Abstract
A major challenge in cell biology is to identify the subcellular distribution of proteins within cells and to characterize how protein localization changes under different cell growth conditions and in response to stress and other external signals. Protein localization is usually determined either by microscopy or by using cell fractionation combined with protein blotting techniques. Both these approaches are intrinsically low throughput and limited to the analysis of known components. Here we use mass spectrometry-based proteomics to provide an unbiased, quantitative, and high throughput approach for measuring the subcellular distribution of the proteome, termed “spatial proteomics.” The spatial proteomics method analyzes a whole cell extract created by recombining differentially labeled subcellular fractions derived from cells in which proteins have been mass-labeled with heavy isotopes. This was used here to measure the relative distribution between cytoplasm, nucleus, and nucleolus of over 2,000 proteins in HCT116 cells. The data show that, at steady state, the proteome is predominantly partitioned into specific subcellular locations with only a minor subset of proteins equally distributed between two or more compartments. Spatial proteomics also facilitates a proteome-wide comparison of changes in protein localization in response to a wide range of physiological and experimental perturbations, shown here by characterizing dynamic changes in protein localization elicited during the cellular response to DNA damage following treatment of HCT116 cells with etoposide. DNA damage was found to cause dissociation of the proteasome from inhibitory proteins and assembly chaperones in the cytoplasm and relocation to associate with proteasome activators in the nucleus.
Many previous studies on organelle proteomics have provided a detailed list of the protein contents of organelles, substructures, or compartments isolated from cells (1–5). Such studies have also used quantitative proteomics in the high throughput assignment of proteins to subcellular compartments using methods such as protein correlation profiling (3, 6), recording the number of ions detected per protein (1, 2), or localization of organelle proteins by isotope tagging (7, 8). However, interpretation of the resulting protein inventory is complicated by the dynamic nature of organelle proteomes and by the fact that many proteins are not exclusive to one compartment but instead partition between separate subcellular locations (9, 10). This is illustrated by our previous studies of the human nucleolar proteome that have identified over 4,000 proteins that can co-purify reproducibly with nucleoli isolated from human cells but many of which are either present in low abundance in nucleoli and/or also have functions in other cellular locations (11). This highlights the importance of not only identifying the presence of a protein in any specific cellular organelle or structure but also measuring its relative abundance in different locations and assessing how this subcellular localization can change between different compartments under different cell growth and physiological conditions.
Stable isotope labeling with amino acids in cell culture (SILAC)1 is the use of stable isotopic atoms along with mass spectrometry for quantitative mass spectrometry analysis (12, 13). This method allows quantitative analyses of proteins by comparison of the mass of light and heavier forms of the same peptide from a given protein, arising from the presence of heavier, stable isotopes such as 13C, 2H, and 15N. These stable isotopes are incorporated in proteins by in vivo labeling, i.e. growing the cells in specialized media where specific amino acids, typically arginine and lysine, are replaced with corresponding heavy isotope-substituted forms in which either all carbons or carbons, hydrogens, or nitrogens are isotope-labeled (14). Cleavage at the substituted arginine or lysine by trypsin generates a peptide with a shift in mass relative to the control (i.e. unsubstituted) peptide, and this can easily be resolved by mass spectrometry. The ratio of intensities of the “light” and “heavy” peptide signals identified by mass spectrometry directly correlates with the relative amount of the cognate protein from each sample. This method has been widely used for both relative quantification of protein levels after exposure of cells to drugs and inhibitors and for the identification of specific protein interaction partners (15–18).
Here we used a quantitative and high throughput MS-based approach we term “spatial proteomics,” which both measures the relative intracellular localization of proteins and facilitates a comparison of changes in their subcellular localization under different conditions. This approach allows the rapid assignment of the cellular localization of proteins using common fractionation techniques. The major advantage of such a technique over other MS-based localization techniques such as protein correlation profiling or localization of organelle proteins by isotope tagging is that it provides a direct quantitative measurement of what fraction of each protein is localized to each cellular compartment, whereas the other techniques associate proteins showing similar profiles in a density centrifugation gradient while not describing the relative fraction of proteins in all locations. The spatial proteomics approach thus facilitates the comparison of protein localization under different conditions. We applied this spatial proteomics technique to determine the subcellular localization of over 2,000 proteins in HCT116 cells and then compared changes in localization following exposure to the topoisomerase inhibitor etoposide.
MATERIALS AND METHODS
Cell Culture
The human colon carcinoma cell line HCT116 was cultured as adherent cells in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, custom order) depleted of arginine and lysine. The DMEM was supplemented with 10% fetal bovine serum dialyzed with a cutoff of 10 kDa (Invitrogen, 26400-044), 100 units/ml penicillin/streptomycin, 2 mm l-glutamine. Arginine and lysine were added in either light (Arg0, Sigma, A5006; Lys0, Sigma, L5501), medium (Arg6, Cambridge Isotope Laboratories, CNM-2265; Lys4, Cambridge Isotope Laboratories, DLM-2640), or heavy (Arg10, Cambridge Isotope Laboratories, CNLM-539; Lys8, Cambridge Isotope Laboratories, CNLM-291) form to a final concentration of 28 μg/ml for arginine and 49 μg/ml for lysine. Cells were tested for full incorporation of the label after six passages.
Cell Fractionation
Cytoplasm, nuclei, and nucleoli were prepared from HCT116 cells using a method originally described in Andersen et al. (4). Briefly, cells were washed three times with PBS, resuspended in 5 ml of buffer A (10 mm HEPES-KOH (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT), and Dounce homogenized 10 times using a tight pestle. Dounce homogenized nuclei were centrifuged at 228 × g for 5 min at 4 °C. The supernatant represents the cytoplasmic fraction. The nuclear pellet was resuspended in 3 ml of 0.25 m sucrose, 10 mm MgCl2; layered over 3 ml of 0.35 m sucrose, 0.5 mm MgCl2; and centrifuged at 1,430 × g for 5 min at 4 °C. The clean, pelleted nuclei were resuspended in 3 ml of 0.35 m sucrose, 0.5 mm MgCl2 and sonicated for 6 × 10 s using a microtip probe and a Misonix XL 2020 sonicator at power setting 5. The sonication was checked using phase-contrast microscopy, ensuring that there were no intact cells and that the nucleoli were readily observed as dense, refractile bodies. The sonicated sample was then layered over 3 ml of 0.88 m sucrose, 0.5 mm MgCl2 and centrifuged at 2,800 × g for 10 min at 4 °C. The pellet contained the nucleoli, whereas the supernatant consisted of the nucleoplasmic fraction. The nucleoli were then washed by resuspension in 500 μl of 0.35 m sucrose, 0.5 mm MgCl2 followed by centrifugation at 2,000 × g for 2 min at 4 °C. Proteins were quantified using the Quant-IT protein assay (Invitrogen) and measured using a Qubit (Invitrogen). Equal amounts of total protein from each fraction were then recombined to recreate a whole cell extract but with cytoplasm (Cyto), nuclei (Nuc), and nucleoli (No) arising from cells with different isotopic labels.
Immunofluorescence
Cells were grown on glass coverslips and fixed with 1% paraformaldehyde in PBS for 10 min. Cells were then permeabilized in PBS containing 0.5% Triton X-100 for 10 min and then labeled with antibodies recognizing α-tubulin (Sigma), lamin B (Abcam), or RNA polymerase I large subunit (RPA194, Santa Cruz Biotechnology, Inc.). After washing with PBS containing 0.1% Triton X-100 and with PBS, cells were then labeled with a secondary antibody coupled to Alexa Fluor 488 (Molecular Probes) and mounted on slides with Vectashield (Vector Laboratories Inc.) containing 4′,6-diamidino-2-phenylindole. Fluorescence imaging was performed on a DeltaVision Spectris wide field deconvolution microscope (Applied Precision) with a CoolMax charge-coupled device camera (Roper Scientific). Cells were imaged using a 60× numerical aperture 1.4 Plan-Apochromat objective (Olympus) and the appropriate filter sets (Chroma Technology Corp.) with 20 optical sections of 0.5 μm each acquired. SoftWorX software (Applied Precision) was used for both acquisition and deconvolution.
Gel Electrophoresis and In-gel Digestion
The reconstituted cell fractions were reduced in 10 mm DTT and alkylated in 50 mm iodoacetamide prior to being boiled in loading buffer and then separated by one-dimensional SDS-PAGE (4–12% Bis-Tris Novex minigel, Invitrogen) and visualized by colloidal Coomassie staining (Novex, Invitrogen). The entire protein gel lanes were excised and cut into eight slices each. Every gel slice was subjected to in-gel digestion with trypsin (19). The resulting tryptic peptides were extracted by 1% formic acid, acetonitrile; lyophilized in a SpeedVac; and resuspended in 1% formic acid.
LC-MS/MS
Trypsin-digested peptides were separated using an Ultimate U3000 (Dionex Corp.) nanoflow LC system consisting of a solvent degasser, micro- and nanoflow pumps, flow control module, UV detector, and thermostated autosampler. 10 μl of sample (a total of 2 μg) was loaded with a constant flow of 20 μl/min onto a PepMap C18 trap column (0.3-mm inner diameter × 5 mm, Dionex Corp.). After trap enrichment, peptides were eluted off onto a PepMap C18 nanocolumn (75 μm × 15 cm, Dionex Corp.) with a linear gradient of 5–35% solvent B (90% acetonitrile with 0.1% formic acid) over 65 min with a constant flow of 300 nl/min. The HPLC system was coupled to an LTQ Orbitrap XL (Thermo Fisher Scientific Inc.) via a nano-ES ion source (Proxeon Biosystems). The spray voltage was set to 1.2 kV, and the temperature of the heated capillary was set to 200 °C. Full-scan MS survey spectra (m/z 335–1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 500,000 ions. The five most intense peptide ions from the preview scan in the Orbitrap were fragmented by collision-induced dissociation (normalized collision energy, 35%; activation Q, 0.250; and activation time, 30 ms) in the LTQ after the accumulation of 10,000 ions. Maximal filling times were 1,000 ms for the full scans and 150 ms for the MS/MS scans. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 90 s and a relative mass window of 10 ppm. The lock mass option was enabled for survey scans to improve mass accuracy (20). Data were acquired using the Xcalibur software.
Quantification and Bioinformatics Analysis
Quantitation was performed using the program MaxQuant version 1.0.13.8 (21, 22). The derived peak list generated by Quant.exe (the first part of MaxQuant) was searched using Mascot (Matrix Sciences, London, UK) as the database search engine for peptide identifications against the International Protein Index human protein database version 3.37 containing 69,290 proteins to which 175 commonly observed contaminants and all the reversed sequences had been added. The initial mass tolerance was set to 7 ppm, and MS/MS mass tolerance was 0.5 Da. Enzyme was set to trypsin with no proline restriction (trypsin/p) with three missed cleavages. Carbamidomethylation of cysteine was searched as a fixed modification, whereas N-acetyl protein and oxidation of methionine were searched as variable modifications. Identification was set to a false discovery rate of 1%. To achieve reliable identifications, all proteins were accepted based on the criteria that the number of forward hits in the database was at least 100-fold higher than the number of reverse database hits, thus resulting in a false discovery rate of 1%. A minimum of two peptides was quantified for each protein. Protein isoforms and proteins that were not able to be distinguished based on the peptides identified are grouped and displayed on a single line with multiple International Protein Index numbers (see the supplemental tables). Alternatively, quantitation was performed using MSQuant version 1.4.3 for clustering analysis with Cluster using complete linkage clustering and visualized using Treeview (23).
RESULTS AND DISCUSSION
The key feature of the spatial proteomics method is to establish a quantitative map of the relative subcellular localization of each protein. This will both annotate the proteome and serve as the basis for a comparison of changes in proteome localization under different conditions. A map of the spatial distribution of the proteome is achieved by creating a whole cell extract where separate compartments are differentially isotope-labeled through recombining separate subcellular fractions derived from cells labeled with amino acids containing distinct heavy isotopes (Fig. 1A). For these experiments, we combined the use of established cell fractionation procedures to generate separate cytoplasmic, nuclear, and nucleolar fractions and SILAC (14) to label the proteins in each fraction with isotopes that can be distinguished using mass spectrometry (Fig. 1B). SILAC provides a convenient and efficient means of metabolically labeling cell proteins with amino acids that incorporate heavy isotopes (for a review, see Mann (13)). The SILAC approach has been successfully used for a range of proteomics studies on cultured cells that rely upon differential mass tagging of proteins (24). In principle, the spatial proteomics approach could be used to analyze protein localization in either tissue samples or in cells where metabolic SILAC labeling is not possible, using alternative quantitative mass spectrometric methods (25, 26). Furthermore, although here we analyzed specifically nuclear, nucleolar, and cytoplasmic fractions, the approach can also be extended to compare the distribution of proteins between other combinations of subcellular compartments so long as a reproducible fractionation procedure is available. Because our major aim was to generate a reproducible map of relative protein distribution within the cell that can be compared under different conditions, we chose to generate the whole cell extract by recombining equal amounts of protein isolated from each subcellular fraction. Alternative schemes for recombining the separate labeled fractions could also be used.
Fig. 1.
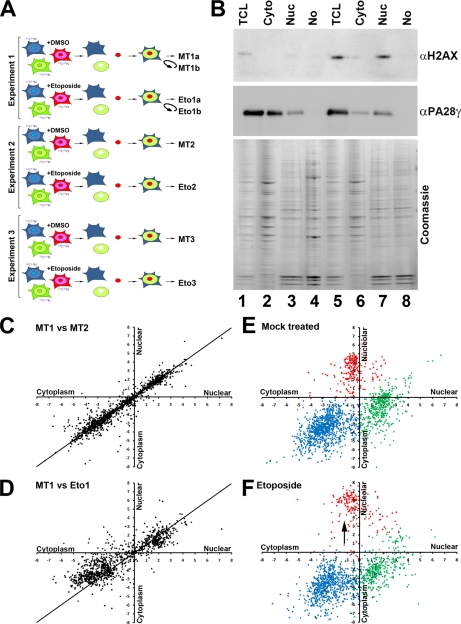
A, human colon carcinoma HCT116 cells were grown in DMEM containing either the normal light isotopes of carbon and nitrogen (i.e. 12C14N) (light), l-[13C6,14N4]arginine and l-[2H4]lysine (medium), or l-[13C6,15N4]arginine and l-[13C6,15N2]lysine (heavy). Separate cytoplasmic, nuclear, and nucleolar fractions were isolated from each labeled cell population. B, equal amounts of total protein from each fraction were then recombined to recreate a whole cell extract but with Cyto, Nuc, and No arising from cells with different isotope labels. The recombined whole cell extract mixture was solubilized with loading buffer; proteins were separated using SDS-PAGE; and the resulting gel was cut into eight equal pieces, trypsin-digested, and analyzed by LC-MS/MS using an LTQ Orbitrap. Triplicate fractions and LC-MS/MS resulted in a proteome of 2,536 proteins.
Human colon carcinoma HCT116 cells were grown in three different SILAC media containing arginine and lysine either with the normal light isotopes of carbon, hydrogen, and nitrogen (i.e. 12C14N) (light), l-[13C6,14N4]arginine and l-[2H4]lysine (medium), or l-[13C6,15N4]arginine and l-[13C6,15N2]lysine (heavy). Separate cytoplasmic, nuclear, and nucleolar fractions were isolated from each labeled cell population as described previously (4). Equal amounts of total protein from each fraction were then recombined to recreate a whole cell extract but with Cyto, Nuc, and No arising from cells with different isotope labels (Fig. 1A). This represent approximately a 1:1:2.4 (cytoplasmic:nuclear:nucleolar) relative distribution of the proteins. Note that any external protein contaminants, such as keratins, will only appear in the light fraction because the heavy isotopes occur at very low levels in the environment. Therefore, because our major biological interest was on the analysis of nuclear proteins, we chose to use the light label for the cytoplasmic fraction. However, the fractions can be recombined in any order as best suits the objective of the analysis, and if sufficient resources and analysis time are available, each permutation of isotope combinations could be analyzed separately, which would confirm the identity of external contaminants (data not shown). There are also a wash step of the nuclear fraction following the hypotonic Dounce homogenization and a wash step of the nucleolar fraction that are excluded from the analysis, which may result in a loss of some proteins. Our quantification of those wash fractions indicated that less than 5% of the total proteins were removed at those steps, although we cannot exclude that this includes a small number of specific proteins. The recombined whole cell extract mixture was solubilized with loading buffer; proteins were separated using SDS-PAGE; and the resulting gel was cut into eight equal pieces, trypsin-digested, and analyzed by LC-MS/MS using an LTQ Orbitrap (27). The resulting ratios between light, medium, and heavy isotopic forms for each peptide identified were quantified using MaxQuant (21). The separate ratio values for each peptide in a given protein were averaged to provide a measure of the relative distribution for the protein between the respective cytoplasmic, nuclear, and nucleolar compartments (Figs. 1, 2, and 3). Three independent experiments of the whole spatial proteomics procedure were performed using separate preparations of isotope-labeled HCT116 cells. A total of 29,541 peptides were quantified, corresponding to 2,427 proteins, and a resulting distribution ratio between the three cellular compartments was derived for each protein, calculated as a mean of the values for all peptides from the protein.
Fig. 2.
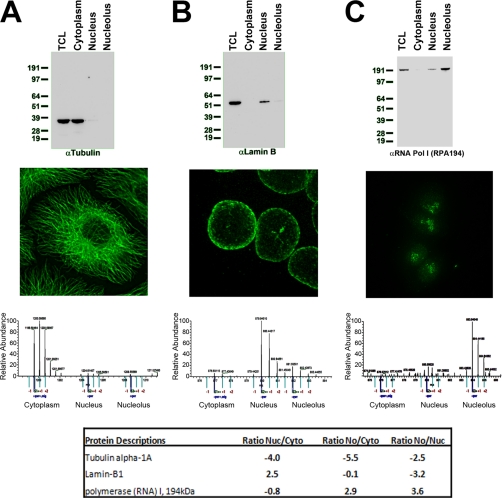
Equal amounts (5 μg) of extract from total cell lysate (TCL) or from cytoplasmic, nuclear, or nucleolar fractions of HCT116 cells were separated by SDS-PAGE and transferred to nitrocellulose, and Western blotting was performed using antibodies recognizing α-tubulin (A), lamin B (B), or RPA194 (C). Alternatively, HCT116 cells were grown on coverslips, fixed, and processed for fluorescence microscopy using the same antibodies used for Western blotting. An example of a spectrum from each of the proteins derived from the spatial proteomics mass spectrometry run is also shown. The table shows the ratios of enrichment of Nuc/Cyto, No/Cyto, and No/Nuc.
Fig. 3.
A, hierarchical clustering was performed using the localization ratios (a) No/Nuc, (b) No/Cyto, and (c) Nuc/Cyto and represented as a tree. In each case, high ratios are shown in red, low ratios are in green, and a 1:1 ratio is in black. B, visualization of the spatial proteomics data by graphical representation, plotting the log base 2 nuclear/cytoplasmic ratio on the x axis and log base 2 nucleolar/cytoplasmic ratio on the y axis. C, the Sm proteins associated with the small nuclear RNP subunits of spliceosomes (blue), proteins associated with small nucleolar RNPs (snoRNPs; red), and protein subunits of the 26 S proteasome cluster (green) are shown as examples. PEP, posterior error probability.
To validate the approach, examples are shown comparing the respective distributions of proteins commonly used as markers for cytoplasmic (tubulin; Fig. 2A), nuclear (lamin B; Fig. 2B), and nucleolar (RNA polymerase I subunit RPA194; Fig. 2C) fractions. Here the localization was independently measured by fluorescence microscopy, protein blotting, and spatial proteomics mass spectrometry, in the latter case illustrated with a representative peptide spectrum from each protein (Fig. 2, A–C). This shows close agreement in the relative protein distribution observed using each method. However, of these three methods, only the spatial proteomics is readily amenable to high throughput analysis, and the accuracy in this case is amplified because multiple peptides from each protein are typically identified and quantified, providing separate, independent measurements. The spatial proteomics method also allows the identification and localization of previously uncharacterized proteins and/or proteins for which no antibodies are currently available. Detailed mass spectrometry analysis could also potentially distinguish the localization of separate spliced isoforms and/or correlate differential localization of proteins with distinct post-translational modifications.
The ratio values derived from the spatial proteomics experiments enable clustering analysis (23), providing an objective approach for unbiased grouping of proteins that show similar subcellular distribution (28). Hierarchical clustering was performed using the localization ratios (a) No/Nuc, (b) No/Cyto, and (c) Nuc/Cyto and represented as a tree (Fig. 3A). In each case, high ratios are shown in red, low ratios are in green, and a 1:1 ratio is in black. The individual ratio values for each protein identified are provided in supplemental Table 1. Visualization of the spatial proteomics data by graphical representation, plotting the log base 2 nuclear/cytoplasmic ratio on the x axis and log base 2 nucleolar/cytoplasmic ratio on the y axis (Fig. 3B), shows that most proteins are enriched within one of the fractions by at least 2-fold. Relatively few proteins show equal distributions between two or three compartments (Fig. 3B), suggesting that, at steady state, the proteome is predominantly partitioned into specific subcellular locations.
The spatial proteomics data can be analyzed to correlate co-localization of either protein families or of separate subunits of a multiprotein complex. For example, the Sm proteins associated with the small nuclear RNP subunits of spliceosomes cluster in the nuclear fraction, proteins associated with small nucleolar RNPs cluster in the nucleolar fraction, and protein subunits of the 26 S proteasome cluster in the cytoplasmic fraction (Fig. 3C). These quantification values were reproducible across independent repeat experiments (see below). We conclude, therefore, that the spatial proteomics method provides a useful new tool for annotating the subcellular localization of the proteome.
The spatial proteomics analysis correctly assigned most known proteins to the expected subcellular fraction according to their gene ontology cellular component. Of the 1,004 proteins with a log base 2 ratio of Nuc/Cyto of 1 and No < Cyto < −1, we found 659 having an annotation. Of those, 578 had a cytosol, cytoplasm, extracellular, or cytoskeleton annotation. 63 proteins were annotated as intracellular with no further annotations, and 18 had a nuclear annotation. For the nuclear proteins with a ratio of Nuc/Cyto > 1 and No/Nuc < −1, we found 464 proteins of which 334 had a gene ontology annotation for their localization. Of those proteins, 193 had a nuclear annotation, 98 had a mitochondrial annotation, and 43 had an endoplasmic reticulum annotation. Finally, 221 proteins were found as nucleolar with No/Cyto > 1 and No/Nuc > 1. Of those proteins, 127 had a gene ontology cellular component annotation of nucleolar, 16 were assigned to other compartments, and 84 had no specific annotation for localization. The fractionation demonstrated that soluble nucleoplasmic proteins are prone to leakage into the cytoplasmic fraction, and mitochondrial proteins as well as endoplasmic reticulum-associated proteins were detected in the nuclear fraction. This is a result of the fractionation protocol and is not a limitation in the SILAC quantitation. For example, under the hypotonic conditions used, any unlysed mitochondria will pellet with the nuclear fraction, whereas unbound nuclear proteins can be partially extracted into the cytoplasm.
We observed that nuclear retention of proteins during fractionation is positively correlated with known association to either chromatin or other dense nuclear structures, such as the nuclear lamina and nucleolus. This provides a convenient assay that can be exploited to detect changes in protein binding to nuclear structures under different conditions. Essentially all fractionation procedures can result in some degree of leakage and mislocalization of proteins during extraction. The spatial proteomics approach provides an excellent internal control for such effects, however, because of the many thousands of peptides whose localization is measured in parallel. This contrasts with the situation in protein blotting experiments where typically only one or two representative markers are used as controls (see Fig. 2, A–C). Here we show that with the fractionation procedure used the majority of proteins were accurately mapped to the respective nuclear and cytoplasmic compartments, although a subset of soluble nuclear proteins, e.g. PA28γ (29), leak into the cytoplasmic fraction (PSME3; supplemental Tables 1 and 2). This underlines how critical the choice of marker protein can be when used as a control for fractionation in a blotting experiment and shows the limitation of relying on a limited number of internal controls. Using spatial proteomics, thousands of control measurements are included within each experiment. We suggest therefore that the spatial proteomics can help to assess the accuracy of a wide range of cell fractionation procedures.
Next, we used spatial proteomics to compare the cellular localization of the proteome in HCT116 cells following exposure to the topoisomerase II inhibitor etoposide (Fig. 4A), which induces DNA damage by generating double strand breaks. HCT116 cells incubated in either light, medium, or heavy SILAC labeling media were either mock-treated (MT) or else exposed to a final concentration of 50 μm etoposide (Eto) for 1 h, which caused the expected increase in phosphorylation of H2AX (Fig. 4B). This was a dose inducing a high level of phosphorylation of the histone H2AX as visualized by immunofluorescence and sufficient to subsequently kill most cells if left to incubate for 48 h after the exposure to etoposide. Cells were then harvested, and separate nuclear, cytoplasmic, and nucleolar fractions were prepared as before (Fig. 4B). The entire experiment was repeated three times, and a whole cell extract recombining cytoplasmic, nuclear, and nucleolar fractions with separate light, medium, and heavy isotopes was prepared independently for the MT and Eto 1, 2, and 3 samples (Fig. 4A). The isotope-labeled extracts were dissolved in sample buffer and separated by SDS-PAGE, the resulting gel was cut into eight slices and trypsin-digested, and peptides were analyzed by mass spectrometry on an LTQ Orbitrap. Each peptide was quantified using MaxQuant (Fig. 4, C and D, and supplemental Table 2).
Fig. 4.
Changes in protein localization were determined using spatial proteomics by comparing cells that were either MT or treated with 50 mm etoposide for 1 h. A, the entire experiment was repeated three times, and a whole cell extract recombining cytoplasmic, nuclear, and nucleolar fractions with separate light, medium, and heavy isotopes was prepared independently for the MT and Eto 1, 2, and 3 samples. Experiment 1 was run twice on the mass spectrometer (MT1a and -b and Eto1a and -b). B, HCT116 cells were either MT (lanes 1–4) or else exposed to a final concentration of 50 μm Eto (lanes 5–8) for 1 h, which caused the expected nuclear increase in phosphorylation of H2AX. C, scatter plot showing the comparison of the cytoplasmic versus nuclear distribution of MT1 and MT2. D, scatter plot showing the comparison of the cytoplasm versus nuclear distribution of MT1 versus Eto1. E, scatter plot showing the cytoplasm versus nuclear (x axis) and the cytoplasm versus nucleolar (y axis) distribution of proteins following mock treatment. F, scatter plot showing the cytoplasm versus nuclear (x axis) and the cytoplasm versus nucleolar (y axis) distribution of proteins following etoposide treatment. Cytoplasmic proteins are shown in blue, nuclear proteins are shown in green, and nucleolar proteins are shown in red. TCL, total cell lysate.
To check whether any differences observed in proteome localization under different conditions arose because of biologically relevant changes and not simply because of experimental variation (e.g. in reproducibility of mass spectrometry or fractionation procedures), statistical evaluation of the repeat data sets was carried out. The data show a Pearson correlation of 0.90 ± 0.01 between samples MT1, MT2, and MT3 with a Pearson correlation of 0.86 ± 0.01 between the respective MT and Eto samples. This is illustrated in the respective scatter plots showing the comparison of MT1 versus MT2 (Fig. 4C) and MT1 versus Eto1 (Fig. 4D) data where a clear increase in the number of proteins deviating from a straight line plot is evident following etoposide treatment. Furthermore, we found similar Pearson correlations whether we compared samples from either the same pair or from different pairs (for example, the correlation between MT1 versus Eto1 is 0.86684 and between MT1 and Eto2 is 0.86623, whereas the correlation between MT1 and MT2 is 0.90906), confirming that the observed differences are largely due to the treatment with etoposide and not due to variation between repeats of the experiment. In addition to the three replicate experiments, one sample (i.e. MT1 and Eto1) was also analyzed twice to assess the reproducibility of the mass spectrometry (defined as samples MT1a and -b and Eto1a and -b). The Pearson correlation is 0.96 for data from the same sample analyzed twice in the mass spectrometer (e.g. MT1a versus MT1b).
To determine whether there are variations due to the different growth media or due to repeats of the fractionation technique, we analyzed each fraction originating from the different growth media (supplemental Fig. 1). Analysis of an equal amount of protein shows an undistinguishable pattern between each of the growth media as visualized by Coomassie staining of each fraction (supplemental Fig. 1A). We mixed equal amounts of cytoplasmic, nuclear, and nucleolar fractions and analyzed the variation resulting from the different growth media, analyzing cytoplasmic fractions from light, medium, and heavy at 1:1:1 (supplemental Fig. 1, B–D), nuclear fractions at 1:1:1 (supplemental Fig. 1, E–G), and nucleolar fractions at 1:1:1 (supplemental Fig. 1, H–J). We found no specific bias resulting from the different culture media (supplemental Fig. 1, B–J). Each protein had a similar localization in the three different culture media. Next, we analyzed whether there was variation due to the repeats of the fractionation protocols. In each case, we found a minor deviation from the expected value of 1 (i.e. 15% for the cytoplasm, 20% for the nucleus, and 1% for the nucleolus). We thus conclude that proteins showing an increase in localization in one compartment by more than those values likely represent a significant change. We also found an inverse correlation between the posterior error probability score, which estimates the probability of a wrong assignment of a spectrum to a peptide sequence and the variation of the data. This is not surprising because the probability of a wrong assignment decreases with the number of peptides identified, which also results in more robust data for the quantification. This can thus be used when trying to determine the reliability of the observed changes (i.e. the ratio of a protein for which more peptides were identified is more reliable).
We conclude that experimental variations, both directly in the mass spectrometer and from the fractionation procedure, can have a small effect on the values measured. Nonetheless, the data demonstrate that the spatial proteomics technique can determine differences in proteome localization directly resulting from inhibitor treatment. The effect of etoposide on proteome localization was specific because most of the proteome did not change in localization after DNA damage (Fig. 4D). However, there was a general effect on nucleolar proteins, which became more enriched in the nucleolus (Fig. 4F, arrow). This increased segregation of nucleolar proteins within the nucleolus following stress is consistent with previous observations (30).
Proteins with similar gene ontology annotations that showed relocalization following treatment with etoposide were analyzed to find protein complexes changing compartments following DNA damage. We found that proteins with the gene ontology annotation “proteolysis” had a change in their cytoplasmic localization, suggesting a partial relocalization of those proteins toward the nucleus (Fig. 5A). As a control, proteins with the gene ontology annotation “ribosomal” did not display any change in their nuclear/cytoplasmic ratio (Fig. 5B). Most of the proteins identified with the proteolysis annotation were components of the 20 S proteasome and the associated activating complexes. The 20 S proteasome (illustrated in Fig. 5C) forms the core of a larger 26 S protease complex that catalyzes the ATP-dependent degradation of ubiquitinated proteins (Fig. 5F) (31). Alternatively, the 20 S proteasome can associate with the PA28 activator (or 11 S proteasome), which is involved in antigen presentation (Fig. 5D) (32). The proteasome is present in the cytoplasm and in the nuclei of all eukaryotic cells (33). Each of the subunits of the 20 S proteasome had a similar relocalization toward the nucleus following etoposide treatment (Fig. 5C). The 11 S proteasome also displayed a relocalization to the nucleus (Fig. 5D) as well as the base and lid of the 19 S proteasome (Fig. 5, E and F). Interestingly, some proteins with the gene ontology annotation proteolysis did not show a change in localization. Of those proteins, the proteasome inhibitor PSMF1 (PI31) showed the same cytoplasmic accumulation as in control cells as did the proteasome assembly chaperone PSMG1 proteins (Fig. 5G). The three proteins that displayed the largest association with a nuclear proteasome after etoposide treatment are all proteasome activators (Fig. 5G). These data demonstrate a dissociation of the proteasome from inhibitory proteins and assembly chaperones in the cytoplasm and relocation to associate with proteasome activators in the nucleus in response to DNA damage.
Fig. 5.
A, proteins with the gene ontology annotation proteolysis are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (blue) or etoposide-treated (red). B, proteins with the gene ontology annotation ribosomal are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (blue) or etoposide-treated (red). C, proteins PSMA1–7 and PSMB1–7 forming the 20 S proteasome are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (blue) or etoposide-treated (red). D, proteins PSME1–3 forming the 11 S proteasome are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (blue) or etoposide-treated (red). E, proteins PSMC1–6 and PSMD1–2 forming the base of the 19 S proteasome are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (blue) or etoposide-treated (red). F, proteins PSMD3–14 forming the lid of the 19 S proteasome are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (blue) or etoposide-treated (red). G, proteins showing the highest relocalization to the nucleus after etoposide and proteins showing no relocalization to the nucleus are displayed with the log base 2 ratio of their enrichment in each compartment.
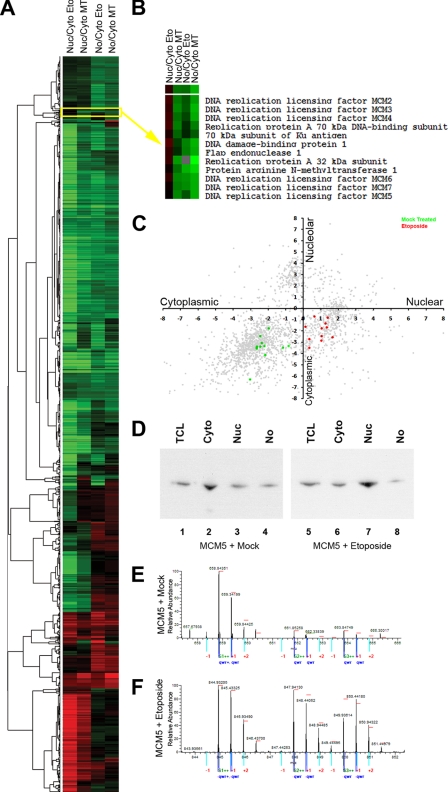
Protein complexes that showed a specific change in localization after etoposide treatment were identified by clustering proteins with similar values of localization and relocalization following etoposide treatment (Fig. 6A). For example, the replication protein A complex proteins and the minichromosome maintenance (MCM) complex proteins MCM2–7 all showed a similar specific change in their spatial proteome (Fig. 6B). All these proteins became concentrated in the nuclear compartment following DNA damage (Fig. 6C), and this was confirmed by Western blotting (Fig. 6D). This most likely reflects a change in the MCM complex going from a soluble nucleoplasmic to a chromatin-bound form after DNA damage. The cytoplasmic location of the MCM proteins prior to DNA damage is thus resulting from leakage during fractionation as discussed above, which appears to affect predominantly nuclear proteins that are not associated stably with either chromatin or other subnuclear structures. Indeed, immunofluorescence microscopy showed the MCM proteins to be easily extracted by treatment with a mild detergent (0.5% Triton X-100) prior to fixation (data not shown).
Fig. 6.
A, protein complexes that showed a specific change in localization after etoposide treatment were clustered according to the log base 2 ratios and displayed as a heat map where green represents negative values and red represents positive values. B, a cluster containing the replication protein A complex proteins and the MCM complex proteins MCM2–7 all showed a similar specific change in their spatial proteome. C, the proteins displayed in B are plotted by their cytoplasmic/nuclear ratios versus their cytoplasmic/nucleolar ratio as either mock-treated (green) or etoposide- treated (red). D, Western blotting of the total cell lysate (TCL), Cyto, Nuc, and No fractions of either mock-treated (lanes 1–4) or etoposide-treated (lanes 5–8) cells with an MCM5 antibody. E and F, spectra for peptides from MCM5 derived from either mock-treated cells (E) or etoposide-treated cells (F) are displayed.
These spatial proteomics data provide unexpected new evidence showing that DNA damage alters the properties of the MCM complex. The specificity of this effect is underlined by the fact that most of the proteome was not affected by the etoposide treatment, whereas each protein analyzed from the MCM complex (and each peptide analyzed for each of these proteins) independently revealed a similar shift in localization (Fig. 6, E and F). The MCM proteins, which act as a replicative helicase during DNA synthesis, are required for processive DNA replication and are a target of S-phase checkpoints (34). In yeast, temperature-sensitive MCM cells at restrictive temperature contain numerous foci recognized by the phosphorylated histone H2AX antibody (34), suggesting a role in the detection and repair of DNA double strand breaks. The spatial proteomics data are consistent with a role for MCM proteins in DNA repair in mammalian cells. Interestingly, we also found three known DNA repair proteins (i.e. Ku70, DDB1, and PRMT1) clustered with a redistribution profile similar to that of the MCM and replication protein A complex replication proteins, which suggests a possible mechanism linking DNA replication and DNA repair. This demonstrates how spatial proteomics can be used to not only look at individual protein responses under different conditions but also to look at how different groups of proteins can be analyzed to have a better understanding of underlying functional interactions.
The ubiquitin-dependent proteasome pathway degrades misfolded and damaged proteins and regulates cellular processes such as cell cycle, apoptosis, and DNA repair by targeting specific regulatory proteins (35). Substrates degraded by proteasomes are covalently linked with a polyubiquitin chain through the coordinated actions of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). The proteasome recognizes target proteins through receptors by specifically interacting with ubiquitinated proteins to degrade them. The proteasome has been involved in the regulation of several DNA repair pathways such as the nucleotide excision repair and homologous recombination (for a review, see Ref. 36). Our data demonstrate a dissociation of at least a subset of the proteasome from inhibitory proteins and assembly chaperones in the cytoplasm and relocation to associate with proteasome activators in the nucleus in response to DNA damage. It remains to be established whether the primary mechanism following DNA damage involves an active transport of the proteosomal proteins into the nucleus, a change in affinity of the proteasome for nuclear proteins, or both. It will be interesting to determine how the proteasome is activated and recruited to nuclear protein targets involved in the DNA repair pathways as our data suggest.
Interpretation of protein inventories using proteomics to identify proteins in purified organelles is complicated by the fact that many proteins are not exclusive to one compartment but instead partition between separate subcellular locations (9, 10). In our previous studies of the human nucleolar proteome, we have identified over 4,000 proteins that can co-purify reproducibly with nucleoli isolated from human cells (11). By comparing the proteins we categorized as being cytoplasmic, nuclear, and nucleolar from our spatial proteomics experiments with the nucleolar proteome, we found that 94% (197 of 210) of the proteins we found enriched in the nucleolus are present in the nucleolar database compared with 68% (299 of 437) and 48% (436 of 911) of proteins from the nucleus and the cytoplasm, respectively. Our measurements using the spatial proteomics method allow us to now classify proteins as enriched in the nucleolus compared with other compartments or as low abundance proteins in that organelle. This highlights the importance of not only identifying the presence of a protein in any specific cellular organelle or structure but also measuring its relative abundance in different locations and assessing how this subcellular localization can change between different compartments under different cell growth and physiological conditions.
In this study, we have introduced spatial proteomics, which represents a “second generation” approach to MS-based proteomics that not only identifies proteins but also provides an unbiased and quantitative measurement of protein properties, in this case, the annotation of subcellular proteome localization under different conditions. Although we have illustrated this here using cells exposed to etoposide, it could just as well be applied to assess other changes in the localization of proteins, for example following gene knock-outs, following metabolic perturbations, after activation of signaling pathways, after viral infection, or following any other altered conditions in cells. This provides information at a proteome-wide level that complements other high throughput approaches and could be combined with mRNA expression, protein-protein interaction, and other databases for bioinformatics analysis to provide insights into differences between cells under specific conditions. The spatial proteomics approach provides information that can be further analyzed and independently verified using complementary methods such as microscopy and molecular studies that are not readily applicable in high throughput. We envisage that spatial proteomics can be used to characterize a wide range of different cell types and can be combined with alternative fractionation techniques to analyze multiple subcellular compartments and structures. As the method is scalable and is meant to study the distribution of all cellular proteins, with increasing sensitivity of mass spectrometry-based protein detection, it should in the future be possible to extend the spatial proteomics to provide a comprehensive annotation of the localization of cell proteomes that can form an objective basis for comparison and analysis in both human cells and other model organisms.
Supplementary Material
Acknowledgments
We thank Matthias Mann and Jurgen Cox for providing us access to MaxQuant and for providing helpful discussions. We thank other members of the Lamond laboratory and colleagues with The Wellcome Trust Gene Regulation and Expression Centre for advice and suggestions.
* This work was supported in part by Proteomics Specification in Time and Space (PROSPECTS) and by Wellcome Trust Program Grant 073980/Z/03/Z.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1 and 2, cluster Figs. 3 and 6, and Tables 1–6.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1 and 2, cluster Figs. 3 and 6, and Tables 1–6.
1 The abbreviations used are:
- SILAC
- stable isotope labeling with amino acids in cell culture
- DMEM
- Dulbecco's modified Eagle's medium
- Cyto
- cytoplasm
- Nuc
- nuclei
- No
- nucleoli
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- LTQ
- linear trap quadrupole
- RNP
- ribonucleoprotein
- MT
- mock-treated
- Eto
- etoposide
- MCM
- minichromosome maintenance.
REFERENCES
- 1.Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J., Hallett M., Paiement J., Kearney R. E., Nilsson T., Bergeron J. J. (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 2.Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 3.Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., Meyer H. E., Just W., Azevedo J. E., Wanders R. J., Warscheid B. (2007) Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell. Proteomics 6, 2045–2057 [DOI] [PubMed] [Google Scholar]
- 4.Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 5.Andersen J. S., Mann M. (2006) Organellar proteomics: turning inventories into insights. EMBO Rep 7, 874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 7.Dunkley T. P., Watson R., Griffin J. L., Dupree P., Lilley K. S. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteomics 3, 1128–1134 [DOI] [PubMed] [Google Scholar]
- 8.Sadowski P. G., Dunkley T. P., Shadforth I. P., Dupree P., Bessant C., Griffin J. L., Lilley K. S. (2006) Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat. Protoc 1, 1778–1789 [DOI] [PubMed] [Google Scholar]
- 9.Gauthier D. J., Lazure C. (2008) Complementary methods to assist subcellular fractionation in organellar proteomics. Expert Rev. Proteomics 5, 603–617 [DOI] [PubMed] [Google Scholar]
- 10.Hall S. L., Hester S., Griffin J. L., Lilley K. S., Jackson A. P. (2009) The organelle proteome of the DT40 Lymphocyte cell line. Mol. Cell. Proteomics 8, 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad Y., Boisvert F. M., Gregor P., Cobley A., Lamond A. I. (2009) NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res 37, D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 13.Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 14.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 15.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 16.Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittler G., Butter F., Mann M. (2009) A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res 19, 284–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vermeulen M., Hubner N. C., Mann M. (2008) High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr. Opin. Biotechnol 19, 331–337 [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 20.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 21.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 22.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 23.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong S. E., Foster L. J., Mann M. (2003) Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130 [DOI] [PubMed] [Google Scholar]
- 25.Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 26.Lill J. (2003) Proteomic tools for quantitation by mass spectrometry. Mass Spectrom. Rev 22, 182–194 [DOI] [PubMed] [Google Scholar]
- 27.Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 28.Kumar C., Mann M. (2009) Bioinformatics analysis of mass spectrometry-based proteomics data sets. FEBS Lett 583, 1703–1712 [DOI] [PubMed] [Google Scholar]
- 29.Soza A., Knuehl C., Groettrup M., Henklein P., Tanaka K., Kloetzel P. M. (1997) Expression and subcellular localization of mouse 20S proteasome activator complex PA28. FEBS Lett 413, 27–34 [DOI] [PubMed] [Google Scholar]
- 30.Rubbi C. P., Milner J. (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldberg A. L. (1995) Functions of the proteasome: the lysis at the end of the tunnel. Science 268, 522–523 [DOI] [PubMed] [Google Scholar]
- 32.Seeger M., Ferrell K., Dubiel W. (1997) The 26S proteasome: a dynamic structure. Mol. Biol. Rep 24, 83–88 [DOI] [PubMed] [Google Scholar]
- 33.Wójcik C., DeMartino G. N. (2003) Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol 35, 579–589 [DOI] [PubMed] [Google Scholar]
- 34.Bailis J. M., Forsburg S. L. (2004) MCM proteins: DNA damage, mutagenesis and repair. Curr. Opin. Genet. Dev 14, 17–21 [DOI] [PubMed] [Google Scholar]
- 35.Demartino G. N., Gillette T. G. (2007) Proteasomes: machines for all reasons. Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 36.Motegi A., Murakawa Y., Takeda S. (2009) The vital link between the ubiquitin-proteasome pathway and DNA repair: impact on cancer therapy. Cancer Lett 283, 1–9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.