Abstract
A number of Arabidopsis (Arabidopsis thaliana) lesion-mimic mutants exhibit alterations in both abiotic stress responses and pathogen resistance. One of these mutants, constitutive expresser of PR genes22 (cpr22), which has a mutation in two cyclic nucleotide-gated ion channels, is a typical lesion-mimic mutant exhibiting elevated levels of salicylic acid (SA), spontaneous cell death, constitutive expression of defense-related genes, and enhanced resistance to various pathogens; the majority of its phenotypes are SA dependent. These defense responses in cpr22 are suppressed under high-humidity conditions and enhanced by low humidity. After shifting plants from high to low humidity, the cpr22 mutant, but not the wild type, showed a rapid increase in SA levels followed by an increase in abscisic acid (ABA) levels. Concomitantly, genes for ABA metabolism were up-regulated in the mutant. The expression of a subset of ABA-inducible genes, such as RD29A and KIN1/2, was down-regulated, but that of other genes, like ABI1 and HAB1, was up-regulated in cpr22 after the humidity shift. cpr22 showed reduced responsiveness to ABA not only in abiotic stress responses but also in germination and stomatal closure. Double mutant analysis with nahG plants that degrade SA indicated that these alterations in ABA signaling were attributable to elevated SA levels. Furthermore, cpr22 displayed suppressed drought responses by long-term drought stress. Taken together, these results suggest an effect of SA on ABA signaling/abiotic stress responses during the activation of defense responses in cpr22.
Plants have evolved a large number of defense systems to protect themselves against pathogen invasion. Whether these defenses are successful depends on the speed and intensity of their activation. The first line of defense is the basal immune system that is activated by molecules that are conserved among many pathogens (microbe-associated molecular patterns). Pathogens in turn have evolved a number of effector molecules that can block the basal resistance response (Jones and Dangl, 2006; Bent and Mackey, 2007). A second, stronger response to pathogen infection is mediated by resistance (R) genes that can interact with particular effectors (previously termed avirulence factors) from the pathogen or that can recognize effector-induced modifications of plant proteins (Flor, 1971; Bent and Mackey, 2007). One defense mechanism activated by R gene-mediated pathogen recognition is the hypersensitive response (HR), which is characterized by apoptosis-like cell death at and around the site of pathogen entry (Hammond-Kosack and Jones, 1996; Heath, 2000). HR development is usually accompanied by an increase in salicylic acid (SA) and the accumulation of defense-related proteins such as the pathogenesis-related (PR) proteins (Vlot et al., 2008). At later times after infection, elevated SA levels and PR gene expression are also detected in the uninoculated leaves, concurrent with the development of systemic acquired resistance (SAR), a long-lasting, broad-based resistance to subsequent infection (Durrant and Dong, 2004; Grant and Lamb, 2006; Vlot et al., 2008).
Many studies have demonstrated that SA is an important signaling molecule in the pathways conferring local and systemic resistance (Dempsey et al., 1999; Vlot et al., 2008). To identify other components in the pathogen resistance signal transduction pathway, many Arabidopsis (Arabidopsis thaliana) mutants with altered resistance to pathogens have been isolated. One class exhibits constitutively increased SA levels and PR gene expression as well as heightened resistance to pathogen infection. This group includes dnd1, dnd2/hlm1, copine1 (cpn1), constitutive expresser of PR genes22 (cpr22), and ssi4 (Yu et al., 1998; Jambunathan et al., 2001; Yoshioka et al., 2001; Shirano et al., 2002; Balague et al., 2003; Jurkowski et al., 2004). The majority of these mutants share similar phenotypes such as spontaneous HR-like lesions and thus are categorized as lesion-mimic mutants (Moeder and Yoshioka, 2008). Interestingly, it has been reported that some lesion-mimic mutants are environmentally sensitive (i.e. their resistance phenotypes are conditional; Moeder and Yoshioka, 2009). For instance, under high-humidity conditions such as on agar plates or when grown at high temperature, both the spontaneous HR and the enhanced pathogen resistance are suppressed (Jambunathan et al., 2001; Yoshioka et al., 2001; Jambunathan and McNellis, 2003; Xiao et al., 2003; Zhou et al., 2004; Noutoshi et al., 2005). On the other hand, relatively low humidity or cold temperature enhances their SA-related phenotypes, including HR-like cell death (Jambunathan et al., 2001; Zhou et al., 2004).
Some of these lesion-mimic phenotypes are caused by mutations in R genes, such as SSI4 and SLH1 (Shirano et al., 2002; Noutoshi et al., 2005), or by the overexpression of an R gene, such as RPW8 (Xiao et al., 2003), indicating the involvement of environmental factors on R gene-mediated signaling pathway(s). Indeed, similar environmental effects were also reported for the response of wild-type R genes. It is well known that the HR induced by the recognition of Tobacco mosaic virus by the N protein can be completely suppressed when plants are kept above 28°C. When plants are shifted back to 22°C, the HR starts to develop, indicating that there is a temperature-sensitive step in the signaling pathway (Samuel, 1931). Both basal and R gene-mediated resistance against the bacterial pathogen, Pseudomonas syringae, is attenuated by a moderate increase in temperature (Wang et al., 2009). It has also been reported that high humidity (greater than 95% relative humidity [RH]) delayed or reduced the HR and other resistance responses induced by the interaction of the Cladosporium fulvum avirulence factors Avr2, Avr4, and Avr9 and their cognate tomato R proteins Cf-2, Cf-4, and Cf-9, respectively (Hammond-Kosack et al., 1996; May et al., 1996; Wang et al., 2005). These findings suggest that there is a universal factor(s) in defense signaling that is environmentally sensitive.
Abscisic acid (ABA) controls various environmental (abiotic) stress responses, including drought, salinity, and temperature stress, and many components involved in these responses have been identified (Shinozaki et al., 2003). Additionally, it is becoming clear that ABA is also involved in biotic stress responses in a complex manner. For instance, treatment with exogenous ABA prior to pathogen infection induces enhanced susceptibility in various plant species (Mauch-Mani and Mauch, 2005). Mohr and Cahill (2003, 2006) suggested that the mechanisms behind this phenomenon are likely related to the antagonistic effect of ABA on SA signaling. Similarly, several groups have reported that virulent P. syringae DC3000 enhances the production of ABA during pathogenesis (Schmelz et al., 2003; de Torres-Zabala et al., 2007). Furthermore, Yasuda et al. (2008) suggested the antagonism between SA and ABA signaling in SAR. These studies suggest that ABA plays a negative role in pathogen resistance. In contrast, Melotto and colleagues (2006) reported that ABA-dependent stomata closure is part of plant innate immunity against bacterial invasion and that SA is required for this response. They also reported that aba3-1, an ABA-deficient mutant, was more susceptible to P. syringae DC3000, suggesting a positive role of ABA in innate immunity (Melotto et al., 2006).
Here, we attempt to characterize the effects of humidity on pathogen resistance responses using the lesion-mimic mutant cpr22. Previously, we reported that most phenotypes of cpr22, such as spontaneous lesion formation, SA accumulation, and constitutive PR gene expression, were suppressed under high RH (Yoshioka et al., 2001). cpr22 contains a deletion that fuses two cyclic nucleotide-gated ion channel (CNGC)-encoding genes, AtCNGC11 and AtCNGC12, generating the novel chimeric AtCNGC11/12 (Yoshioka et al., 2006). We proposed that the expression of AtCNGC11/12 activates pathogen resistance responses through the same signal transduction pathway used by R genes and that cell death induced by the expression of AtCNGC11/12 is HR-like programmed cell death (Yoshioka et al., 2006; Urquhart et al., 2007). Here, we report intriguing alterations in ABA-related phenotypes in cpr22. Our data demonstrate that elevated SA accumulation is the cause of these alterations, suggesting complex SA-ABA cross talk during lesion formation.
RESULTS
Humidity Shift Influences cpr22 Phenotypes
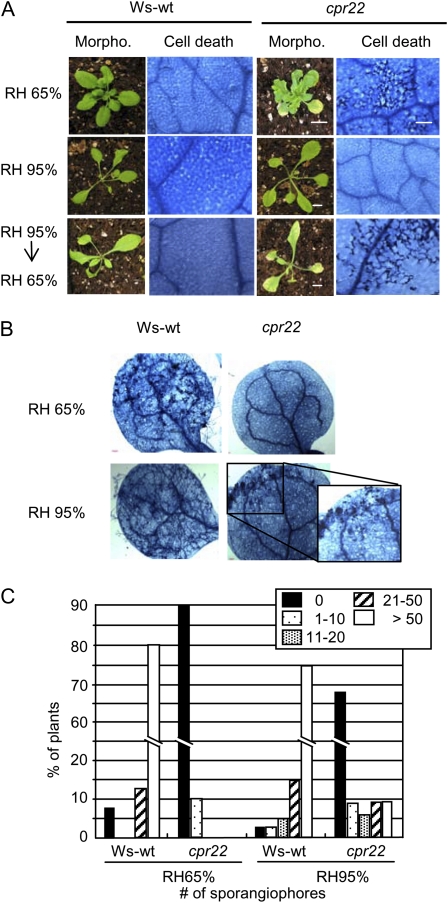
Under moderate RH (approximately 65%), heterozygous cpr22 plants display stunted growth, HR-like cell death, and induction of PR genes. Furthermore, cpr22 in the homozygous state is lethal (Yoshioka et al., 2001). However, when grown under high RH (greater than 90%), all SA-related phenotypes including spontaneous cell death and lethality were suppressed (Fig. 1A, top row; Yoshioka et al., 2001). To test the effects of environmental conditions further, plants grown under high RH on soil were shifted to moderate RH. HR-like cell death appeared approximately 2 d after shifting and gradually became more visible and severe (Fig. 1A). Quantitative analysis using ion conductivity measurements also supported this observation (Supplemental Fig. S1A). When grown under slightly cooler temperature conditions (16°C versus 22°C), the cpr22 phenotypes were enhanced and plants were even more stunted. Additionally, plants shifted from 22°C to 16°C showed more intense cell death (Supplemental Fig. S1, A and B). In contrast, Wassilewskija (Ws; the background ecotype of cpr22) wild-type plants (Ws-wt) that underwent the same treatments did not exhibit cell death (Fig. 1A; Supplemental Fig. S1). Similar phenomena have also been observed in other lesion-mimic mutants, such as ssi4 and cpn1 (Jambunathan et al., 2001; Zhou et al., 2004).
Figure 1.
Effect of humidity on cpr22 phenotypes. A, Morphology, chlorotic phenotypes, and spontaneous cell death formation of 4-week-old cpr22 and Ws-wt plants grown under 65% RH (top row), under 95% RH (middle row), or under 95% RH and shifted to 65% RH for 2 d (bottom row) at 22°C. B and C, Growth of H. arabidopsidis Emwa1 on Ws-wt and cpr22 plants grown under 65% or 95% RH conditions. Cotyledons of 7-d-old seedlings were inoculated with H. arabidopsidis Emwa1 (106 spores mL−1). At 7 d post infection, infected cotyledons were stained by trypan blue to visualize pathogen growth (B) or the number of sporangiophores on two cotyledons per plant was determined (C). Experiments were done three times with similar results. Bars = 1 cm (plants) and 125 μm (trypan blue staining).
Zhou et al. (2004) reported that in addition to HR-like cell death, enhanced pathogen resistance is also affected by RH in ssi4. To test if this is also the case in cpr22, Ws-wt and cpr22 plants that had been grown under moderate and high RH were infected with the oomycete pathogen Hyaloperonospora arabidopsidis isolate Emwa1, which is virulent on the Ws ecotype. While cpr22 plants grown under moderate RH displayed strong resistance compared with wild-type plants, cpr22 plants grown under high RH displayed a partial breakdown of its enhanced resistance phenotype (Fig. 1, B and C). The reason we observe only a partial breakdown is likely related to the fact that our high-humidity condition does not suppress cpr22 phenotypes perfectly (e.g. we have to open the growth chamber's door for irrigation). Slight accumulation of SA and up-regulation of defense-related genes were observed under this condition in cpr22 (see Fig. 3B below; Supplemental Table S1).
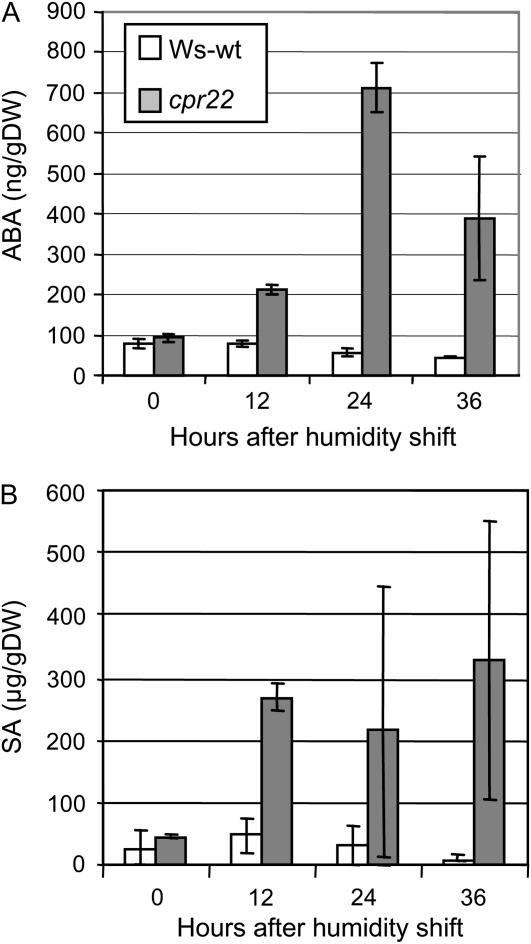
Figure 3.
ABA and SA increase after humidity shift in cpr22. ABA (A) and SA (B) levels in Ws-wt (white bars) and cpr22 (gray bars) at 0, 12, 24, and 36 h after the shift from 95% RH to 65% RH conditions were measured. The data represent mean levels of three extracts. Each extract was made from three to five plants. Experiments were repeated four times with similar results. DW, Dry weight.
In order to test whether the suppression of HR in high RH can be generalized to natural pathogen resistance responses, Arabidopsis wild-type plants were grown under moderate RH and shifted to high RH for 7 d prior to pathogen infection. Plants that had been shifted to high RH exhibited less visual AvrRpt2-induced HR formation as shown by trypan blue staining (Supplemental Fig. S2A). Quantitative ion leakage also supported this observation (Supplemental Fig. S2B). Bacterial growth was also enhanced in high-humidity plants, indicating an attenuated resistance response under high-humidity conditions (Supplemental Fig. S2C; note that bacterial inoculum was infiltrated, so altered stomata conductance is not the cause of the observed difference.). These data support the notion that an environmentally sensitive factor(s) is part of at least some R gene signal transduction pathways.
Expression of a Subset of ABA Signaling Genes Is Altered in cpr22 after Humidity Shift
To determine how humidity alters defense responses, microarray analysis was performed to screen for altered gene expression. cpr22 and wild-type plants were grown in high RH, and samples were taken before and 24 h after the plants had been shifted to moderate RH. Two independent experiments were performed, and more than 10 plants were pooled for each RNA extraction. Comparison of gene expression between the mutant and the wild type at 0 and 24 h after the shift identified genes that showed more than 2-fold changes (induced or repressed) in both experiments. Many up-regulated genes were related to pathogen defense, such as PR-1, EDS1, PAD4, WRKY70, and various R genes (Uknes et al., 1992; Falk et al., 1999; Jirage et al., 1999; Li et al., 2004), suggesting that the shift from high to low humidity induced resistance responses in cpr22 as we expected (Supplemental Table S1). Similar results were obtained in the microarray analysis of another defense mutant, ssi4 (Supplemental Table S1). In addition, the SA biosynthesis genes, ISC1 and EDS5, were up-regulated in ssi4 after the shift. However, in cpr22, these genes were already up-regulated before the shift. This could be due to the imperfect suppression of defense responses as mentioned above.
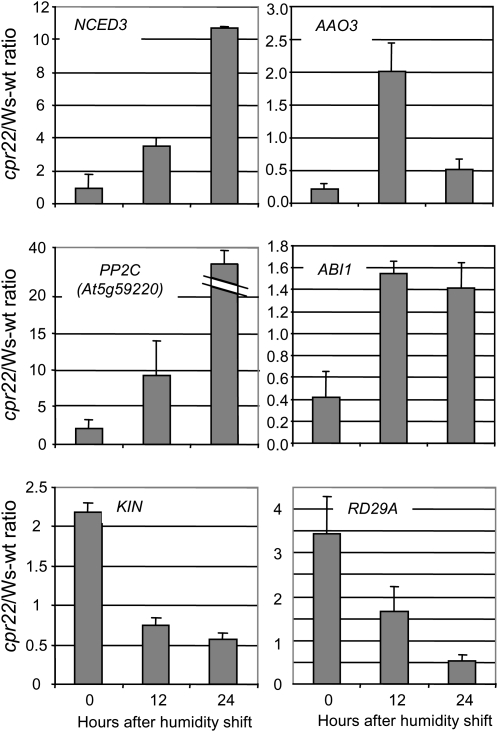
Interestingly, a number of genes related to ABA biosynthesis and signaling were also altered in cpr22 (Supplemental Table S1). For example, the ABA biosynthetic genes 9-cis-epoxycarotenoid dioxygenase3 (NCED3) and abscisic aldehyde oxidase3 (AAO3) were up-regulated. One ABA 8′-hydroxylase isoform, CYP707A4, was also induced. ABA 8′-hydroxylases are considered to be the key catabolitic enzymes for ABA metabolism (Nambara and Marion-Poll, 2005). Several protein phosphatases of the 2C class (PP2C) including ABA-insensitive1 (ABI1), HAB1, and At5g59220 (PP2C) were also induced. Other induced ABA signaling components included RD26, ERA1, and RD20. These genes have previously been shown to be induced by ABA and drought stress (Shinozaki et al., 2003). Although the expression of the transcription factors AtMYC2 and ABF3/4, which are in the ABA signaling pathway, was not significantly altered, a number of components that are under the control of these transcription factors, such as RD22, RD29A, KIN1/2, and ERD3, were down-regulated after the shift. Additionally, the newly identified ABA receptor genes, Pyrabactin resistance1 (PYR1) and some PYR1-like (PYL) genes (Park et al., 2009), were suppressed (Supplemental Table S1). Suppression of PYR1/PYL genes by exogenous ABA has been reported by Park et al. (2009). The expression patterns of NCED3, AAO3, PP2C, ABI1, RD29A, and KIN1/2 were monitored by quantitative real-time PCR, including an additional time point (12 h after the shift; Fig. 2). As observed in the microarray analysis, NCED3, AAO3, PP2C, and ABI1 were induced while RD29A and KIN1/2 were down-regulated after the humidity shift. Again, similar results were obtained in the microarray analysis in ssi4 (Supplemental Table S1).
Figure 2.
Quantitative real-time PCR analysis of NCED3, AAO3, PP2C, ABI1, RD29A, and KIN1/2 expression in cpr22 at 0, 12, and 24 h after shift from 95% RH to 65% RH conditions. Transcript levels were normalized to the expression of 18S RNA. Shown is the ratio of cpr22 to Ws-wt. Each bar represents the mean level of three replicates ± se. The experiment was repeated three times with similar results.
Increased ABA Levels Were Observed in cpr22 after Humidity Shift
Since ABA biosynthetic genes were up-regulated after humidity shift, the endogenous levels of ABA were analyzed. At 0 h, the levels of ABA were identical in mutant and wild-type plants. At 24 h after the shift, ABA levels were significantly increased in cpr22, whereas in Ws-wt they did not change substantially (Fig. 3A). These data, together with the microarray/quantitative PCR results, indicate that the humidity shift induces the ABA biosynthetic pathway and leads to heightened accumulation of ABA in cpr22 mutants. The increased level of ABA was transient, gradually decreasing by 36 h post humidity shift. Although the absolute amount of ABA increase was variable from experiment to experiment, we observed the same trend in four independent experiments. Endogenous SA levels also were analyzed to determine whether the humidity shift altered SA accumulation. Similar to ABA, although the absolute amount was variable in each experiment, we observed increased SA levels in cpr22 in four independent experiments and also in ssi4 (Fig. 3B; Supplemental Fig. S3).
cpr22 Plants Display Partial Insensitivity to ABA
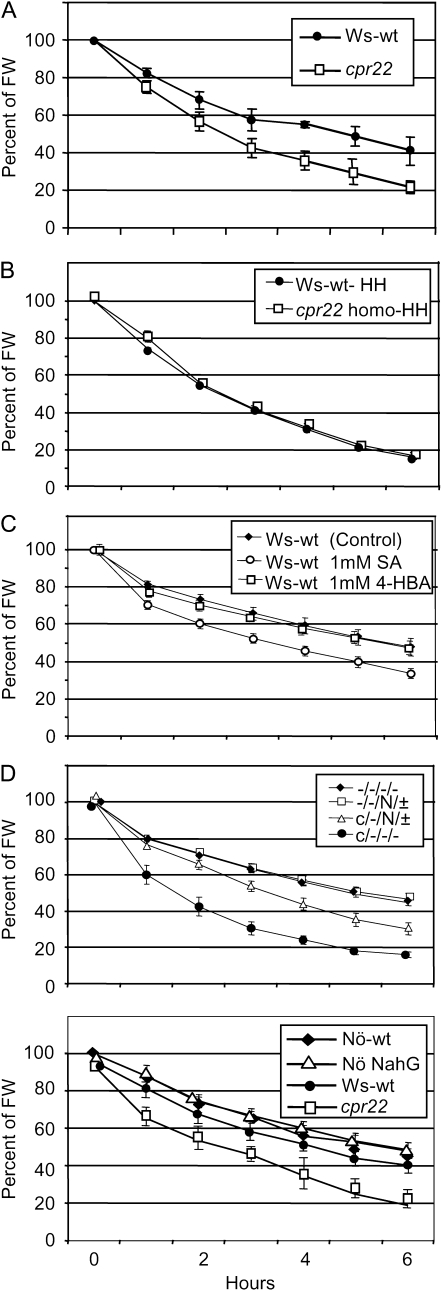
Since the microarray/quantitative PCR data indicated an alteration in ABA signaling in cpr22, we suspected that cpr22 may have altered sensitivity to ABA. To address this point, the rate of water loss was analyzed using a well-established method (Nambara et al., 1998). Interestingly, cpr22 showed enhanced water loss compared with wild-type plants (Fig. 4A), indicating reduced responsiveness to dehydration. Similar results were also obtained with the other humidity-sensitive lesion-mimic mutants, ssi4 and cpn1 (Supplemental Fig. S4A).
Figure 4.
cpr22 displays enhanced dehydration. Plants were weighed at various times after detachment from their roots. A, Ws-wt (black circles) and cpr22 heterozygous (white squares) plants grown under ambient humidity conditions. B, Ws-wt (black circles) and cpr22 homozygous (white squares) plants grown under 95% RH conditions. C, Ws-wt plants 16 h after soil drenching with 1 mm SA (white circles), 4-hydroxybenzoic acid (4-HBA; white squares), or control solution (0.1% methanol in water, the same solution in which SA was dissolved; black diamonds). D, F2 progeny of cross-pollination between nahG transgenic (Nö background) and cpr22 (Ws background) plants. −/−/−/− plants carry neither cpr22 nor nahG (black diamonds), −/−/N/± plants carry nahG but not cpr22 (white squares), c/−/N/± plants are heterozygous for cpr22 and carry nahG (white triangles), and c/−/−/− plants are heterozygous for cpr22 but do not carry nahG (black circles). All plants were genotyped prior to experiments by PCR-based molecular markers (see “Materials and Methods”). Each bar represents the mean level of five plants ± se. All experiments were repeated three times with similar results. FW, Fresh weight.
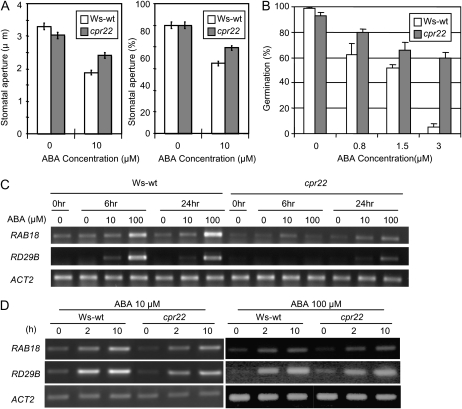
The increased water loss in cpr22 plants suggested that guard cell response in cpr22 might be attenuated. Therefore, the responsiveness of cpr22 guard cells to exogenous ABA was assessed by monitoring stomotal closure. As expected, guard cells in cpr22 plants were significantly less responsive to ABA than those of wild-type plants (Fig. 5A). We also tested whether ABA-induced inhibition of seed germination was altered. Sixty percent of cpr22 seeds were able to germinate in the presence of 3 μm ABA compared with only 5% of wild-type seeds, indicating reduced sensitivity to ABA (Fig. 5B). However, with increasing ABA concentrations, germination of cpr22 seeds was gradually suppressed (Supplemental Fig. S5). To further investigate this ABA insensitivity at the molecular level, the expression of several ABA marker genes upon ABA treatment was analyzed in soil-grown plants. The induction of RAB18 and RD29B was attenuated in cpr22 after ABA treatment under ambient humidity (Fig. 5C). In summary, all of the above analyses indicate a reduced sensitivity of cpr22 to ABA.
Figure 5.
cpr22 plants display reduced sensitivity to ABA. A, Impairment in ABA-induced stomatal closure of Ws-wt (white bars) and cpr22 (gray bars) plants in the presence or absence of 10 μm ABA. Stomatal aperture is shown in μm (left panel) and in percentage of untreated plants (right panel); n = 3 experiments, 30 stomata per condition per experiment. Student's t test shows significant difference between the wild type and cpr22. B, Effects of ABA on germination of Ws-wt (white bars) and cpr22 homozygous (gray bars) seeds. The experiment was conducted with approximately 100 seeds per plate. Each bar represents the mean level of three plates ± se. Student's t test shows significant difference between Ws-wt and cpr22. The experiment was repeated more than three times with similar results. C, Four-week-old plants grown on soil were sprayed with 0, 10, and 100 μm ABA. Samples were taken at 0, 6, and 24 h after treatment, and gene expression was analyzed by semiquantitative RT-PCR. D, Three-week-old plants grown on MS agar plates were soaked with 10 or 100 μm ABA solution (for details, see “Materials and Methods”). Samples were taken at 0, 2, and 10 h after treatment, and gene expression was analyzed by semiquantitative RT-PCR. Actin (ACT2) served as a loading control in both experiments. All experiments were repeated three times with similar results.
Elevated Levels of SA Are the Cause of the Alterations in ABA-Related Phenotypes in cpr22
A major change in cpr22 after humidity shift is an increase in its endogenous SA levels. Therefore, we assessed whether increased accumulation of SA in cpr22 causes the observed partial insensitivity to ABA. Since cpr22 does not accumulate SA on Murashige and Skoog (MS) agar plates (Yoshioka et al., 2001) and this condition suppresses cpr22 phenotypes even more efficiently than a high-RH growth chamber, we tested how plants grown on MS agar plates respond to ABA treatment with respect to ABA-responsive genes. While cpr22 plants that were grown on soil under ambient RH displayed a clear attenuation in RD29B and RAB18 gene expression upon ABA treatment (Fig. 5C), cpr22 plants that were grown on MS agar plates showed almost no difference in the induction of these genes (Fig. 5D), indicating that heightened SA levels may result in ABA insensitivity.
These results led us to test whether the enhancement of water loss in cpr22 is also due to SA accumulation. As shown in Figure 4B, high-RH growth conditions suppressed the enhanced water loss observed in cpr22 plants. This observation led us to assess the effect of SA treatment on water loss in wild-type plants. Strikingly, pretreatment with 1 mm SA by soil drenching increased water loss in wild-type plants. This increased water loss was not observed in wild-type plants treated with control solution or the biologically inactive SA analog 4-hydroxybenzoic acid (Fig. 4C). These data indicated that SA has a strong effect on the water loss stress responses in general and that elevated levels of SA are the likely cause of enhanced water loss in cpr22. We also investigate if ABA can rescue this water loss phenotype in cpr22. Strikingly, 10 μm ABA treatment partially rescued the phenotype, suggesting an SA-ABA antagonism (Supplemental Fig. S4B). To further test whether ABA is required for the SA-induced water loss phenotype, we used the ABA biosynthesis mutant aba2-2 (Nambara et al., 1998). As shown in Supplemental Figure S4C, we did not observe any difference between control solution-or SA-treated aba2-2 mutants, suggesting that the SA-inducible enhanced water loss phenotype of wild-type plants (Fig. 4C) in our experimental setting requires functional ABA biosynthesis. Alternatively, since aba2-2 plants lose water much faster than wild-type plants, SA effects may not be seen (Supplemental Fig. S4C). Collectively, the data suggested that the elevated level of SA is responsible for the observed attenuation of ABA responses in cpr22.
To further confirm this possibility, we asked whether removal of SA would reverse the observed ABA-related phenotypes. To test this, we took a genetic approach utilizing cpr22 plants carrying the nahG gene, which degrades SA into biologically inactive catechol (Gaffney et al., 1993; Yoshioka et al., 2001). We first tested the effect of nahG on SA accumulation after humidity shift. As shown in Supplemental Figure S6A, humidity shift induced a significant increase of SA in cpr22 plants (Ws and Nössen [Nö] mixed background; c/−/−/−) but not in cpr22/nahG plants (Ws and Nö mixed background; c/−/N/±), indicating that the nahG transgene effectively suppresses the accumulation of SA. Cell death enhancement after humidity shift was also suppressed in cpr22/nahG plants (Supplemental Fig. S6B). The suppression of cell death was further supported by quantitative ion leakage analysis (Supplemental Fig. S6C). These results indicate that the nahG transgene suppresses humidity shift-induced cell death formation in cpr22 through the suppression of SA accumulation.
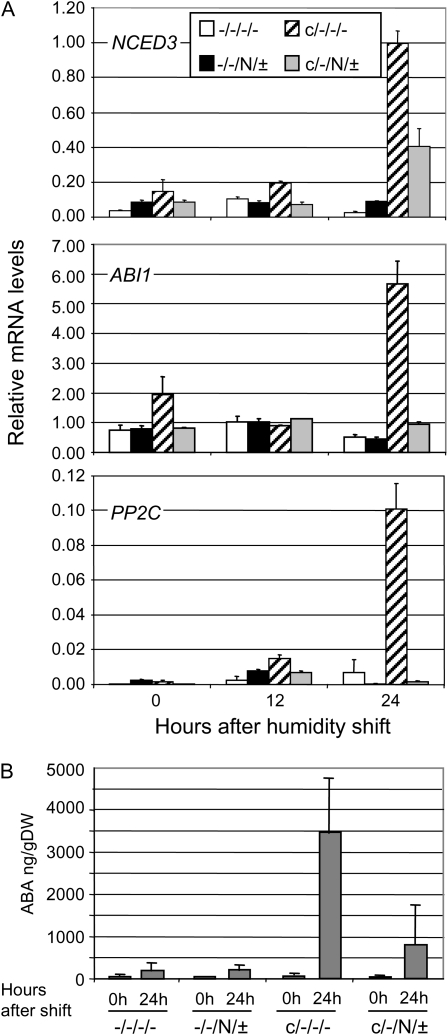
To further confirm the role of SA, ABA-related phenotypes in cpr22/nahG were determined. As expected, cpr22/nahG (Ws and Nö mixed background; c/−/N/± in Fig. 4D) plants displayed significantly less water loss than cpr22. Moreover, induction of the ABA biosynthetic gene NCED3 as well as the ABA signaling genes ABI1 and PP2C after humidity shift was significantly suppressed in cpr22/nahG double mutant plants (Ws and Nö mixed background; c/−/N/± in Fig. 6A) compared with cpr22 (Ws and Nö mixed background; c/−/−/− in Fig. 6A).
Figure 6.
The nahG transgene rescues ABA-related phenotypes in cpr22. A, Quantitative real-time PCR analysis of NCED3, ABI1, and PP2C expression in F2 progeny of cross-pollination between nahG transgenic and cpr22 plants at 0, 12, and 24 h after shift from 95% RH to 65% RH. Transcript levels were normalized to the expression of 18S rRNA (multiplied by 1,000 for clarity). Each bar represents the mean level of three replicates ± se. Experiments were repeated three times with similar results. B, Elevated SA levels cause ABA accumulation in cpr22. F2 progeny of cross-pollination of nahG transgenic (Nö background) and cpr22 (Ws background) plants were used: −/−/−/− plants carry neither cpr22 nor nahG, −/−/N/± plants carry nahG but not cpr22, c/−/−/− plants are heterozygous for cpr22 but do not carry nahG, and c/−/N/± plants are heterozygous for cpr22 and carry nahG. All plants were grown under 95% RH conditions and then shifted to 65% RH conditions. ABA levels at 0 and 24 h after the shift from 95% RH to 65% RH conditions were measured. All plants were genotyped prior to experiments (see “Materials and Methods”). The data represent mean levels of three extracts. Each extract was made from three to five plants. Student's t test shows significant difference between cpr22 and cpr22/nahG plants. DW, Dry weight.
In addition, endogenous levels of ABA were significantly suppressed in cpr22/nahG (Ws and Nö mixed background; c/−/N/± in Fig. 6B) compared with cpr22 (Ws and Nö mixed background; c/−/−/− in Fig. 6B). Taken together, these data suggest that elevated SA levels are the cause of alterations of ABA-related phenotypes and enhanced ABA accumulation in cpr22. Note that we do not know whether reduced amounts of basal SA can affect ABA signaling. That is beyond the scope of this study. However, we have not seen any significant change in the water loss phenotype in nahG plants, which have a slightly lower basal level of SA compared with wild-type plants.
cpr22 Exhibits an Attenuated ABA Response upon Moderate Drought Stress
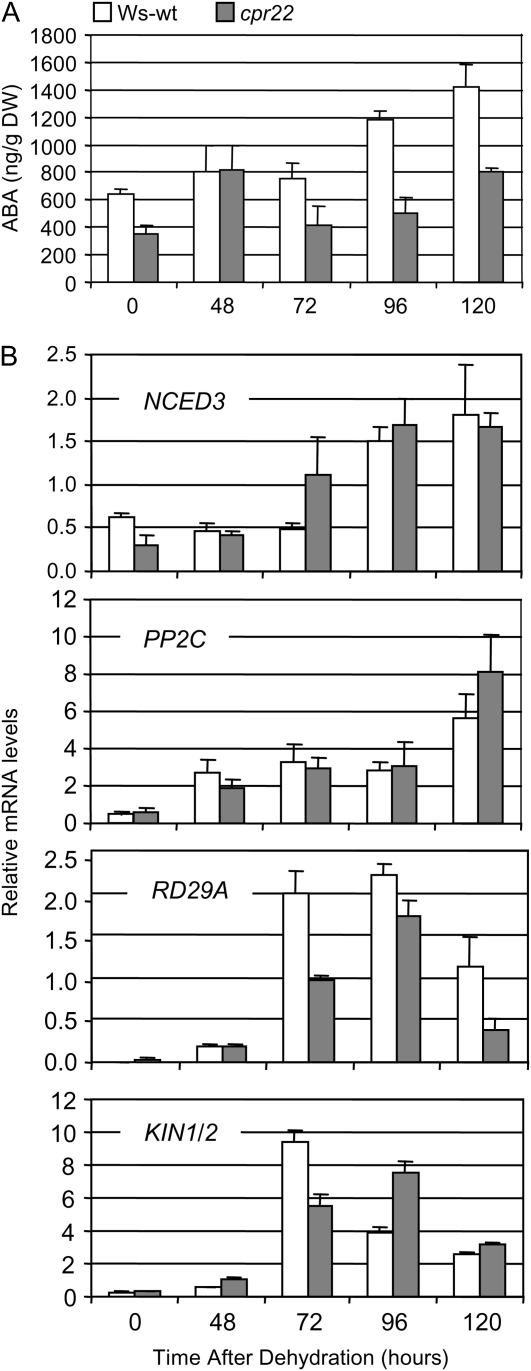
In order to subject plants to a more natural drought situation, we mimicked drought stress by terminating irrigation and monitored ABA levels over a time course of 120 h. Interestingly, while ABA levels in cpr22 plants increased initially and then returned to control levels, those of wild-type plants increased continuously over the time period analyzed (Fig. 7A). In addition, while the transcript levels of NCED3 and PP2C in cpr22 were mostly comparable to those of the wild type, the expression of KIN1/2 and RD29A showed a delayed and lower peak than in wild-type plants (Fig. 7B). These data suggest that under ambient humidity conditions, when SA levels are already increased in cpr22 plants (Supplemental Fig. S7), both ABA biosynthesis and downstream gene expression are attenuated. This attenuation likely prevents cpr22 from responding properly to this abiotic stress, suggesting another node of antagonism between SA and ABA in moderate long-term drought stress responses in addition to the one observed in acute stress induced by humidity shift.
Figure 7.
cpr22 plants display an attenuated drought stress response. Moderate drought stress was applied by terminating irrigation using 4-week-old plants. The first sample was taken when the soil surface became dry (0 h). A, ABA levels in Ws-wt (white bars) and cpr22 heterozygous (gray bars) plants. The data represent mean levels of three extracts. Each extract was made from three to five plants. B, Quantitative real-time PCR analysis of NCED3, PP2C, RD29A, and KIN1/2 expression in Ws-wt (white bars) and cpr22 (gray bars) plants. Transcript levels were normalized to the expression of 18S rRNA (multiplied by 1,000 for clarity) measured in the same samples. Each bar represents the mean level of three replicates ± se. Experiments were repeated three times with similar results. DW, Dry weight.
DISCUSSION
It has been known for a long time that HR and pathogen resistance are influenced by temperature and humidity (Samuel, 1931; Hammond-Kosack et al., 1996; May et al., 1996). Here, we also show that the HR induced by avirulent P. syringae (avrRpt2) is suppressed when Arabidopsis plants are kept under high RH. However, when analyzing the effect of environmental conditions on pathogenesis, difficulties arise if both plants and microorganisms are used, because we cannot conclude whether the plant, the microorganism, or both are being affected by environmental factors. Furthermore, it has been reported that some effectors of microbes manipulate plant hormonal signaling (de Torres-Zabala et al., 2007; Grant and Jones, 2009). Therefore, it is difficult to study the environmental effect on the plant defense response using pathogens from a hormonal point of view. Interestingly, spontaneous HR-like cell death and pathogen resistance in some Arabidopsis mutants/transgenic plants have also been revealed to be sensitive to temperature, humidity, and light conditions, as in the aforementioned plant-pathogen systems (Moeder and Yoshioka, 2009). Therefore, the use of these environmentally sensitive mutants can be an effective way to analyze these phenomena.
Here, we used a well-studied environmentally sensitive lesion-mimic mutant, cpr22, whose conditional phenotype is useful to analyze the induction of HR and other defense responses (Moeder and Yoshioka, 2009). Our study revealed alterations in ABA signaling and ABA-related phenotypes in this mutant. In contrast to the wild type, cpr22 transiently increased endogenous ABA after being shifted from high RH to ambient RH, coincident with an increase in SA. cpr22 also exhibited ABA-insensitive phenotypes, such as reduced stomatal closure and enhanced water loss. Some of these findings were also confirmed with other lesion-mimic mutants such as ssi4 and cpn1 (Supplemental Fig. S4A). Both the ABA increase and ABA insensitivity are due to enhanced SA levels. Why ABA levels transiently increase during HR development in cpr22 is currently not known. There are three likely possibilities: (1) it is the consequence of the plants sensing increased dehydration caused by acute cell death; (2) it is the consequence of the suppression of downstream ABA signaling by SA (possible positive feedback mechanism or loss of negative feedback); or (3) ABA may play an active role to promote HR cell death in these mutants. Since cold shift treatment also enhanced HR development (Supplemental Fig. S1) and such conditions generally induce ABA accumulation, it is tempting to favor the third possibility. Interestingly, Dong et al. (1999, 2005) reported that HrpN, a protein that is produced by the plant pathogenic bacterium Erwinia amylovora and stimulates pathogen resistance responses including HR development in a SA-dependent manner, also increased plant ABA levels. This suggests a connection between the increased amounts of ABA and SA accumulation and HR development. To address this question, we tested the effect of ABA treatment on various environmentally sensitive lesion-mimic mutants. However, so far we have not obtained conclusive results. This could be due to difficulties in experimentally recreating the fine balance and timing of endogenous levels of ABA and SA by exogenous treatment.
The observed partial insensitivity to ABA in cpr22 is intriguing, since there are many reports suggesting that ABA-insensitive mutants display enhanced resistance against various pathogens. For example, the ABA-deficient tomato (Solanum lycopersicum) mutant sitiens shows both an increase in SA-mediated responses and greater resistance to P. syringae DC3000 and Botrytis cinerea (Audenaert et al., 2002). Furthermore, the ABA-insensitive Arabidopsis mutants abi1-1 and abi2-1, as well as a 35S:HAB1 transgenic line, were 20- to 80-fold more resistant to virulent P. syringae than wild-type plants (de Torres-Zabala et al., 2007). These reports and our data strongly suggest the importance of the suppression of ABA signaling to enhance pathogen resistance. Additionally, our data indicate that this partial ABA insensitivity is correlated with the suppression of a subset of genes, such as RD29A and KIN1/2, in the ABA signaling pathway through the action of SA.
There was a significant induction of several PP2C genes, including ABI1, in cpr22 as well as ssi4 after humidity shift. The products of the ABA-inducible group of PP2C genes have been reported to antagonize the action of some kinases that are regulated by ABA, through dephosphorylation of their targets, and to play a significant role in ABA signaling (Schweighofer et al., 2004; Yoshida et al., 2006; Park et al., 2009). Therefore, it is tempting to think that SA, induced by humidity shift, activates these phosphatase genes in order to suppress the subset of genes that play a role in abiotic stress responses in ABA signaling, such as RD29A. In this scenario, SA may play a role to shift plants from abiotic stress to biotic stress response mode. Further analysis is required for complete understanding of the role of PP2C genes.
We also have observed different responses in experiments applying acute stress (humidity shift) and moderate long-term stress (drought) in cpr22. Under long-term drought stress, accumulation of ABA was suppressed in cpr22 compared with the wild type, and the mutant showed less tolerance to drought stress. A major difference that may have affected the results of the two types of experiments is the SA level at the start of the experiment. Plants used for the drought experiment were grown in ambient humidity; thus, cpr22 plants already contain high SA levels, which mimic a state of pathogen infection. On the other hand, in the humidity shift experiment, plants started with relatively low levels of SA, which then increased greatly over the course of the experiment. Mohr and Cahill (2003, 2006) suggested that the levels of ABA at the time of pathogen challenge may be crucial for resistance phenotypes. Our data suggest that the same may be true in the opposite direction; that is, the levels of SA at the time of an abiotic stress challenge may be crucial for the abiotic stress response. Alternatively, since our analysis could not detect ABA derivatives, it is possible that under drought stress responses, ABA derivatives accumulated but could not be detected. This possibility should be addressed in the future.
Regarding this, Yasuda et al. (2008) reported that SAR activators that can induce SA accumulation suppress salt-induced expression of ABA-inducible genes, indicating that SA may negatively affect ABA signaling. Our drought stress experiment supports their findings; however, the humidity shift experiments contradict their data, indicating that the SA-ABA cross talk could be more complex than previously thought. Further detailed studies are required to address this discrepancy.
The observed antagonistic effects of ABA and SA on the water loss phenotype is intriguing. It has been reported that some phenolic compounds, including SA, reverse ABA-induced stomatal closure (Rai et al., 1986), thus supporting our results. Although the biological significance of this phenomenon in defense responses is not known, our data further indicate antagonism between ABA and SA in response to water stress. Interestingly, Noutoshi et al. (2005) analyzed drought stress responses of the Arabidopsis mutant sensitive to low humidity1 (slh1). This mutant has a mutation in a TIR-NBS-LRR-WRKY-type R gene and shows typical lesion-mimic phenotypes, such as heightened levels of SA and enhanced pathogen resistance resembling those of cpr22. However, slh1 exhibits normal responses to drought stress and ABA (Noutoshi et al., 2005). Therefore, the comparison of ABA-related phenotypes between slh1 and cpr22 could provide further evidence for signaling complexity.
Recently, a suppressor screen for snc1, another gain-of-function R gene mutant like ssi4, identified mos8, which is a novel allele of ENHANCED RESPONSE TO ABA1 (ERA1; Goritschnig et al., 2008). ERA1 encodes the β-subunit of a protein farnesyltransferase that plays an important role in ABA responses (Cutler et al., 1996). This indicates that hypersensitivity to ABA may suppress the snc1 enhanced pathogen resistance phenotypes, including SA accumulation. This correlates well with ABA insensitivity in the pathogen-resistant mutants cpr22 and ssi4. Additionally, Wawrzynska et al. (2008) also conducted a suppressor screen of enhanced disease resistance1 and identified a suppressor that has a mutation in the Keep on going gene, a regulator of ABA (Stone et al., 2006). Although in both cases the mechanism of suppression of pathogen resistance by these ABA-related mutations is not yet clear, this type of forward genetics approach could be a powerful method to investigate ABA-SA cross talk.
Hormonal cross talk is essential for living organisms to survive under natural conditions. This is especially important for plants, since they must continuously adjust their physiology to the ever-changing environment throughout their life cycle. The primary focus of this study is to understand the effect of environmental conditions on pathogen resistance. An increasing body of evidence suggests that the environmental conditions affect defense responses against pathogens and that these effects are likely related to hormonal cross talk. The data presented here demonstrate, to our knowledge for the first time, both SA-induced overinduction of ABA and ABA insensitivity in lesion-mimic mutants. A future challenge is to uncover the key elements that act as nodes during this cross talk and how these elements work at the molecular level.
MATERIALS AND METHODS
Plant Growth Conditions and Pseudomonas syringae and Hyaloperonospora arabidopsidis Infections
Arabidopsis (Arabidopsis thaliana) plants were grown on Pro-Mix soil (Premier Horticulture; http://www.premierhort.com/) in a growth chamber under RH 65% or RH 95% under a 14-h/10-h light/dark regimen at 22°C. Pathogen infections were conducted as described previously (Yoshioka et al., 2006).
Genotyping, RNA Extraction, and Reverse Transcription-PCR
PCR-based marker analysis for cpr22 and its homozygosity and the nahG gene was performed as described previously (Yoshioka et al., 2001, 2006; Baxter et al., 2008). RNA extraction and reverse transcription-PCR were performed as described previously (Baxter et al., 2008). For primer sequences, see Supplemental Table S2.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed using a Bio-Rad Chromo4 real-time PCR detector. The amplification reaction mixture was created using Applied Biosystems Power SYBR Green PCR Master Mix (http://www3.appliedbiosystems.com/) according to the manufacturer's instructions. Thermal cycling conditions consisted of 3 min at 95°C and 40 cycles of 10 s at 95°C, 20 s at 52°C, and 20 s at 72°C. The results were obtained and analyzed using the Opticon Monitor version 3.1 (Bio-Rad). Transcript levels were normalized to the expression of 18S rRNA measured in the same samples (Sakuma et al., 2006; for primer sequences, see Supplemental Table S2).
Microarray Analysis
All plants were grown in high RH, and samples were taken before (0 h) and 24 h after the plants had been shifted to moderate RH. Two independent experiments were performed (biological replicates), and at least 10 plants were pooled for each RNA extraction. Total RNA was extracted using the RNeasy kit (Qiagen; http://www.qiagen.com/). The microarray procedure was conducted as described previously (Bassel et al., 2008). Microarrays were preprocessed together using GC-robust multiarray analysis (Wu et al., 2004). Expression data were filtered to remove probe sets with low expression and low variance across all arrays (genefilter; Gentleman et al., 2004; minimum intensity of 100 on a minimum of 25% of arrays, and minimum interquartile range of 0.5 on the log2 scale). Genes that showed more than 2-fold changes between mutant and the wild type (induced or repressed) in their expression levels in both experiments were identified. Annotations of Arabidopsis genes based on the probe set identifiers were obtained from The Arabidopsis Information Resource (www.arabidopsis.org).
Microarray data are available from the Bio-Array Resource for Plant Functional Genomics (BAR) Web site under Project 50 of BAR's project browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_proj_browser.cgi) using the following identifiers: Ws-0h, bot0848, bot0926; Ws-24h, bot0849, bot0927; cpr22-0h, bot0850, bot0928; cpr22-24h, bot0851, bot0929; Nö-0h, bot0934, bot0930; Nö-24h, bot0935, bot0931; ssi4-0h, bot0936, bot0932; ssi4-24h, bot0937, bot0933. Affymetrix CEL files are available upon request.
Extraction and Determination of ABA and SA
Levels of endogenous ABA were measured as described (Priest et al., 2006) with some modifications (Supplemental Materials and Methods S1).
Loss of Fresh Weight during Desiccation
Aerial parts of 4-week-old plants were detached from their roots and were dehydrated on paper towels for 6 h under ambient humidity conditions unless otherwise stated (Nambara et al., 1998). Plants were weighed at various times up to 6 h. Hormone treatments were accomplished by drenching the soil with 100 to 125 mL of 1 mm SA or 10 μm ABA 16 h prior to analysis.
Drought Treatment
Arabidopsis plants were grown in a growth chamber under RH 65% conditions. The samples were created by terminating irrigation once plants were 4 weeks old. Samples were taken for RNA and ABA analysis at 0, 48, 72, 96, and 120 h.
Germination Assays
Approximately 50 seeds were placed on each of three replicate MS agar plates containing the indicated concentrations of ABA unless otherwise stated. After stratification at 4°C for 5 d, the plates were transferred to light racks and germination was scored on day 5 for root radical emergence.
Trypan Blue Staining
Trypan blue staining was performed as described previously (Bowling et al., 1994).
Ion Leakage
Plant ion leakage was measured using an Oakton CON5 ACORN series conductivity meter as described before (Urquhart et al., 2007).
Stomatal Response to ABA
Four-week-old plants of wild-type and cpr22 heterozygous plants were grown under ambient humidity conditions, and their leaves were incubated for 2 h in stomatal opening buffer (5 mm KCl, 50 μm CaCl2, and 10 mm MES-Tris, pH 6.15) and then treated with 0 or 10 μm ABA for 2 h. Data in Figure 5A were determined as single-blind experiments in which the ABA concentration was unknown to the experimenter (n = 3 experiments, 30 stomata per condition and experiment). All data are presented as means ± se.
ABA Treatment for MS Agar-Grown Plants
Plants were grown on MS agar plates vertically for about 3 weeks and preincubated for 2 h in water prior to each treatment. Plants were then soaked with 10 or 100 μm ABA solution. Samples were taken at 0, 2, and 10 h after treatment, and gene expression was analyzed by semiquantitative reverse transcription-PCR.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1.
Supplemental Figure S2.
Supplemental Figure S3.
Supplemental Figure S4.
Supplemental Figure S5.
Supplemental Figure S6.
Supplemental Figure S7.
Supplemental Table S1. Fold differences in cpr22/ssi4 of ABA- and defense/SA-related genes at high humidity and 24 h after shift to ambient humidity.
Supplemental Table S2. Primer sequences.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Ms. T. Nguyen, Ms. E. Zhao, and Ms. C. To for their technical assistance. We thank Dr. T.W. McNellis for providing cpn1 seeds, Dr. N.J. Provart for his assistance with transcriptome analysis, and Dr. Y. Kamiya for hormonal analysis. We also thank Drs. S. Lumba and P. McCourt for fruitful discussions.
References
- Audenaert K, De Meyer GB, Höfte MM. (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balague C, Lin B, Alcon C, Flottes G, Malmstrom S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D. (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Fung P, Chow TFF, Foong JA, Provart NJ, Cutler SR. (2008) Elucidating the germination transcriptional program using small molecules. Plant Physiol 147: 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Moeder W, Urquhart W, Shahinas D, Chin K, Christendat D, Kang HG, Angelova M, Kato N, Yoshioka K. (2008) Identification of a functionally essential amino acid for Arabidopsis cyclic nucleotide gated ion channels using the chimeric AtCNGC11/12 gene. Plant J 56: 457–469 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. (1996) A protein farnesyl transferase involved in ABA transduction in Arabidopsis. Science 273: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Dempsey D, Shah J, Klessig DF. (1999) Salicylic acid and disease resistance in plants. Crit Rev Plant Sci 18: 547–575 [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Delaney TP, Bauer DW, Beer SV. (1999) Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J 20: 207–215 [DOI] [PubMed] [Google Scholar]
- Dong HP, Yu H, Bao Z, Guo X, Peng J, Yao Z, Chen G, Qu S, Dong H. (2005) The ABI2-dependent abscisic acid signalling controls HrpN-induced drought tolerance in Arabidopsis. Planta 221: 313–327 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost JN, Jones JDJ, Daniels MJ, Parker JE. (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor H. (1971) Current status of gene-for-gene concept. Annu Rev Phytopathol 9: 275–296 [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritschnig S, Weihmann T, Zhang Y, Fobert P, McCourt P, Li X. (2008) A novel role for protein farnesylation in plant innate immunity. Plant Physiol 148: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Lamb C. (2006) Systemic immunity. Curr Opin Plant Biol 9: 414–420 [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JD. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. (1996) Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Silverman P, Raskin I, Jones JDG. (1996) Race-specific elicitors of Cladosporium fulvum induce changes in cell morphology and the synthesis of ethylene and salicylic acid in tomato plants carrying the corresponding Cf disease resistance gene. Plant Physiol 110: 1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Jambunathan N, McNellis TW. (2003) Regulation of Arabidopsis COPINE 1 gene expression in response to pathogens and abiotic stimuli. Plant Physiol 132: 1370–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N, Siani JM, McNellis TW. (2001) A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13: 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDJ, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK, Yu IC, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF. (2004) Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact 17: 511–520 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8: 409–414 [DOI] [PubMed] [Google Scholar]
- May MJ, Hammond-Kosack KE, Jones JDG. (1996) Involvement of reactive oxygen species, glutathione metabolism, and lipid peroxidation in the Cf-gene-dependent defense response of tomato cotyledons induced by race-specific elicitors of Cladosporium fulvum. Plant Physiol 110: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K. (2008) Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav 3: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K. (2010) Environmental sensitivity in pathogen resistant Arabidopsis mutants. Yoshioka K, Shinozaki K, , Signal Crosstalk in Plant Stress Responses. Wiley-Blackwell, Ames, IA, pp 113–135 [Google Scholar]
- Mohr PG, Cahill DM. (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30: 461–469 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. (2006) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct Integr Genomics 7: 181–191 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S. (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K. (2005) A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fuji H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest DM, Ambrose SJ, Vaistij FE, Elias L, Higgins GS, Ross ARS, Abrams SR, Bowles DJ. (2006) Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J 46: 492–502 [DOI] [PubMed] [Google Scholar]
- Rai VK, Sharma SS, Sharma S. (1986) Reversal of ABA-induced stomatal closure by phenolic compounds. J Exp Bot 37: 129–134 [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel G. (1931) Some experiments on inoculating methods with plant viruses, and on local lesions. Ann Biol (Ludhiana) 18: 494–507 [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, O'Donnell P, Sammons M, Toshima H, Tumlinson JH. (2003) Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc Natl Acad Sci USA 100: 10552–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Shirano Y, Kachroo P, Shah J, Klessig DF. (2002) A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart W, Gunawardena AHLAN, Moeder W, Ali R, Berkowitz G, Yoshioka K. (2007) The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol Biol 65: 747–761 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park SW. (2008) Systemic acquired resistance: the elusive signal(s). Curr Opin Plant Biol 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Wang C, Cai X, Zheng Z. (2005) High humidity represses Cf-4/Avr4-and Cf-9/Avr9-dependent hypersensitive cell death and defense gene expression. Planta 222: 947–956 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J. (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Wawrzynska A, Christiansen KM, Lan Y, Rodibaugh NL, Innes RW. (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol 148: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Xiao S, Brown S, Patrick E, Brearley C, Turner JG. (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. (2006) ABA-Hypersensitive Germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Kachroo P, Tsui F, Sharma SB, Shah J, Klessig DF. (2001) Environmentally-sensitive, SA-dependent defense response in the cpr22 mutant of Arabidopsis. Plant J 26: 447–459 [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. (2006) The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IC, Parker J, Bent AF. (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Menke FLH, Yoshioka K, Moder W, Shirano Y, Klessig DF. (2004) High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J 39: 920–932 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.