Abstract
Clinical resistance to epidermal growth factor receptor (EGFR) inhibition in lung cancer has been linked to the emergence of the EGFR T790M resistance mutation or amplification of MET. Additional mechanisms contributing to EGFR inhibitor resistance remain elusive. By applying combined analyses of gene expression, copy number, and biochemical analyses of EGFR inhibitor responsiveness, we identified homozygous loss of PTEN to segregate EGFR-dependent and EGFR-independent cells. We show that in EGFR-dependent cells, PTEN loss partially uncouples mutant EGFR from downstream signaling and activates EGFR, thereby contributing to erlotinib resistance. The clinical relevance of our findings is supported by the observation of PTEN loss in 1 out of 24 primary EGFR-mutant non–small cell lung cancer (NSCLC) tumors. These results suggest a novel resistance mechanism in EGFR-mutant NSCLC involving PTEN loss.
Introduction
Activating mutations in the epidermal growth factor receptor (EGFR) are present in ~10% of non–small cell lung cancers (NSCLC) in Caucasian patients and in up to 40% of East-Asian patients. By contrast, EGFR mutations are much more rare in African Americans. These mutations lead to the “addiction” of mutant cells to the oncogenic signals driven by mutant EGFR. This dependency is thought to be the cause of the clinical observations that EGFR-mutant tumors shrink when treated with EGFR inhibitors (1, 2). Eventually, these tumors recur; in ~60% to 70% (3) of cases, this has been linked to the emergence of either the T790M resistance mutation of EGFR or amplification of MET (2-4). However, a mechanistic explanation for acquired resistance in the remaining cases is lacking.
Here, we used a large collection of genomically characterized NSCLC cell lines in order to derive genomic features that segregate EGFR-dependent from EGFR-independent EGFR-mutant lung tumor cells. We combined computational, biochemical, and cellular approaches to identify novel, clinically relevant mechanisms uncoupling EGFR-dependent tumors from downstream signaling.
Materials and Methods
A detailed description of all methods is given in the Supplementary Methods. As part of a larger effort to characterize the genomes of NSCLC, we have collected 84 NSCLC cell lines, which we analyzed for chromosomal gene copy number alterations, mutations, as well as transcriptional changes. The detailed description of this collection will be published elsewhere. Here, a subset of 53 of these cell lines was studied (Supplementary Table S1). Hierarchical clustering was performed using dCHIP. Genomic lesions differentiating between erlotinib-sensitive and erlotinib-insensitive cells were analyzed by inferring the mean copy number of chromosomal windows from five contiguous loci. Statistical analyses were performed using R.
Results and Discussion
In order to analyze oncogene dependencies in lung cancer, we used a collection of 84 NSCLC cell lines that we have recently characterized in-depth genomically and phenotypically (Supplementary Table S1).14
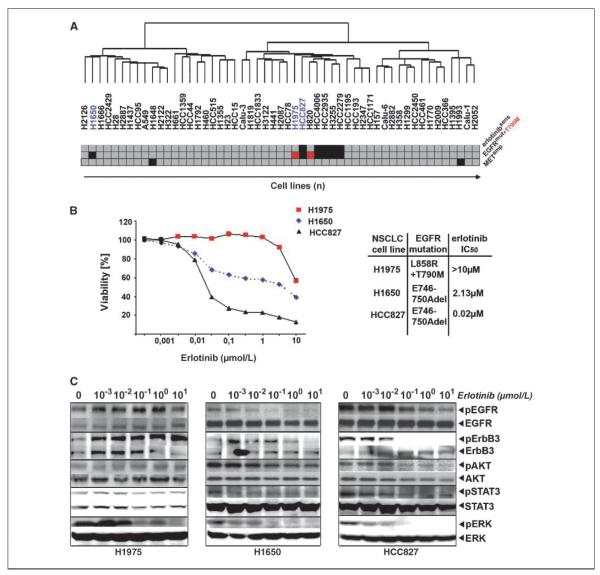
We performed hierarchical clustering of gene expression data of 53 of these lines. In this analysis, the EGFR-mutant cell line, H1650, did not share a cluster with all other EGFR-mutant cell lines (Fig. 1A). This cell line has previously been reported to be erlotinib-resistant, despite lacking known resistance mechanisms (Fig. 1A; ref. 5).
Figure 1.
An EGFR independence signature in H1650 cells. A, hierarchical clustering of 53 NSCLC cells according to gene expression. Erlotinib sensitivity (IC50 < 1 μmol/L, red; IC50 > 1 μmol/L, gray) and EGFR mutations (EGFR-mutant, black; T790M, red; EGFR wild-type, gray) as well as MET amplification (black). B, left, cellular viability as a function of erlotinib dose for all three cell lines studied. Right, mutation status and IC50 values. C, cells were treated with different doses of erlotinib. Activation of EGFR and downstream signaling pathways was determined by analyzing the amount of phosphorylated versions of the respective proteins in comparison with their total levels using phosphorylation-specific antibodies.
Confirming these observations, H1650 cells were erlotinib-resistant with a half-maximal inhibitory concentration (IC50)of 2.13 μmol/L (Fig. 1B). As previously reported, EGFR-mutant HCC827 cells were erlotinib-sensitive (IC50, 0.02 μmol/L), whereas H1975 cells expressing both the erlotinib-sensitizing L858R mutation and the T790M resistance mutation were resistant (IC50 > 10 μmol/L; Fig. 1B; refs. 5, 6). Treatment with 100 nmol/L of erlotinib led to the dephosphorylation of EGFR in H1650 and HCC827 but not in H1975 cells (Fig. 1C). However, although the dephosphorylation of EGFR was accompanied by a reduction in p-Akt levels in erlotinib-sensitive HCC827 cells, H1650 cells retained high levels of p-Akt despite inhibition of EGFR (Fig. 1C). By contrast, erlotinib-mediated inhibition of known signal transducers of the EGFR such as ErbB3, STAT3, and ERK was similar to the levels observed in HCC827, consistent with the uncoupling of mutant EGFR from downstream survival signaling at the level of Akt (Fig. 1C).
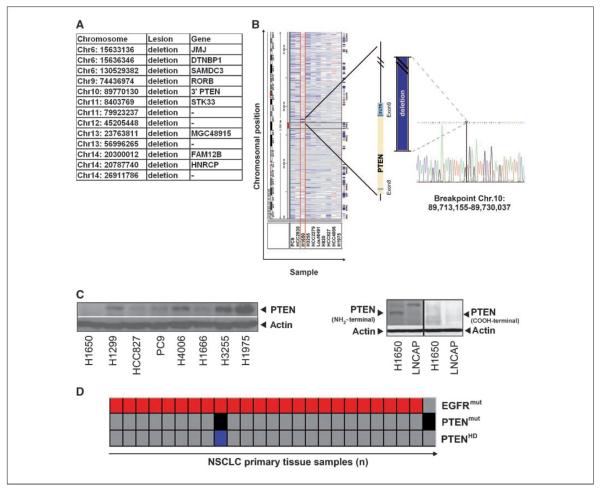
We speculated that chromosomal aberrations might be causatively involved in this phenotype and sought for chromosomal regions displaying differential copy numbers between H1650 cells and the EGFR-mutant and erlotinib-sensitive cell lines. We identified 13 H1650-specific chromosomal loci harboring nine known genes, including a chromosomal region affected by homozygous deletion 3′ to the locus containing the tumor suppressor gene PTEN (Fig. 2A; ref. 7). Furthermore, when analyzing the transcription of IGFBP2, a marker predictive of PTEN loss in glioblastoma (8), H1650 was the highest scoring line in our panel (data not shown). PTEN counteracts Akt activation by dephosphorylating phosphatidylinositol-3,4,5-triphosphate (PIP3), the product of class I phosphoinositide-3-kinases (7, 9). Because PTEN loss has been shown to be involved in EGFR inhibitor resistance in some tumor cell lines (10, 11) and in glioblastoma patients (12), we reasoned that PTEN loss might also be involved in the EGFR-independent phenotype of H1650. Furthermore, lack of PTEN protein expression has previously been speculated to be involved in erlotinib resistance in H1650 cells (13, 14).
Figure 2.
Genomic characterization of PTEN loss in H1650 cells. A, list of genes affected by differential lesions between H1650 cells and EGFR-mutant and erlotinib-sensitive cell lines. B, left, screenshot showing chromosomal aberrations at chromosome 10 (Integrative Genomics Viewer; http://www.broad.mit.edu/igv/) of all EGFR-mutant cells. Middle, 3′-region mapping of PTEN using quantitative PCR reveals a homozygous deletion deleting parts of exon 8 and the entire exon 9. Right, the sequence bridging the breakpoint. C, left, PTEN protein status determined using immunoblotting in different NSCLC cell lines. Right, NH2-terminal and COOH-terminal PTEN detection by immunoblotting. LNCAP cells, known to express a truncated version of PTEN, served as controls. D, analysis of EGFR mutations (red) and homozygous deletions of PTEN (black) and PTEN mutations (blue) in 140 lung cancer biopsy specimens.
To determine whether loss of PTEN protein in H1650 cells (13, 14) might be caused by genomic loss, we mapped the PTEN locus by quantitative PCR. Fine-mapping followed by long-distance PCR revealed that the homozygous deletion (spanning 16.8 kb) leads to the deletion of the 3′ part of exon 8 and the entire exon 9 (Fig. 2B). The deletion results in a COOH-terminally truncated protein that could only be detected using antibodies against NH2-terminal epitopes (Fig. 2C). Previous functional genetics experiments have shown a critical role of the COOH-terminal part of PTEN (15). Thus, the COOH-terminal deletion in H1650 cells might be causally involved in uncoupling mutant EGFR from downstream Akt survival signaling.
We next analyzed a panel of 140 primary lung adenocarcinomas (predominantly Caucasian patients), annotated for copy number alterations and mutations in 623 genes, for the presence of cooccurring lesions in PTEN and EGFR (16, 17). We found cooccurrence of homozygous deletion of PTEN and EGFR mutation in 1 out of 24 samples with EGFR mutations (Fig. 2D). Thus, primary resistance of EGFR-mutant NSCLC might, in rare cases, be due to homozygous loss of PTEN. Furthermore, we found hemizygous loss of chromosome 10 to be significantly enriched in EGFR-mutant patients in the cohort of 140 primary samples (P = 0.012; data not shown). Loss of the other allele by mutation might thus confer acquired resistance in patients initially responding to EGFR inhibition. This notion is also supported by a previous study reporting favorable survival of EGFR-mutant patients with high expression of PTEN (18).
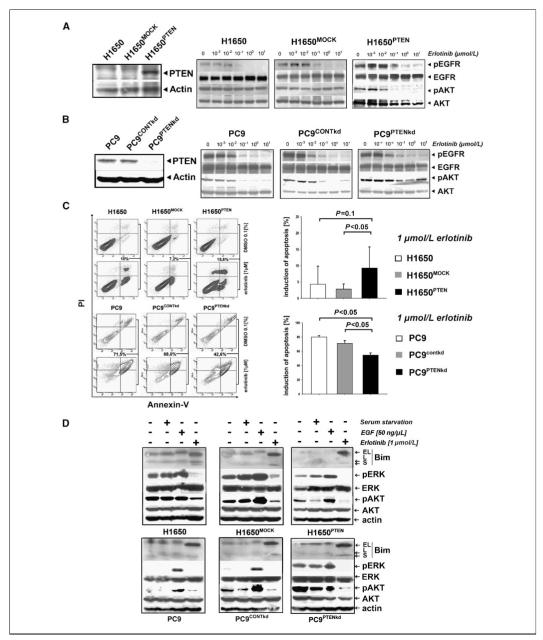
We reconstituted wild-type PTEN in H1650 cells by stable retroviral expression (Fig. 3A). Reconstitution of PTEN restored coupling of the EGFR signal to downstream Akt signaling as evidenced by dephosphorylation of both EGFR and Akt upon erlotinib treatment (Fig. 3A). Cellular proliferation of H1650PTEN cells treated with erlotinib was virtually identical to that seen in the parental cells (data not shown) but combinatorial treatment of H1650 cells with erlotinib and an AKT inhibitor led to a reduction of viability when compared with cells treated with erlotinib alone (Supplementary Fig. S1). However, when analyzing the fraction of cells undergoing apoptosis upon treatment with erlotinib, we observed an increase of apoptotic H1650PTEN cells when compared with the parental and the mock-transduced cells (Fig. 3C). Thus, PTEN reconstitution increases the susceptibility to erlotinib-induced apoptosis in H1650 cells.
Figure 3.
Erlotinib resistance in EGFR-mutated NSCLC with PTEN loss. A, left, in H1650PTEN cells, PTEN levels were determined by immunoblotting. Right, levels of phospho-EGFR and phospho-AKT were assessed by immunoblotting in H1650, H1650MOCK, and H1650PTEN cells treated with erlotinib. B, left, in PC9PTENkd cells, PTEN levels were determined by immunoblotting. Right, levels of phospho-EGFR and phospho-AKT were assessed in PC9, PC9CONTkd, and PC9PTENkd cells treated with erlotinib. C, left, percentage of apoptotic cells (in %, analyzed by measuring the fraction of cells positive for Annexin V and/or propidium iodide by flow cytometry) after treatment with either erlotinib (1 μmol/L) or control. Right, cumulative histograms of apoptosis induction. D, levels of Bim (EL, extra long; L, long; S, short), phospho-ERK, phospho-pAKT, and actin were measured after serum starvation (serum starvation “+”), EGF stimulation (EGF “+”), or treatment with erlotinib (1 μmol/L erlotinib “+”) for 24 h.
We next silenced PTEN in EGFR-mutant and erlotinib-sensitive PC9 cells by lentiviral short hairpin RNAs (Fig. 3B). Similar to our observation in the parental H1650 cells, PTEN loss in PC9 cells (PC9PTENkd) induced the uncoupling of EGFR and downstream Akt signaling as shown by continuous Akt phosphorylation under erlotinib treatment (Fig. 3B). Again, recapitulating our observations in H1650 cells, silencing of PTEN expression in PC9 cells led to a significant decrease in the fraction of apoptotic cells when treated with erlotinib (Fig. 3C). Induction of apoptosis in both PTEN-proficient and PTEN-deficient cells was paralleled by activation of the proapoptotic protein Bim, recently shown to play a key role in erlotinib-induced apoptosis in EGFR-mutant NSCLC (refs. 19, 20; Fig. 3D). Thus, the differential induction of apoptosis is not mediated through modulation of Bim levels. Interestingly, in PC9PTENkd cell lines, we observed the activation of Erk under steady-state and serum-starved conditions, whereas PTEN-proficient cells hardly showed Erk activity (Fig. 3D). Thus, PTEN loss partially uncouples EGFR signaling from downstream Akt survival signaling, activates ERK, and contributes to EGFR inhibitor resistance.
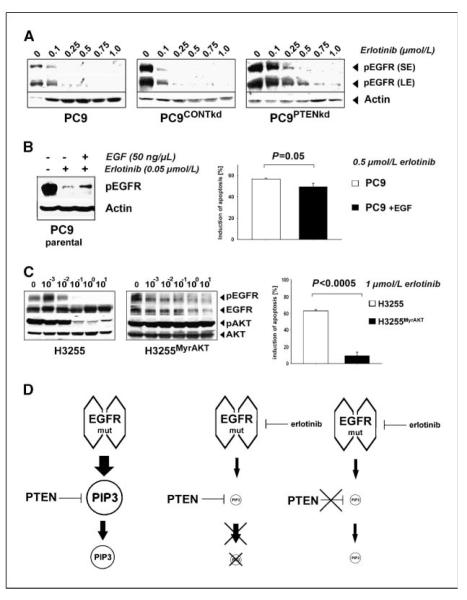
While analyzing the activity of Akt in PTEN-deficient H1650 and PC9PTENkd EGFR-mutant cells, we observed an increase in phospho-EGFR when compared with PTEN-proficient cells. In PC9PTENkd cells, complete deactivation of EGFR was achieved at 750 nmol/L of erlotinib, whereas in parental and control PC9 cells, 250 nmol/L of erlotinib was sufficient to fully dephosphorylate the receptor (Fig. 4A). Thus, the resistance phenotype observed in PTEN-deficient H1650 cells may be partially explained by the prolonged activation of EGFR under treatment with EGFR tyrosine kinase inhibitors. To test whether PTEN loss–induced EGFR activation may be mimicked by stimulation of EGFR in PTEN-proficient PC9 cells, we treated parental PC9 cells with a combination of erlotinib and EGF (Fig. 4B). We observed an induction of phospho-EGFR by dual EGF stimulation and EGFR inhibition resembling the situation in PTEN-deficient cells (Fig. 4B). Confirming the functional relevance of PTEN loss– induced EGFR activation, this treatment also led to a reduction of the fraction of apoptotic cells (Fig. 4B).
Figure 4.
PTEN loss activates EGFR. A, phospho-EGFR was detected by immunoblotting after short exposure (SE) and long exposure (LE) in PC9, PC9CONTkd, and PC9PTENkd cells. Actin levels served as a loading control. B, left, levels of phospho-EGFR of PC9PTENkd and PC9 cells treated with erlotinib were determined (+/− EGF) under serum starvation. Right, apoptosis (%) after erlotinib treatment (0.5 μmol/L) in the given cells. C, left, phospho-EGFR and phospho-AKT in H3255 and H3255MyrAKT cells were assessed by immunoblotting. Right, the fraction of apoptotic cells (in %) in the given cells. D, a simplified model explaining our observations: in EGFR-mutant cells, EGFR is the sole input for production of PIP3. Inhibiting EGFR dramatically reduces the input into PIP3 production. Therefore, the lack of negative regulation of PIP3 production by loss of PTEN is limited.
Finally, we asked whether survival signaling activated by loss of PTEN is equivalent to immediate activation of Akt. We introduced a constitutively active allele of Akt (MyrAkT) into EGFR-mutant and erlotinib-sensitive H3255 cells. As expected, levels of phospho-Akt but not of phospho-EGFR levels remained elevated in H3255MyrAKT cells under erlotinib treatment (Fig. 4C). Furthermore, this pronounced Akt activity was associated with erlotinib resistance (P < 0.0005) of H3255MyrAKT cells when measuring apoptosis (Fig. 4C). Thus, immediate and constitutive activation of Akt is more effective than PTEN loss to induce erlotinib resistance in EGFR-mutant NSCLC cells.
Others have recently shown that PTEN loss leads to robust EGFR inhibitor resistance in cells lacking EGFR mutations (10, 11). Our findings in EGFR-mutant NSCLC cells differ from these observations, as the phenotype elicited by PTEN loss was less dominant. This discrepancy may be explained by the fact that EGFR-mutant NSCLC cells are exclusively dependent on EGFR signaling for their survival. Thus, erlotinib-mediated inhibition of EGFR as the sole input of PIP3 production may only partially be rescued by PTEN loss (Fig. 4D).
In summary, we have shown that in-depth genomic and phenotypic analyses of large cell line collections can be applied to identify a novel cell biology phenotype. Here, computational genomic analyses implied homozygous deletion of PTEN as a candidate for EGFR inhibitor resistance. Functional studies revealed that PTEN loss induces a significant reduction in apoptosis sensitivity in EGFR-mutant cells by activation of Akt and EGFR. We speculate that activation of Erk in PTEN-deficient cells (Fig. 3D) may lead to transcriptional up-regulation of EGFR ligands, such as amphiregulin (21). Moreover, PTEN loss and EGFR mutation co-occurred in 1 out of 24 EGFR-mutant patients in a genomic analysis of 140 lung adenocarcinomas, thus confirming the clinical relevance of our findings. Thus, PTEN loss may represent an additional mechanism of initial or acquired resistance to erlotinib-induced apoptosis in EGFR-mutant NSCLC.
Supplementary Material
Acknowledgments
Grant support: R.K. Thomas is a fellow of the International Association for the Study of Lung Cancer; and is supported by the Deutsche Krebshilfe (107954), the Fritz-Thyssen-Stiftung (10.08.2.175), and the NGFN-Plus Program of the German Ministry of Science and Education (BMBF, 01GS08100). J.D. Minna is supported by grants from the Specialized Programs of Research Excellence P50CA70907, DOD PROSPECT, and the Longenbaugh Foundation.
We thank Dr. Ingo Mellinghoff for sharing unpublished results.
Footnotes
M.L. Sos et al., under revision.
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Sharma SV, Fischbach MA, Haber DA, Settleman J. “Oncogenic shock”: explaining oncogene addiction through differential signal attenuation. Clin Cancer Res. 2006;12:4392–5s. doi: 10.1158/1078-0432.CCR-06-0096. [DOI] [PubMed] [Google Scholar]
- 2.Thomas RK, Greulich H, Yuza Y, et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:73–81. doi: 10.1101/sqb.2005.70.056. [DOI] [PubMed] [Google Scholar]
- 3.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 4.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo A, Villen J, Kornhauser J, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–7. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sos ML, Zander T, Thomas RK, Staratschek-Jox A, Claasen J, Wolf J. Expression of signaling mediators downstream of EGF-receptor predict sensitivity to small molecule inhibitors directed against the EGF-receptor pathway. J Thorac Oncol. 2008;3:170–3. doi: 10.1097/JTO.0b013e3181622c05. [DOI] [PubMed] [Google Scholar]
- 7.Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 8.Mehrian-Shai R, Chen CD, Shi T, et al. Insulin growth factor-binding protein 2 is a candidate biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc Natl Acad Sci U S A. 2007;104:5563–8. doi: 10.1073/pnas.0609139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 10.Yamasaki F, Johansen MJ, Zhang D, et al. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res. 2007;67:5779–88. doi: 10.1158/0008-5472.CAN-06-3020. [DOI] [PubMed] [Google Scholar]
- 11.She QB, Solit DB, Ye Q, O’Reilly KE, Lobo J, Rosen N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005;8:287–97. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 13.McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci U S A. 1999;96:10182–7. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endoh H, Yatabe Y, Kosaka T, Kuwano H, Mitsudomi T. PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol. 2006;1:629–34. [PubMed] [Google Scholar]
- 19.Gong Y, Somwar R, Politi K, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng J, Shimamura T, Perera S, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–75. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- 21.Toulany M, Baumann M, Rodemann HP. Stimulated PI3K-AKT signaling mediated through ligand or radiation-induced EGFR depends indirectly, but not directly, on constitutive K-Ras activity. Mol Cancer Res. 2007;5:863–72. doi: 10.1158/1541-7786.MCR-06-0297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.