Abstract
Iron (Fe) deficiency is counteracted by a suite of responses to ensure maintenance of vital processes for which Fe is essential. Here, we report on transcriptional changes upon Fe deficiency, investigated in two Arabidopsis (Arabidopsis thaliana) accessions, Columbia (Col-0) and C24. Functional modules of the Arabidopsis Fe deficiency syndrome were inferred from clustering of Fe-responsive genes according to their coexpression. It was found that the redistribution of transition metals is an integral part of the reduction-based response to Fe starvation. The differential expression of metal transporters under the control of the FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR appeared to reflect an anticipated reaction rather than a response to the actual change in metal distribution. In contrast, the regulation of the zinc transporters ZRT/IRT-LIKE PROTEIN2 (ZIP2), ZIP3, ZIP4, and ZIP9 was dependent on the cellular zinc level, and their regulation by Fe was a secondary effect. Cellular Fe homeostasis was found to be closely coupled to Fe-related processes in the plastids. Using clustered genes as bait in gene-fishing experiments, we were able to attribute potentially important roles for gene candidates that have no previously described function in the Fe deficiency response. These results demonstrate a conceptually novel and integrative view into the regulation and interactions that allow Arabidopsis to adapt to suboptimal Fe availability.
As an essential element for virtually all organisms, iron (Fe) plays a crucial role in many vital processes such as respiration and photosynthesis. Although the availability of Fe is generally restricted in most soils due to its tendency to form complexes of poor solubility, the concentration of Fe can vary considerably in both time and space and can reach toxic amounts in soils with low oxygen levels. The variability in Fe availability led to the evolution of a variety of acclimatory responses, most of which are conserved among species and mediated by proteins encoded by close homologs. All nongraminaceous plants investigated acquire Fe by reducing ferric Fe in stable but soluble ferric chelates by a FERRIC OXIDASE REDUCTASE (FRO) homolog, first described in Arabidopsis (Arabidopsis thaliana; Robinson et al., 1999). The ferrous Fe liberated from the complex is then taken up by an IRON-REGULATED TRANSPORTER (IRT) of the ZRT/IRT-LIKE PROTEIN (ZIP) family, named IRT1 in Arabidopsis (Eide et al., 1996). In grasses, the reductive step is circumvented by the secretion of phytosiderophores with high affinity to ferric Fe and subsequent uptake of the Fe-loaded complex into the cell by an oligopeptide transporter, YELLOW-STRIPE1 (Curie et al., 2001). The reduction-based strategy is supported by electrogenic transport of protons across the plasma membrane, mediated by P-type ATPases (Santi and Schmidt, 2009). Exudation of low-molecular-weight compounds upon Fe deficiency was described for many species, but a clear function has not yet been attributed to this part of the Fe deficiency response (Jin et al., 2007).
The substrate specificity of the Arabidopsis IRT1 protein is low, allowing other transition metals such as zinc (Zn), manganese (Mn), and cobalt (Co) to be transported in addition of Fe (Korshunova et al., 1999). Arabidopsis irt1 mutants show enhanced resistance to cadmium (Cd) toxicity, suggesting that undesired metals can also accumulate within the cell during Fe deficiency (Vert et al., 2002). To balance the distribution of these metals according to the plant's demand and to avoid potential toxicity, their cellular homeostasis needs to be strictly controlled. Rapid regulation of Zn and copper transporters upon changes in the Fe regime was reported recently, suggesting that the regulation of the homeostasis of essential nutrients other than Fe is linked to the acquisition of Fe (Buckhout et al., 2009). Similarly, vacuolar sequestration of nickel (Ni) and Co by the IRON-REGULATED PROTEIN2 (IREG2) transporter has been described as part of the Fe deficiency response in Arabidopsis (Schaaf et al., 2006; Morrissey et al., 2009). How the distribution of transition metals is regulated under Fe-deficient conditions has not yet been sufficiently elucidated.
Fe deficiency is associated with alterations in several metabolic pathways in both roots and shoots that lead to, among other alterations in metabolic profiles, an increase in CO2 dark fixation and an accumulation of organic acids in roots, in particular citrate (Thimm et al., 2001; Buckhout and Thimm, 2003; Zocchi et al., 2007; López-Millán et al., 2009). Whether these changes represent adaptations to Fe deficiency or merely reflect imbalances between carbon metabolism and energy-generating reactions remains obscure. Some metabolic responses, such as the increase in amino acid biosynthesis, can be interpreted as the result of a higher demand for S-adenosyl Met (SAM) as a precursor for nicotianamine, an important Fe chelator that plays a crucial role in Fe transport and homeostasis (Klatte et al., 2009). Citrate accumulation in roots may be an important prerequisite for Fe transport to the leaves. Many facets of the surprisingly complex metabolic processes that are triggered upon Fe deficiency, however, still await elucidation.
The physiological responses to Fe shortage are associated with developmental alterations such as the development of shorter lateral roots, formation of transfer cells in peripheral cell layers, and changes in the density, positions, and characteristics of root hairs (Landsberg, 1986; Schmidt and Schikora, 2001). In Arabidopsis, the major morphological response induced by Fe deficiency is the formation of root hairs that are branched at their base and a weakening of the strict pattern of hair cells and nonhair cells that is typical of Arabidopsis (Müller and Schmidt, 2004). Similar to the physiological processes that are triggered by Fe deficiency, changes in postembryonic development are thought to adapt the plant to low availability of Fe.
Knowledge regarding the regulation of reduction-based Fe deficiency responses is derived from the Fe-inefficient tomato (Solanum lycopersicum) mutant fer (T3238fer; Brown and Chaney, 1971; Brown and Ambler, 1974). fer plants do not show any of the responses described above, which led to the assumption that the fer mutant is defective in a crucial Fe deficiency regulator. FER encodes a basic helix-loop-helix (bHLH) transcription factor that controls the expression of genes with key functions in Fe acquisition, including the tomato orthologs of IRT1 and FRO2 (Ling et al., 2002). The Arabidopsis FER ortholog, named FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT), was identified almost synchronously by three different groups and was shown to regulate a relatively large subset of genes under Fe-deficient conditions (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). FIT forms heterodimers with two other bHLH proteins, bHLH38 and bHLH39 (Yuan et al., 2008). All three bHLH genes are Fe responsive, indicating the existence of upstream components in the cascade that mediates the sensing and signaling of Fe deficiency (Wang et al., 2007).
Arabidopsis accessions from different habitats provide a means to study qualitative and quantitative traits. Physiological traits such as nutrient uptake strategies differ among accessions, providing a vast potential resource for studying the control of environmental responses in addition to forward genetics (Alonso-Blanco et al., 2009). By comparing the Fe deficiency responses of two widely used Arabidopsis accessions, Columbia (Col-0) and C24, we sought to identify the conserved core responses and those that are specific to either genotype. The two accessions differ in their flowering time, presumably reflecting different ecological strategies that may affect the regulation of processes associated with nutrient acquisition. We demonstrate that while a relatively large number of genes differ in their expression level between the two accessions, their responses to Fe deficiency were surprisingly similar. By clustering differentially expressed genes on the basis of their coexpression using publicly available expression profiling experiments, we were able to identify gene networks that could be organized into functional modules.
RESULTS
Transcriptional Profiling Reveals a Core Group of Genes Responsive to Fe Deficiency
To identify genes that are differentially expressed upon Fe deficiency in roots of the two accessions, we performed transcriptional profiling experiments using ATH1 GeneChips. The transcriptional Fe deficiency response of the two accessions was remarkably similar (Fig. 1A; Supplemental Table S1). Some of the genes that were found to be responsive to the Fe regime in only one of the accessions were just below the given threshold in the other accession. Therefore, we defined a gene as being responsive to Fe deficiency in both accessions if the expression change was greater than 2-fold in at least one of the accessions and greater than 1.5-fold in the other accession (Supplemental Table S1).
Figure 1.
Venn diagrams summarizing the microarray analysis between Col-0 and C24. The comparison was performed using the parameters of treatment (A) and accession (B). The numbers in the intersections represent the extent of the overlap between two analyses. Gene transcripts were tallied whose P values were was less than 0.05 and that were up- or down-regulated by greater than 2-fold. Plants were grown for 3 d on media deprived of Fe before harvest. [See online article for color version of this figure.]
Under Fe-deficient conditions, a set of FIT-regulated genes showed a 50% reduction in transcript level in C24 roots when compared with Col-0, among them the genes encoding the copper transporter COPT2, the Fe transporters IRT1 and IRT2, an oxidoreductase (At3g12900), a cytochrome P450 (At4g31940), and an unknown gene (At3g61930). The group also included bHLH38, which has been shown to form a heterodimer with FIT, thereby regulating the transcription of a set of genes that are responsive to the Fe regime (Yuan et al., 2008). Since FRO2 is not present on the ATH1 GeneChip but is reportedly closely coregulated with IRT1 and IRT2, we further determined FRO2 expression levels by real-time reverse transcription (RT)-PCR. Similar to IRT1, the up-regulation of FRO2 in C24 roots upon Fe deficiency was about half of that found in Col-0 (22.0-fold ± 5.4-fold in C24 and 52.0-fold ± 10.3-fold in Col-0).
We found a notable overlap of Fe-responsive genes in Col-0 and C24 with genes that show rapid responses to Fe starvation in a previous study using hydroponically grown Landsberg erecta plants (Buckhout et al., 2009). All but one of the rapidly (6 h of Fe deficiency) responding genes in the Landsberg erecta background were also found to be among the most differentially expressed genes in Col-0 and C24 roots (Supplemental Table S1). A relatively small subset of genes was responsive to Fe deficiency in only one of the accessions (Table I). A comparison of transcript levels of genes that do not respond to Fe deficiency but differ between the two accessions revealed a relatively large group of genes (Fig. 1B; Supplemental Table S2). The largest difference was observed for the MADS box protein encoded by FLOWERING LOCUS C, a repressor of flowering that has been shown to be highly expressed in C24 compared with Col-0 (Sheldon et al., 2000).
Table I. Genes that are Fe deficiency responsive in either Col-0 or C24.
Genes were selected that are more than 2-fold up- or down-regulated by Fe deficiency in only one of the accessions under investigation. Plants were grown for 3 d on media deprived of Fe before harvest.
| Gene Locus | Gene Name | Fold Change, Col-0 | Fold Change, C24 |
| At5g50590/At5g50690 | HYDROXYSTEROID DEHYDROGENASE4, AtHSD4/AtHSD7 | 0.94 | 0.40 |
| At5g38820 | Amino acid transporter family protein | 16.31 | 1.06 |
| At5g04730 | Expressed protein | 5.66 | 1.37 |
| At2g01880 | Purple acid phosphatase (PAP7) | 3.61 | 1.04 |
| At1g49820 | 5-MTK family | 2.55 | 1.05 |
| At1g71400 | RECEPTOR LIKE PROTEIN12, AtRLP12 | 2.30 | 0.88 |
We performed validation of the microarray data by real-time RT-PCR. A set of genes was selected that showed differential expression between C24 and Col-0 according to the GeneChip results (Supplemental Fig. S1). We did not observe major deviations of the RT-PCR results from the microarray data, demonstrating good consistency between the two approaches.
Coexpression Analysis Allows Prediction of Functional Modules in Fe Homeostasis
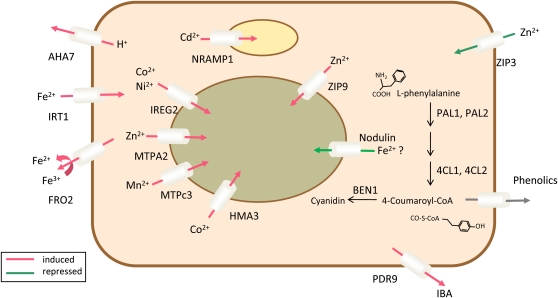
In order to uncover regulatory networks from the transcriptional profiles, we conducted coexpression analysis of genes that are affected by Fe deficiency in both accessions. To measure the similarity between expression patterns of any pair of genes, we downloaded a set of 300 root-related GeneChip experiments from the NASCarrays database, performed a robust multiarray average adjustment using the R software package, and searched for clusters of closely coexpressed genes. Interactions were analyzed for each pair of genes. Coexpressed genes were grouped into a module in which the nodes represent the genes that are responsive to Fe deficiency and the edges represent all pairs of genes whose expression similarity has a Pearson correlation greater than 0.7. To validate the robustness of the analysis, we compared the cluster obtained by this analysis with a separate analysis using a larger set of transcriptional profiling experiments not restricted to root samples (for details, see “Materials and Methods”). Using the root-related experiments as a data source, two functional modules could be deduced from the data, comprising 56 and 14 genes named cluster 1300 and 2300, respectively (Fig. 2; Supplemental Table S3). Three smaller clusters consisting of two and three genes were considered as being too small to represent functional modules and were not further discussed. The first module consists of two subclusters, comprising 47 genes (cluster 1A300; Fig. 2; Supplemental Table S3) and nine genes (cluster 1B300), the genes in each subcluster being more closely related to each other than to the rest of the cluster. In cluster 1A300, 17 genes were regulated by the bHLH transcription factor FIT (Colangelo and Guerinot, 2004). The “transportome” of this module comprises eight metal transporters, the P-type proton ATPases AHA7 and HMA3, and the ATP-binding cassette (ABC) transporter PLEIOTROPIC DRUG RESISTANCE9 (PDR9; Fig. 3). The FRO2 gene is not present on the ATH1 GeneChip and has been added to the module because of its well-documented coexpression with IRT1 and AHA7 (Eide et al., 1996; Vert et al., 2003; Colangelo and Guerinot, 2004). A nodulin (At3g25190) with unknown function was also found in this cluster. Based on the similarity with the yeast CCC1 transporter, which has been shown to function in Fe transport, a role of this nodulin in intracellular Fe transport, potentially in sequestering Fe into the vacuole or intracellular vesicles, may be inferred.
Figure 2.
Coexpression-based clustering of Fe-responsive genes. Genes were clustered on the basis of their expression patterns in 300 root-related microarray experiments (for details, see “Materials and Methods”). Similar colors represent genes that were clustered together when a larger set of microarrays was applied. White nodes represent genes for which no overlap was observed, and gray nodes indicate genes that correlate with small (fewer than four genes) clusters in the larger data set.
Figure 3.
Graphical representation of the transportome of cluster 1300. Localizations and functions of the genes were inferred from the literature or, when no information was available, assumed. Red arrows represent genes that were up-regulated upon Fe deficiency, and green arrows indicate down-regulation. An exporter for phenolic compounds has not yet been identified. The yellow compartment indicates plastids, and the green compartment indicates the vacuole. [See online article for color version of this figure.]
A total of five genes in this cluster are related to the flavonoid pathway (Fig. 3). Two Phe ammonia lyase (PAL) genes were up-regulated by Fe deficiency. PAL catalyzes the initiation of the phenylpropanoid pathway, ultimately leading to the formation of lignin, sinapic acid esters, and flavonoids. Two coumarate:CoA ligases, encoded by At4CL1 and At4CL2, mediate the last step of the general phenylpropanoid pathway. Since no genes encoding enzymes that mediate later steps in lignin synthesis were found to be induced upon Fe deficiency, it can be assumed that these gene products may play a role in the production of phenolics, a well-documented response of strategy I plants to Fe deficiency.
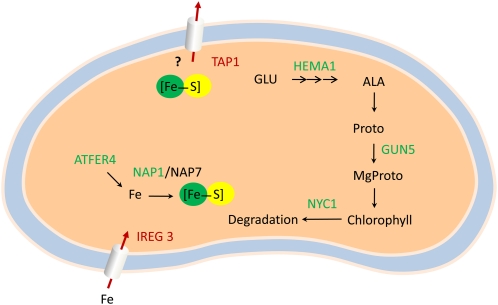
The second cluster (2300) comprises 14 genes, of which nine have been attributed with specific functions in the plastid (Fig. 4). One of the strongly induced genes of this cluster encodes an l-Gal dehydrogenase involved in ascorbate synthesis. The glutamyl-tRNA reductase HEMA1 catalyzes the second step in 5-aminolevolinic acid synthesis, the rate-limiting step in chlorophyll biosynthesis. GENOMES UNCOUPLED5 (GUN5) encodes the largest subunit of Mg chelatase, CHLH. Both HEMA1 and GUN5 were significantly down-regulated upon Fe deficiency. The ABC transporter NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) is a homolog of the prokaryotic SufB protein, which plays a role in Fe-S cluster assembly/repair (Xu et al., 2005). Another ABC transporter, TRANSPORTER ASSOCIATED WITH ANTIGEN PROCESSING PROTEIN1 (TAP1), might function in the export of Fe-S clusters from the plastid, similar to the ABC transporter STARIK in mitochondria (Kushnir et al., 2001). IREG3 represents another gene in this cluster that is up-regulated upon Fe deficiency. A sequence homolog from mouse, IREG1, was shown to mediate Fe efflux in the basolateral membrane of polarized epithelial cells. It is thus tempting to speculate that IREG3 is involved in Fe import from the cytosol. The expression of the ferritin gene FER4 was found to be repressed by Fe deficiency. NON-YELLOW COLORING1, a gene involved in chlorophyll degradation, was also found to be associated with this cluster.
Figure 4.
Putative locations and functions of the genes in cluster 2300. NAP1 may act as a Fe sensor, down-regulating the assembly of Fe-S clusters upon Fe deficiency. To counteract Fe shortage, Fe is released from ferritin and imported from the cytosol by IREG3. The expression of the ABC transporter TAP1, putatively involved in the export of Fe-S clusters, is up-regulated upon Fe deficiency, compensating for the reduced assembly. To prevent the accumulation of MgProto, the expression of HEMA1 and GUN5 is reduced. Green protein names indicate processes that are down-regulated upon Fe deficiency, and red arrows and protein names indicate up-regulation of the respective processes. [See online article for color version of this figure.]
Transporters Involved in Detoxification of Transition Metals Are Differentially Regulated
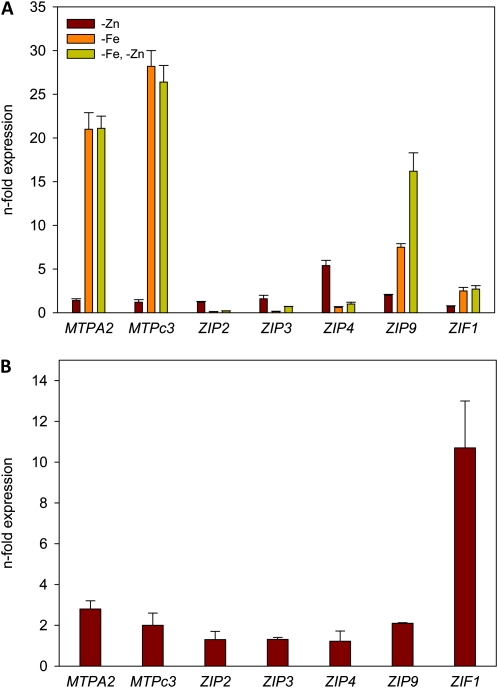
A surplus of transition metal ions is transported into the cell along with IRT1 under conditions of Fe deficiency and need to be sequestered to avoid the accumulation of these metal ions to toxic levels (Korshunova et al., 1999). A number of Zn transporters were affected by Fe deficiency, suggesting that Zn homeostasis is affected when IRT1 is up-regulated. To prove whether the detoxification response is triggered by a surplus of transition metals taken up during Fe deficiency or solely regulated by the availability of Fe, we measured the transcript abundance of Zn transporters under various Fe and Zn regimes by real-time RT-PCR. Upon Fe deficiency, the level of induction of METAL TOLERANCE PROTEIN A2 (MTPA2), MTPc3, ZIP2, ZIP3, ZIP4, ZIP9, and ZINC-INDUCED FACILITATOR1 (ZIF1) was comparable to the microarray experiments (Fig. 5A). In media deprived of Zn, the expression levels of the vacuolar Zn transporters MTPA2, MTPc3, and ZIF1 were essentially the same as under –Fe/+Zn conditions, suggesting that the up-regulation is caused by the Fe status of the plants and independent on the presence of Zn in the media (Fig. 5A). A different pattern was found for the expression of the other Zn transporter genes. In the absence of both Fe and Zn, the transcript levels of ZIP2, ZIP3, and ZIP9 were markedly higher when compared with a –Fe/+Zn regime, indicating that the transcription of these genes is mainly dependent on the Zn status of the cell. It thus appears that the change in transcript abundance of the FIT-regulated transporters MTPA2 and MTPc3 is solely regulated by Fe availability, but the expression of ZIP2, ZIP3, and ZIP9 is controlled by the availability of Zn and the regulation upon Fe deficiency occurs because of the surplus of Zn under these conditions.
Figure 5.
Expression patterns of transition metal transporters. Transcript levels were determined by real-time RT-PCR in plants that were grown on media deprived of Zn, Fe, or both nutrients for 3 d (A) or in the presence of Fe and a surplus of 10 μm Zn (B). Data are expressed as ΔΔCT. All data are presented as means from three independent biological replicates, and error bars represent se. [See online article for color version of this figure.]
When Zn was omitted from the media in the presence of Fe, only ZIP4 showed a significant (greater than 2-fold) increase in transcript abundance (Fig. 5A), indicating a function in Zn uptake under Zn-deficient conditions. In contrast, a surplus of Zn (10 μm Zn) caused a marked (greater than 2-fold) induction in the expression of MTPA2, MTPc3, ZIP9, and ZIF1, the most pronounced increase being observed for the latter gene. This result suggests a role in detoxifying a surplus of Zn brought into the cells by either Fe deficiency or high external Zn concentrations (Fig. 5B).
Robust Gene Clustering Using Coexpression Analysis
To investigate how the clustering is affected when different databases are used, we analyzed our data on the basis of a larger set of 1,436 microarray experiments, comprising experiments conducted with leaves, roots, and seedlings (Supplemental Fig. S2). Similar to the root-related data set, genes could be differentiated into two major clusters. Cluster 11436 contains 38 genes, 29 of which overlap with the cluster generated based on the root-related microarray experiments, including most of the transporters. This cluster also contains IRT2, which was unexpectedly absent in cluster 1300 and could be interpreted as being less closely related to the other genes in roots compared with other tissues. The two PAL genes and one of the 4-coumarate:CoA ligases, At4CL1, were also clustered separately. The second major cluster comprised genes encoding proteins localized in the plastids, including HEMA1, TAP1, GUN5, and NAP1. Interestingly, the cluster contained NICOTIANAMINE SYNTHASE4, a gene not related to chloroplasts but with a well-known function in Fe homeostasis.
While cluster 1A300 and cluster 2300 show relatively large overlaps with the respective clusters resulting from the larger data set, this was not the case for cluster 1B300, which may thus represent a module that is specific to roots. Interestingly, the only gene that has been associated with the formation of root hairs, COBRA-LIKE PROTEIN 6 PRECURSOR, was associated with this cluster (Fig. 2).
Genome-Wide Fishing for Coexpressed Genes Identifies Candidates That Play Potential Roles in Fe Homeostasis
To identify genes closely related to the clustered genes but not considered as being affected by Fe deficiency, we performed a whole genome-fishing analysis using the coexpressed genes from the two bigger modules as bait. We searched for genes that are closely coexpressed in the Arabidopsis genome, using strict parameters (Pearson correlation > 0.90). Applying these criteria, only six genes were found to be closely related to genes in clusters 1A/B300. Using the genes from cluster 2300 as bait, we found two clusters consisting of 24 and 89 genes (Supplemental Fig. S3). In the smaller of these two clusters, GUN5 was the only bait gene and all fished genes were localized in the chloroplast. The second cluster revealed a close correlation of an unknown, Fe deficiency-responsive gene to a set of genes mostly localized in chloroplasts and peroxisomes. The only other bait gene, CYP71B5, was strongly induced by Fe deficiency and the only FIT-regulated gene in cluster 2, representing a link between the general response to Fe deficiency regulated by the FIT-bHLH38/39 heterodimer and the responses that are triggered in the plastids during Fe deficiency. Using the genes from cluster 11436 as bait, we obtained two clusters comprising 58 and 14 genes (Supplemental Fig. S4). While in the first cluster several bait genes were included, the only Fe deficiency-responsive gene in the second cluster was the Fe transporter IRT2. Interestingly, the unknown gene At5g40510, only weakly regulated by Fe deficiency, was found in a central position in the first cluster, pointing to a putatively important function in this module.
DISCUSSION
The Accumulation of Transition Metals upon Fe Deficiency Is Anticipated and Counteracted by a Concerted Response
The clustering analysis indicates that the transport of Fe into the cell is coupled to a concerted detoxification response, mediating the sequestration of Zn, Mn, and Ni and, under natural conditions, probably also nonessential metals such as Cd and Co. This response is conserved in both accessions under investigation; the relatively subtle differences probably reflect different ecological preferences and/or habitat requirements. The evolution of this detoxification response is most likely the result of the low specificity of the IRT1 transporter (Korshunova et al., 1999; Vert et al. 2002), which increased the concentrations of transition metals upon Fe deficiency, a phenomenon well described in plants. The accumulation of metal ions positively correlates with the severity of Fe deficiency (Schmidt et al., 1997; Li and Kaplan, 1998), necessitating a fast and robust redistribution of metals that are transported into the cell alongside of Fe. The Fe deficiency-responsive transportome consists of several transporters that regulate metal influx and sequestration. The FIT-dependent Zn detoxification appears to be mainly mediated by MTPA2, which was even more strongly expressed in our microarrays than IRT1 upon Fe deficiency. MTPA2 (MTP3) is a vacuolar Zn transporter induced under the condition of excess Zn or Fe deficiency (Arrivault et al., 2006). The highest induction upon Fe deficiency was observed for MTPc3 (MTP8), a transporter of the cation diffusion facilitator family. A close sequelog of MTPc3 is MTP11, which has been shown to be involved in Mn sequestration into the vacuole (Delhaize et al., 2007; Peiter et al., 2007). The gene encoding MTPc3 was strongly induced by Fe deficiency in both Col-0 and C24 plants and was found to be down-regulated upon Mn deficiency (Yang et al., 2008), pointing to a potential role in Mn detoxification upon Fe deficiency. The role of NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN1 (NRAMP1) in Fe homeostasis remains obscure, but it appears likely that NRAMP1 localizes to an intracellular compartment, probably to the plastids (Curie et al., 2000). In support of this assumption, the expression of NRAMP1 was found to be slightly higher in plants defective in the plastid-localized Fe transporter PERMEASE IN CHLOROPLASTS1 (Duy et al., 2007). The increase in NRAMP1 expression was relatively weak under our conditions, and the differential expression upon Fe deficiency was restricted to Col-0 roots and clearly less pronounced when compared with other transport components of this cluster. It has been shown that NRAMP1 can transport Cd (Thomine et al., 2000), which is also taken up along with Fe by IRT1 from the soil. It thus appears that NRAMP1 might represent a further component of the detoxification response, sequestering Cd into an intracellular compartment. NRAMP1 can complement yeast mutants defective in Mn uptake (Curie et al., 2000; Thomine et al., 2000), which may indicate the existence of an alternative or additional pathway in sequestering Mn. The P-type ATPase HMA3 has been associated with the vacuolar sequestration of several metals (Morel et al., 2009). Ectopic expression of HMA3 conferred tolerance to Co and Cd, suggesting that HMA3 is involved in sequestering nonessential elements. Lastly, the gene encoding the vacuolar Ni transporter IREG2 was highly induced upon Fe deficiency, suggesting that Ni sequestration is crucial (Schaaf et al., 2006). More recently, IREG2 has been associated with Co and Fe transport (Morrissey et al., 2009), implying a more complex role of this transporter in metal homeostasis. The fact that all these genes were strictly coregulated, induced to a similar extent, and affected early after transfer to Fe-deficient media (Buckhout et al., 2009) suggests that changes in metal homeostasis occur in anticipation of the actual changes in ion distribution. This hypothesis is supported by experiments that showed that the expression levels of MTPA2 and MTPc3 were essentially unchanged when plants were grown on media deprived of Zn in addition to Fe (Fig. 5). Similar results have been obtained when Mn was omitted from the media in addition to Fe. Furthermore, the increased concentration of Ni in the media was shown not to affect the expression of IREG2 (Schaaf et al., 2006), supporting such a scenario. It can thus be postulated that plants developed a response to avoid imbalances in ion distribution by inducing a suite of transporters that compensate for the surplus of undesired metals. Interestingly, the expression of the transition metal transporters in cluster 1A300 was more closely related to IRT1 in terms of expression profiles than the expression of its close homolog, IRT2, for which a coregulation has been demonstrated during Fe deficiency (Vert et al., 2009).
The part of the transportome that is not FIT regulated consists of a putative nodulin, the ABC transporter PDR9, and the metal transporters ZIP3 and ZIP9. Interestingly, ZIP3 expression was reported to be dependent on the function of the cell wall-associated kinase WAKL4 (Hou et al., 2005), a gene that was induced in our arrays by Fe deficiency. WAKL4 might thus be a likely candidate responsible for the regulation of the FIT-independent part of the Zn detoxification response. Another Zn transporter gene, ZIP2, which is regulated by WAKL4, was found to have a similar expression pattern under our conditions (Fig. 5B).
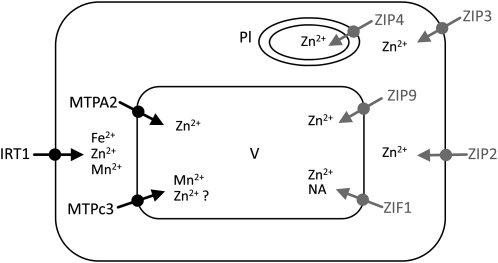
The expression of the Zn transporter ZIF1 is not regulated by FIT and deviates from the pattern of the ZIPs in that its expression under Fe deficiency was not affected by the presence of Zn in the media. ZIF1 transcript abundance is strongly increased by a surplus of Zn under Fe-sufficient conditions (Fig. 5B), indicating a role in Zn detoxification. Interestingly, ZIF was suggested to transport a ligand of Zn (and possibly of Fe), either as a sole substrate or in addition to Zn (Haydon and Cobbett, 2007), suggesting an important role of this transporter in both Zn and Fe homeostasis. The effects of Fe deficiency on Zn and Mn homeostasis are depicted in Figure 6.
Figure 6.
Putative regulation of Zn and Mn homeostasis under Fe-deficient conditions. Black symbols denote transporters mainly regulated by Fe and independent of the Zn concentration, and gray symbols indicate transporters regulated by Zn. Among other cations, Zn accumulates upon Fe starvation, due to a low substrate specificity of IRT1. While IRT1, MTPA2, and MTPc3 are regulated by Fe in anticipation of a surplus of transition metals, ZIP2, ZIP3, ZIP4, and ZIP9 are regulated by the Zn concentration. ZIF1 plays a role in both Zn and Fe homeostasis, probably by transporting a metal chelator, nicotianamine (NA). Pl, Plastid; V, vacuole.
Similar to the other ZIPs analyzed in this study, the expression of both ZIP3 and ZIP9 was strongly affected by the presence of Zn in the media, suggesting that their regulation depends on the cellular availability of Zn rather than on the Fe concentration. It can be further deduced that the regulation of the ZIP transporters by Fe deficiency might be a secondary effect. Therefore, it is perhaps not surprising that a separate Zn detoxification response has evolved that is apparently under the control of FIT.
PDR9, a member of the PDR subfamily of ABC transporters, is expressed predominantly in the lateral root cap and in epidermal cells at the root tip (Ito and Gray, 2006). PDR9 and a close homolog, PDR8, were shown to be plasma membrane-localized exporters of the auxin storage precursor indole-3-butyric acid (Strader et al., 2008; Strader and Bartel, 2009). Indole-3-butyric acid increases indole-3-acetic acid levels via β-oxidation in peroxisomes. Increased expression of PDR9 would suggest decreased indole-3-acetic acid levels, but a clear difference in auxin levels upon Fe deficiency could not be inferred from the analysis of DR5-GUS plants. The role of PDR9 in Fe homeostasis thus remains elusive. From the ability of PDR9 to transport the synthetic auxin analog 2,4-dichlorophenoxyacetic acid, an alternative role for PDR9 may be inferred for the export of phenolic compounds such as caffeic acid or chlorogenic acid, which improves Fe nutrition by helping to reutilize apoplasic Fe (Jin et al., 2007).
An interesting side aspect is related to ethylene production of the two accessions in response to Fe deficiency. Ethylene has been suggested to modulate some of the Fe deficiency responses and exerts a great impact on root hair development, a trait that is enhanced upon Fe deficiency (Masucci and Schiefelbein, 1996; Schmidt and Schikora, 2001; Waters et al., 2007). Methylthioribose kinase (MTK) is required for the recycling of methylthioadenosine during ethylene production (Sauter et al., 2004). The gene encoding this enzyme was found to be induced by Fe deficiency in Col-0 roots but not in C24 (Table I). Clustering revealed that MTK is closely related to SAM1 in cluster 1A300 (data not shown). It is thus tempting to assume that ethylene production differs between the two accessions. MTK is regulated by the FIT transcription factor (Colangelo and Guerinot, 2004) and is a potential marker enzyme for Fe deficiency-induced induction in ethylene production.
A Role for Plastids in Cellular Fe Homeostasis in Roots?
Unexpectedly, the majority of genes in the second cluster (2300) were localized in the plastids (Fig. 4). In leaves, the majority of Fe is sequestered in PSI and the Fe-storage protein ferritin in the chloroplasts. All four FER genes were markedly down-regulated under Fe-deficient conditions, suggesting an important role of plastidic Fe in root Fe homeostasis in root cells. In plastids, both heme biosynthesis and Fe-S cluster assembly can take place. Decreased Fe-S cluster assembly is reflected by a decrease in NAP1 (AtABC1) expression, a plastid-localized homolog of bacterial SufB. NAP1 interacts with NAP6 and NAP7 to form a NAP1-NAP7-NAP6 complex in chloroplasts that is associated with the repair of oxidatively damaged Fe-S clusters and/or transfer of Fe-S clusters to apoproteins (Xu et al., 2005). NAP1 is transcriptionally and posttranscriptionally regulated by Fe, suggesting a potential role as a Fe sensor, adapting the repair or assembly of Fe-S clusters to the available Fe (Xu et al., 2005). To compensate for the reduced Fe level in plastids, IREG3 is induced. This gene, also known as MAR1, was shown to encode a Fe-regulated chloroplast envelope protein with putative roles in Fe homeostasis. Ectopic expression of IREG3 causes chlorosis, consistent with a function in Fe import (Conte et al., 2009). Although its precise function and localization are unknown, TAP1 might function in the export of Fe-S clusters from the plastid, similar to the ABC transporter STARIK in mitochondria, which has been linked to intracellular Fe homeostasis (Kushnir et al., 2001). Enhanced expression of TAP1 may compensate for the reduced assembly of Fe-S clusters in the plastids upon Fe starvation.
Genes involved in chlorophyll synthesis and degradation were found to be down-regulated upon Fe deficiency. Since chlorophyll is not produced in roots and is reduced in leaves during Fe deficiency, it is tempting to speculate that down-regulated HEMA1 and GUN5 activities prevent the accumulation of MgProtoIX, an important signaling molecule (Mochizuki et al., 2001; Strand, 2004). The gene with the highest Fe responsiveness and the only FIT-dependent gene in this cluster, the cytochrome CYP71B5, was also shown to be highly induced upon Fe deficiency in hydroponically grown plants (Buckhout et al., 2009). The regulation of chlorophyll biosynthesis genes could be explained by the loss of activity of Fe metalloproteins that catalyze chlorophyll biosynthesis.
Gene Fishing Highlights the Importance of Individual Genes
Using a larger set of microarray experiments for gene clustering, the majority of genes were found to be assembled in two major clusters in a size comparable to that of the clusters generated by only comparing coexpression in root-related microarrays. While this clearly helps to prove the robustness of the clustering method used in this study, fishing for co-regulated genes that were not, or were only slightly, affected by Fe deficiency resulted in markedly different results when different data sets were used as a basis. Using the root-related experiments, fishing with clusters 1A300 and cluster 1B300 resulted only in a few genes under our threshold values. With cluster 2300 as bait, genes related to the plastids and the peroxisomes were fished. Interestingly, only two genes were found to be efficient as baits. The gene At5g55620, encoding an unknown protein, had a close relation to all genes in the fishing cluster, implying a central role in plastid-related processes that has not been described yet. The only other bait gene, GUN5, was associated with an independent cluster also composed of chloroplast-related genes (Supplemental Fig. S3).
Gene fishing with cluster 11436, derived from the larger set of microarray experiments, resulted in the formation of two clusters composed of non-Fe-regulated genes (Supplemental Fig. S4). In the first cluster, cell wall-related genes associated with the response to oxidative damage, cell wall synthesis, and cell wall alterations were clearly overrepresented. A total of six genes encoding mineral nutrient transporters were found, indicating a response to alterations in the overall nutrient status of the plant. Overall, this module appears to be related to detoxification during abiotic stress. The second cluster is related to the bait gene IRT2, a FIT-regulated Fe transporter. Unexpectedly, all genes in this module have been found to play a role related to the formation of root hairs. Namely, all genes are part of the “root hair elongation transcriptome” (Jones et al., 2006). Furthermore, 11 out of 14 genes possess cis-motifs that have been attributed to root hair-related genes (Won et al., 2009), pointing to a putative function of IRT2 in root hair formation.
In conclusion, we show in this study novel interactions of genes that are differentially regulated upon Fe deficiency. Our results suggest that a detoxification response, regulating the cellular homeostasis of several transition metals, is integrally coupled to the induction of the Fe acquisition response and predominately regulated by the FIT-bHLH38/39 transcription factors. A conceptually novel finding is that the differential regulation of a suite of metal transporters is not the direct result of an imbalance in metal distribution but a concerted, anticipated response in order to minimize the effect of such an imbalance. Fe stored in plastids may significantly affect cellular Fe homeostasis, and genes involved in Fe-S cluster assembly may have sensing or signaling functions that could be important for the regulation of Fe-related processes in the plant. Finally, our data analysis uncovers potentially important roles of genes that have not been described to function in Fe acquisition, distribution, or transport. While most of the processes described seem to be conserved among accessions and probably among most strategy I species, we demonstrated that the expression of genes upon Fe deficiency differs both quantitatively and qualitatively among Arabidopsis accessions, which may provide a starting point to unravel differences in Fe efficiency and elucidate regulatory mechanisms in the adaptation to a suboptimal supply of Fe.
MATERIALS AND METHODS
Plants
Arabidopsis (Arabidopsis thaliana) plants were grown in a growth chamber on an agar-based medium as described by Estelle and Somerville (1987). Seeds of the accessions Col-0 and C24 were obtained from the Arabidopsis Biological Resource Center. Seeds were surface sterilized by immersing them in 5% (v/v) NaOCl for 5 min and 96% ethanol for 7 min, followed by four rinses in sterile water. Seeds were placed onto petri dishes and kept for 1 d at 4°C in the dark, before the plates were transferred to a growth chamber and plants were grown at 21°C under continuous illumination (50 μmol m−2 s−1; Phillips TL lamps). The medium was composed of (mm) KNO3 (5), MgSO4 (2), Ca (NO3)2 (2), KH2PO4 (2.5), and (μm) H3BO3 (70), MnCl2 (14), ZnSO4 (2), CuSO4 (0.5), NaCl (10), Na2MoO4 (0.2), and FeEDTA (40) solidified with 0.3% Phytagel (Sigma-Aldrich). Suc (43 mm) and 4.7 mm MES were included, and the pH was adjusted to 5.5. After 10 d of precultivation, plants were transferred to fresh agar medium with 40 μm FeEDTA (+Fe plants), without Fe and with 100 μm 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate (−Fe plants), without Zn, or with a surplus of Zn (10 μm) for 3 d.
Real-Time Quantitative RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and DNase treated with the Turbo DNA-free Kit (Ambion) following the manufacturer's instructions. cDNA was synthesized using 4 μL of DNA-free-RNA with oligo(dT)20 primer and the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer's instructions. The cDNA was then used as a template in a 20-μL reaction using Power SYBR Green PCR Master Mix (Applied Biosystems), and subsequent experiments were carried out on an Applied Biosystems 7500 Fast RealTime PCR System with programs recommended by the manufacturer. All gene expression analyses were performed with three independent biological replicates. Three independent technical replicates were performed for each sample. The samples were normalized first to a selected internal control gene, and then the relative target gene expression was determined by performing a comparative ΔΔCT.
The following primers were used for the validation of the microarray results and the expression analysis of transition metal transporters. 5′-TCCAGAGAATTCGCCAGAGTTT-3′ and 5′-ATGAAGTTTCTTCCTGCACAAGTAAC-3′ were used for At5g20700; 5′-CGAGCTGGATTATCCGAATGA-3′ and 5′-CTCCAGTTTCAGCTCCCATGA-3′ for At5g35940; 5′-AGCTTTTGAACACTGAGAGGAAGAG-3′ and 5′-CCTTACTGCGCTGGACGAA-3′ for At5g45070; 5′-TTGGCCACCTTCATTTGATA-3′ and 5′-TCTGCAGAACCAAACTCAGG-3′ for At2g42250; 5′-CCATTGCCTCATCTTACAACTCTTATA-3′ and 5′-CCCCGCAGAGAGAGAATCG-3′ for At4g33020 (ZIP9); 5′-CGCACCCTAAAATGCAATCC-3′ and 5′-AGGCCAGCACCAAGAAGGA-3′ for At2g32270 (ZIP3); 5′-GGAACAAATCAGCGACCATAATG-3′ and 5′-AGGCCACTGCAGTTCCAATC-3′ for At1g10970 (ZIP4); 5-GTTTGTAGCGGCTGGGAGTAA-3′ and 5′-GTCATCATCCTCCCTCGACTCT-3′ for At5g59520 (ZIP2); 5′-TGAACAAAGCTGTGCCTCAGA-3′ and 5′-ACATCGCGGTCATAGAAATGC-3′ for At5g13740 (ZIF1); 5′-TGCATATACCTTCGGTGTTCTTTATT-3′ and 5′-CGCATGAGCTTCTTTCAATGG-3′ for At3g58060 (MTPc3); 5′-TGCTTGAAAAGGGAGTGTGTGA-3′ and 5′-ACAGTAATCGCCCAAATGTGAA-3′ for At3g58810 (MTPA2); 5′-CACCATTCGGAATAGCGTTAGG-3′ and 5′-CCAGCGGAGCATGCATTTA-3′ for At4g19690 (IRT1); 5′-GGCCACCACATATCCGAAGAT-3′ and 5′-CGACGTGGAGGACAAAGAAGAG-3′ for At1g01580 (FRO2); and 5′-AACACGAAGACCGAACGAAT-3′ and 5′-GTGCTGAAGGTGGAGACGAT-3′ for At5g19770 (tubulin).
Microarray Experiments
The Affymetrix GeneChip Arabidopsis ATH1 Genome Array was used for microarray analysis. Total RNA from roots of control and Fe-deficient plants was isolated as described above. All RNA samples were quality checked using the Agilent Bioanalyzer 2100 (Agilent). Complementary RNA synthesis was performed by use of the GeneChip One-Cycle Target Labeling Kit (Affymetrix). GeneChips were hybridized with 15 μg of fragmented complementary RNA. Hybridization, washing, staining, and scanning procedures were performed as described in the Affymetrix technical manual.
Microarray Data Analysis
Data from the microarray experiments were imported directly into GeneSpring (version 10; Agilent). The software was used to normalize the data on a per chip basis, to the 50th percentile and per gene to the control samples. The data were then filtered using the following: (1) by removing genes that were flagged as absent in at least three replicates; (2) by expression level to remove those genes that were deemed to be unchanging between log values 0.8 and 1.2 (greater than 1.5-fold difference); and (3) by confidence using a one-sample t test and a P value cutoff of 0.05, so that any gene with a P value of 0.05 or less when compared with the normalized control was regarded as statistically significant (i.e. up- or down-regulated compared with the expression baseline of 1). To determine those genes that were statistically differentially expressed between groups of samples, GeneSpring's ANOVA statistical analysis function (a Welch t test) was applied to the filtered data set. The microarray data are available in Supplemental Table S4.
Gene Clustering and Fishing
Gene clustering was performed based on the expression correlations of Fe-responsive genes. We downloaded robust multiarray averaged microarray data of 1,436 ATH1 slides from The Arabidopsis Information Resource Web site (ftp://ftp.arabidopsis.org/home/tair/Microarrays/analyzed_data/affy_data_1436_10132005.zip) and constructed a look-up table that records the normalized levels of every probe in the 1,436 slides. For every pair of Fe-responsive genes, their expression correlation was assigned the Pearson correlation of their normalized levels. For the gene clustering, we developed a software package that is available with a thorough description at http://maccu.openfoundry.org/. A graph was constructed whose nodes represent the Fe-responsive genes, and edges are all pairs of genes whose expression correlation is greater than 0.7. In order to capture the coexpression relationships specifically in roots, we collected 300 ATH1 slides from the NASCarrays database (Craigon et al., 2004), where the sample of each slide was derived nonexclusively from roots. After robust multiarray averaging of these 300 slides, we replicated all aforementioned procedures once. A list of 300 root slides is included in Supplemental Table S5.
In addition to gene clustering, we also performed gene fishing to find potentially coexpressed partners of a gene cluster. Taking a gene cluster as bait, a gene not in the given list was fished if it had an expression correlation greater than 0.9 with the bait gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of microarray data.
Supplemental Figure S2. Coexpression-based clustering.
Supplemental Figure S3. Clusters resulting from genome-wide fishing (300 root-related experiments).
Supplemental Figure S4. Clusters resulting from genome-wide fishing (1,436 experiments).
Supplemental Table S1. Differentially expressed genes (+Fe/−Fe).
Supplemental Table S2. Differentially expressed genes (Col-0/C24).
Supplemental Table S3. Clustering of Fe-responsive genes.
Supplemental Table S4. GeneChip data.
Supplemental Table S5. List of the microarray experiments.
Supplementary Material
Acknowledgments
We thank the bioinformatics core facility at the Institute of Plant and Microbial Biology for proficient support, Chung-Wen Lin for skillful technical assistance, and Prof. Thomas Buckhout for valuable comments on the manuscript. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/), supported by Academia Sinica.
References
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault S, Senger T, Kramer U. (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Brown JC, Ambler JE. (1974) Iron-stress response in tomato (Lycopersicon esculentum). 1. Sites of Fe reduction, absorption and transport. Physiol Plant 31: 221–224 [Google Scholar]
- Brown JC, Chaney RL. (1971) Effect of iron on the transport of citrate into the xylem of soybeans and tomatoes. Plant Physiol 47: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckhout TJ, Thimm O. (2003) Insights into metabolism obtained from microarray analysis. Curr Opin Plant Biol 6: 288–296 [DOI] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJ, Schmidt W. (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte S, Stevenson D, Furner I, Lloyd A. (2009) Multiple antibiotic resistance in Arabidopsis thaliana is conferred by mutations in a chloroplast-localized transport protein. Plant Physiol 151: 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. (2004) NASCarrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32: 575–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347: 749–755 [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE. (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51: 198–210 [DOI] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wiren N, Soll J, Philippar K. (2007) PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19: 986–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- Haydon MJ, Cobbett CS. (2007) A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol 143: 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Tong H, Selby J, Dewitt J, Peng X, He ZH. (2005) Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol 139: 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gray WM. (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol 142: 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Jin CW, You GY, He YF, Tang C, Wu P, Zheng SJ. (2007) Iron deficiency-induced secretion of phenolics facilitates the reutilization of root apoplastic iron in red clover. Plant Physiol 144: 278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P. (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, De Rycke R, Engler G, Stephan UW, Lange H, Kispal G, et al. (2001) A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg EC. (1986) Function of rhizodermal transfer cells in the Fe stress response mechanism of Capsicum annuum L. Plant Physiol 82: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kaplan J. (1998) Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem 273: 22181–22187 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Gogorcena Y, Abadia A, Abadia J. (2009) Metabolic responses in iron deficient tomato plants. J Plant Physiol 166: 375–384 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. (2009) AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol 149: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML. (2009) The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21: 3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Schmidt W. (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, Montanini B, Gobert A, Pedas P, Husted S, Maathuis FJ, Blaudez D, Chalot M, Sanders D. (2007) A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc Natl Acad Sci USA 104: 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Sauter M, Cornell KA, Beszteri S, Rzewuski G. (2004) Functional analysis of methylthioribose kinase genes in plants. Plant Physiol 136: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Honsbein A, Meda AR, Kirchner S, Wipf D, von Wiren N. (2006) AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J Biol Chem 281: 25532–25540 [DOI] [PubMed] [Google Scholar]
- Schmidt W, Bartels M, Tittel J, Fühner C. (1997) Physiological effects of copper on iron acquisition processes in Plantago. New Phytol 135: 659–666 [Google Scholar]
- Schmidt W, Schikora A. (2001) Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol 125: 2078–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Monroe-Augustus M, Rogers KC, Lin GL, Bartel B. (2008) Arabidopsis iba response5 suppressors separate responses to various hormones. Genetics 180: 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A. (2004) Plastid-to-nucleus signalling. Curr Opin Plant Biol 7: 621–625 [DOI] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ. (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol 127: 1030–1043 [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Barberon M, Zelazny E, Seguela M, Briat JF, Curie C. (2009) Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta 229: 1171–1179 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C. (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Baumlein H, Weisshaar B, Bauer P. (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Waters BM, Lucena C, Romera FJ, Jester GG, Wynn AN, Rojas CL, Alcantara E, Perez-Vicente R. (2007) Ethylene involvement in the regulation of the H(+)-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiol Biochem 45: 293–301 [DOI] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. (2009) Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Adams S, Chua NH, Moller SG. (2005) AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J Biol Chem 280: 6648–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Perry PJ, Ciani S, Pandian S, Schmidt W. (2008) Manganese deficiency alters the patterning and development of root hairs in Arabidopsis. J Exp Bot 59: 3453–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zocchi G, De Nisi P, Dell'Orto M, Espen L, Gallina PM. (2007) Iron deficiency differently affects metabolic responses in soybean roots. J Exp Bot 58: 993–1000 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.