Abstract
With the aim of determining the genetic basis of metabolic regulation in tomato fruit, we constructed a detailed physical map of genomic regions spanning previously described metabolic quantitative trait loci of a Solanum pennellii introgression line population. Two genomic libraries from S. pennellii were screened with 104 colocated markers from five selected genomic regions, and a total of 614 bacterial artificial chromosome (BAC)/cosmids were identified as seed clones. Integration of sequence data with the genetic and physical maps of Solanum lycopersicum facilitated the anchoring of 374 of these BAC/cosmid clones. The analysis of this information resulted in a genome-wide map of a nondomesticated plant species and covers 10% of the physical distance of the selected regions corresponding to approximately 1% of the wild tomato genome. Comparative analyses revealed that S. pennellii and domesticated tomato genomes can be considered as largely colinear. A total of 1,238,705 bp from both BAC/cosmid ends and nine large insert clones were sequenced, annotated, and functionally categorized. The sequence data allowed the evaluation of the level of polymorphism between the wild and cultivated tomato species. An exhaustive microsynteny analysis allowed us to estimate the divergence date of S. pennellii and S. lycopersicum at 2.7 million years ago. The combined results serve as a reference for comparative studies both at the macrosyntenic and microsyntenic levels. They also provide a valuable tool for fine-mapping of quantitative trait loci in tomato. Furthermore, they will contribute to a deeper understanding of the regulatory factors underpinning metabolism and hence defining crop chemical composition.
Tomato (Solanum species) fruits constitute one of the most important sources of minerals, vitamins, fibers, and antioxidants in the human diet. Genomic approaches focused on determining the genetic basis of pathway regulation underlying quantitative variation in the production of these nutrients will likely provide information to facilitate the identification of cardinal genes. The cumulative body of data concerning developmental and metabolic shifts coupled with those recently acquired using postgenomic tools has prompted the adoption of this species as a model system for plants bearing fleshy fruits (Mueller et al., 2005). Such a model is required, because although many of these processes might be shared by many different plant species, this should not be straightly assumed, since remarkable metabolic differences have been observed even between closely related species (Fernie and Willmitzer, 2001).
The facts that the domesticated species Solanum lycopersicum can be crossed with a number of its wild relatives and that these species have shown tremendous variation in metabolite content both in leaves and fruits (Schauer et al., 2006) render wild germplasm as an important source for metabolic gene discovery (Zamir, 2001). For this reason, extensive germplasm collections, including numerous natural and induced mutants and interspecific populations, have been made publicly available by the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu/). An example of this is the collection of 76 introgression lines (ILs; Eshed and Zamir, 1995) comprising single overlapping introgressions of the Solanum pennellii (accession no. LA716) genome within the S. lycopersicum genome (cv M82). These lines are an excellent source of the stable genetic variation used worldwide by different researchers to map 2,795 quantitative trait loci (QTLs) to date, including those affecting plant biomass, yield, drought tolerance, morphology, gene expression, and metabolism (Lippman et al., 2007). The success of the S. pennellii ILs in establishing new principles for plant breeding and for resolving the molecular basis of complex traits has been demonstrated by the cloning of the first QTLs in this species: FW2.2 (a fruit size gene; Frary et al., 2000) and Brix9-2-5 (a sugar yield gene; Fridman et al., 2000, 2004). The broad phenotypic diversity inherent in this population alongside its simple mapping framework has also led to the identification of heterotic QTLs for biomass production, a major step toward isolating heterosis genes (Lippman et al., 2007). A clone-based physical map of S. pennellii, however, is necessary to fully exploit this population and to produce a unique resource for Solanaceae genomics. Such a map would also provide previous information for comparative analyses, map-based cloning, and validation of the sequence assemblies of the tomato whole genome (Solanaceae Genome Network [SGN]; www.sgn.cornell.edu).
The aim of our current research was to perform a large-scale comparative genome analysis of the S. pennellii wild tomato and S. lycopersicum. Five genomic regions (BINs) comprising more than 100 recently identified QTLs associated with fruit carbon primary metabolism (Schauer et al., 2006, 2008) were chosen. These QTLs include fruit color (Liu et al., 2003), volatile content (Tieman et al., 2006), and yield traits linked to metabolite variations found in the fruits (Eshed and Zamir, 1995; Schauer et al., 2006; Semel et al., 2006). The physical map, built by anchoring 374 clones from bacterial artificial chromosome (BAC) and cosmid S. pennellii genomic libraries to the tomato genetic map, revealed a consistent pattern of coaligning regions, thus suggesting that the two genomes can largely be considered as colinear. Targeted sequencing of 1,238,705 bp allowed the annotation of 407 genes and uncovered a high degree of microsynteny between the two species. To a lesser extent, microsynteny was also observed with the Arabidopsis genome. However, this colinearity was somewhat perturbed by differential transposable element (TE) insertion patterns, different intergenic region lengths, and extensive single nucleotide polymorphism (SNP) and insertion-deletion (InDel) polymorphism. The time of divergence between species was estimated to be 2.7 million years ago (MYA). Furthermore, the large data set presented here would constitute a useful tool for QTL fine-mapping and relatively easy screening of target clones in map-based cloning approaches.
RESULTS
Integration of Genetic and Physical Maps of S. pennellii and S. lycopersicum Genomic Regions Spanning QTLs
The S. pennellii IL population has allowed the association of more than 2,700 QTLs (for review, see Lippman et al., 2007), including many related to fruit compositional quality (Schauer et al., 2006, 2008). Based on this data set repository and with the intention of resolving the genetic determinants responsible for the observed phenotypic variations, five genomic regions were selected: BINs 1C, 2B, 4I, 7H, and 11C. These regions are associated with a total of 104 QTLs comprising traits related to fruit primary metabolite content, yield, and fruit volatile content (Eshed and Zamir, 1995; Schauer et al., 2006; Tieman et al., 2006; Table I). In order to reach the goal proposed above, an integration of the genetic and physical maps of S. pennellii and S. lycopersicum was performed. By screening two S. pennellii genomic libraries, 614 seed clones were identified using pooled overgo probes for the five selected BINs. Over 70% of the overgo probes hit at least one BAC or cosmid clone, and no major differences were observed between libraries (Supplemental Table S1). Two-dimensional pooling hybridization allowed the anchoring of 374 clones to their corresponding map positions (Table I). In addition, the map positions of 24% of these clones were confirmed by independent hybridizations to closely linked or cosegregating markers (Supplemental Table S2A).
Table I. S. pennellii BAC and cosmid clones anchored onto five genomic regions spanning fruit quantitative metabolic loci (QML) and yield-associated loci (YAL).
Metabolite QTLs and YALs listed here were previously reported by Eshed and Zamir (1995), Liu et al. (2003), Schauer et al. (2006), Semel et al. (2006), and Tieman et al. (2006).
| Genomic Region (BIN) | No. of Markers | Seed Clones | Clones Anchored by Individual Hybridization | Average Clone No. per Marker | Average Clone No. per cM | No. of End Sequences | No. of Contigs | Total Length of Nonredundant End Sequences | Clones with Confirmed Position by End Sequencing | Clones with Single Map Positions | Associated QML/YAL |
| bp | |||||||||||
| 1C (19.4 cM) | 19 | 323 (for 1C and 2B) | 49 | 2.6 | 2.5 | 78 | 7 | 51,372 | 19 | 8 | 13a |
| 2B (13.0 cM) | 25 | 132 | 5.3 | 10.1 | 173 | 21 | 91,196 | 51 | 23 | 10b/5c | |
| 4I (11.7 cM) | 19 | 81 | 55 | 2.8 | 4.7 | 71 | 6 | 43,361 | 31 | 21 | 37d/10e |
| 7H (36.0 cM) | 18 | 100 | 68 | 3.8 | 1.8 | 105 | 13 | 65,200 | 41 | 31 | 7f/2g |
| 11C (19.0 cM) | 23 | 110 | 70 | 3.0 | 3.7 | 92 | 7 | 60,966 | 19 | 17 | 12h/8i |
| Total | 104 | 614 | 374 | 519 | 54 | 312,095 | 161 | 100 | 104 | ||
| Average | 3.5 | 4.6 |
Ala, Asp, Cys, Met, oxoproline-5, S-methyl-Cys, isomaltose, phosphate, Glc-6-P, sorbitol, quinate, succinate, threonate.
Glu, S-methyl-Cys, Fru, Gal, Glc, Man, glycerol-3-phosphate, dehydroascorbate, gluconate, isocitrate.
Flowers per plant, pericarp, seed number per fruit, seed number per plant, leaf morphology.
Asn, Asp, β-Ala, γ-aminobutyric acid, Glu, Gln, Ile, Leu, Lys, Met, Pro, Fru, Fuc, Glc, Suc, phosphate, Fru-6-P, Glc-6-P, glycerol-3-phosphate, inositol-1-phosphate, stearate, palmitate, citrate, dehydroascorbate, gluconate, glycerate, isocitrate, 3-methylbutanal, 3-methylbutanol, 2-methylbutanal, 2-methylbutanol, isovaleronitrile, isobutyl acetate, pentanal, trans-2-pentenal, 1-penten-3-one, cis-2-penten-1-ol.
Plant weight, Brix, fruit color, fruit length per width, fruit width, harvest index, pericarp, seed width, style length, green fruit at harvest time.
γ-Aminobutyric acid, homoserine, Ile, Leu, Met, Ser, Glc-6-P.
Fruit length, fruit weight.
Asp, γ-aminobutyric acid, Leu, Glc, dehydroascorbate, fumarate, pentanal, 3-methylbutanal, 3-methylbutanol, 2-methylbutanal, 2-methylbutanol, isovaleronitrile.
Anther length, Brix yield, flowers per plant, fruit number, inflorescence number, pericarp per width, seed number per plant, seed weight per plant.
The average insert size of anchored clones, as estimated by pulsed-field gel electrophoresis, was 108.3 kb (125.8 and 90.8 kb for BAC and cosmid, respectively). Since the S. pennellii genome has been estimated to be approximately 1.2 Gb (Arumuganathan and Earle, 1991) and considering the total number of clones of each library (see “Materials and Methods”), we estimate that genome coverage is 5.5 and 3.8 for the BAC and cosmid libraries, respectively, with an average of 4.7×. Thus, the anchored 374 BAC/cosmid clones represent around 1% of the wild tomato genome and approximately 10% of the physical distance of the selected regions. However, these values should be taken with caution, since they rely on a genetic map constructed with less than 100 individuals (Tomato-EXPEN 2000, S. lycopersicum LA925 × S. pennellii LA716; http://www.sgn.cornell.edu/).
To support the integration of the wild species clones with both the S. lycopersicum genomic sequence and the available genetic map, the 374 anchored BAC and cosmid clones were end sequenced. Both end sequences were obtained for 233 clones, only one end sequence reached a suitable quality criteria in 53 of the clones, and no high-quality sequence was produced for 88 clones. Thus, after quality and vector trimming, a total of 519 end sequences, representing 76% of all anchored clones, were obtained and subjected to further analyses. After masking repetitive elements, the sequences from clones anchored with the same marker were assembled into 54 contigs (Table I; Supplemental Table S2B). These analyses rendered a total of 436 nonredundant end sequences (312,095 bp).
To support the above analysis, S. pennellii clones were selected for full-length sequencing using the following criteria: (1) PCR confirmation for the presence of candidate genes previously identified by our group (Bermúdez et al., 2008); (2) confirmation of physical map position by end sequence anchoring; and (3) availability of information from orthologous S. lycopersicum genomic regions. On the basis of these criteria, nine clones were sequenced: one clone anchored to BIN 1C, two to BIN 2B, one to BIN 4I, four to BIN 7H, and one to BIN 11C. Insert lengths ranged from 50,564 to 148,978 bp. Map positions and clone identifiers are available in Supplemental Table S2A.
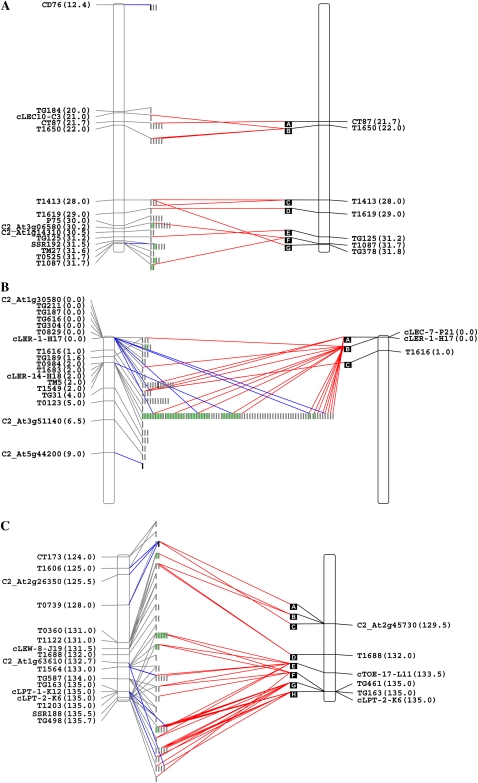
The full-length sequencing of these clones resulted in a total of 926,691 bp of sequence, which, together with the end sequences described above, encompasses a total of 1,238,786 bp. This sequence volume represents 1.5% of the five genomic regions under study and about 0.1% of the entire S. pennellii genome. The complete data set, including hybridization anchored clones, masked nonredundant end sequences, and large insert clone sequences, was used to build a physical map. Figure 1 shows a graphical representation of the relationships found between the S. lycopersicum genetic map and the anchored clones of the wild species. The identity of S. pennellii sequences to mapped markers (blue lines) and to S. lycopersicum genomic sequence (red lines) allowed the determination of orthologous regions between the two species for 43% of the anchored clones.
Figure 1.
Integrated genetic and physical maps of selected genomic regions of S. pennellii and S. lycopersicum. Genetic markers and their corresponding map positions (according to the Tomato-EXPEN 2000 map) are shown on the left and right sides of the S. pennellii and S. lycopersicum chromosome fragment representations, respectively. S. pennellii anchored BAC/cosmid clones are represented by gray bars. Clones belonging to the same contig are grouped into green boxes. Full-length sequenced clones are represented by black bars. S. lycopersicum clones are represented as black squares ordered alphabetically. Gray lines link S. pennellii BAC/cosmid clones with their corresponding hybridizing genetic markers. Blue lines indicate the S. pennellii BAC/cosmid end sequences containing the corresponding marker sequences. Red lines link S. pennellii BAC/cosmid end sequences matching S. lycopersicum BAC clone sequences. Black lines link S. lycopersicum BAC clones to their corresponding markers. The entire data set is available at the SGN Web site in the Tomato Physical Map interface (http://www.sgn.cornell.edu/cview/). A, BIN 1C. B, BIN 2B. C, BIN 4I. D, BIN 7H. E, BIN 11C. [See online article for color version of this figure.]
At first glance, the topology of the maps of these five genomic regions showed that the S. pennellii genome can be considered colinear with that of the domesticated tomato. This can clearly be observed in Figure 1 by the density of the lines linking S. pennellii with S. lycopersicum clones and their positions on the map. However, it is possible to identify a few regions where red lines connect distant S. pennellii clones to a single S. lycopersicum one and vice versa (i.e. the interval 93–108 centimorgan [cM] in BIN 7H; Fig. 1D). These features suggest the occurrence of rearrangements such as chromosomal inversions and/or InDels. Another interesting feature of the map topology is apparent in the junction of the red lines linking several distant S. pennellii clones to narrower intervals within the S. lycopersicum genome. The S. pennellii clones anchored to a 10.7-cM interval (T1606-TG498 markers) on BIN 4I matched S. lycopersicum BACs (A–H) that were anchored with markers covering 5.5 cM (C2_At2g45730-cLPT-2-K6) according to the Tomato-EXPEN 2000 map (Fig. 1C). Similarly, on BIN 11C, several S. pennellii BAC clones anchored within an interval of 3.0 cM (SSR76-C2_At1g44790) matched a single S. lycopersicum BAC (A) anchored with the T1161 marker at 38.0 cM (Fig. 1E). Although this result is suggestive of a genome expansion event in S. pennellii, as reflected by the relative genome sizes of these species, it is not possible to formally exclude that this colinearity break reflects the low resolution of the map and/or the current relative paucity of information concerning the S. lycopersicum genomic clones in the regions studied here.
The number of anchored clones per marker (3.5; Table I) is in general agreement with the libraries' redundancy (4.7×). However, the cLER-14H18 marker anchored 76 S. pennellii clones (BIN 2B; Fig. 1B), while only eight S. lycopersicum clones were identified (Supplemental Table S1). End sequence analyses and contig assemblies of S. pennellii clones revealed that most of them span the anchoring marker or a cosegregating one, thus confirming their map positions. Furthermore, these clones hit only two contiguous S. lycopersicum BACs and harbor ribosomal genes and TEs. Taken together, these results suggest that a local amplification may well have occurred in the S. pennellii genome.
Annotation and Functional Categorization of S. pennellii Coding Sequences
The significant number of sequences generated, together with the fact that they are linked to physiologically relevant QTLs, renders this information an untapped resource for exposing known and novel genes that could be involved in controlling these traits. For this reason, an exhaustive annotation was attempted for the 436 nonredundant end sequences and the nine BAC/cosmid clones. From the nonredundant end sequences, a total of 298 putative genes were identified and assigned to functional categories (Supplemental Table S3). From the fully sequenced BAC/cosmid clones, 109 complete open reading frames were identified by two approaches: (1) utilizing the FGENESH (Salamov and Solovyev, 2000) and Augustus (Stanke et al., 2004) prediction softwares, and (2) manual confirmation by the cross-match interface of the phrap program and BLAST comparison (Supplemental Table S4). A detailed analysis of the large-insert sequence annotation revealed comparable gene density values of 0.1 genes kb−1 in BINs 1C, 2B, 7H, and 11C. However, BIN 4I displayed 0.2 genes kb−1 and the lowest density of TEs (Supplemental Tables S4 and S5). The annotated sequences are representative of unigenes derived from cDNA libraries from a broad spectrum of tomato tissues and physiological conditions. Moreover, 103 of the 109 predicted open reading frames through the five genomic regions matched Arabidopsis genes, revealing a high degree of conservation in the gene repertories of the two species. Furthermore, with the exception of the BAC analyzed from chromosome 1, in all other clones, 13 microsyntenic segments were identified between the wild tomato species and Arabidopsis (Arabidopsis thaliana), which are highlighted in color in Supplemental Table S4. These regions encompass 54 genes that span 218,060 nucleotides representing about 25% of the annotated large-insert clone sequences.
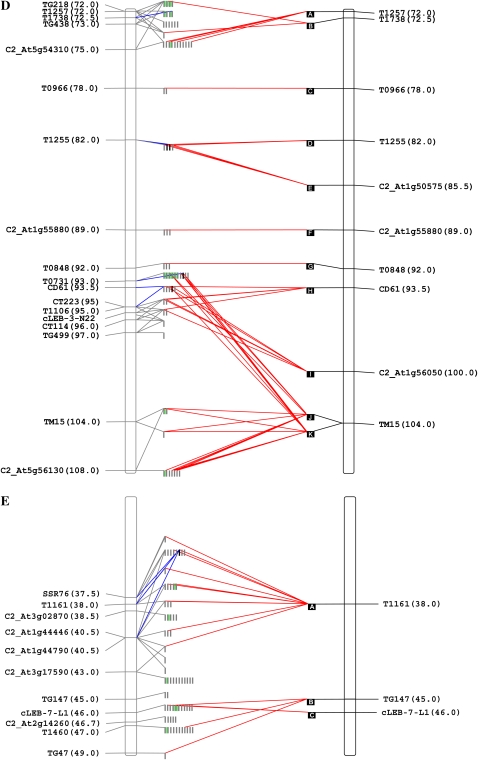
The 407 annotated genes were further subjected to functional categorization using the MapMan ontology adapted for solanaceous species by Urbanczyk-Wochniak et al. (2006). These genes fell into 23 of the 35 gene categories, and their distribution along the five genomic regions is shown in Figure 2. Total sequence percentage per BIN that fell into any functional category varied between 51% and 84% for BINs 11C and 4I, respectively. The number of gene classes per BIN ranged from nine in BIN 11C to 19 in BIN 7H, revealing differences in gene diversity among BINs (Fig. 2). With the exception of region B on chromosome 2, “unknown function” (category 35) was the most represented category for all BINs. The second two most abundant categories, “DNA modifiers” (category 28) and “protein modification” (category 29), are those comprising ribosomal proteins and TEs, respectively. In all the BINs analyzed, a significant percentage of predicted coding regions fell into the “RNA regulation” category (category 27), which includes regulatory proteins such as MYB, WRKY, and zinc finger transcription factors. Several MapMan functional classes were found exclusively in certain genomic regions. For example, genes falling into the “major carbohydrates” category (category 2) were found only in regions of chromosomes 2 and 4. “TCA [tricarboxylic acid] cycle/organic acid transformations” and “cofactor and vitamin synthesis” categories (categories 8 and 18) were found only in BIN 1C, while “redox regulation” (category 21) was found exclusively in BINs 4I and 7H. Genes related to “transport” (category 34) were found only in BINs 4I, 7H, and 11C. The same was found for genes related to “lipid metabolism” (category 11) and “tetrapyrrole synthesis” (category 19), which were present only in BIN 7H and 11C, respectively.
Figure 2.
Functional categorization of S. pennellii genomic sequences. BAC/cosmid end (singletons and contigs) and full-length sequences were categorized according to MapMan software (http://mapman.mpimp-golm.mpg.de). Categories are as follows: 1, photosynthesis; 2, major carbohydrates; 3, minor carbohydrates; 7, oxidative pentose phosphate pathway; 8, tricarboxylic acid cycle/organic acid transformations; 10, cell wall; 11, lipid metabolism; 13, amino acid metabolism; 16, secondary metabolism; 17, hormones; 18, cofactor and vitamin synthesis; 19, tetrapyrrole synthesis; 20, stress; 21, redox regulation; 26, miscellaneous enzyme families; 27, RNA regulation; 28, DNA modifiers; 29, protein modifications; 30, signaling; 31, cell; 33, development; 34, transport; 35, unknown function. [See online article for color version of this figure.]
SNP and InDel Discovery
Due to the importance of SNP and InDel discovery in genetic mapping, the number and density of these differences were evaluated within the 436 nonredundant end sequences and their corresponding S. lycopersicum orthologous regions (Supplemental Table S2C). Polymorphisms along each identified alignment were analyzed following stringent criteria: (1) nucleotide variations were regarded as reliable SNPs only if the quality of the base call achieved a quality value (Qv) ≥ 20; and (2) end sequences with only one S. lycopersicum matching region were used. This allowed the comparison of 59,510 bp between both species. SNPs varied among genomic regions, ranging from 2.1 per 100 bp in BIN I of chromosome 4 to 9.7 per 100 bp in BIN B of chromosome 2, with an average of 4.3 (Supplemental Table S2C), according to the values expected for these plant species (L. Mueller, personal communication). In the same regions, the distribution of InDels also varied between 0.37 per 100 bp in BIN 4I and 0.5 per 100 bp in BIN 1C. A graphical overview of SNP and InDel mapping revealed that there were no particular patterns of polymorphism within the analyzed chromosomal regions (Supplemental Fig. S1). A high proportion of the SNPs and InDels identified were detected within predicted coding regions (Supplemental Table S3). In the case of the analyzed region of chromosome 2 (which showed the highest percentage of polymorphisms), all the SNPs and InDels detected fell within three gene categories: 29 (protein modification), 35 (unknown function), and 28 (DNA modifiers).
Microsynteny and Evolutionary Analyses of S. pennellii and S. lycopersicum Orthologous Regions
Large-insert clones from S. pennellii were compared with the corresponding conserved syntenic segments of S. lycopersicum in order to explore both the differences in genome structure and the sequence divergence between these closely related species. Although this work embraces the study of five genomic regions, the following analyses were focused solely on BINs 4I, 7H, and 11C because they are the only ones that do not span repetitive sequences and for which unambiguous orthologous sequences from S. lycopersicum were available.
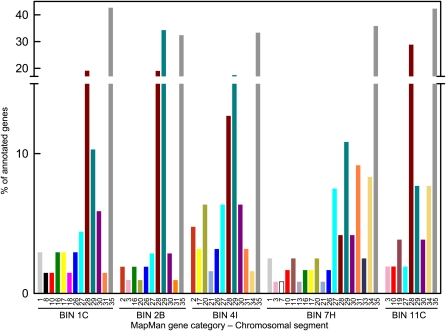
We observed that the syntenic regions contain 38 distinct genes showing conserved genomic ordering, orientation, and gene structure (exon/intron) between both species and that this nearly perfect colinearity is slightly altered by intergenic regions of variable size and a differential pattern of TE insertions (Fig. 3; Supplemental Table S5). An analysis of the distribution of polymorphisms revealed, as expected, a higher percentage of InDels in intergenic regions than in genic ones (Fig. 3) and in introns as opposed to exons (Table II). The degree of conservation for each gene was evaluated by estimating the rate of synonymous and nonsynonymous substitution distances (ds/dn). This analysis was carried out for all genes that fulfilled the following criteria: (1) they displayed full-length coding sequences (thus, uncompleted genes located on the BAC/cosmid ends were excluded); (2) they exhibited no frameshifts or stop codons (gene Sp07gCP5C04.8 was excluded); (3) they had Arabidopsis orthologs; and (4) they showed homogeneous relative substitution rates between S. pennellii and S. lycopersicum, considering Arabidopsis as an outgroup. For all 27 analyzed genes, the ratio ds/dn was greater than 1, thus indicating the absence of positive selection (Table II). A test of selection was performed in order to assess whether genes were undergoing neutral (ds = dn) or purifying (ds > dn) selection, revealing that both cases were present. Interestingly, all the genes that displayed nonstatistically significant purifying selection between S. pennellii and S. lycopersicum also displayed amino acid polymorphisms. It is important to note that the ds/dn ratio may be subject to misinterpretation if the codon usage is restricted; however, for the data set used in this work, no codon bias was observed for any gene in all three species analyzed.
Figure 3.
Comparative analysis between S. lycopersicum and S. pennellii orthologous genomic regions. Genes are indicated as blue arrows and coded according to Table II. Green blocks depict transposable elements. Green blocks squared in black correspond to retrotransposons, and their LTRs are indicated as black bars at the ends of the elements. The percentage identity and InDels (in parentheses) between genotypes along genic (including untranslated regions) and intergenic regions are indicated. Compared orthologous genomic regions of BIN 4I (A), 7H (B), and 11C (C) are presented. [See online article for color version of this figure.]
Table II. Comparative analysis between S. lycopersicum and S. pennellii orthologous genomic regions.
Sl, S. lycopersicum; Sp, S. pennellii; At, Arabidopsis; CB, total of compared bases; ND, not determined; UD, undetermined because dn = 0.
| BIN | IDa | Nameb | Identity (InDels)c CB: XXX |
Exonsd | Lengthe (Amino Acids) | Amino Acid Polymorphismsf |
ds/dng |
|||
| Exons | Introns | Sl-Sp | Sl-At | Sp-At | ||||||
| 41 | Sp04gBP4C04.1 | Diphthine synthase | 99 (0) CB: 100 | 1h | 33i | 0 | ||||
| Sp04gBP4C04.2 | Putative chaperone protein DNAJ-related | 99 (0) CB: 480 | 95.3 (2.7) CB: 2,224 | 5 | 159 | 2 | 5.3 | 7.2j | 7.5j | |
| Sp04gBP4C04.3 | Expressed gene 1 | 98.6 (0) CB: 633 | 94.9 (1.1) CB: 2,077 | 7 | 179 | 4 | ||||
| Sp04gBP4C04.4 | Abscisic acid-responsive HVA22 family protein | 98.4 (0.0) CB: 966 | 96.0 (1.5) CB: 1,202 | 6 | 321 | 9 | 1.83 | 2.23j | 2.15j | |
| Sp04gBP4C04.5 | E2F transcription factor | 98.8 (0) CB: 996 | 94.3 (3.3) CB: 5,294 | 10 | 331 | 8 | ||||
| Sp04gBP4C04.6 | Putative eukaryotic initiation factor 3 γ-subunit family protein | 98.7 (0) CB: 1,341 | 95.3 (3.1) CB: 4,320 | 13 | 446 | 7 | 3.0 | 3.12j | 3.11j | |
| Sp04gBP4C04.7 | Putative zinc finger (C2H2 type) family protein | 97.6 (0.4) CB: 1,359 | 91.4 (5.0) CB: 1,865 | 4 | 451 | 13 | 3.8j | 3.9j | 3.9j | |
| Sp04gBP4C04.8 | Plastidic hexokinase | 99.2 (0) CB: 1,500 | 88.8 (8.1) CB: 2,111 | 9 | 499 | 2 | 11j | 4.6j | 4.6j | |
| Sp04gBP4C04.9 | Putative clathrin assembly protein AP17-like protein | 100 (0) CB: 429 | 73.1(25.2) CB: 3,113 | 6 | 142 | 0 | UD | 17.8j | 17.8j | |
| Sp04gBP4C04.10 | Expressed gene 2 | 96.5 (0.6) CB: 909 | 1 | ND | ND | |||||
| Sp04gBP4C04.11 | Expressed gene 3 | 98.3 (0.3) CB: 1,851 | 96.7 (1.6) CB: 1,684 | 10 | 608 | 9 | ||||
| Sp04gBP4C04.12 | Putative transcription coactivator-like protein | 99.4 (0) CB: 507 | 96.2 (2.2) CB: 1,604 | 5 | 168 | 0 | UD | 8.7j | 8.7j | |
| Sp04gBP4C04.13 | β-Fructofuranosidase | 99.2 (0) CB: 1,713 | 91.2 (6.1) CB: 2,008 | 4 | 570 | 1 | 26j | 8.0j | 7.7j | |
| Sp04gBP4C04.14 | Putative cytomatrix protein | 99 (0) CB: 1,062 | 94 (3.8) CB: 1,796 | 4 | 353 | 8 | 1.1 | 1.3 | 1.3 | |
| 7H | Sp07gCP5C04.1 | Glycerophosphoryl diester phosphodiesterase family protein | 98.8 (0.0) CB: 162 | 95.9 (1.9) CB: 1,025 | 2h | 54i | 2 | |||
| Sp07gCP5C04.2 | Protease inhibitor/seed storage/lipid transfer protein family protein | 97.8 (0.0) CB: 315 | 1 | 104 | 5 | 1.04 | 1.99j | 1.95j | ||
| Sp07gCP5C04.3 | Polygalacturonase-inhibiting protein 1 (PGIP1) | 97.7 (0.0) CB: 984 | 1 | 327 | 6 | 5.8j | 4.1j | 3.6j | ||
| Sp07gCP5C04.4 | Similar to fimbrin-like protein | 98.8 (0.0) CB: 2,571 | 97.0 (1.5) | 12 | 856 | 0 | UD | 7.3j | 7.4j | |
| Sp07gCP5C04.5 | Expressed gene 1 | 96.9 (1.3) CB: 669 | 1 | ND | ND | |||||
| Sp07gCP5C04.6 | Wax synthase isoform 3 | 86.8 (11.1) CB: 1,053 | 1 | 350 | 53 | 2 | 2.07j | 2.06j | ||
| Sp07gCP5C04.7 | Wax synthase isoform 3 | 98.4 (0.0) CB: 1,035 | 1 | 344 | 10 | 1.9 | 2.1j | 2.0j | ||
| Sp07gCP5C04.8 | Cell division protein FtsZ | 99.0 (0.0) CB: 1,254 | 95.1 (2.2) CB: 3,438 | 6 | 417 | 1 | 15.5j | 6.0j | 6.3j | |
| Sp07gCP5C04.9 | TATA-binding protein-associated factor | 99.3 (0.0) CB: 1,734 | 96.5 (1.8) CB: 2,580 | 12 | 577 | 9 | 0.87 | 2.9j | 2.9j | |
| Sp07gCP5C04.10 | Similar to syntaxin 52 (SYP52) | 94.8 (4.8) CB: 735 | 94.9(2.8) CB: 2,427 | 5 | 243 | 11 | ||||
| Sp07gCP5C04.11 | Ser/Thr protein phosphatase | 99.5(0.0) CB: 918 | 96.1 (1.1) CB: 5,494 | 8 | 305 | 1 | 8.0 | 29.8j | 32.6j | |
| Sp07gCP5C04.12 | Lipid-binding serum glycoprotein family protein | 98.8 (0.0) CB: 1,302 | 95.1 (2.3) CB: 2,731 | 5 | 433 | 8 | 1.9 | 2.1j | 2.2j | |
| Sp07gCP5C04.13 | PHD finger family protein | 98.1 (0.1) CB: 2,184 | 98.2 (0.3) CB: 1,150 | 3 | 727 | 24 | ||||
| Sp07gCP5C04.14 | S-Adenosyl-l-Met:carboxyl methyltransferase family protein | 99.1 (0.0) CB: 1,155 | 80.1 (11.7) CB: 1,222 | 4 | 384 | 3 | 5.5 | 7.0j | 6.6j | |
| Sp07gCP5C04.15 | Hyp-rich glycoprotein family protein | 96.5 (1.2) CB: 964 | 91.8 (3.1) CB: 1,397 | 3h | 295i | 23 | ||||
| 11C | Sp11gBP4E08.1 | Tetratricopeptide repeat-containing protein | 98.7 (0.0) CB: 476 | 97.0 (1.6) CB: 507 | 3h | 158i | 3 | |||
| Sp11gBP4E08.2 | Expressed gene 2 | 99.4 (0.0) CB: 360 | 93.8 (4.3) CB: 5,444 | 2 | 119 | 1 | 2.7 | 2.9j | 3.0j | |
| Sp11gBP4E08.3 | Glc-6-P/phosphate translocator-related | 98.2 (0.0) CB: 1,014 | 93.8 (1.2) CB: 1,376 | 5 | 337 | 9 | 2.6 | 7.03j | 7.04j | |
| Sp11gBP4E08.4 | Peptidoglycan-binding LysM domain-containing protein | 98.9 (0.0) CB: 1,237 | 94.7 (2.2) CB: 1,692 | 5 | 413 | 11 | 1.2 | 4.1j | 4.2j | |
| Sp11gBP4E08.5 | Coiled-coil protein | 98.8 (0.0) CB: 2,013 | 96.0 (2.8) CB: 2,106 | 4 | 670 | 11 | 3.8j | 1.5j | 1.5j | |
| Sp11gBP4E08.6 | CH1 (chlorophyll b biosynthesis); chlorophyllide a oxygenase | 98.9 (0.0) CB: 1,612 | 95.4 (1.5) CB: 2,203 | 9 | 536 | 2 | 16.5j | 6.6j | 6.8j | |
| Sp11gBP4E08.7 | BTB/POZ domain-containing protein | 99.8 (0.0) CB: 987 | 95.1 (2.7) CB: 1,812 | 4 | 329 | 1 | 4.0 | 6.5j | 6.3j | |
| Sp11gBP4E08.8 | F-box family protein | 99.3 (0.0) CB: 990 | 96.5 (1.1) CB: 7,288 | 11 | 330 | 3 | 3.9 | 6.1j | 6.1j | |
| Sp11gBP4E08.9 | Putative replication factor C 37-kD subunit | 98.4 (0.0) CB: 1,020 | 78.6 (16.8) CB: 10,653 | 12 | 340 | 1 | 54.0j | 8.0j | 7.6j | |
Gene identification.
Gene functional identity.
Percentage of identity and InDels between the S. lycopersicum and S. pennellii regions analyzed.
Number of exons compared between both genotypes.
Number of amino acids compared between both genotypes.
Polymorphic amino acids on the predicted protein sequence.
Synonymous distance/nonsynonymous distance.
Exons not included in the comparative evolutionary analysis of S. pennellii and S. lycopersicum orthologous regions.
Only partial sequences were compared because genes were located at the end of the clones for any of the genotypes.
Statistically significant purifying selection (P < 0.05).
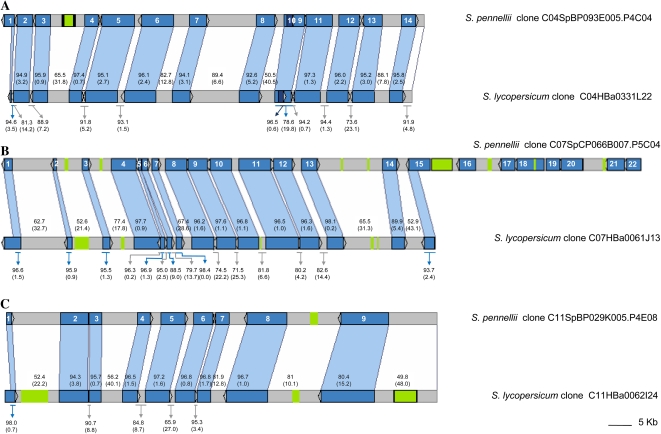
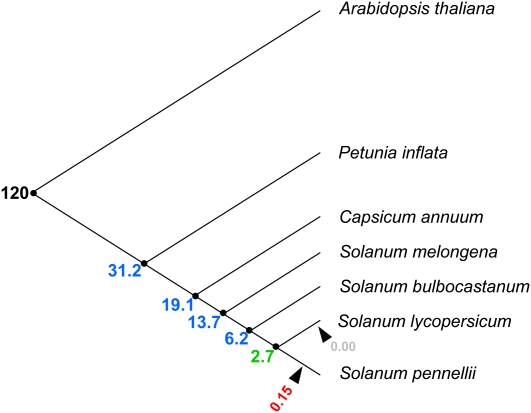
Since in this work, 27 loci belonging to three different chromosomes of S. pennellii were sequenced and the corresponding orthologs from S. lycopersicum were identified, these data represent a unique source to investigate the divergence time between these closely related tomato species. Using the fossil record estimation of 120 MYA for the divergence between Arabidopsis and Solanaceae (Bell et al., 2005; Magallon and Sanderson, 2005) and the ds of the concatenated coding sequences, the genomic region-specific substitution rates and the species divergence time were estimated. Analysis of the substitution rate revealed no significant differences between BINs, suggesting that the three regions analyzed may code for important conserved proteins (Table III). When all three regions are considered together, the species divergence time was estimated at 2.7 MYA (Fig. 4).
Table III. Estimation of divergence time between S. lycopersicum and S. pennellii.
| BIN | S. lycopersicum BACa | S. pennellii BAC/Cosmidb | Substitution Ratec | Divergence Timed |
| 4Ie | C04HBa0331L22 | C04SpBP093E005.P4C04 | 4.26 × 10−09 | 2,936,857 (±690,736) |
| 7Hf | C07HBa0061J13 | C07SpCP066B007.P5C04 | 4.44 × 10−09 | 2,700,421 (±661,591) |
| 11Cg | C1HBa0062I24 | C11SpBP029K005.P4E08 | 4.40 × 10−09 | 2,838,.221 (±667,537) |
| 4I/7H/11C | All S lycopersicum genes | All S. pennellii genes | 4.37 × 10−09 | 2,744,163 (±448,205) |
S. lycopersicum concatenated coding regions.
S. pennellii concatenated coding regions.
Region-specific substitution rate (substitutions per site per year) calculated according to the fossil record estimation of 120 MYA for the divergence between Arabidopsis and Solanaceae (Bell et al., 2005; Magallon and Sanderson, 2005).
Species divergence time in MYA (±confidence interval).
Concatenated coding regions of genes 2, 4, 6, 7, 8, 9, 12, 13, and 14.
Concatenated coding regions of genes 2, 3, 4, 6, 7, 8, 9, 11, 12, and 14.
Concatenated coding regions of genes 2 to 9.
Figure 4.
Species divergence time in MYA according to Bell et al. (2005; black), Magallon and Sanderson (2005; black), and Wang et al. (2008; blue). S. pennellii and S. lycopersicum divergence (green) was estimated using a molecular clock. SHACOP_I_MT (red) and TOPSCOTCH_LP_I (gray) retrotransposon insertions are indicated by arrows. [See online article for color version of this figure.]
Two Copia-like long terminal repeat (LTR) retrotransposons with perfect tandem duplication sites were found among the genomic regions analyzed in this work. A SHACOP_I_MT in BIN 4I was present only in S. pennellii (Supplemental Table S5), and a TOPSCOTCH_LP_I in BIN 11C was found only in S. lycopersicum. Based on the molecular clock, their insertion times were estimated and, as expected, both elements appeared after the species split (Fig. 4), in agreement with the species divergence time estimated above.
DISCUSSION
In the study described herein, we performed a large-scale analysis of the S. pennellii genome and compared it with that of its cultivated tomato counterpart S. lycopersicum. To achieve this goal, different approaches were taken: (1) a macrosynteny analysis via the construction of a physical map; (2) sequencing, annotation, and functional categorization of a portion of the wild tomato genome; (3) polymorphism evaluation between these species; and (4) a microsynteny analysis aimed at exposing the variation that has driven the evolutionary change between these two species. Data compilation was focused on five genomic regions: BINs 1C, 2B, 4I, 7H, and 11C, spanning 104 QTLs associated with fruit carbon primary metabolism (Schauer et al., 2006, 2008), fruit color (Liu et al., 2003), volatile content (Tieman et al., 2006), and yield traits linked to metabolite variations found in the fruits (Eshed and Zamir, 1995; Schauer et al., 2006; Semel et al., 2006). Although the high number of QTLs in the studied regions may merely reflect the effect of a few QTLs on several related traits, rather than the existence of multiple independent QTLs, the analyses of the generated data set will serve as a bridge for linking the genome to phenotypes for basic and applied studies.
Comparative Physical Map between S. pennellii and S. lycopersicum
Due to its versatility and low cost, physical mapping has become a natural component of large genome-sequencing endeavors (Gregory et al., 2002; Wallis et al., 2004). In fact, this strategy was originally adopted for the tomato genome-sequencing project (Mueller et al., 2009). Here, a comparative physical map was built anchoring 374 S. pennellii BAC/cosmid clones, which resulted in approximately 10% of the physical distance coverage of the five selected regions and 1% of the total wild tomato genome. Across the five regions studied, 58 S. lycopersicum BAC clones were fully sequenced and available on the SGN Web site. The physical map presented here anchored 31 of these clones to S. pennellii sequences, representing reliable coverage of 53% of the sequence information available for the reference genome.
Overall, the topology of the physical map built here reveals a high level of colinearity between S. pennelli and the cultivated tomato genomes. However, this colinearity is based on the quality of the S. lycopersicum physical map and on the genetic distances between the markers of the S. lycopersicum × S. pennellii reference map, which are far from perfect, especially considering that the latter was constructed with fewer than 100 individuals. However, two independent results support the accuracy of the S. pennelli physical map. First, 24% of the initially anchored clones were further localized at the same position by hybridization with another closely linked or cosegregating marker. Second, the positions of 43% of the anchored clones were confirmed by sequence identity to both mapped markers and to S. lycopersicum genomic sequences.
The hypothesis of Solanaceae genome colinearity has been proposed several times based on comparative genetic mapping experiments, which yielded evidence for the conservation of gene repertoire and colinear chromosome segments for related species (Wang et al., 2008). Particular features of the physical map presented here suggest the presence of local breaks in the colinearity of the regions analyzed. For example, the instances wherein S. pennelli and S. lycopersicum clones are distant from each other are suggestive of chromosomal inversion and/or InDels. Moreover, some S. pennellii genomic intervals seem to be compressed in S. lycopersicum, hinting at the expansion of specific S. pennellii regions, which is in agreement with the larger genome size estimation for this species (Arumuganathan and Earle, 1991). The results of microsynteny analysis, which revealed larger intergenic regions in S. pennellii than in S. lycopersicum, reinforce this statement. These rearranged regions, together with the screening for SNPs and InDels, will likely facilitate the identification of polymorphic markers.
As reported by Zhao et al. (2002), the sequence of 114 BAC clones from the taxonomically related rice subspecies Oryza sativa indica ‘Guangluai 4’ was particularly useful for gap closing of sequence gaps of rice chromosome 4. Although a significant number of InDels were detected between the two tomato species, the S. pennellii BAC/cosmid clones anchored in this work allowed the detection of 64 extension clones (those matching the S. lycopersicum genome at only one end) and another 21 clones filling in gaps between two S. lycopersicum clones (those matching two nonoverlapping S. lycopersicum clones of the same region). Together with the information obtained from the anchoring results described above, all these sequences provide useful tools for comparative genome analyses, map-based cloning, validation of genome sequence assemblies, and an easier access to the regulatory sequences surrounding important genes.
Annotation of S. pennellii Coding Sequences
After annotation of the 436 nonredundant end sequences and the nine BAC/cosmid clones, 407 genes were identified. In agreement with all large-scale plant genome analyses, our results for large-insert sequences display an inverse distribution between genes and TE-containing regions (Arabidopsis Genome Initiative, 2000; Paterson et al., 2009). In particular, the gene density along BIN 4I (0.2 genes kb−1) doubles that of the rest of the genomic regions analyzed (0.1 genes kb−1), while it displays the lowest density of TEs. Gene identification by end sequence data analysis also revealed BINs 4I and 7H as the most gene-enriched regions. These gene density values are higher than those previously estimated for S. lycopersicum heterochromatin (Van der Hoeven et al., 2002; Wang et al., 2008; Mueller et al., 2009), although it should be noted that they may reflect a bias considering the high abundance of QTLs in these genomic regions.
Focusing on a region of tomato chromosome 7 of about 57 kb, Rossberg et al. (2001) found the same arrangement of five predicted genes as that found in a region of Arabidopsis chromosome 1. This kind of colinearity with Arabidopsis has also been reported for a region of tomato chromosome 2 harboring the major fruit-shape locus ovate with Arabidopsis chromosome 4 (Ku et al., 2001) and between a tomato centromeric BAC of chromosome 6 harboring the FER locus and three distinct regions on Arabidopsis chromosomes 2, 3, and 5 (Guyot et al., 2005). With the exception of the case of BIN 1C, our results from BAC/cosmid full-length sequencing revealed several examples of microsynteny between the S. pennellii and Arabidopsis genomes. These results, together with those recently published from genome-wide approaches in Arabidopsis detecting a few genetic “hot spots” exhibiting major effects on hundreds of QTLs (Lisec et al., 2008; Fu et al., 2009), open new routes to explore further relations between common pathways regulating metabolism in these two species.
Concerning gene categorization, the number of gene classes per BIN ranges between nine for BIN 11C and 19 for BIN 7H, suggesting a functional compartmentalization of the genome, at least for the analyzed regions. Among all the identified genes, several plausible candidates that could directly underlie previously reported metabolic QTLs can be identified. However, the analyzed sequences represent only 10% of these regions, and the participation of these genes defining the metabolic QTLs remain to be functionally proven. A combination of the generation of recombinant lines harboring smaller introgressions of the S. pennellii genome and reverse genetic approaches is under way at present to assess the candidature of these genes.
Microsynteny and Evolutionary Analysis of Orthologous Regions
Domestication of crops is arguably the most dramatic recent event in plant evolution. Evidence indicates that most of the anatomical changes selected during domestication are determined by the modification of gene expression patterns of a few regulatory loci (Doebley et al., 1997). This is the case, for example, of the fw2.2 locus, responsible for the fruit weight variation between wild and cultivated tomato (Frary et al., 2000). Moreover, the large- and small-fruit alleles do not differ in protein sequence but in transcription levels due to promoter polymorphisms (Nesbitt and Tanksley, 2002). With the putative fruit-related loci analyzed in this work, a slightly different panorama is observed when comparing S. pennellii and S. lycopersicum. A total of 277,534 bases were compared between both species. A total of 32 out of the 38 completed genes identified displayed amino acid polymorphisms, and 16 out of the 27 genes with estimated ds/dn displayed neutral selection. Even the eight genes with significant purifying selection showed amino acid polymorphisms. These results indicate that coding regions have accumulated polymorphisms since both species diverged. With the exception of a cytomatrix-encoding protein (Sp04gBP4C04.14), all the coding regions displayed significant purifying selection between tomato and Arabidopsis, thus indicating that the analyzed genes have a conserved function.
Using an estimated substitution rate of 6.03 × 10−9 and four loci, Nesbitt and Tanksley (2002) reported that S. pennellii and S. lycopersicum diverged 7.0 MYA. More recently, Wang et al. (2008) reported the analysis of sequences for an unduplicated conserved syntenic segment in the genomes of five Solanaceae species (petunia [Petunia hybrida], pepper [Capsicum annuum], eggplant [Solanum melongena], potato [Solanum tuberosum], and tomato). The results indicated that the last common ancestor of these species lived approximately 31.2 MYA and that the divergence of tomato and potato had occurred approximately 6.2 MYA. In this study, a more precise calculation using specific substitution rates and genetic distances of 27 different loci along three different chromosomes allows us to estimate the divergence between S. pennellii and S. lycopersicum at 2.7 MYA. Since S. pennellii is one of S. lycopersicum's most distantly related wild species, our results indicate that the lycopersicum species of the genus Solanum began its radiation more recently than previously estimated. Two different Copia-like LTR retrotransposons were found through the analyzed regions. Their corresponding insertion dates were estimated, and they were shown to have occurred after both tomato species diverged. Recently, several reports have demonstrated the impact of TEs on important agronomical traits. As illustrative examples, we can mention the effects of the insertion of retrotransposons in β-carotene accumulation in cauliflower (Brassica oleracea) plastids (Lu et al., 2006) and in gene duplication causing morphological variations of tomato fruit (Xiao et al., 2008). In this direction, work is in progress to further investigate whether the recently inserted retrotransposons identified here could be involved in differential gene expression patterns between S. pennellii and S. lycopersicum.
CONCLUSION
In this work, we integrated physical and genetic maps of selected regions of two tomato species, S. lycopersicum and S. pennellii. Together with BAC/cosmid end sequencing, we detected a number of putative SNPs and InDels that could be particularly useful as a screening tool in fine-mapping approaches and that, following experimental validation, could be of considerable practical relevance in breeding programs. Moreover, they will aid in filling marker-poor segments of the genome and in understanding the relationship between levels of crossing over per physical distance and levels of polymorphism in Solanaceae species. Targeted sequencing of BAC/cosmid clones, gene annotation, and comparative analyses suggest that changes in both coding and noncoding sequences could be implicated in the phenotypic diversity within the genus. In addition, a precise estimation of S. pennellii and S. lycopersicum divergence time indicates that tomato radiation occurred 2.7 MYA.
The framework of the physical map, sequence assembly, and novel candidate genes presented here provides a considerable collection of resources for tomato research, especially considering the 104 QTLs that have previously been mapped to the genomic regions analyzed. Together with previous genomic sequences of S. pennellii candidate genes (Bermúdez et al., 2008), these resources constitute a valuable data set repository of genomic information for this Solanaceae species. Work is in progress to further characterize the functions of several of the novel candidate genes described here.
Note
While this paper was under revision, a draft of the tomato genome sequence was released (December 1, 2009, International Tomato Genome Sequencing Consortium; http://solgenomics.net/tomato/). Thus, the analyses performed in this work were rerun and 82 further S. pennellii BAC/cosmid clones were putatively anchored to S. lycopersicum scaffolds. However, it is important to point out that these scaffolds are not yet physically ordered and, consequently, the expected map positions of these S. pennellii sequences are not confirmed. Although the S. pennellii sequences reported here were compared with a considerable number of BAC sequences and with a draft version of the cultivated tomato genome, 35% of the S. pennellii BAC/cosmid clones remained unanchored. This could be indicative of either a discontinued synteny between the species and/or a lack of corresponding S. lycopersicum genomic sequence information and thus requires further analyses.
MATERIALS AND METHODS
Screening and Anchoring of Solanum pennellii Genomic Clones
The five genomic regions selected for analysis were BINs 1C, 2B, 4I, 7H, and 11C, which were determined by the overlapping of Solanum pennelli IL introgressed fragments according to the Tomato-EXPEN 1992 map (Solanum lycopersicum LA925 × S. pennellii LA716; http://www.sgn.cornell.edu/; Eshed and Zamir, 1995). In order to select markers spanning the regions of interest, Southern-blot hybridizations were performed using the BIN flanking markers as probes (Sambrook et al., 1989). This analysis allowed us to delimit the five selected BINs between markers TG24 and TG80, TG31 and CT106, CT173 and TG464, TG199 and TG499, and TG523 and TG384 for the 1C, 2B, 4I, 7H, and 11C chromosomal segments, respectively (data not shown). The selected regions carry a total of 277 molecular markers mapped on the publicly available Tomato-EXPEN 2000 and Tomato-EXPEN 1992 maps spanning 99.1 cM (BIN 1C, 19.4 cM; BIN 2B, 13.0 cM; BIN 4I, 11.7 cM; BIN 7H, 36.0 cM; BIN 11C, 19.0 cM). A total of 104 markers that map to single loci in S. lycopersicum were used for library screening (Table I). Ninety overgo-derived markers were selected from the collection available at the SGN site. The other 14 were in-house designed with the OligoSpawn software (Zheng et al., 2006). The complete list of the 104 overgoes used in this study is given in Supplemental Table S1.
The S. pennellii (accession LA716, the donor parent of the IL population) genomic libraries were built in pBeloBACII (52,992 clones) and pCLD04541 (50,304 clones) vectors for BAC and cosmid, respectively (Chen et al., 2007). Both libraries were the result of genomic DNA partially digested with HindIII endonuclease. High-density membranes were screened with 15 to 20 pooled overgo probes (first hybridization). A two-dimensional pooling strategy was performed in order to anchor each clone to its corresponding hybridizing marker (second hybridization). Hybridizations were performed according to Sambrook et al. (1989) with minor modifications. BAC/cosmid filters were prehybridized in a solution containing 2× SSC, 0.1% SDS, and 10 mg mL−1 salmon sperm DNA for 3 h at 65°C. Probes were denatured with 0.4 n NaOH added to each hybridization recipient and hybridized at 65°C overnight. Filters were washed in 2× SSC and 0.1% SDS at 65°C for 5 min and then in 1.0× SSC and 0.1% SDS at 65°C for 30 min.
BAC/Cosmid Clone Insert Size Estimation and Sequencing
DNA was purified using the R.E.A.L Prep 96 Plasmid Kit (Qiagen). Clone insert sizes were estimated by pulsed-field gel electrophoresis by digesting 0.5 to 2 μg of purified DNA with NotI endonuclease in order to release the insert. Insert size was calculated with ImageQuant TL 7.0 software (G&E).
For BAC/cosmid end sequencing, 0.2 to 0.5 μg of purified DNA was used, and sequencing reactions were performed with ABI Big Dye Terminator version 3.1. Samples were read in an ABI 3730 sequencer. Vector and low-quality sequences (Qv < 20) were trimmed out, and only reads above 300 nucleotides were considered.
BAC/cosmid full-length sequencing was performed either by shotgun (Macrogen) or pyrosequencing (454 Life Sciences, through a partnership with Roche). Reads were assembled using the Phred+Phrap+Consed software package (Ewing and Green, 1998; Gordon et al., 1998).
Map Building
All the generated sequences were masked using the Repeat Masker program (http://www.repeatmasker.org) before analysis in order to avoid further associations with nonorthologous regions. End reads from BAC/cosmid clones, anchored to the same marker, were subjected to contig assembly with the DNA Baser version 2.60.90 software with default parameters. A Python script (Chapman and Chang, 2000) was used to analyze the nonrepetitive sequences, either from contigs or singletons by BLASTN (Altschul et al., 1990). The databases used were (1) all currently available S. lycopersicum genomic clone sequences and (2) the marker sequence database. Both databases are accessible at the ftp site of http://www.sgn.cornell.edu/. For comparison against S. lycopersicum BAC sequences (version 441, October 5, 2009, containing 1,188 sequences and encompassing 127,592,092 bp), a hit criterion of above 80% identity was considered. For comparison against marker sequences (database containing 9,625 sequences and encompassing 3,947,844 bp), only hits with at least 96% identity across more than 150 bp were considered. Furthermore, the associations between S. lycopersicum genomic clone sequences and marker sequences were identified using the Marker search feature from the SGN Web site with the parameters mentioned above. The complete set of established relationships is presented in Supplemental Table S2A. Anchored clones and end sequences are available at the SGN Web site in the Tomato Physical Map interface (http://www.sgn.cornell.edu/cview/).
Annotation of BAC/Cosmid End Sequences and Polymorphism Analyses
Nonrepetitive singletons and contigs were annotated by comparison against the following databases: nonredundant proteins at the National Center for Biotechnology Information (November 2008 version, 9,487,554 sequences and 3,243,454,420 bp), the unigenes database at SGN (February 2009 version, 123,550 sequences and 92,327,235 bp), and the Arabidopsis (Arabidopsis thaliana) protein database (The Arabidopsis Information Resource 8, 38,963 sequences and 85,192,362 bp) using BLASTN and BLASTX with e-values ≤ 10−6, above 80% identity along ≥300 bp. Repetitive sequences were annotated by comparison against The Institute for Genomic Research (TIGR) plant repeat databases (ftp://ftp.tigr.org/pub/data/TIGR_Plant_Repeats/) using BLASTN with e-values ≤ 10−5 (Altschul et al., 1990). All annotated sequences (including large-insert sequences; see below) were organized in functional classes according to the MapMan ontology software (http://mapman.mpimp-golm.mpg.de).
SNP and InDel analyses were performed with the SeqScape version 2.5 software using a base call value of 20 or greater as true nucleotides, removing bases from the ends until fewer than four bases out of 20 had Qv < 20 and filtering settings by default. Graphical output was performed by a bespoke Python script, which parses the formatted SeqScape output and reads both the position and the kind of polymorphism (SNP or InDel). An arbitrary value of 1 was used for SNPs, while InDels could take a value of 2 or 3 depending of the InDel size. Positive values were used for SNPs and insertions and negative values were used for deletions.
Large-Insert Sequence Annotation
S. pennellii BAC/cosmid full-length sequences as well as three S. lycopersicum BAC clones, C04HBa0331L22, C07HBa0061J13, and C11HBa0062I24, were annotated using two different gene prediction programs: FGENESH (www.softberry.com; Salamov and Solovyev, 2000) and Augustus (http://augustus.gobics.de; Stanke et al., 2004). Each BAC/cosmid was screened independently against the SGN unigene database (http://www.sgn.cornell.edu/; September 2009 version, 123,550 sequences and 92,327,235 bp) using the sequence comparison programs cross-match (www.phrap.org). Predicted genes were hand curated by comparison with the corresponding identified unigene by BLAST (Altschul et al., 1990). When no unigene hit was found at the SGN database, TIGR Plant Transcript Assemblies database was used (http://plantta.tigr.org/). Only genes predicted both by gene prediction programs and matching an mRNA sequence were annotated. The criteria for gene annotation were (1) at least 95% identity and above 90% coverage of the unigene length or (2) at least 70% identity over 85% of sequence length when only Arabidopsis mRNAs were found.
Repetitive sequences were identified using RepeatMasker (http://www.repeatmasker.org) software, and only elements covering at least 70% of the matching repeat sequence were considered. LTR retrotransposons were also identified using LTR finder (http://tlife.fudan.edu.cn/ltr_finder/). Element class and family name, as well as the matching repeat sequence available at Repbase (www.girinst.org), are indicated in Supplemental Table S5.
A new nomenclature to name S. pennellii genes is proposed: SpXXgB(or C)XXXXX.#, where Sp stands for S. pennellii, the two digits indicate the chromosome number, g represents the genomic sequence, B stands for BAC or C for cosmid vectors, the five digits address library coordinates, and # stands for consecutive numbers indicating gene order within the clone.
For comparative analysis between S. lycopersicum and S. pennellii orthologous genomic regions, the S. lycopersicum BAC clone sequence that better spanned the S. pennellii sequence was selected. Sequence comparisons between genotypes for InDel and SNP identification were calculated using the Needleman-Wunsch global alignment algorithm available at http://www.ebi.ac.uk/Tools/emboss/align/.
Evolutionary Analyses
Alignments of coding regions were performed with ClustalW multiple alignment software (version 1.5; Thompson et al., 1994) and hand curated with reference to amino acid alignment. Synonymous (ds) and nonsynonymous (dn) distances and their se values were estimated with MEGA 3.1 (Kumar et al., 2004) using the corrected Nei-Gojobori method (Jukes-Cantor). Codon bias was determined by the effective number of codons value computed using DNAsp version 4.10.9 (Rozas et al., 2003). Test of selection was also performed with MEGA 3.1 (Kumar et al., 2004) using the corrected Nei-Gojobori method. To reject the null hypothesis of neutral selection (ds = dn), P < 0.05 was considered in the Z test. Relative rate tests were performed utilizing HYPHY (http://www.hyphy.org) using the codon model proposed by Muse and Gaut (1994). To reject the homogeneity null hypothesis, P < 0.05 was considered in the χ2 test. Genomic region-specific substitution rates (K) and species divergence time were estimated using K = ds/2T, where ds is the estimated number of synonymous substitutions per site between homologous sequences (synonymous distance) and T is the divergence time. K was estimated using the fossil-supported Arabidopsis/Solanaceae divergence time (120 MYA; Bell et al., 2005; Magallon and Sanderson, 2005), and ds was estimated based on concatenated coding regions per chromosome.
For retrotransposon dating, both LTRs of copies with target site duplications were aligned using the ClustalW multiple alignment software (version 1.5; Thompson et al., 1994). The distance (D) between LTRs and its se were estimated with MEGA 3.1 (Kumar et al., 2004) using Kimura 2-parameter, which corrects for homoplasy and differences in the rates of transition and transversion. The retrotransposon insertion date was estimated using T = D/2K, where K represents the 1.3 × 10−8 substitutions per site per year as proposed by Ma and Bennetzen (2004). Confidence intervals were calculated using the se of the mean distance (D) as estimated with MEGA 3.1.
Sequence data from this article have been deposited within the GenBank data libraries under accession numbers FI277973 to FI278499, FJ809740 to FJ809747, and FJ812349.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SNP and InDel mapping.
Supplemental Table S1. Markers used for S. pennellii BAC/cosmid genomic library screening.
Supplemental Table S2. S. pennellii physical map data, S. pennellii contig sequences, and SNPs and InDels between S. pennellii end sequences and S. lycopersicum BAC clones.
Supplemental Table S3. BAC/cosmid end sequence annotation.
Supplemental Table S4. Annotation of S. pennellii fully sequenced BAC/cosmid clones from BINs 1C, 2B, 4I, 7H, and 11C.
Supplemental Table S5. S. pennellii transposable elements identified in full-length sequenced clones.
Supplementary Material
Acknowledgments
We thank Mariana Conte for technical assistance and Agustín Montenegro and Ignacio Tevez for taking care of the tomato plants. L.K., R.A., and F.C. are members of Consejo Nacional de Investigaciones Científicas y Técnicas.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle E. (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep 9: 208–218 [Google Scholar]
- Bell CD, Soltis DE, Soltis PE. (2005) The age of the angiosperms: a molecular timescale without a clock. Evolution 59: 1245–1258 [PubMed] [Google Scholar]
- Bermúdez L, Urias U, Milstein D, Kamenetzky L, Asis R, Fernie AR, Van Sluys MA, Carrari F, Rossi M. (2008) A candidate gene survey of quantitative trait loci affecting chemical composition in tomato fruit. J Exp Bot 59: 2875–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman BA, Chang JT. (2000) Biopython: Python tools for computational biology. ACM SIGBIO Newsletter 20: 15–19 [Google Scholar]
- Chen KY, Cong B, Wing R, Vrebalov J, Tanksley SD. (2007) Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318: 643–645 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. (1998) Basecalling of automated sequencer traces using phred II: error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Fernie AR, Willmitzer L. (2001) Molecular and biochemical triggers of potato tuber development. Plant Physiol 127: 1459–1465 [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie A, Zamir D. (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D. (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJB, Bouwmeester H, America R, Verstappen FWA, Ward JL, Beale MH, de Vos RCH, Dijkstra M, Scheltema RA, et al. (2009) System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. (1998) Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Gregory SG, Sekhon M, Schein J, Zhao S, Osoegawa K, Scott CE, Evans RS, Burridge PW, Cox TV, Fox CA, et al. (2002) A physical map of the mouse genome. Nature 418: 743–750 [DOI] [PubMed] [Google Scholar]
- Guyot R, Cheng X, Su Y, Cheng Z, Schlagenhauf E, Keller B, Ling HQ. (2005) Complex organization and evolution of the tomato pericentromeric region at the FER gene locus. Plant Physiol 138: 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HM, Liu J, Doganlar S, Tanksley SD. (2001) Exploitation of Arabidopsis-tomato synteny to construct a high-resolution map of the ovate-containing region in tomato chromosome 2. Genome 44: 470–475 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Semel Y, Zamir D. (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr Opin Genet Dev 17: 545–552 [DOI] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D. (2003) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol J 1: 195–207 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O'Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallon SA, Sanderson MJ. (2005) Angiosperm divergence times: the effect of genes, codon positions, and time constraints. Evolution 59: 1653–1670 [DOI] [PubMed] [Google Scholar]
- Mueller L, Tanksley S, Giovannoni JJ, Vaneck J, Stack S, Buels R. (2009) A snapshot of the emerging tomato genome sequence. Plant Genome 2: 78–92 [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al. (2005) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. (1994) A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol 11: 715–724 [DOI] [PubMed] [Google Scholar]
- Nesbitt TC, Tanksley SD. (2002) Comparative sequencing in the genus Lycopersicon: implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Rossberg M, Theres K, Acarkan A, Herrero R, Schmitt T, Schumacher K, Schmitz G, Schmidt R. (2001) Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497 [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch T, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D. (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA 103: 12981–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. (2004) AUGUSTUS: a Web server for gene finding in eukaryotes. Nucleic Acids Res 32: 309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ. (2006) Identification of loci affecting flavour volatile emissions in tomato fruits. J Exp Bot 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Bläsing O, Kowalczyk M, Weicht D, Polinceusz A, et al. (2006) Conversion of MapMan to allow the analysis of transcript data from solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60: 773–792 [DOI] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S. (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JW, Aerts J, Groenen MA, Crooijmans RP, Layman D, Graves TA, Scheer DE, Kremitzki C, Fedele MJ, Mudd NK, et al. (2004) A physical map of the chicken genome. Nature 432: 761–764 [DOI] [PubMed] [Google Scholar]
- Wang Y, Diehl A, Wu F, Vrebalov J, Giovannoni J, Siepel A, Tanksley SD. (2008) Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics 180: 391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, Van der Knaap E. (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Zamir D. (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2: 983–990 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y, Cheng Z, Chen M, Wang S, Feng Q, Huang Y, Li Y, Tang Y, Zhou B, et al. (2002) A fine physical map of the rice chromosome 4. Genome Res 12: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Svensson JT, Madishetty K, Close TJ, Jiang T, Lonardi S. (2006) OligoSpawn: a software tool for the design of overgo probes from large unigene datasets. BMC Bioinformatics 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.