Abstract
A regulatory protein from grass pea (Lathyrus sativus), LS-24, a close homolog of albumin 2 from garden pea (Pisum sativum) that is associated with polyamine biosynthesis, was characterized and the structure of a hemopexin-type fold among plant proteins illustrated. Crystal structure of LS-24 determined at 2.2 Å resolution by multiple isomorphous replacement phasing showed four-bladed β-propeller structure having a pseudo 4-fold molecular symmetry along a metal ion-binding central channel. The structure represents typical mammalian hemopexin fold with discernible features correlated with the possible functional variations. The protein was found to exist in the dimeric state. While LS-24 dimer binds to spermine in the crystal structure as well as in solution, binding of heme in solution resulted in the dissociation of the dimer into monomers with concomitant release of bound spermine. Interactions of heme and spermine with LS-24 bear physiological implications. While binding of spermine to LS-24 can be linked with polyamine biosynthesis that of heme correlates with oxidative stress. Mutually exclusive binding of heme and spermine in different oligomeric states suggest a role for LS-24 in sensing oxidative stress through a ligand-regulated monomer-dimer transition switch.
Plant seeds are a rich source of proteins that have been historically assigned to four different classes: albumins, globulins, prolamines, and glutelins, based on their solubility properties (Osborne and Campbell, 1898). Globulins and albumins are the major legume seed proteins (Higgins et al., 1986) whereas prolamines and glutelins predominantly occur in cereals (Shewry and Tatham, 1990). While globulins comprise of storage proteins, albumins form a heterogeneous class of unrelated proteins and include metabolic, structural, and/or regulatory proteins (Vioque et al., 1999). Many albumins such as lipoxygenases (Clemente et al., 2000), glycosidases (Agrawal and Bahl, 1968), proteinases (Schlereth et al., 2001), proteinase inhibitors (Kochhar et al., 2000), and lectins (Harley and Beevers, 1986) have been demonstrated to exhibit distinct functions. Recently, another protein belonging to albumin class, referred to as albumin 2 from garden pea (Pisum sativum) has been demonstrated to have regulatory role in polyamine metabolism (Vigeolas et al., 2008).
Grass pea (Lathyrus sativus) seeds are a promising source of dietary proteins (Campbell, 1997). Although proteins constitute approximately 20% of the total seed dry weight (Rosa et al., 2000), the grass pea crop has presently been banned due to the presence of various antinutritional substances responsible for neurolathyrism (Grela et al., 2001). Also, grass pea exposure results in occupational allergies leading to asthma and rhinoconjunctivitis in susceptible individuals (Valdivieso et al., 1988; Porcel et al., 2001; Gironés et al., 2005). In light of its unusual properties, we set out for a comparative analysis of grass pea seed proteome with respect to other legumes.
We had earlier isolated LS-24, a protein from the albumin fraction of seed proteins from grass pea (Qureshi et al., 2006). LS-24 shows significant sequence identity with albumin 2 from garden pea (Croy et al., 1984; Harris and Croy, 1985; Gruen et al., 1987; Higgins et al., 1987; Jenne, 1991) and chickpea (Cicer arietinum; Kolberg et al., 1983; Vioque et al., 1998). They are associated with metabolic processes being cytoplasmic in localization (Harris and Croy, 1985), in contrast to the other seed storage proteins that are found in membrane-bound vesicles (Larkins and Hurkman, 1978; Okita and Rogers, 1996). Unlike other seed storage proteins, these proteins persist long after the beginning of seed germination process, indicating their possible functional role during this process (Higgins et al., 1987).
Here we have determined the crystal structure of LS-24 and described the architecture of the first four-bladed β-propellar protein from plants. Further, while the crystal structure showed a molecule of spermine bound to LS-24 dimer, the binding of heme to LS-24 was found to dissociate the protein into monomeric state with subsequent loss of spermine-binding ability as demonstrated using competition assay. Also, the presence of bound spermine in the protein purified directly from the seeds has been established using anti-spermine antibodies. Our structural and biochemical data, in light of the known regulatory roles of spermine and heme, implicate LS-24 in sensing oxidative stress as a regulatory switch.
RESULTS
Protein Characterization, Crystallization, and Structure Determination
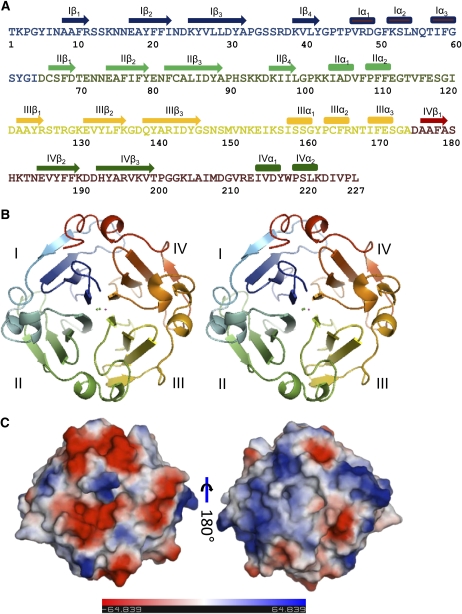
LS-24 was identified as a close homolog of albumin 2 from garden pea, exhibiting 85% sequence identity with it, while functionally screening the grass pea seed proteome (Qureshi et al., 2006; Fig. 1A). The sequences of the internal protein fragments of LS-24 obtained by chemical and enzymatic methods and interpreting electron density map at 2.2 Å resolution calculated from multiple isomorphous replacement (MIR) phasing yielded the complete sequence of LS-24 (Supplemental Fig. S1).
Figure 1.
The overall structure of LS-24 monomer. A, Amino acid sequence of LS-24 along with secondary structure assignment on the basis of crystal structure. B, Stereo view of ribbon diagram highlighting the β-propeller topology of the protein. The blades of the propeller have been differently colored and marked as I, II, III, and IV, from N to C terminus. C, Surface diagrams of LS-24 displaying differential distribution of charge in two opposite views.
Crystallization and preliminary characterization of LS-24 have been reported earlier (Qureshi et al., 2006). LS-24 crystals were obtained in P212121 space group, with the unit cell dimensions a = 83.2 Å, b = 88.1 Å, and c = 154.5 Å, and four molecules in the asymmetric unit accounting for 56.3% solvent content. This crystal form was different in one of the unit cell dimensions in comparison to the one reported earlier (Qureshi et al., 2006). All subsequent crystallographic analyses were carried out using the new crystal form that diffracted at higher resolution. The three-dimensional structure of LS-24 was determined using MIR method of phase determination (Table I). The model was refined to an Rfree of 26.6% and Rwork of 24.6% in the resolution range of 50 to 2.2 Å, with 99.9% of the residues in the allowed region of the Ramachandran plot (Table II).
Table I. Data collection and phasing statistics.
All the datasets were collected at 120 K on Rigaku RU-H3R rotating anode generator using MAR345dtb detector. Phases were determined using MIR. Data in parentheses are for highest resolution bin.
| Dataset | Native | HgCN | Pb(CH3COO)2 | CdCl2 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å; a, b, c) | 83.2, 88.1, 154.5 | 83.4, 89.0, 154.1 | 83.4, 85.9, 154.1 | 82.8, 87.4, 154.0 |
| Resolution (Å) | 50–2.2 (2.32–2.2) | 50-2.8 (2.882.8) | 50–2.5 (2.59–2.5) | 50–2.5 (2.61–2.5) |
| Observed reflections | 185,883 (26,826) | 101,610 (9,155) | 124,744 (11,542) | 182,495 (17,141) |
| Unique reflections | 54,633 (8,105) | 28,804 (2,543) | 38,827 (3,607) | 37,629 (3,586) |
| Rmerge (%)a | 12.9 (49.1) | 13.4 (32.22) | 13.7 (40.6) | 8.7 (25.9) |
| Completion (%) | 93.9 (96.5) | 97.3 (88.0) | 99.0 (93.8) | 97.1 (94.6) |
| Multiplicity | 3.4 (3.3) | 3.5 (3.6) | 3.2 (3.2) | 4.79 (4.78) |
| I/σ | 5.2 (1.9) | 3.0 (1.5) | 2.8 (1.2) | 3.7 (1.8) |
| MIR statistics | ||||
| Riso (%)b | 20.6 | 17.1 | 11.9 | |
| Phasing powerc; centric (acentric) | 0.87 (1.11) | 0.51 (0.61) | 0.24 (0.25) | |
| Rcullis (%)d | 0.66 | 0.71 | 0.83 | |
| No. of sites | 5 | 4 | 1 | |
| Figure of merit/Z score | 0.40/22.2 | |||
Rmerge = Σ[I − (I)]/Σ(I), where I is the integrated intensity of a given reflection.
Riso = Σ|FPH − FP|/ΣFP, where FPH and FP are derivative and native structure factor amplitudes, respectively.
Phasing power = <r.m.s. heavy atom structure factor>/<r.m.s. lack of closure>.
Rcullis = Σ‖FPH ± FP| − FH|/Σ|FPH − FP| for centric reflections.
Table II. Refinement statistics.
Model refinement was carried out with 10% random reflections used for cross validation.
| Refinement Statistics | |
| Resolution range (Å) | 50.0–2.2 |
| No. of reflections | 49,032 |
| Rwork (%)a | 24.6 |
| Rfree (%)b | 26.6 |
| RMSD bond length (Å) | 0.021 |
| RMSD bond angle (°) | 1.984 |
| No. of atoms | |
| Protein | 7,220 |
| Solvent | 565 |
| Ligand/ion | 58 |
| Average B factor (Å2) | |
| Protein | 16.57 |
| Solvent | 16.84 |
| Ligand/ion | 18.40 |
Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|.
Rfree = Σ‖Fo| − |Fc‖/Σ|Fo|.
Overall Structure
The structure of LS-24 momomer is shown in Figure 1. The molecular structure of a single monomer shows β-propeller domain, characteristic of the proteins belonging to the hemopexin superfamily. Each monomer exhibits a pseudo 4-fold axis of symmetry passing through the center of each of the monomer, characterized by a channel (Fig. 1B). The discoid subunit, with an average diameter of 40 Å and a thickness of 30 Å, is characterized by differential distribution of surface charge with predominantly positive electrostatic potential on one face and the negative on the other (Fig. 1C).
The four blades of the β-propeller polypeptide are arranged around a pseudo 4-fold axis of symmetry such that the blade I, toward the N terminus, comes in close proximity to the blade IV, toward the C-terminal end. Each blade comprises of four stranded twisted β-sheet. The innermost strand, toward the central channel, is most regular whereas the outer one is not as regular. The β-strands of the adjacent blades together form a β-sandwich enclosing a hydrophobic cavity. The adjacent blades are connected by a linker region consisting of three short α-helices. A region consisting of α-helices extends from the blade IV toward the N terminus of the subunit, thereby maintaining cementing interactions between blade I and blade IV similar to those at the other three interfaces (Fig. 1B).
The four blades of LS-24 superimpose with root mean square deviations (RMSDs) varying between 0.9 Å to 1.2 Å. The fourth strand in the case of blade III and IV has a β-bulge, disrupting the extended β-strand conformation. The other hemopexin domains also show the presence of β-bulge in the blades. The structural symmetry of the monomer is consistent with the 4-fold tandem repeats in the sequence (Fig. 1A). Interestingly, the sequences among the four blades of the monomer are more identical in LS-24 than the hemopexin fold of mammalian origin (Supplemental Fig. S2).
The central channel is formed by the innermost strands of the blades, which run parallel to each other. The channel is 4.3 Å in diameter toward the N-terminal opening and 7.24 Å on the opposite side with basic residues lining the wider opening and acidic residues lining the narrower opening, producing a net dipole inside the channel. Based on the electron density level, chemical environment, and the B factors, Ca2+, Na+, Cl−and a water molecule have been identified within the central channel. The ions provide stability to the structure by minimizing electrostatic repulsion through neutralization of the interiorly facing backbone carbonyl and amide groups in the channel (Fig. 1B).
Oligomerization and Spermine Binding
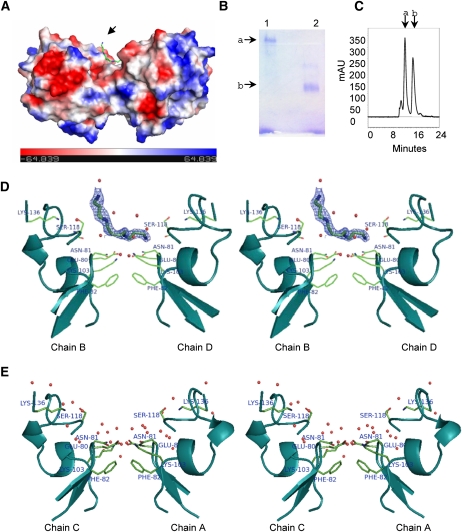
The crystallographic asymmetric unit contains four monomeric chains, A, B, C, and D organized into two dimers, A:C and B:D, one of which is shown in Figure 2A. The oligomerization has been analyzed in the solution state as well. While LS-24 migrates as a 24-kD protein on SDS-PAGE (Qureshi et al., 2006), a comparison in the mobility of heat-denatured and native forms of the protein by PAGE indicated oligomerization (Fig. 2B). This was further confirmed by size-exclusion chromatography. Two distinct peaks were evident, indicating the presence of monomeric and dimeric species, with dimer constituting the major molecular population in solution (Fig. 2C; Supplemental Fig. S3).
Figure 2.
LS-24 dimer and spermine binding. A, Surface topology of LS-24 dimer along with bound spermine molecule (indicated by an arrow), showing surface charge distribution. Spermine is shown as stick. B, Native gel profile showing the oligomeric state of LS-24. Bands a and b correspond to dimer and monomer, respectively. Lanes 1 and 2 have been loaded with native and heat denatured proteins, respectively. C, Size-exclusion chromatogram of LS-24. Peaks a and b correspond to dimeric and monomeric forms of LS-24, respectively. mAU, Milli absorbance units. D, Stereo view showing interactions of bound spermine at the dimer interface. Bias removed Fo-Fc map highlighting the bound spermine molecule at 1.5 σ cutoff. E, Stereo view showing the interactions at dimer interface without spermine, in another dimer in the asymmetric unit. Chains A, B, C, and D refer to four monomers in the asymmetric unit organized into two dimers, A:C and B:D.
The two monomers of the dimer are held together by noncovalent interactions. The last strand of blade II and the interblade linker, connecting blade I and blade II of both the monomers are involved in dimerization. The hydrophobic interactions, largely mediated by Phe, Tyr, and Ile of both the chains, dominate at the center of the dimeric interface, while the salt bridges are at the periphery of the interface. The interacting surfaces of the two monomers at the dimer interface are equivalent and are related by a noncrystallographic 2-fold axis. The average surface area of LS-24 monomer involved in dimerization is 640.0 Å2. A water molecule is present at the center of the dimer interface tightly held in position by forming a hydrogen bond with the backbone amide of Ile-58 of both the monomers with an average B factor of 6.5 Å2.
A molecule of spermine, in an extended conformation, has been identified bound to one of the two dimers (B:D) with an average B factor of 18.90 Å2 (Fig. 2D). The site of spermine interaction is contributed by both the monomers of the dimer. The residues Glu-80, Asn-81, and Ser-118 of both the monomers are crucial for spermine binding. The bound spermine makes two direct contacts with Asn-81D and Ser-118D and 14 van der Waals interactions with the protein. It makes three water-mediated contacts with Glu-80B, Asn-81B, and Ser-118B of LS-24. The water molecules involved in bridging spermine with LS-24 are held tightly by well-defined protein structure. One of these water molecules is held in position by a chelating network formed by the side chains of residues Glu-80B and Asn-81B with a B factor of 14.24 Å2. The other water molecule is held in position by interaction with the main chain carbonyl of Ser-118B with a B factor of 17.84 Å2. Thus, out of the four polar amines of the spermine molecule, three are interacting either directly with the protein molecule or through highly structured protein-bound water molecules. These interactions, contributed by both the subunits, provide a flat hydrophilic surface facilitating spermine binding at one of the two dimer interfaces (B:D) in the crystallographic asymmetric unit (Fig. 2D). The area of dimer involved in interaction with the spermine was calculated to be 102.7 Å2. The other dimer (A:C), devoid of spermine, was found to have a water network at the corresponding spermine interaction site (Fig. 2E). Spermine binding resulted in an increase of the area of interaction between the two subunits by 30.8 Å2, marginally enhancing the association between the two monomers.
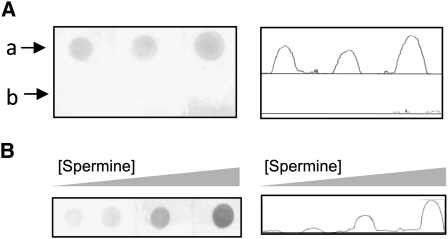
The presence of spermine in the protein purified directly from the source was confirmed using anti-spermine antibody in a dot-blot assay, with bovine serum albumin (BSA) as negative control (Fig. 3A). Furthermore, the spermine-binding potential of the LS-24 was confirmed from the concomitant increase in the ability of binding anti-spermine antibody, when the protein was incubated with increasing amounts of exogenous spermine (Fig. 3B). This dose-dependent increase in signal is indicative of the fact that under physiological conditions, not all the spermine-binding sites are occupied, consistent with the observed stoichiometry of one spermine molecule as against two independent dimers of LS-24 in the crystallographic asymmetric unit.
Figure 3.
Interaction of spermine with LS-24. A, Dot-blot analysis showing the ability of LS-24 (lane a) to interact with anti-spermine antibody. BSA (lane b) was used as negative control. Relative intensities of signals obtained during dot-blot analysis have been presented. B, Dot-blot analysis presenting an increase in the anti-spermine antibody signal when LS-24 was preincubated with increasing concentration of exogenous spermine.
Heme Binding and Monomerization
Heme-binding potential of LS-24 was analyzed in light of the structural resemblance of this protein with the mammalian serum hemopexin. The protein was subjected to mobility shift assay, size-exclusion chromatography, and the quantitative measurements of the affinity parameters in the presence of hemin (Fig. 4).
Figure 4.
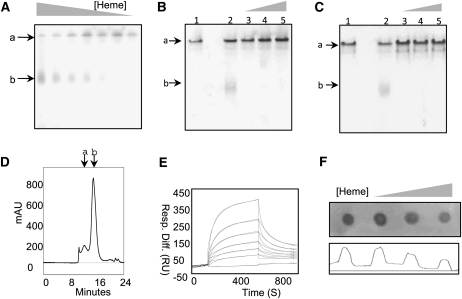
Analysis of hemin binding with LS-24. A to C, Mobility shift assay of LS-24 in the presence of decreasing concentrations of hemin (A), increasing concentrations of protoporphyrin IX (B), and increasing concentrations of hematoporphyrin (C). Bands a and b correspond to LS-24 dimer and monomer, respectively. Lanes 1 and 2 correspond to native LS-24 and LS-24 preincubated with hemin (B and C). D, Size-exclusion chromatogram of heme-bound LS-24, elucidating the monomerization of LS-24 on heme binding. Peaks a and b correspond to dimeric and monomeric forms of LS-24, respectively. mAU, Milli absorbance units. E, Binding analysis by SPR of LS-24 interaction with hemin. Resp. Diff., Response difference; RU, response units. F, Dot-blot analysis presenting a decrease in anti-spermine antibody binding when LS-24 is preincubated with increasing concentration of hemin.
LS-24 exhibited lower mobility on the native polyacrylamide gel than that of LS-24 in the presence of hemin (Fig. 4A). However, no change in mobility was observed with other porphyrin analogs (Fig. 4, B and C). To dissect out the mechanism involved in the shift observed in the mobility of LS-24 incubated with hemin, the samples were analyzed over gel-exclusion chromatography. Gel-exclusion chromatography of LS-24 preincubated with hemin showed loss of the peak corresponding to dimeric molecular species of LS-24 (Figs. 2C and 4D; Supplemental Fig. S3), indicating that hemin binding causes dimer to dissociate into monomers.
The kinetics of hemin binding was quantitatively studied using surface plasmon resonance (SPR) on BIAcore 2000. The SPR data recorded with hemin flowing over LS-24 immobilized on the biosensor chip revealed a dose-dependent binding of heme to LS-24 at physiological pH of 7.4. The sensogram could be fitted to the 1:1 Langmuir binding model. Further analysis gave association (ka) and dissociation (kd) rate constants of 4.41 × 103 m−1 s−1 and 7.73 × 10−3 s−1, respectively, showing an equilibrium dissociation constant (KD) of 1.75 ± 0.61 μm (Fig. 4E). Other porphyrin analogs, protoporphyrin IX and hematoporphyrin, were also assessed by SPR and they did not show interaction with LS-24 (data not shown), consistent with the data from gel-shift assay. Thus, LS-24 exhibited specificity for heme binding.
On one hand, spermine binding, as observed in the crystal structure, involves both the monomers simultaneously and, on the other, heme binding results in dissociation of the dimer into monomers. These data when considered together suggest that heme binding should be accompanied by disruption of the spermine-binding site. This would, in turn, imply that the two ligands should interact with LS-24 in mutually exclusive manner. Indeed, that was the case since a loss in spermine-binding potential was observed in dot-blot assay after incubation of the protein with increasing heme concentrations (Fig. 4F).
DISCUSSION
It was intriguing to find that LS-24 belongs to the hemopexin structural family. More than 500 proteins are designated to contain hemopexin-like domain(s), from simple organisms such as prokaryotes to the complex organisms including plants and mammals (Piccard et al., 2007). While, on one hand, mammalian hemopexins are involved in scavenging biologically active ligands (Hrkal et al., 1974), on the other, they are involved in modulating the enzymatic activities of the associated domains (Gomis-Rüth et al., 1996; Jozic et al., 2005; Iyer et al., 2006). In the context of LS-24 homologs being involved in polyamine biosynthetic pathway (Vigeolas et al., 2008) and stress physiology (Huang et al., 2006), the structural correlation of mammalian hemopexins with LS-24 would shed light on the physiological role of this protein.
Screening of grass pea seed proteome for allergenic proteins led to the identification of LS-24, a close homolog of albumin 2 from garden pea. The four-bladed propeller having a pseudo 4-fold molecular symmetry along the central channel, characteristic of the hemopexin fold, was apparent in the structure of LS-24. While sharing the fold, hemopexin domains of plant and mammalian origin exhibit distinctive tertiary structural features. The prominent structural differences are in the length and spatial arrangement of the loops connecting the outermost β-strands and the linker region between the blades (Supplemental Fig. S4). The entire LS-24 structure is folded stable with the N and C termini in close proximity by hydrophobic interactions alone, as against the stabilization through a disulphide bond in the case of mammalian hemopexins. Presence of better 4-fold internal structural symmetry indicates plant hemopexins may be structurally more primitive in comparison to the corresponding mammalian proteins. LS-24 structure illustrates the first protein with four-bladed hemopexin fold, although the propeller structures with higher number of blades have often been observed, among the plant proteins.
The dimeric state of LS-24 is evident both in the crystal structure as well as in solution. Since the asymmetric unit contains spermine-bound and unbound dimers, with small structural differences, it may indicate that the dimer may exist in both these states under the physiological conditions. The spermine-binding site of LS-24 is contributed by both the monomers of a dimer and dimerization would thus be required for spermine binding as also evident from the biochemical studies. Spermine and other polyamines have been implicated in a wide range of biological processes in plants. They include growth, development, and stress responses (Galston and Sawhney, 1990). Plant cells have their intrinsic machinery to synthesize polyamines (Martin-Tanguy, 2001). It has been shown that the intracellular levels of spermine and spermidine are under tight regulatory control (Bassie et al., 2000; Lepri et al., 2001; Capell et al., 2004). Polyamine metabolism is modulated by various physical stimuli and stress conditions (Palavan-Ünsal, 1995; Bouchereau et al., 1999). In fact, albumin 2 from garden pea, an LS-24 homolog, has been linked with the regulation of polyamine biosynthesis. A decreased amount of spermidine and spermine during the later stages of seed development has been observed in mutant lines lacking albumin 2. This decrease was attributed to a decrease in the activities of enzymes of polyamine biosynthesis, Arg decarboxylase, and spermidine synthase (Vigeolas et al., 2008). Thus, the association of plant hemopexins with this regulatory mechanism is further strengthened by our demonstration of LS-24-spermine interaction.
Being a hemopexin fold protein, it is not surprising that LS-24 binds heme with physiological affinity. In fact, a homologous protein from chickpea has been previously demonstrated to interact with hemin (Pedroche et al., 2005). However, the mode of heme binding appears to be different in LS-24 compared to that of the mammalian serum hemopexin (Paoli et al., 1999). The mammalian serum hemopexin is a two-domain protein, both the domains folded with the hemopexin topology. The heme-binding pocket is formed by the juxtaposition of the two domains and the interdomain linker. Such an architecture for heme binding is absent in LS-24 since it has a single domain monomer without any linker region. Also, binding of heme competes with the LS-24 dimerization, implying that the protein binds to heme in the monomeric state. Other hemopexin domains lacking the architecture of mammalian serum hemopexin are also known to show heme-binding ability (Geurts et al., 2008).
Although the pathway for heme biosynthesis has been well defined in plants (Papenbrock and Grimm, 2001), the transportation of heme from the site of its synthesis to its insertion into hemoproteins residing in cytoplasm or other subcellular compartments remains poorly understood (Hamza, 2006). Due to its poor solubility and toxicity, heme is unlikely to exist in free state in cytoplasm. Therefore the existence of protein-based intracellular heme transportation is plausible. The cytoplasmic localization of LS-24 homologs (Harris and Croy, 1985) along with their ability to interact with heme at physiological affinities make LS-24 to be an ideal candidate to serve as a heme carrier protein within the plant cell.
It is important to note that LS-24 binds spermine in dimeric state and monomerizes while binding to heme. Since heme and spermine are two molecules associated with stress among plants, their association with LS-24 indicated an involvement of this protein in the regulatory pathways related to stress. Indeed, LS-24 homolog has been shown to be involved in the seed stress physiology (Huang et al., 2006). The function of LS-24 may be modulated by different oligomeric states in the context of relative availability of two different regulatory molecules. Under normal conditions, the LS-24 dimer binds polyamine, as observed in this study, and regulates activities of key enzymes of the polyamine biosynthetic pathway as was shown in the case of albumin 2 (Vigeolas et al., 2008). Under conditions of oxidative stress, the levels of free heme reportedly increases (Nagai et al., 2007), which could possibly dissociate LS-24 dimer into monomers, thereby releasing and further increasing the levels of free spermine. On the other hand, the free spermine has been shown to induce nitric oxide synthesis (Tun et al., 2006), which in turn, regulates the expression of genes involved in the plant stress physiology.
In conclusion, we have determined the structure of LS-24, a regulatory plant seed protein exhibiting hemopexin-like fold. Our data demonstrate that while spermine binds to LS-24 dimer, binding of heme results in the monomerization of the protein. Essentially, LS-24 may be involved in regulating the activity of other specific enzymes associated with, for example, polyamine biosynthetic pathway and management of oxidative stress by acting as a scaffold for the recognition of corresponding substrates.
MATERIALS AND METHODS
Protein Purification
Mature seeds of grass pea (Lathyrus sativus) were purchased from the Indian Agricultural Research Institute Regional Station, Karnal, India. The seeds were ground to a fine powder and defatted with petroleum ether and subsequently homogenized with 10 mm phosphate buffer, pH 7.2 containing 140 mm NaCl by continuous stirring for 4 h at 277 K. The crude extract was prepared by centrifugation at 48,384g for 30 min and then subjected to ammonium sulfate fractionation. LS-24 was purified from 95% ammonium sulfate fraction using PI/M weak anion-exchange column (Applied Biosystems), preequilibrated with 50 mm Tris-HCl, pH 7.5. Elution was carried out using a gradient of 0 to 1 m NaCl in 50 mm Tris-HCl buffer, pH 7.5 for 35 min.
Protein Sequencing
Protein sequencing was carried out by subjecting the purified protein to limited proteolyses by using endoproteinase Lys-C, endoproteinase Arg-C, endoproteinase Asn-N, and CNBr to obtain internal protein fragments. The internal proteolytic fragments thus obtained were transferred onto polyvinylidene difluoride membrane and subjected to N-terminal sequencing using Edman chemistry on a Procise protein sequencer (Applied Biosystems). All the sequence searches were performed with BLAST (Altschul et al., 1997) and sequence alignments were carried out using ClustalW (Thompson et al., 1994).
Heme-Binding Analysis
Hemin binding was initially screened by gel-retardation assay and gel-exclusion chromatography. For gel-retardation assay, LS-24 was incubated with increasing concentrations of hemin with [LS-24]/[hemin] molar ratios ranging from 1:1 to 1:50 at room temperature for 1 h in 50 mm mm Tris-HCl, pH 7.5. As negative controls, protoporphyrin IX and hematoporphyrin were used with [LS-24]/[ligand] molar ratios ranging from 1:1 to 1:50. The samples were run on 7.5% native PAGE in nondenaturing and nonreducing conditions. A total of 0.192 m Gly, 25 mm Tris-HCl, pH 8.3 was used as the running buffer. Protein samples were prepared by mixing with the running buffer containing 10% glycerol and 0.01% bromphenol blue in a ratio of 1:1 (v/v). Gel staining was carried out in 0.25% Coomassie Brilliant Blue G-250 in water/methanol/acetic acid (4:5:1) for 1 h and washing in water/methanol/acetic acid (8:1:1). To demarcate the positions of dimeric and monomeric protein species in negative control experiments, LS-24 incubated with hemin in a molar ratio of 1:5 ([LS-24]/[hemin]) was used, to allow its partial monomerization. LS-24 preincubated with hemin in a molar ratio of 1:50 ([LS-24]/[hemin]) was loaded onto S-300 TSK-GEL SW(XL) gel filtration column (Tosoh Corporation), preequilibrated with 50 mm Tris-HCl, pH 7.5 at a flow rate of 1 mL min−1. Low Mr gel filtration calibration kit (Amersham Biosciences) was used for plotting standard curve: Mr against Kav ([Ve − Vo]/[Vc − Vo]; Supplemental Fig. S3). Ve is the elution volume, Vo is the void volume of the column (10.8 mL), and Vc is the total bed volume of the column (26 mL). Vo was determined using blue dextran. Stock solutions of 100 mm, each of hemin, protoporphyrin IX, and hematoporphyrin in 100 mm NaOH were prepared and protein was used at a concentration of 5 mg mL−1 in 50 mm Tris-HCl, pH 7.5.
LS-24-hemin interaction analysis was performed using a BIAcore 2000 biosensor system (Amersham Biosciences). LS-24 was immobilized onto CM4 (carboxymethylated)-certified grade sensor chips using an equal mixture of N-ethyl-N-(dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide in 10 mm sodium acetate buffer, pH 4.5. Approximately 500 response units (RU) of LS-24 were immobilized, where 1,000 RU corresponds to an immobilization level of approximately 1 ng mm−2. The nonreacted activated sites were blocked with 1 m ethanolamine. The reference surface was treated in the same way except that no protein was passed over this. Binding studies were conducted in 10 mm HEPES, pH 7.4, containing 150 mm NaCl, 3.0 mm EDTA, and 0.005% surfactant P20 at 25°C. For the determination of association rate constant (ka; m−1 s−1), various concentrations of heme in binding buffer (varied between 0.1 μm to 3.0 μm) were allowed to bind to the immobilized LS-24 at a flow rate of 30 μL min−1. The dissociation rate constant (kd; s−1) was measured by replacing with binding buffer. Regeneration was conducted using 100 mm NaOH. Analysis was done by BIAevaluation 3.2 software.
Dot-Blot Assay
Dot-blot assay was carried out to demonstrate the presence of bound spermine with LS-24, purified directly from the seeds of grass pea, and the loss of spermine-binding potential by LS-24 on heme binding. BSA was used as the negative control. Polyclonal anti-spermine antibody (Abcam) and horseradish peroxidase conjugated goat anti-rabbit IgG antibodies (Abcam) were used as the primary (1:1,200 dilution) and secondary antibodies (1:3,000 dilution), respectively. For blotting, grids were drawn on nitrocellulose membrane (Millipore) using a pencil to demarcate the regions for blotting. Two microliters of the sample (BSA or LS-24) was applied to the membrane at a concentration of 5 mg mL−1 in 50 mm Tris-HCl, pH 7.5. For demonstrating binding ability of the native protein to spermine, protein was incubated with increasing concentrations of spermine in [LS-24]/[spermine] molar ratios ranging from 1:1 to 1:10. After applying the samples, membrane was dried and the nonspecific sites were blocked with 5% lactogen in Tris-buffered saline (TBS)-T (0.05% Tween 20 in TBS buffer: 20 mm Tris-HCl, 150 mm NaCl, pH 7.5). The membrane was incubated with primary antibody for 30 min at room temperature followed by washing in TBS-T. The membrane was then incubated with secondary antibody for 30 min at room temperature. Colorimetric method employing 3,3′-diaminobenzidine (Sigma) and chemiluminescence method (BIORAD) were used to assess the reaction catalyzed by horseradish peroxidase.
Crystallization
Crystallizations were performed using a protein concentration of 5 mg mL−1 in 50 mm Tris-HCl, pH 7.5 at room temperature using hanging drop vapor diffusion method. The final crystallization conditions were optimized to be 0.1 m MES pH 6.5 containing 10% (w/v) polyethylene glycol 20,000. Isomorphous heavy atom derivatives were prepared by soaking the native crystals with heavy atom compounds at concentrations ranging from 2.5 mm to 5.0 mm prepared in 33% (v/v) glycerol made in mother liquor.
Data Collection and Structure Solution
Data collection for both the native and the heavy atom derivatized crystals was carried out at 120 K on Rigaku RU-H3R rotating anode x-ray generator equipped with Osmic focusing mirrors using MAR345dtb detector (MAR Research), using 33% glycerol (v/v) in the mother liquor as cryo-protectant. The program automar was used for data processing. The crystal structure was determined by the MIR method using SOLVE (Terwilliger, 2003) for heavy atom site determination and phase calculation. Initial phases and heavy atom sites were determined using three heavy atom derivatives: HgCN, Pb(CH3COO)2, and CdCl2, at the resolution of 18 to 3.3 Å, which were further refined using CHECKSOLVE at the resolution of 18 to 2.8 Å.
Model Building, Refinement, and Quality of Model
Phase extension, density modification, and model building were carried out using PHENIX (Adams et al., 2002) at a resolution of 18 to 2.2 Å. CNS (Brünger et al., 1998) was used for the calculation of noncrystallographic symmetry rotation and translation matrices and structure refinement. Ten percent of the reflections, assigned randomly, were used for the calculation of Rfree. TLS refinement was carried out using REFMAC5 (Murshudov et al., 1999; Winn et al., 2001). The quality of structure was assessed using PROCHECK (Laskowski et al., 1993). Structure analysis was carried out using PyMol. Most of the superpositions were carried out using secondary structure superposition and least square method of superposition in COOT (Emsley and Cowtan, 2004) and UCSF Chimera (Pettersen et al., 2004). The dimer interface was analyzed using PISA (Krissinel and Henrick, 2007).
The protein sequence data reported in this article will appear in the UniProt Knowledgebase (http://www.ebi.ac.uk/swissprot/) under the accession number P86190. The coordinates of LS-24 structure and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with accession number 3LP9.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence of LS-24.
Supplemental Figure S2. Comparison of the blades within LS-24 and other hemopexins.
Supplemental Figure S3. Gel filtration chromatography standard curve for Mr determination.
Supplemental Figure S4. Structure-based multiple sequence alignment of repeat sequences.
Supplementary Material
Acknowledgments
We thank Drs. J.K. Batra and D. Mohanty for critically reading the manuscript.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Agrawal KM, Bahl OP. (1968) Glycosidases of Phaseolus vulgaris. II. Isolation and general properties. J Biol Chem 243: 103–111 [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassie L, Noury M, Lepri O, Lahaye T, Christou P, Capell T. (2000) Promoter strength influences polyamine metabolism and morphogenic capacity in transgenic rice tissues expressing the oat adc cDNA constitutively. Transgenic Res 9: 33–42 [DOI] [PubMed] [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J. (1999) Polyamines and environmental challenges: recent development. Plant Sci 140: 103–125 [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Campbell CG. (1997) Grass Pea Lathyrus sativus L.: Promoting the Conservation and Use of Underutilized and Neglected Crops. 18. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome, pp 1–91 [Google Scholar]
- Capell T, Bassie L, Christou P. (2004) Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA 101: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente A, Olías R, Olías JM. (2000) Purification and characterization of broad bean lipoxygenase isoenzymes. J Agric Food Chem 48: 1070–1075 [DOI] [PubMed] [Google Scholar]
- Croy RRD, Hoque MS, Gatehouse JA, Boulter D. (1984) The major albumin proteins from pea (Pisum sativum L): purification and some properties. Biochem J 218: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Galston AW, Sawhney RK. (1990) Polyamines in plant physiology. Plant Physiol 94: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts N, Martens E, Van Aelst I, Proost P, Opdenakker G, Van den Steen PE. (2008) Beta-hematin interaction with the hemopexin domain of gelatinase B/MMP-9 provokes autocatalytic processing of the propeptide, thereby priming activation by MMP-3. Biochemistry 47: 2689–2699 [DOI] [PubMed] [Google Scholar]
- Gironés MA, de la Hoz Caballer B, Martín TM, Agustín MC, Sánchez-Cano M. (2005) Occupational rhinoconjunctivitys and asthma by exposure to Lathyrus sativus flour. Allergol Immunopathol (Madr) 33: 326–328 [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Gohlke U, Betz M, Knäuper V, Murphy G, López-Otín C, Bode W. (1996) The helping hand of collagenase-3 (MMP-13): 2.7 Å crystal structure of its C-terminal haemopexin-like domain. J Mol Biol 264: 556–566 [DOI] [PubMed] [Google Scholar]
- Grela ER, Studziòski T, Matras J. (2001) Antinutritional factors in seeds of Lathyrus sativus cultivated in Poland. Lathyrus Lathyrism Newsletter 2: 101–104 [Google Scholar]
- Gruen LC, Guthrie RE, Blagrove RJ. (1987) Structure of a major pea seed albumin: implication of a free sulphydryl group. J Sci Food Agric 41: 167–178 [Google Scholar]
- Hamza I. (2006) Intracellular trafficking of porphyrins. ACS Chem Biol 1: 627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley SM, Beevers H. (1986) Lectins in castor bean seedlings. Plant Physiol 80: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N, Croy RRD. (1985) The major albumin protein from pea (Pisum sativum L.). Planta 165: 522–526 [DOI] [PubMed] [Google Scholar]
- Higgins TJV, Beach LR, Spencer D, Chandler PM, Randall PJ, Blagrove RJ, Kortt AA, Guthrie RE. (1987) cDNA and protein sequence of a major pea seed albumin (PA 2 : Mr ˜ 26 000). Plant Mol Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Higgins TJV, Chandler PM, Randall PJ, Spencer D, Beach LR, Blagrove RJ, Kortt AA, Inglis AS. (1986) Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J Biol Chem 261: 11124–11130 [PubMed] [Google Scholar]
- Hrkal Z, Vodrážka Z, Kalousek I. (1974) Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur J Biochem 43: 73–78 [DOI] [PubMed] [Google Scholar]
- Huang B, Chu CH, Chen SL, Juan HF, Chen YM. (2006) A proteomics study of the mung bean epicotyl regulated by brassinosteroids under conditions of chilling stress. Cell Mol Biol Lett 11: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Visse R, Nagase H, Acharya KR. (2006) Crystal structure of an active form of human MMP-1. J Mol Biol 362: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne D. (1991) Homology of placental protein 11 and pea seed albumin 2 with vitronectin. Biochem Biophys Res Commun 176: 1000–1006 [DOI] [PubMed] [Google Scholar]
- Jozic D, Bourenkov G, Lim NH, Visse R, Nagase H, Bode W, Maskos K. (2005) X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding. J Biol Chem 280: 9578–9585 [DOI] [PubMed] [Google Scholar]
- Kochhar S, Gartenmann K, Juillerat MA. (2000) Primary structure of the abundant seed albumin of Theobroma cacao by mass spectrometry. J Agric Food Chem 48: 5593–5599 [DOI] [PubMed] [Google Scholar]
- Kolberg J, Michaelsen TE, Sletten K. (1983) Properties of a lectin purified from the seeds of Cicer arietinum. Hoppe Seylers Z Physiol Chem 364: 655–664 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Larkins BA, Hurkman WJ. (1978) Synthesis and deposition of zein in protein bodies of maize endosperm. Plant Physiol 62: 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291 [Google Scholar]
- Lepri O, Bassie L, Safwat G, Thu-Hang P, Trung-Nghia P, Hölttä E, Christou P, Capell T. (2001) Over-expression of a cDNA for human ornithine decarboxylase in transgenic rice plants alters the polyamine pool in a tissue-specific manner. Mol Genet Genomics 266: 303–312 [DOI] [PubMed] [Google Scholar]
- Martin-Tanguy J. (2001) Metabolism and function of polyamines in plants: recent development (new approaches). Plant Growth Regul 34: 135–148 [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Nagai S, Koide M, Takahashi S, Kikuta A, Aono M, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T. (2007) Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol 144: 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita TW, Rogers JC. (1996) Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol 47: 327–350 [DOI] [PubMed] [Google Scholar]
- Osborne TB, Campbell GF. (1898) Proteids of the pea. J Am Chem Soc 20: 348–362 [Google Scholar]
- Palavan-Ünsal N. (1995) Stress and polyamine metabolism. Bulg J Plant Physiol 21: 3–14 [Google Scholar]
- Paoli M, Anderson BF, Baker HM, Morgan WT, Smith A, Baker EN. (1999) Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains. Nat Struct Biol 6: 926–931 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Grimm B. (2001) Regulatory network of tetrapyrrole biosynthesis—studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta 213: 667–681 [DOI] [PubMed] [Google Scholar]
- Pedroche J, Yust MM, Lqari H, Megías C, Girón-Calle J, Alaizn M, Millán F, Vioque J. (2005) Chickpea pa2 albumin binds hemin. Plant Sci 4: 1109–1114 [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Piccard H, Van den Steen PE, Opdenakker G. (2007) Hemopexin domains as multifunctional liganding modules in matrix metalloproteinases and other proteins. J Leukoc Biol 81: 870–892 [DOI] [PubMed] [Google Scholar]
- Porcel S, León F, Calderín PM, Valero A, Botello A, Cuesta EA. (2001) Occupational asthma caused by grass pea used in the industrial processing of parquet. Allergol Immunopathol (Madr) 29: 207–211 [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Sethi DK, Salunke DM. (2006) Purification, identification and preliminary crystallographic studies of an allergenic protein from Lathyrus sativus. Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 869–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa MJS, Ferreira RB, Teixeira AR. (2000) Storage proteins from Lathyrus sativus seeds. J Agric Food Chem 48: 5432–5439 [DOI] [PubMed] [Google Scholar]
- Schlereth A, Standhardt D, Mock HP, Müntz K. (2001) Stored cysteine proteinases start globulin mobilization in protein bodies of embryonic axes and cotyledons during vetch (Vicia sativa L.) seed germination. Planta 212: 718–727 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. (1990) The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J 267: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374: 22–37 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Floh EI, Scherer GF. (2006) Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol 47: 346–354 [DOI] [PubMed] [Google Scholar]
- Valdivieso R, Quirce S, Sainz T. (1988) Bronchial asthma caused by Lathyrus sativus flour. Allergy 43: 536–539 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Chinoy C, Zuther E, Blessington B, Geigenberger P, Domoney C. (2008) Combined metabolomic and genetic approaches reveal a link between the polyamine pathway and albumin 2 in developing pea seeds. Plant Physiol 146: 74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque J, Clemente A, Sánchez-Vioque R, Pedroche J, Bautista J, Millán F. (1998) Comparative study of chickpea and pea Pa2 albumins. J Agric Food Chem 46: 3609–3613 [Google Scholar]
- Vioque J, Sánchez-Vioque R, Clemente A, Pedroche J, Bautista J, Millán F. (1999) Purification and partial characterization of chickpea 2S albumin. J Agric Food Chem 47: 1405–1409 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.