an illustrated manual for identification. Pennsylvania State University Press, University Park, PA.
- 25.Nirenberg, H. I. 1990. Recent advances in the taxonomy of Fusarium. Stud. Mycol. 32:91-101. [Google Scholar]
- 26.Nirenberg, H. I., and K. O'Donnell. 1998. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90:434-458. [Google Scholar]
- 27.Nucci, M., and E. Anaissie. 2002. Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: implications for diagnosis and management. Clin. Infect. Dis. 35:909-920. [DOI] [PubMed] [Google Scholar]
- 28.Nucci, M., and E. Anaissie. 2007. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20:695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odds, F. C., and M. D. Jacobsen. 2008. Multilocus sequence typing of pathogenic Candida species. Eukaryot. Cell 7:1075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Donnell, K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919-938. [Google Scholar]
- 31.O'Donnell, K., E. Cigelnik, and H. I. Nirenberg. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465-493. [Google Scholar]
- 32.O'Donnell, K., C. Gueidan, S. Sink, P. R. Johnston, P. W. Crous, A. Glenn, R. Riley, N. C. Zitomer, P. Colyer, C. Waalwijk, T. van der Lee, A. Moretti, S. Kang, H.-S. Kim, D. M. Geiser, J. H. Juba, R. P. Baayen, M. G. Cromey, S. Bithel, D. A. Sutton, K. Skovgaard, R. Ploetz, H. C. Kistler, M. Elliott, M. Davis, and B. A. J. Sarver. 2009. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 46:936-948. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell, K., H. C. Kistler, E. Cigelnik, and R. C. Ploetz. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U. S. A. 95:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Donnell, K., B. A. J. Sarver, M. Brandt, D. C. Chang, J. Noble-Wang, B. J. Park, D. A. Sutton, L. Benjamin, M. Lindsley, A. Padhye, D. M. Geiser, and T. J. Ward. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J. Clin. Microbiol. 45:2235-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Donnell, K., D. A. Sutton, M. G. Rinaldi, K. C. Magnon, P. A. Cox, S. G. Revankar, S. Sanche, D. M. Geiser, J. H. Juba, J.-A. H. van Burik, A. Padhye, E. J. Anaissie, A. Francesconi, T. J. Walsh, and J. S. Robinson. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 42:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Donnell, K., D. A. Sutton, A. Fothergill, D. McCarthy, M. G. Rinaldi, M. E. Brandt, N. Zhang, and D. M. Geiser. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46:2477-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Donnell, K., D. A. Sutton, M. G. Rinaldi, C. Gueidan, P. W. Crous, and D. M. Geiser. 2009. A novel MLST scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 47:3851-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prados-Rosales, R. C., C. Serena, J. Delgado-Jarana, J. Guarro, and A. Di Pietro. 2006. Distinct signalling pathways coordinately contribute to virulence of Fusarium oxysporum on mammalian hosts. Microbes Infect. 8:2825-2831. [DOI] [PubMed] [Google Scholar]
- 39.Pujol, I., J. Guarro, J. Gené, and J. Sala. 1997. In-vitro antifungal susceptibility of clinical and environmental Fusarium spp. strains. J. Antimicrob. Chemother. 39:163-167. [DOI] [PubMed] [Google Scholar]
- 40.Romano, C., C. Miracco, and E. M. Difonzo. 1998. Skin and nail infections due to Fusarium oxysporum in Tuscany, Italy. Mycoses 41:433-437. [DOI] [PubMed] [Google Scholar]
- 41.Schroers, H.-J., R. P. Baayen, J. P. Meffert, J. de Gruyter, M. Hooftman, and K. O'Donnell. 2004. Fusarium foetens, a new species pathogenic to begonia elatior hybrids (Begonia × hiemalis) and the sister taxon of the Fusarium oxysporum complex. Mycologia 96:393-406. [PubMed] [Google Scholar]
- 42.Schroers, H.-J., K. O'Donnell, S. C. Lamprecht, P. L. Kammeyer, S. Johnson, D. A. Sutton, M. G. Rinaldi, D. M. Geiser, and R. C. Summerbell. 2009. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia 101:44-70. [DOI] [PubMed] [Google Scholar]
- 43.Simonetti, O., M. L. Bernardini, D. Arzeni, A. Cellini, F. Barchiesi, and A. Offidani. 2004. Epidemiology of onychomycosis and paronychia in the area of Ancona (Italy) over a period of 5 years. Mycopathologia 158:271-274. [DOI] [PubMed] [Google Scholar]
- 44.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 45.Taylor, J. W., D. J. Jacobson, and M. C. Fisher. 1999. The evolution of asexual fungi: reproduction, speciation and classification. Annu. Rev. Phytopathol. 37:197-246. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, J. W., D. J. Jacobson, S. Kroken, T. Kasuga, D. M. Geiser, D. S. Hibbett, and M. C. Fisher. 2000. Phylogenetic species recognition and species concepts in Fungi. Fungal Genet. Biol. 31:21-32. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, J. W., and M. C. Fisher. 2003. Fungal multilocus sequence typing—it's not just for bacteria. Curr. Opin. Microbiol. 6:351-356. [DOI] [PubMed] [Google Scholar]
- 48.Tortorano, A. M., A. Prigitano, G. Dho, M. C. Esposto, C. Gianni, A. Grancini, C. Ossi, and M. A. Viviani. 2008. Species distribution and in vitro antifungal susceptibility patterns of 75 clinical isolates of Fusarium spp. from northern Italy. Antimicrob. Agents Chemother. 52:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Townsend, J. P. 2007. Profiling phylogenetic informativeness. Syst. Biol. 56:222-231. [DOI] [PubMed] [Google Scholar]
- 50.Walsh, T. J., A. Groll, J. Hiemenz, R. Fleming, E. Roilides, and E. Anaissie. 2004. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 10:48-66. [DOI] [PubMed] [Google Scholar]
- 51.Ward, T. J., R. M. Clear, A. P. Rooney, K. O'Donnell, D. Gaba, S. Patrick, D. E. Starkey, J. Gilbert, D. M. Geiser, and T. W. Nowicki. 2008. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 45:473-484. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, N., K. O'Donnell, D. A. Sutton, F. A Nalim, R. C. Summerbell, A. A. Padhye, and D. M. Geiser. 2006. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 44:2186-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]