Abstract

The objective of this review is to enable researchers to use the software package Rosetta for biochemical and biomedicinal studies. We provide a brief review of the six most frequent research problems tackled with Rosetta. For each of these six tasks, we provide a tutorial that illustrates a basic Rosetta protocol. The Rosetta method was originally developed for de novo protein structure prediction and is regularly one of the best performers in the community-wide biennial Critical Assessment of Structure Prediction. Predictions for protein domains with fewer than 125 amino acids regularly have a backbone root-mean-square deviation of better than 5.0 Å. More impressively, there are several cases in which Rosetta has been used to predict structures with atomic level accuracy better than 2.5 Å. In addition to de novo structure prediction, Rosetta also has methods for molecular docking, homology modeling, determining protein structures from sparse experimental NMR or EPR data, and protein design. Rosetta has been used to accurately design a novel protein structure, predict the structure of protein−protein complexes, design altered specificity protein−protein and protein−DNA interactions, and stabilize proteins and protein complexes. Most recently, Rosetta has been used to solve the X-ray crystallographic phase problem.
Rosetta is a unified software package for protein structure prediction and functional design. It has been used to predict protein structures with and without the aid of sparse experimental data, perform protein−protein and protein−small molecule docking, design novel proteins, and redesign existing proteins for altered function. Rosetta allows for rapid tests of hypotheses in biomedical research which would be impossible or exorbitantly expensive to perform via traditional experimental methods. Thereby, Rosetta methods are becoming increasingly important in the interpretation of biological findings, e.g., from genome projects and in the engineering of therapeutics, probe molecules, and model systems in biomedical research.
Rosetta, like all structure prediction algorithms, must perform two tasks. First, Rosetta must explore or sample the relevant conformational space, and in the case of design, sequence space. Second, Rosetta must accurately rank or evaluate the energy of the resulting structural models. For this purpose, Rosetta implements (mostly) knowledge-guided Metropolis Monte Carlo sampling approaches coupled with (mostly) knowledge-based energy functions. Knowledge-based energy functions assume that most molecular properties can be derived from available information, in this case the Protein Data Bank (PDB) (1,2).
While other reviews have focused on a specific Rosetta functionality, this work briefly reviews the approaches to sampling and scoring used by each of the major Rosetta protocols (protein structure prediction, protein docking, ligand docking, and protein design). Additionally, in the Supporting Information, we provide tutorials demonstrating six of the protocols introduced in this review. The tutorials we provide are intended as starting points and are therefore as basic as possible. Rosetta provides experienced users the option of extending and tailoring these protocols to their biomedical research question. Some of the calculations described below can be run on a standard workstation in a reasonable time, while others require small computer clusters found at most universities.
Rosetta Conformational Sampling Strategies
The majority of conformational sampling protocols in Rosetta use the Metropolis Monte Carlo algorithm to guide sampling. Gradient-based minimization is often employed for the last step of refinement of initial models. Since each Rosetta protocol allows degrees of freedom specific for the task, Rosetta can perform a diverse set of protein modeling tasks (3).
Sampling Strategies for Backbone Degrees of Freedom
Rosetta separates large backbone conformational sampling from local backbone refinement. Large backbone conformational changes are modeled by exchanging the backbone conformations of nine or three amino acid peptide fragments. Peptide conformations are collected from the PDB for homologous stretches of sequence (4) that capture the structural bias for the local sequence (5). For local refinement of protein models, Rosetta utilizes Metropolis Monte Carlo sampling of ϕ and ψ angles that are calculated not to disturb the global fold of the protein. Rohl (6) provides a review of the fragment selection and backbone refinement algorithms in Rosetta.
Sampling Strategies for Side Chain Degrees of Freedom
Systematic sampling of side chain degrees of freedom of even short peptides quickly becomes intractable (7). Rosetta drastically reduces the number of conformations sampled through the use of discrete conformations of side chains observed in the PDB (8,9). These “rotamers” capture allowed combinations between side chain torsion angles, as well as the backbone ϕ and ψ angles, and thereby reduce the amount of conformational space (9). A Metropolis Monte Carlo simulated annealing run is used to search for the combination of rotamers occupying the global minimum in the energy function (8,10). This general approach is extended to protein design via replacement of a rotamer of amino acid A with a rotamer of amino acid B in the Monte Carlo step.
Rosetta Energy Function
Simulations with Rosetta can be classified on the basis of whether amino acid side chains are represented by super atoms or centroids in the low-resolution mode or at atomic detail in the high-resolution mode. Both modes come with tailored energy functions that have been reviewed previously by Rohl (6).
Rosetta Knowledge-Based Centroid Energy Function
The low-resolution energy function treats the side chains as centroids (4,11). The energy function models solvation, electrostatics, hydrogen bonding between β strands, and steric clashes. Solvation effects are modeled as the probability of seeing a particular amino acid with a given number of α carbons within an amino acid-dependent cutoff distance. Electrostatic interactions are modeled as the probability of observing a given distance between centroids of amino acids. Hydrogen bonding between β strands is evaluated on the basis of the relative geometric arrangement of strand fragments. Backbone atom and side chain centroid overlap is penalized and thus provides the repulsive component of a van der Waals force. A radius of gyration term is used to model the effect of van der Waals attraction. All probability profiles have been derived using Bayesian statistics on crystal structures from the PDB. The lower resolution of this centroid-based energy function smoothes the energy landscape at the expense of its accuracy. The smoother energy landscape allows structures that are close to the true global minima to maintain a low energy even with structural defects that a full atom energy function would stiffly penalize.
Knowledge-Based All Atom Energy Function
The all atom high-resolution energy function used by Rosetta was originally developed for protein design (8,12). It combines the 6-12 Lennard-Jones potential for van der Waals forces, a solvation approximation (13), an orientation-dependent hydrogen bonding potential (14), a knowledge-based electrostatics term, and a knowledge-based conformation-dependent amino acid internal free energy term (9). An important consideration in the construction of this potential was that all energy terms are pairwise decomposable. The pairwise decomposition of each of the terms limits the total number of energy contributions to 1/2N(N − 1) when N is the number of atoms within the system. This limitation allows precomputation and storage of many of these energy contributions in the computer memory, which is necessary for rapid execution of the Metropolis Monte Carlo sampling strategies employed by Rosetta during protein design and atomic-detail protein structure prediction.
Protein Structure Prediction
The central tenet of structural biology is that structure determines function. Thus, the structure of a protein is critical for a full understanding of its function. Experimental structure determination techniques such as X-ray crystallography, nuclear magnetic resonance, electron paramagnetic resonance, and electron microscopy require significant investments of effort to produce structures of a molecule. Conversely, the advent of the genomic revolution created an explosion in the number of known sequences for biopolymers. For example, from October 2008 to March 2009, approximately 8 million (!) new, nonredundant sequences were added to the BLAST database. Rosetta remedies the shortfall in structural information by predicting high-probability structures for a given amino acid sequence.
De Novo Folding Simulation
The “protein folding problem” (given an amino acid sequence, predict the tertiary structure into which it folds) is considered the greatest challenge in computational structural biology. The Rosettade novo structure prediction algorithm has been reviewed and described in detail elsewhere (4,6,11,15). Briefly, Rosetta begins with an extended peptide chain. Insertion of backbone fragments rapidly “folds” the protein using the low-resolution energy function and sampling approaches (Figure 1). Rosetta attempts approximately 30000 nine-residue fragment insertions followed by another 10000 three-residue fragment insertions to generate a protein model (6). Usually, 20000−50000 models are folded for each individual protein (15). The resulting models can undergo atomic-detail refinement, or if computational expense is an issue, clustering based on the Cα root-mean-square deviation (rmsd) (16,17) can reduce the number of models before performing refinement. The clustering parameters are chosen by the user to generate clusters that maintain the same overall fold (i.e., Cα rmsd < 5 Å) while maximizing coverage of the structure space sampled. The lowest-energy models and the structures at the center of the clusters enter atomic-detail refinement (read below). The “Protein Folding” and “Refinement” tutorials in the supplement cover many aspects of this protocol.
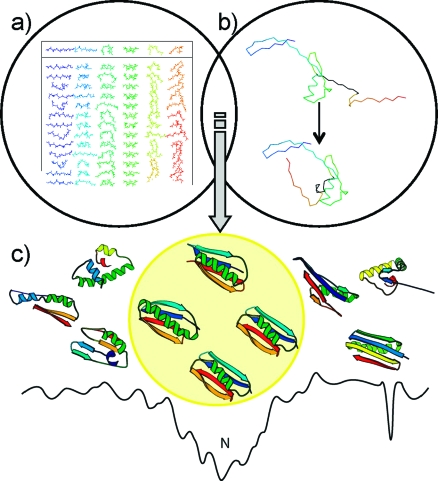
Figure 1.
De novo folding algorithm. Rosetta starts from (a) fragment libraries with sequence-dependent (φ and ψ) angles that capture the local conformational space accessible to a sequence. (b) Combining different fragments from the libraries folds the protein through optimization of non-local contacts. The low-resolution energy function depicted in panel c smoothes the rough energy surface, resulting in a deep, broad minimum for the native conformation. Metropolis Monte Carlo minimization drives the structure toward the global minimum.
In 2009, Das et al. implemented an addition to the existing de novo protein folding protocol that allowed for accurate prediction of homomeric proteins (18). They combined elements of Rosettade novo structure prediction (19) with protein−protein docking (20) to develop RosettaFold-and-dock. Fold-and-dock alternates between cycles of symmetric fragment insertion as in Rosettade novo prediction and rigid-body docking between the partially assembled monomers. Following complex assembly, the entire complex undergoes full atom refinement.
Fold-and-dock assumes that secondary structural elements of a homomer are symmetric and inserts the same fragments into every subunit. As the interface between a homomer is largely buried, docking while folding allows this region to be protected and stabilized during the folding simulation, which greatly improved the accuracy. Using this method, the structure of a K138A mutant of the Rab6-GTP·GCC185 Rab binding domain complex (PDB entry 3bbp) (21) was predicted within 1 Å rmsd of the experimental structure in a blind prediction test.
To further improve resolution, sparse NMR-derived chemical shift restraints were added, yielding models with rmsd values of <1 Å. Typically, structure elucidation for homomers with NMR-derived restraints would have required extensive data sets of RDCs, NOEs, chemical shifts, and scalar couplings.
Comparative Modeling
Comparative modeling in Rosetta starts after the alignment of a target sequence with a template protein, using sequence−sequence or sequence−structure alignment tools as described by Raman et al. (19). The quality of the alignment determines the aggressiveness of the sampling in Rosetta(19). In a case with a high degree of sequence homology (>50% identical sequence), the protein backbone is only rebuilt in regions surrounding insertions and deletions in the sequence alignment (19,22). Consequently, Rosetta starts with the template structure and builds in missing loops using fragment insertion or randomization of ϕ and ψ angles followed by one of the loop closure algorithms such as cyclic coordinate descent or kinematic closure (23−25). In the case of a medium to low degree of sequence identity between the template and target, Raman et al. applied a more aggressive iterative stochastic rebuild and refine protocol that allowed the complete rebuilding of large regions of the protein, which in some cases included entire secondary structure elements (19).
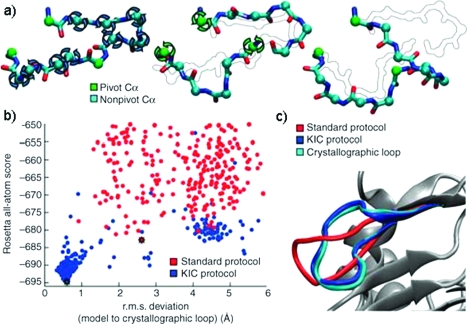
Mandell et al. (24) recently developed a Loop Closure algorithm in Rosetta that achieves rmsd values of better than 1 Å. Their adaptation of Kinematic closure (KIC) first selects three Cα atoms as pivots. Next, nonpivot (φ and ψ) torsion angles are sampled, leading to a chain break at the middle pivot. Finally, KIC is used to determine φ and ψ torsion angles for the pivot atoms that close the loop. For a data set of 25 loops containing 12 residues each, Rosetta achieved a median accuracy of 0.8 Å rmsd (see Figure 2). This demonstrates an improvement over both the standard Rosetta cyclic coordinate descent protocol and a state-of-the-art molecular dynamics protocol (median accuracies of 2.0 and 1.2 Å rmsd, respectively). The “Loop Modeling” tutorial demonstrates the kinematic loop closure protocol.
Figure 2.
Kinematic loop closure. (a) The kinematic loop closure algorithm samples φ and ψ angles at the cyan Cα spheres from a residue specific Ramachandran map. The φ and ψ angles at green Cα spheres are determined analytically to close the loop. (b) The energy vs rmsd plot shows accuracies for the prediction of loop conformation better than 1 Å achieved through the improved sampling offered by the kinematic closure protocol. (c) The kinematic closure prediction (blue) closely resembles the crystallographic conformation (cyan). From (24) reprinted with permission from Nature Methods.
Model Relaxation and Refinement
After construction of a protein backbone via de novo protein folding or comparative modeling, the model enters atom-detail refinement (15,26,27). During the iterative relaxation protocol, ϕ and ψ angles of the backbone are perturbed slightly while the overall global conformation of the protein is maintained. The side chains of the protein are adjusted using a simulated annealing Metropolis Monte Carlo search of the rotamer space. Finally, gradient minimization is applied to all torsional degrees of freedom (ϕ, ψ, ω, and χ). The repulsive portion of van der Waals potential is increased incrementally, moving the structure to the nearest energy minimum. Extensive use of the all atom model refinement has proven integral to the success of Rosetta in the recent Critical Assessment of Structure Prediction (CASP) experiments. A basic refinement protocol is introduced in the “Refinement” tutorial.
Recently, Qian et al. applied the refinement protocol to protein structures determined de novo, via comparative modeling, or using NMR-derived restraints (26). In this protocol, protein models derived from NMR constraints or comparative modeling were used as a basis for solving the crystallographic phase problem. The model was initially minimized using Rosetta’s all atom Monte Carlo Refinement protocol. The results of this refinement were used to identify regions likely to deviate from the native structure. In this context, it was demonstrated that regions of high variability between refined models often correlate to areas of deviation from the native structure. The conformational space in these areas was sampled extensively using the fragment replacement approach used by Rosetta’s de novo structure prediction protocol. The resulting models are then subjected to another round of all atom refinement. This cycle of refinement and conformational sampling is performed iteratively, each time using only the lowest-energy models from the previous round of refinement, until the system converges. The final models were then used in molecular replacement to solve the crystallographic phase problem. In a blind test, this ab initio phase solution method resulted in significant improvement in structural resolution compared to that of the original unrefined models. The protocol can also be applied for the refinement of models derived from medium-resolution NMR data.
Rosetta’s Performance in the CASP Experiment
Rosetta has displayed remarkable success in de novo structure prediction in the last several blind CASP experiments; this is evidenced by the method’s ranking among the top structure prediction groups (16,19,22,28−31). During CASP, sequences of proteins not yet reported in the PDB are distributed among participating laboratories. Within a given time limit, predictions are collected and assessed on the basis of the experimental structure that is typically available shortly after the CASP experiment (http://www.predictioncenter.org). Generally, Rosetta has superseded competing approaches at predicting the structure of small to moderately sized protein domains with fewer than 200 amino acids de novo. Shortly after the CASP6 (held in 2004), Bradley et al. showed that for a benchmark of small proteins Rosettade novo structure prediction found models at atomic detail accuracy in an encouraging eight of 16 cases (15,29). In that same year, Misura et al. found that homology models built with Rosetta can be more accurate than their templates (15). During CASP 7, with the assistance of the Rosetta@Home distributed computer network, several moderately sized domains were predicted to atomic-detail accuracy within 2 Å of the experimental structure for the first time (16). On the basis of the performance of Rosetta in improving models over the best template structures available (see Figure 3) (19), Raman et al. suggest that the limitation of the Rosetta structure prediction protocol remains in the sampling algorithms rather than the energy function. For this reason, prediction of larger domains becomes possible upon introduction of experimental data which restricts the conformational space.
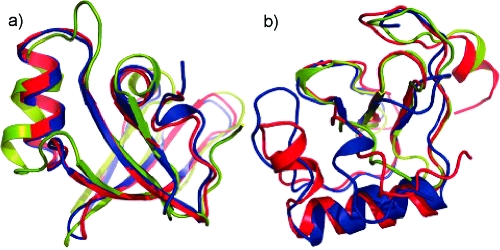
Figure 3.
Comparative modeling CASP performance. Raman and colleagues demonstrated that comparative models refined with Rosetta improved upon the best template structure available for several CASP targets, for example, (a) T0492 and (b) T0464. The native structure is colored blue, the best submitted Rosetta model red, and the best template structure green. The Rosetta models for T0492 resulted in atomic-level accuracy for side chains in the core of the protein. For T0464, a 25-residue insertion was predicted which resulted in models that were significantly improved over the best templates available. One of the models was further improved in the model refinement category. From (19) reprinted with permission from Proteins.
Rosetta Leverages Sparse Experimental Data from NMR and EPR Experiments
Rosetta allows incorporation of many types of experimental restraints. Rosetta’s ability to deal with restraints derived from nuclear magnetic resonance (NMR) spectroscopy is the most developed (32). NMR restraints have two entry points into the Rosetta protein structure prediction routine. Chemical shift assignments for backbone atoms can be converted to ϕ and ψ backbone angle restraints (33) and are used during the selection of the fragment libraries. Distance and orientation restraints [e.g., nuclear Overhauser effect (NOE) restraints and residual dipolar couplings (RDCs), respectively] are incorporated into the scoring function used during folding. Bowers et al. showed that a sparse mixture of short- and long-range NOE restraints (approximately one restraint per residue) in addition to the backbone chemical shifts enables Rosetta to build models at atomic-detail accuracy (34). Rohl and Baker (35) likewise demonstrated that limited RDC measurements (approximately one per residue) in combination with backbone chemical shifts identify the correct fold. Meiler and Baker presented a protocol that uses unassigned NMR restraints for rapid protein fold determination (36). More recently, Shen et al. showed the use of a modified fragment selection protocol in Rosetta to generate structures of a quality comparable to those from traditional NMR structure determination methods (37). Furthermore, Shen found that Rosetta sampling can compensate for the incomplete and incorrect NMR restraints (38). A major point to note is that in each of these examples Rosetta is used to complement structure restraints obtained early in the structure determination process. Consistently, Rosetta models are accurate at the atomic level of detail that would only be apparent from either significantly more or higher-resolution experimental information. For example, Rohl and Baker found that Rosetta produced ubiquitin models with an rmsd of <4 Å of the experimental structure using RDC restraints that were also consistent with models that have an rmsd of >20 Å (35).
Beyond NMR restraints, any experimental data suitable to represent the distance between atoms can be used in the simulation. Through site-directed spin labeling (SDSL), such distance restraints can be obtained from electron paramagnetic resonance (EPR) spectroscopy (39). Alexander et al. generated accurate atomic-detail models of T4 lysozyme (see Figure 4) and the heat shock protein α-crystallin using SDSL-EPR data using as few as one distance restraint for every four residues. Similar approaches can be used to model multimeric complexes from monomers, as Hanson et al. showed for the Arrestin tetramer in solution (40). Both the “Protein Folding” and “Loop Modeling” tutorials demonstrate the use of distance restraints.
Figure 4.
De novo protein structure prediction from sparse EPR data. Alexander et al. demonstrated that approximately one low-resolution distance restraint for every four residues is sufficient to determine a model of T4 lysozyme that is accurate at an atomic level of detail. The distance restraints had been determined using SDSL-EPR experiments. The native T4 lysozyme structure is colored gray, while the model with an rmsd of 1.0 Å is shown with a rainbow coloring scheme. Side chain conformations in the core of the protein are accurately represented in the model. From (39) reprinted with permission from Structure.
Rosetta Models Assist in the Determination of Molecular Structures from Electron Diffraction Data
De novo-predicted models have also been used to assist in phasing of X-ray diffraction data (see Figure 5) (26,41,42). Das and Baker found that for 15 of 30 benchmark cases, Rosettade novo models successfully solved the phase problem by molecular replacement (43). Das and Baker suggest that approximately one in six X-ray diffraction data sets for proteins with 100 or fewer residues can be solved via molecular replacement using Rosetta-generated de novo models. In a subsequent study, Ramelot et al. showed that refinement of NMR ensembles using Rosetta results in higher-quality molecular replacement solutions to X-ray diffraction data than direct use of the NMR ensembles (42). DiMaio et al. extended Rosetta to directly build models from electron density (44). Both Lindert and DiMaio have obtained atomic accuracy models via cryo-electron microscopy density maps at resolutions of 4−7 Å using Rosetta44−46. In both cases, resulting models have a higher resolution than the density from which they were built.
Figure 5.
Crysallographic phase problem. Qian et al. demonstrated that Rosetta-predicted protein models can be used in conjunction with automated molecular replacement algorithms to determine phases for electron density maps. The coordinates of BH3980 from Bacillus halodurans [PDB entry 2hh6 (unpublished), colored red] fit well into the isosurface of the electron density determined by molecular replacement using a Rosetta prediction for T0283 at CASP 7. From (26) reprinted with permission from Nature.
Protein Structure Prediction Servers
Large parts of the Rosetta protein structure prediction protocol, including generation of fragments, de novo folding, and comparative modeling, have been replicated in an automated server Robetta30,47,48. Chivian found that comparative models built with early versions of Robetta generally did not improve upon templates from close homologues; however, recently, Robettaperformed well in fold recognition and produced models that serve as good starting points for further refinement (48). In the most recent CASP, however, Robetta produced several models with accuracy comparable to that of the best human predictions (19). For instance, Robetta’s top model for the server only target, T0513 domain 2, had an rmsd of 0.84 Å. In general, the performance of Robetta compared to that of other servers increases as the quality of the template structures decreases (19). Robetta is publicly accessible at http://robetta.bakerlab.org.
Protein−Protein Docking
While protein function is often determined by interactions with other proteins, most structures found in the PDB contain single chains. Because of the difficulties in determining structures of protein complexes, computational methods for predicting protein−protein interactions are important. RosettaDock provides tools for predicting the interaction of two proteins (49). RosettaDock employs first a low-resolution rigid-body docking. The second high-resolution refinement stage provides for side chain conformational sampling and backbone relaxation.
The RosettaDock algorithm begins with random reorientation of both proteins (49). Next, one protein slides into contact with the other. The following low-resolution docking conformational search involves 500 Monte Carlo rigid-body movements. These moves rotate and translate one protein around the surface of the other with movements chosen from a Gaussian distribution centered at 5° and 0.7 Å. Each conformation is scored using the low-resolution energy function based on residue pair interaction statistics, residue environment statistics, and van der Waals attractive and repulsive terms. In this low-resolution step, side chains are represented by their centroids.
Next, 50 cycles of high-resolution refinement at the atomic level of detail are performed. Each cycle consists of a 0.1 Å random rigid-body translation, Monte Carlo-based side chain rotamer sampling (packing), and gradient-based rigid-body minimization to find a local energy minimum. Finally, backbone flexibility is introduced around the protein interface. The “Protein−Protein Docking” tutorial demonstrates the entire protocol. RosettaDock is available as a web server (http://rosettadock.graylab.jhu.edu) but is limited to complexes for which the approximate binding orientation is known. The server produces 1000 structures and returns details for the 10 lowest-energy models (50).
RosettaDock successfully recovered the native structures of 42 of 54 benchmark targets from which side chains had been removed (49). Starting with randomly placed proteins, RosettaDock predicts more than 50% of the interface contacts for 23 of 32 benchmark targets (49). These results have improved with the addition of backbone flexibility (3) and conformational sampling (51).
RosettaDock has been used to predict the structures of anthrax protective antigen (52) and epidermal growth factor (53) bound to monoclonal antibodies. Both docking studies led to predicted interface structures consistent with known mutant binding properties and were used to select residues for site-directed mutagenesis. The antibody modeling protocol has been made accessible through a web server (http://antibody.graylab.jhu.edu).
RosettaDock was benchmarked in the Critical Assessment of PRediction of Interactions (CAPRI) experiment (Figure 6), where it achieved full-atom rmsds of better than 1.6 Å for most targets (54). Its success has been attributed to advances such as the inclusion of gradient-based energy minimization of side chain torsion angles (54), incorporation of biochemical data (55), and coupling of docking with loop modeling (55).
Figure 6.
Protein interface prediction. High-resolution CAPRI prediction of the colicin D−immunity protein D interface. Both rigid-body orientation and side chain conformation were modeled. The crystal structure is colored red and orange, and the Rosetta model is colored blue. (a) Whole protein complex. (b) The interface shows the side chains of catalytic residue H611 and additional positively charged residues that are thought to bind to the RNA, as well as their matching negatively charged residues in the immunity protein. From (54) reprinted with permission from Proteins: Structure, Function, and Bioinformatics.
Sircar and Gray recently reported on an extension of the RosettaDock algorithm that allows for accurate modeling of antibody−antigen complexes in the absence of an antibody crystal structure (56). SnugDock simultaneously samples the rigid-body antibody−antigen positions, the orientation of antibody light and heavy chains, and the conformations of the six complementary determining loops. Additionally, antibody conformational ensembles can be provided to mimic conformational selection. As in RosettaDock, side chain rotamers are sampled during high-resolution refinement.
SnugDock was compared with RosettaDock in a blind prediction of human MCP-1 binding 11k2 antibody (PDB entry 2bdn) (57). While the lowest-energy structure produced by RosettaDock is incorrect, the model produced by SnugDock meets the CAPRI acceptable criterion of having more than 30% of the residue−residue contacts predicted correctly. When combined with ensemble sampling, five of the 10 lowest-energy models meet the CAPRI medium-quality criterion of correctly predicting more than 50% residue−residue contacts. Similar results were seen in a benchmark of 15 antibody−antigen complexes.
Protein−Ligand Docking
Ligand docking seeks to predict the interaction between a protein and a small molecule. Most ligand docking applications struggle to correctly predict conformational selection or induced-fit effects (58) resulting from ligand and protein flexibility. As applications were originally designed for protein−ligand docking, flexibility is often a feature added as an afterthought. On the other hand, Rosetta was originally developed for de novo structure prediction. As such, it was designed from its inception to efficiently model flexibility. While protein flexibility is well-defined by side chain rotamers and backbone ϕ and ψ angle changes, small molecule flexibility was newly introduced into Rosetta(59). Modeling ligand flexibility using knowledge-based score functions is especially challenging since the available small molecule crystal structures pale in comparison to the possible chemical diversity available to small molecules.
RosettaLigand is an application for docking a small molecule in the binding pocket of a protein that considers both ligand and protein flexibility (60). The RosettaLigand algorithm is a modification of the RosettaDock algorithm. First, a ligand conformer is chosen randomly from a user-provided ligand conformational ensemble. Second, the ligand is moved to a user-defined putative binding site. A low-resolution shape complementarity search translates and rotates the ligand optimizing attractive and repulsive score terms. In the high-resolution phase, cycles of Monte Carlo minimization perturb the ligand pose and optimize amino acid side chain rotamers and ligand conformers. Lastly, all torsion degrees of freedom in the ligand and protein undergo gradient minimization, and the model is output. The “Small Molecule Docking” tutorial demonstrates this protocol.
In a benchmark, RosettaLigand successfully recovered the native structure of 80 of 100 protein−ligand complexes with an rmsd better than 2.0 Å. When docking ligands into experimental protein structures determined with different binding partners (cross-docking), RosettaLigand recovered the native structure in 14 of 20 cases. Comparing binding energy predictions with 229 experimentally determined binding energies from the Ligand-Protein Database (http://lpdb.chem.lsa.umich.edu) (61), RosettaLigand achieved an overall correlation coefficient of 0.63, which is comparable to the best scoring functions available for protein−ligand interfaces (62).
Recently, backbone flexibility was added to the docking algorithm which led to improved predictions, including lower rmsds among top scoring ligands (63). Backbone flexibility allows prediction of induced-fit effects that occur upon ligand binding. When RosettaLigand was tested in a blind study on a set of lead-like compounds, its performance was comparable to those of other commercially available docking programs (64). The authors caution, however, that current docking programs fail 70% of the time on at least one of the receptors in the test set.
Often researchers seek to understand the interaction of a small molecule with a target protein whose structure has not yet been determined. In such cases, docking studies utilize comparative models. RosettaLigand was recently used by Kaufmann et al. (65) to dock serotonin into comparative models of human and Drosophila serotonin transporters (hSERT and dSERT, respectively). The models were based on the leucine transporter (LeuTAa) structure reported by Yamashita et al. (66) which has an overall sequence similarity of 17% and a binding site similarity of 50% with SERT. Using these models alone, RosettaLigand predicted a binding mode that places serotonin deep in the binding pocket of SERT (see Figure 7). This binding mode is consistent with site-directed mutagenesis studies and substituted cysteine accessibility method (SCAM) data and retains the amine placement seen in the LeuTAa structure. Additionally, binding energy predictions of serotonin analogues agree with experimental data (R = 0.72).
Figure 7.
Complex of the human serotonin transporter with its substrate. The color scheme of serotonin displays the differential sensitivity of human and Drosophila serotonin transporter (SERT) for serotonin derivatives as dervied from a QSAR study. Blue indicates a higher sensitivity in dSERT, while red indicates a higher sensitivity in hSERT. The QSAR data indicate that the docking pose predicted by RosettaLigand is plausible. From (65) reprinted with permission from Proteins.
Kaufmann and Meiler find that RosettaLigand successfully docks a variety of small molecules into comparative models (unpublished results). RosettaLigand identified the binding mode within 2 Å rmsd for six of nine protein−ligand complexes in which models had been submitted in the eighth CASP competition. In seven additional examples, Kaufmann and Meiler observe that RosettaLigand samples the correct binding mode in at least one template for most ligands, yielding an overall success rate of better than 70%. This success rate is comparable to RosettaLigand's performance with an experimental structure for the protein partner and can be attributed to RosettaLigand's ability to sample protein conformational changes.
Protein Design
All protocols discussed up to this point relate to protein structure prediction and seek to determine the position of amino acid atoms in space. Protein design, on the other hand, seeks to determine an amino acid sequence that folds into a given protein structure or performs a given function. In this context, the protein design problem (to find a sequence that folds into a given tertiary structure) is also known as the “inverse protein folding problem”. The RosettaDesign algorithm (12) is an iterative process that energetically optimizes both the structure and sequence of a protein. RosettaDesign alternates between rounds of fixed backbone sequence optimization and flexible backbone energy minimization (12). During the sequence optimization step, a Monte Carlo simulated annealing search is used to sample the sequence space. Every amino acid is considered at each position in the sequence, and rotamers are constrained to the Dunbrack Library (67). After each round of Monte Carlo sequence optimization, the backbone is relaxed to accommodate the designed amino acids (12). The practical uses of RosettaDesign can be divided into five basic categories: design of novel folds (12), redesign of existing proteins (68), protein interface design (69), enzyme design (70), and prediction of fibril-forming regions in proteins (71). The “De Novo Protein Design” tutorial demonstrates the complete redesign of the protein ubiquitin.
De Novo Protein Design
The RosettaDesign method has been used for the de novo design of a fold that was not (yet) represented in the PDB. A starting backbone model consisting of a five-strand β sheet and two packed α helices was constructed with the Rosettade novo protocol using distance constraints derived from a two-dimensional sketch (12). The sequence was iteratively designed with five simulation trials of 15 cycles each. The final sequence was expressed, and the structure was determined using X-ray crystallography. The experimental structure has an rmsd with respect to the computational design of <1.1 Å (see Figure 8) (12).
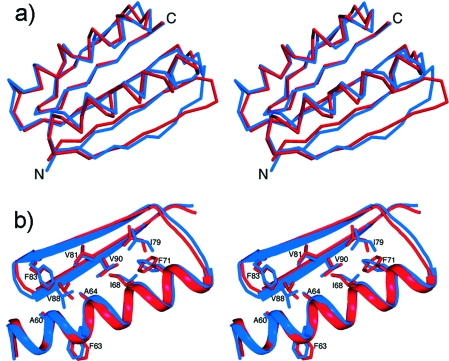
Figure 8.
Design of a novel protein fold. (a) The experimentally determined structure of the Top7 (red) fold displays an rmsd of 1.17 Å with respect to the model that had been previously designed for this protein (blue). (b) In the core of the protein, side chain conformations have been designed to atomic-detail accuracy. From (12) reprinted with permission from AAAS.
Similarly, a molecular switch that folded into a trimeric coiled coil in the absence of zinc, and a monomeric zinc finger in the presence of zinc, was designed by extending RosettaDesign to simultaneously optimize a sequence in two different folds. The sequence of an existing zinc finger domain was aligned with a coiled-coil hemaglutinin domain. During the design protocol, the sequence was optimized to fold into both tertiary structures (72).
Redesign of Existing Proteins
When nine globular proteins were stripped of all side chains and then redesigned using RosettaDesign, the average sequence recovery was 35% for all residues (73). In four of nine cases, the protein stability improved as measured by chemical denaturation. The structure of a redesigned human procarboxypeptidase (PDB entry 1aye) (74) was determined experimentally. RosettaDesign was then used to systematically identify mutations of procarboxypeptidase that would improve the stability of the protein. All of the tested mutants were more stable than the wild-type protein, with the top-scoring mutant having a reduction of free energy of 5.2 kcal/mol (75).
The RosettaDesign server (http://rosettadesign.med.unc.edu) (76) is a Web-based interface to the fixed backbone design module of Rosetta that allows design of proteins with up to 200 residues. The average design takes 5−30 min to complete.
Interface Design
Computational design techniques have been used to engineer an endonuclease with altered specificity. A 1400 Å2 interface was designed between individual domains of two homodimeric endonucleases (I-DomI and I-CreI). The design retained specificity and catalytic activity and crystallized with an rmsd of 0.8 Å with respect to the model (77). Similarly, a highly effective specificity switch was designed into the colicin E7 DNase−Im7 immunity complex through the design of a novel hydrogen bond network (Figure 9). This designed network exhibited a 300-fold increase in specificity (78). Rosetta’s alanine scanning application simulates experimental alanine scanning in silico. Each residue in the protein complex is iteratively mutated to an alanine, and the change in binding free energy is calculated. In silico alanine scanning has been implemented in the current version of Rosetta and is available through a Web-based interface (http://robetta.bakerlab.org) (69). More recently, multispecific designs have been generated in which a single protein interface sequence is simultaneously optimized to bind to multiple targets, producing a so-called “hub” protein (79).
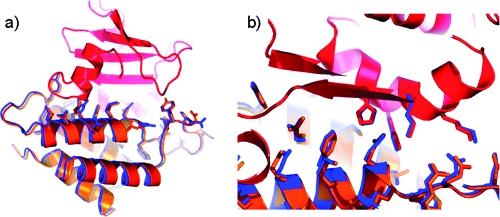
Figure 9.
Design of a novel protein interface. Comparison of the designed specificity switch in the colicin E7 DNase−Im7 immunity complex with the experimentally determined structure. (a) Experimentally determined coordinates, including a density map for computationally designed residues. (b) The computational design (yellow) is superimposed on an experimental structure (orange). (c and d) Side-by-side comparison of the designed and experimentally determined hydrogen bond networks. (e) Hydrogen bonding connectivity in the context of the interface region. From (78) reprinted with permission from Journal of Molecular Biology.
Protein design approaches have enhanced our knowledge of how protein sequence relates to protein structure. For instance, the finding that designed protein sequences are highly similar to the native sequence suggests that native protein sequences are optimal for their structure (8). Recently, Babor and Kortemme investigated the antibody sequence−structure relationship using Rosetta protein design. They demonstrated that native sequences of antibody H3 loops are optimal for conformational flexibility (80). The authors collected pairs of unbound and antigen-bound antibody structures. They used multiconstraint design to find low-scoring sequences that were consistent with both unbound and bound structures. The sequences predicted by multiconstraint design were more similar to the native sequences than the sequences predicted to preferentially bind either the unbound or bound conformations. Next, they collected pairs of antibody structures differing only in their degree of maturation. They used protein design in Rosetta to show that mature antibody sequences are optimized for the bound conformation.
A current major challenge in protein interface design is the de novo design of a novel protein−protein interface. So far, the most successful attempts at de novo interface design have been relatively modest, focusing on small proteins and yielding micromolar affinity (20,81).
Enzyme Design
The RosettaMatch algorithm (82) starts from the protein backbone and attempts to build toward the specified transition state geometry. In this method, all possible active site positions are defined for the protein scaffold, and rotamers from the Dunbrack library (67) are placed at each sequence position in the catalytic site. The sequence of the area surrounding the catalytic site is then designed using the RosettaDesign method (82).
Recently, the RosettaMatch algorithm was used to design enzymes that catalyze the retro-aldol reaction (70). The degrees of freedom in the transition state, the orientation of the active site side chains, and the conformations of the active site side chains were simultaneously optimized. Of 72 models tested, a total of 32 were found to have catalytic activity as much as four orders of magnitude greater than that of an uncatalyzed reaction. Two of the active enzymes were crystallized. The experimental structures share a high degree of similarity with the computational design (rmsd better than 1.1 Å), although the loop regions surrounding the catalytic site show significant variance from the model (70).
Röthlisberger et al. have computationally designed functional Kemp elimination catalysts using RosettaMatch. Quantum chemical predictions were used to generate an idealized transition state model, and RosettaMatch was used to search for backbone configurations that would support the predicted transition sate. The resulting designs were expressed and found to have kcat/Km values between 6 and 160 M−1 s−1. Directed evolution was then performed on the designed enzymes to produce an optimized enzyme with a kcat/Km of 2600 M−1 s−1(83).
The RosettaDesign Algorithm Can Be Used To Identify Structurally Similar Peptide Fragments
A method for predicting peptides capable of forming amyloid fibrils was recently developed using the RosettaDesign protocol (71). The most well understood fibril-forming fragment, the NNQQNY peptide, was used as a template, and the sequence space was searched for alternative fibril-forming sequences. This method was then used to predict fibril-forming regions in proteins known to form amyloids.
Caveats of Modeling
Despite Rosetta’s success in producing accurate and precise models, some of its predictions will necessarily be incorrect, whether due to imperfections in the statistically derived energy function or to practical limits on exhaustive sampling. The following four strategies must be employed by the skeptical researcher to reject incorrect models and validate the low-energy predictions.
(1) Model precision is a necessary prerequisite for model accuracy. Hence, it is an important strategy to ensure precision by insisting upon convergence of multiple independent trials toward a single low-energy solution. This strategy is employed, for example, during the analysis of pairwise rmsd values of low-energy models in an “energy funnel”.
(2) A modification of the precision analysis is clustering. If more than one low-energy solution is found, clustering assesses whether independent trajectories converged to a limited number of low-energy solutions. For example, clustering of models and ranking by cluster size is commonly used in de novo structure prediction, based on the observation that the deepest energy well is frequently also the widest (84). This is important because even using Monte Carlo search, adequate sampling is expensive to achieve due to the extreme roughness of the energy landscape (15).
(3) Every mode of Rosetta described in this review has been benchmarked on a set of test cases. Before these protocols are applied to a system that falls outside the scope of the test cases, it is necessary to apply the protocol to a closely related system of known structure. This experiment serves as a positive control for the method. Even if the application falls within the scope of the original benchmark, it is advisible to reproduce the benchmark results to ensure the operator-independent performance of the respective version of the software and accurate application of the protocol.
(4) It is insufficient to rely solely on the Rosetta energy function to discriminate good models from bad. The reliability of the result can be improved by incorporating the scores from disparate structure evaluators such as PROCHECK (85) and the DOPE scoring function implemented in Modeler (86).
All of the preceding avenues are available without a departure from purely computational methodology. However, the most powerful and only conclusive method to ensure the reliability of computational models is the incorporation of experimental data. There are three strategies to incorporate experimental data into a modeling project: (a) Experimental restraints can be applied during the simulation (compare protein structure determination from NMR/EPR restraints); (b) Experimental restraints can discriminate inaccurate models in a post-simulation filtering; (c) Experimental restraints can be recorded to verify a computational model or hypothesis. More broadly, Rosetta is most valuable as an integrated component of a research program in which initial structural models are used to guide hypothesis generation, and then data from experimental testing of these predictions are used to select and refine supported models in an iterative process.
Conclusion
The Rosetta protein modeling suite provides a variety of tools for protein structure prediction and functional design. These techniques have been used in conjunction with traditional molecular and biochemical techniques to make predictions that would be prohibitively expensive or time-consuming via noncomputational methods. The quality of predictions has reached atomic-detail accuracy in many examples and is a practical tool for biochemical and biomedical research. Rosetta’s de novo folding protocol is applicable if the protein of interest has no detectable homologues in the PDB and is fewer than 100 residues in length. For comparative models based on medium to distant homologues (25 and 50% identical sequence), Rosetta’s comparative modeling protocols offer the ability of remodeling variable regions and regions of poor alignment. Rosetta’s knowledge-based energy function and large-scale sampling strategies allow for construction of models from incomplete or limited experimental data sets. Rosetta shows the capability of supplying structural detail in regions of the models underdetermined by the experimental data. Rosetta’s protein−protein and protein−ligand docking protocols have proven to be particularly helpful if induced fit and conformational selection play a critical role in the interaction. Specialized protocols make Rosetta an attractive option for antibody modeling. While de novo protein design remains a challenging problem, Rosetta can routinely be applied when searching for thermo-stabilizing mutations, when redesigning protein−protein interfaces, and when performing in silico mutagenesis studies such as alanine scanning.
Installation and Licensing
The Rosetta licenses are available at http://www.rosettacommons.org/software free of charge for noncommercial use. Rosetta is compatible with most Unix-based operating systems and is distributed as source code. A user manual describing compilation, installation, and usage for the current release can be found at http://www.rosettacommons.org/manuals/rosetta3_user_guide. Interested developers can join the RosettaCommons setup to contribute to the Rosetta software package.
Additional Features
Several Rosetta Methods under Development Have Been Excluded from This Review
In addition to the protocols described above, several additional methods are currently in development. These methods have been excluded from this review as they are not yet fully implemented in the release version of the software. RosettaMembrane is a transmembrane helix scoring potential that allows Rosetta to predict and design membrane bound proteins at atomic detail. In 2007, Barth et al. used this potential to predict the structure of small transmembrane helices at up to 2.5 Å rmsd (87). The RosettaDesign protocol has also been adapted to model DNA−protein interactions. In 2002, Chevalier et al. used a DNA−protein interaction scoring function in combination with RosettaDesign to produce a novel endonuclease with high specificity (77). In addition to DNA−protein interactions, scoring potentials have been developed to score RNA−RNA interactions and allow for de novo prediction of RNA tertiary structure. This method was developed by Das et al. and uses the Rosetta fragment-based design approach in conjunction with a knowledge-based potential for modeling RNA interactions. Through the use of this method, RNA structures have been predicted with a 4.0 Å rmsd with respect to the backbone (16). Sheffler et al. implemented a space filling VDW model called RosettaHoles that detects voids and packing errors in protein structures (88). Extensions to the experimental modes available for docking small molecule ligands are also under development. These extensions will allow users to simultaneously dock multiple ligands and perform fragment-based design based on a scaffold and a library of small chemical fragments.
Rosetta Interfaces
Rosetta provides several optional user interfaces for interacting with the Rosetta library. In addition to the standard command line interface, pyRosetta (http://pyrosetta.org) has been developed. It contains a set of Python bindings to the Rosetta libraries which integrates many aspects of Rosetta into Python scripts. A simple XML-based scripting language is available which allows users without programming experience to quickly generate custom protocols consisting of existing Rosetta movers and filters. In addition to these conventional interfaces, the “FoldIt” game has been developed in which the player manually alters the protein conformation to identify energy minima using the Rosetta scoring function (http://www.fold.it).
Acknowledgments
We thank Laura Mizoue for discussions about the manuscript.
Supporting Information Available
We provide six tutorials that demonstrate basic usage of Rosetta: (1) protein folding, (2) refinement, (3) loop modeling, (4) protein−protein docking, (5) small molecule docking, and (6) protein design. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Berman H. M.; Battistuz T.; Bhat T. N.; Bluhm W. F.; Bourne P. E.; Burkhardt K.; Feng Z.; Gilliland G. L.; Iype L.; Jain S.; Fagan P.; Marvin J.; Padilla D.; Ravichandran V.; Schneider B.; Thanki N.; Weissig H.; Westbrook J. D.; Zardecki C. (2002) The Protein Data Bank. Acta Crystallogr. D58, 899–907. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C.; Koetzle T. F.; Williams G. J.; Meyer E. F. Jr.; Brice M. D.; Rodgers J. R.; Kennard O.; Shimanouchi T.; Tasumi M. (1977) The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542. [DOI] [PubMed] [Google Scholar]
- Wang C.; Bradley P.; Baker D. (2007) Protein-protein docking with backbone flexibility. J. Mol. Biol. 373, 503–519. [DOI] [PubMed] [Google Scholar]
- Simons K. T.; Kooperberg C.; Huang E.; Baker D. (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268, 209–225. [DOI] [PubMed] [Google Scholar]
- Bystroff C.; Simons K. T.; Han K. F.; Baker D. (1996) Local sequence-structure correlations in proteins. Curr. Opin. Biotechnol. 7, 417–421. [DOI] [PubMed] [Google Scholar]
- Rohl C. A.; Strauss C. E.; Misura K. M.; Baker D. (2004) Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93. [DOI] [PubMed] [Google Scholar]
- Levinthal C. (1968) Are there pathways for protein folding. J. Chim. Phys. Phys.-Chim. Biol. 65, 44–45. [Google Scholar]
- Kuhlman B.; Baker D. (2000) Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. U.S.A. 97, 10383–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack R. L. Jr.; Karplus M. (1993) Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J. Mol. Biol. 230, 543–574. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A.; Kuhlman B.; Snoeyink J. (2005) Rotamer-Pair Energy Calculations Using a Trie Data Structure. Lect. Notes Comput. Sci. 3692, 389–400. [Google Scholar]
- Simons K. T.; Ruczinski I.; Kooperberg C.; Fox B. A.; Bystroff C.; Baker D. (1999) Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins 34, 82–95. [DOI] [PubMed] [Google Scholar]
- Kuhlman B.; Dantas G.; Ireton G. C.; Varani G.; Stoddard B. L.; Baker D. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368. [DOI] [PubMed] [Google Scholar]
- Lazaridis T.; Karplus M. (1999) Effective energy function for proteins in solution. Proteins 35, 133–152. [DOI] [PubMed] [Google Scholar]
- Kortemme T.; Morozov A. V.; Baker D. (2003) An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 326, 1239–1259. [DOI] [PubMed] [Google Scholar]
- Bradley P.; Misura K. M.; Baker D. (2005) Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871. [DOI] [PubMed] [Google Scholar]
- Das R.; Qian B.; Raman S.; Vernon R.; Thompson J.; Bradley P.; Khare S.; Tyka M. D.; Bhat D.; Chivian D.; Kim D. E.; Sheffler W. H.; Malmstrom L.; Wollacott A. M.; Wang C.; Andre I.; Baker D. (2007) Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins 69, Suppl. 8118–128. [DOI] [PubMed] [Google Scholar]
- Bonneau R.; Strauss C. E.; Rohl C. A.; Chivian D.; Bradley P.; Malmstrom L.; Robertson T.; Baker D. (2002) De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322, 65–78. [DOI] [PubMed] [Google Scholar]
- Das R.; Andre I.; Shen Y.; Wu Y.; Lemak A.; Bansal S.; Arrowsmith C. H.; Szyperski T.; Baker D. (2009) Simultaneous prediction of protein folding and docking at high resolution. Proc. Natl. Acad. Sci. U.S.A. 106, 18978–18983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S.; Vernon R.; Thompson J.; Tyka M.; Sadreyev R.; Pei J.; Kim D.; Kellogg E.; Dimaio F.; Lange O.; Kinch L.; Sheffler W.; Kim B. H.; Das R.; Grishin N. V.; Baker D. (2009) Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77, Suppl. 989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell D. J.; Kortemme T. (2009) Computer-aided design of functional protein interactions. Nat. Chem. Biol. 5, 797–807. [DOI] [PubMed] [Google Scholar]
- Burguete A. S.; Fenn T. D.; Brunger A. T.; Pfeffer S. R. (2008) Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell 132, 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl C. A.; Strauss C. E.; Chivian D.; Baker D. (2004) Modeling structurally variable regions in homologous proteins with Rosetta. Proteins 55, 656–677. [DOI] [PubMed] [Google Scholar]
- Canutescu A. A.; Dunbrack R. L. Jr. (2003) Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 12, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell D. J.; Coutsias E. A.; Kortemme T. (2009) Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6, 551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutsias E. A.; Seok C.; Jacobson M. P.; Dill K. A. (2004) A kinematic view of loop closure. J. Comput. Chem. 25, 510–528. [DOI] [PubMed] [Google Scholar]
- Qian B.; Raman S.; Das R.; Bradley P.; McCoy A. J.; Read R. J.; Baker D. (2007) High-resolution structure prediction and the crystallographic phase problem. Nature 450, 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura K. M.; Chivian D.; Rohl C. A.; Kim D. E.; Baker D. (2006) Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc. Natl. Acad. Sci. U.S.A. 103, 5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R.; Tsai J.; Ruczinski I.; Chivian D.; Rohl C.; Strauss C. E.; Baker D. (2001) Rosetta in CASP4: Progress in ab initio protein structure prediction. Proteins 5, Suppl.119–126. [DOI] [PubMed] [Google Scholar]
- Bradley P.; Malmstrom L.; Qian B.; Schonbrun J.; Chivian D.; Kim D. E.; Meiler J.; Misura K. M.; Baker D. (2005) Free modeling with Rosetta in CASP6. Proteins 61, Suppl. 7128–134. [DOI] [PubMed] [Google Scholar]
- Chivian D.; Kim D. E.; Malmstrom L.; Bradley P.; Robertson T.; Murphy P.; Strauss C. E.; Bonneau R.; Rohl C. A.; Baker D. (2003) Automated prediction of CASP-5 structures using the Robetta server. Proteins 53, Suppl. 6524–533. [DOI] [PubMed] [Google Scholar]
- Bradley P.; Chivian D.; Meiler J.; Misura K. M.; Rohl C. A.; Schief W. R.; Wedemeyer W. J.; Schueler-Furman O.; Murphy P.; Schonbrun J.; Strauss C. E.; Baker D. (2003) Rosetta predictions in CASP5: Successes, failures, and prospects for complete automation. Proteins 53, Suppl. 6457–468. [DOI] [PubMed] [Google Scholar]
- Rohl C. A. (2005) Protein structure estimation from minimal restraints using Rosetta. Methods Enzymol. 394, 244–260. [DOI] [PubMed] [Google Scholar]
- Cornilescu G.; Delaglio F.; Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Bowers P. M.; Strauss C. E.; Baker D. (2000) De novo protein structure determination using sparse NMR data. J. Biomol. NMR 18, 311–318. [DOI] [PubMed] [Google Scholar]
- Rohl C. A.; Baker D. (2002) De novo determination of protein backbone structure from residual dipolar couplings using Rosetta. J. Am. Chem. Soc. 124, 2723–2729. [DOI] [PubMed] [Google Scholar]
- Meiler J.; Baker D. (2003) Rapid protein fold determination using unassigned NMR data. Proc. Natl. Acad. Sci. U.S.A. 100, 15404–15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Lange O.; Delaglio F.; Rossi P.; Aramini J. M.; Liu G.; Eletsky A.; Wu Y.; Singarapu K. K.; Lemak A.; Ignatchenko A.; Arrowsmith C. H.; Szyperski T.; Montelione G. T.; Baker D.; Bax A. (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Vernon R.; Baker D.; Bax A. (2009) De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR 43, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander N.; Bortolus M.; Al-Mestarihi A.; McHaourab H.; Meiler J. (2008) De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure 16, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S. M.; Dawson E. S.; Francis D. J.; Van Eps N.; Klug C. S.; Hubbell W. L.; Meiler J.; Gurevich V. V. (2008) A model for the solution structure of the rod arrestin tetramer. Structure 16, 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.; Baker D. (2008) Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 77, 363–382. [DOI] [PubMed] [Google Scholar]
- Ramelot T. A.; Raman S.; Kuzin A. P.; Xiao R.; Ma L. C.; Acton T. B.; Hunt J. F.; Montelione G. T.; Baker D.; Kennedy M. A. (2009) Improving NMR protein structure quality by Rosetta refinement: A molecular replacement study. Proteins 75, 147–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.; Baker D. (2009) Prospects for de novo phasing with de novo protein models. Acta Crystallogr. D65, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F.; Tyka M. D.; Baker M. L.; Chiu W.; Baker D. (2009) Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 392, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert S.; Staritzbichler R.; Wotzel N.; Karakas M.; Stewart P. L.; Meiler J. (2009) EM-fold: De novo folding of α-helical proteins guided by intermediate-resolution electron microscopy density maps. Structure 17, 990–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert S.; Stewart P. L.; Meiler J. (2009) Hybrid approaches: Applying computational methods in cryo-electron microscopy. Curr. Opin. Struct. Biol. 19, 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. E.; Chivian D.; Baker D. (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D.; Kim D. E.; Malmstrom L.; Schonbrun J.; Rohl C. A.; Baker D. (2005) Prediction of CASP6 structures using automated Robetta protocols. Proteins 61, Suppl. 7157–166. [DOI] [PubMed] [Google Scholar]
- Gray J. J.; Moughon S.; Wang C.; Schueler-Furman O.; Kuhlman B.; Rohl C. A.; Baker D. (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299. [DOI] [PubMed] [Google Scholar]
- Lyskov S.; Gray J. J. (2008) The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 36, W233–W238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S.; Gray J. J. (2008) Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J. Mol. Biol. 381, 1068–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian A.; Maynard J. A.; Gray J. J. (2008) Modeling the structure of mAb 14B7 bound to the anthrax protective antigen. Proteins 70, 218–230. [DOI] [PubMed] [Google Scholar]
- Sivasubramanian A.; Chao G.; Pressler H. M.; Wittrup K. D.; Gray J. J. (2006) Structural model of the mAb 806-EGFR complex using computational docking followed by computational and experimental mutagenesis. Structure 14, 401–414. [DOI] [PubMed] [Google Scholar]
- Schueler-Furman O.; Wang C.; Baker D. (2005) Progress in protein-protein docking: Atomic resolution predictions in the CAPRI experiment using RosettaDock with an improved treatment of side-chain flexibility. Proteins: Struct., Funct., Bioinf. 60, 187–194. [DOI] [PubMed] [Google Scholar]
- Chaudhury S.; Sircar A.; Sivasubramanian A.; Berrondo M.; Gray J. J. (2007) Incorporating biochemical information and backbone flexibility in RosettaDock for CAPRI rounds 6−12. Proteins 69, 793–800. [DOI] [PubMed] [Google Scholar]
- Sircar A.; Gray J. J. (2010) SnugDock: Paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput. Biol. 6, e1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid C.; Rushe M.; Jarpe M.; van Vlijmen H.; Dolinski B.; Qian F.; Cachero T. G.; Cuervo H.; Yanachkova M.; Nwankwo C.; Wang X.; Etienne N.; Garber E.; Bailly V.; de Fougerolles A.; Boriack-Sjodin P. A. (2006) Structure activity relationships of monocyte chemoattractant proteins in complex with a blocking antibody. Protein Eng., Des. Sel. 19, 317–324. [DOI] [PubMed] [Google Scholar]
- Taylor R. D.; Jewsbury P. J.; Essex J. W. (2002) A review of protein-small molecule docking methods. J. Comput.-Aided Mol. Des. 16, 151–166. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Glab K., Mueller R., and Meiler J. (2008) Small Molecule Rotamers Enable Simultaneous Optimization of Small Molecule and Protein Degrees of Freedom in ROSETTALIGAND Docking. German Conference on Bioinformatics (Beyer A., and Schroeder M., Eds.) pp 148−157, Dresden. [Google Scholar]
- Meiler J.; Baker D. (2006) ROSETTALIGAND: Protein-small molecule docking with full side-chain flexibility. Proteins 65, 538–548. [DOI] [PubMed] [Google Scholar]
- Roche O.; Kiyama R.; Brooks C. L. III (2001) Ligand-protein database: Linking protein-ligand complex structures to binding data. J. Med. Chem. 44, 3592–3598. [DOI] [PubMed] [Google Scholar]
- Ferrara P.; Gohlke H.; Price D. J.; Klebe G.; Brooks C. L. III (2004) Assessing scoring functions for protein-ligand interactions. J. Med. Chem. 47, 3032–3047. [DOI] [PubMed] [Google Scholar]
- Davis I. W.; Baker D. (2009) RosettaLigand docking with full ligand and receptor flexibility. J. Mol. Biol. 385, 381–392. [DOI] [PubMed] [Google Scholar]
- Davis I. W.; Raha K.; Head M. S.; Baker D. (2009) Blind docking of pharmaceutically relevant compounds using RosettaLigand. Protein Sci. 18, 1998–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K. W.; Dawson E. S.; Henry L. K.; Field J. R.; Blakely R. D.; Meiler J. (2009) Structural determinants of species-selective substrate recognition in human and Drosophila serotonin transporters revealed through computational docking studies. Proteins 74, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A.; Singh S. K.; Kawate T.; Jin Y.; Gouaux E. (2005) Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223. [DOI] [PubMed] [Google Scholar]
- Dunbrack R. L.; Karplus M. (1993) Backbone-Dependent Rotamer Library for Proteins: Application to Side-Chain Prediction. J. Mol. Biol. 230, 543–574. [DOI] [PubMed] [Google Scholar]
- Korkegian A.; Black M. E.; Baker D.; Stoddard B. L. (2005) Computational thermostabilization of an enzyme. Science 308, 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T.; Joachimiak L. A.; Bullock A. N.; Schuler A. D.; Stoddard B. L.; Baker D. (2004) Computational redesign of protein-protein interaction specificity. Nat. Struct. Mol. Biol. 11, 371–379. [DOI] [PubMed] [Google Scholar]
- Jiang L.; Althoff E. A.; Clemente F. R.; Doyle L.; Röthlisberger D.; Zanghellini A.; Gallaher J. L.; Betker J. L.; Tanaka F.; Barbas C. F.; Hilvert D.; Houk K. N.; Stoddard B. L.; Baker D. (2008) De novo computational design of retro-aldol enzymes. Science 319, 1387–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. J.; Sievers S. A.; Karanicolas J.; Ivanova M. I.; Baker D.; Eisenberg D. (2006) The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 4074–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroggio X. I.; Kuhlman B. (2006) Computational design of a single amino acid sequence that can switch between two distinct protein folds. J. Am. Chem. Soc. 128, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Dantas G.; Kuhlman B.; Callender D.; Wong M.; Baker D. (2003) A large scale test of computational protein design: Folding and stability of nine completely redesigned globular proteins. J. Mol. Biol. 332, 449–460. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez I.; Reverter D.; Vendrell J.; Aviles F. X.; Coll M. (1997) The three-dimensional structure of human procarboxypeptidase A2. Deciphering the basis of the inhibition, activation and intrinsic activity of the zymogen. EMBO J. 16, 6906–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas G.; Corrent C.; Reichow S. L.; Havranek J. J.; Eletr Z. M.; Isern N. G.; Kuhlman B.; Varani G.; Merritt E. A.; Baker D. (2007) High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J. Mol. Biol. 366, 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Kuhlman B. (2006) RosettaDesign server for protein design. Nucleic Acids Res. 34, W235–W238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B. S.; Kortemme T.; Chadsey M. S.; Baker D.; Monnat R. J.; Stoddard B. L. (2002) Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell 10, 895–905. [DOI] [PubMed] [Google Scholar]
- Joachimiak L. A.; Kortemme T.; Stoddard B. L.; Baker D. (2006) Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J. Mol. Biol. 361, 195–208. [DOI] [PubMed] [Google Scholar]
- Humphris E. L.; Kortemme T. (2007) Design of multi-specificity in protein interfaces. PLoS Comput. Biol. 3, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babor M.; Kortemme T. (2009) Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins 75, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S.; Love J. J.; Mayo S. L. (2007) A de novo designed protein protein interface. Protein Sci. 16, 2770–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini A.; Jiang L.; Wollacott A. M.; Cheng G.; Meiler J.; Althoff E. A.; Röthlisberger D.; Baker D. (2006) New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 15, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röthlisberger D.; Khersonsky O.; Wollacott A. M.; Jiang L.; DeChancie J.; Betker J.; Gallaher J. L.; Althoff E. A.; Zanghellini A.; Dym O.; Albeck S.; Houk K. N.; Tawfik D. S.; Baker D. (2008) Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195. [DOI] [PubMed] [Google Scholar]
- Shortle D.; Simons K. T.; Baker D. (1998) Clustering of low-energy conformations near the native structures of small proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 11158–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A.; Macarthur M. W.; Moss D. S.; Thornton J. M. (1993) Procheck: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Shen M. Y.; Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P.; Schonbrun J.; Baker D. (2007) Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U.S.A. 104, 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler W.; Baker D. (2009) RosettaHoles: Rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 18, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.