Abstract
Mammalian genomes encode numerous natural antisense transcripts, but the function of these transcripts is not well understood. Functional validation studies indicate that antisense transcripts are not a uniform regulatory RNA group, but instead belong to multiple categories with some common features. Recent evidence indicates that antisense transcripts are frequently functional and use diverse transcriptional and post-transcriptional gene regulatory mechanisms to carry out a wide variety of biological roles.
Natural antisense transcripts are RNA molecules that are transcribed from the opposite DNA strand compared with other transcripts and overlap in part with sense RNA. Both sense and antisense RNAs can encode proteins or be non-protein-coding transcripts; however, the most prominent form of antisense transcription in the mammalian genome is a non-protein-coding antisense RNA partner of a protein-coding gene1. The presence of non-protein-coding sense and antisense transcript pairs implies that natural antisense transcripts may also regulate non-protein-coding sense RNAs.
Various techniques have been used to identify natural antisense transcripts, including large-scale sequencing of cDNA clones1, tiling arrays2, bioinformatics analysis of RefSeq and EST databases3, hybridization techniques4, SAGE libraries5, strand-specific microarrays6 and, most recently, asymmetric strand-specific analysis of gene expression (ASSAGE)7 and global run-on sequencing (GRO-seq)8. These studies have demonstrated widespread antisense transcription in mammalian genomes. The mammalian transcriptomes are still not sequenced deeply enough to provide the organization of all low copy number antisense transcripts. Nevertheless, the majority of transcription units (TUs; see Glossary box) appear to contain antisense transcripts1. Moreover, antisense transcripts have been documented for many active promoters8-10. Therefore, the presence of antisense transcripts is no longer a curiosity but rather a pervasive feature of mammalian genomes. For instance, the FANTOM-3 mouse transcriptome sequencing consortium identified natural antisense transcripts for more than 70% of TUs, the majority of which represent non-protein-coding RNA 1.
Characteristics of antisense RNAs
Antisense transcripts are not evenly distributed across the genome. Both ends of protein-coding genes have a propensity for natural antisense transcription7,11; specifically, antisense transcription is enriched 250 nucleotides upstream of the transcription start site (TSS)8,9 and 1.5 kilobases downstream of sense genes8,12. The basal expression levels of sense and antisense transcripts in different tissues and cell lines may be either positively or negatively correlated1,13. Moreover, antisense transcript expression in different human cell lines is not always linked to expression of the sense gene, which suggests the use of alternative transcriptional regulatory elements7. Antisense RNAs have a tendency to undergo fewer splicing events and typically show lower abundance compared with sense transcripts7. Interestingly, knockdown or blockade of endogenous antisense transcripts can have multiple outcomes, with the corresponding sense transcript concentration showing either an increase (discordant regulation) or a decrease (concordant regulation)14. It has been proposed that discordant de-repression of sense transcript expression, resulting in upregulation of sense RNA expression, can be achieved by removal or steric blockade of many but not all antisense transcripts1,14. These and other variable intrinsic properties imply that antisense-mediated regulation of gene expression must operate through a variety of mechanisms, and further suggest that antisense transcripts are a heterogeneous group of regulatory RNAs.
Supplementary information S1 (table) contains all reported functional antisense transcripts in mammalian genomes characterized to date. This growing list of validated sense–antisense transcript pairs includes many important developmental genes, as well as genes known to be involved in a variety of human disorders. Natural antisense transcripts must therefore be assumed to exert positive or negative regulation at different levels of gene expression invoking diverse and largely uncharacterized co-regulatory factors. Hence, potential involvement of natural antisense transcripts should be considered in a wide range of future studies on the regulation of gene expression.
There are a number of proposed mechanisms for antisense-mediated regulation of sense mRNA. Here, we categorize the mechanisms into four main groups: mechanisms related to transcription, RNA–DNA interactions, RNA–RNA interactions in the nucleus and RNA–RNA interactions in the cytoplasm. Each type of mechanism will be discussed in detail below.
Transcription-related modulation
According to this mechanism, the act of transcription in the antisense direction, but not the antisense RNA molecule itself, modulates transcription of sense RNA. Transcription in the antisense direction is suggested to cause alterations in sense RNA, as in the case of transcriptional collision, and is also proposed to be involved in genomic rearrangements (Figure 1).
Figure 1. Modulation of transcription.
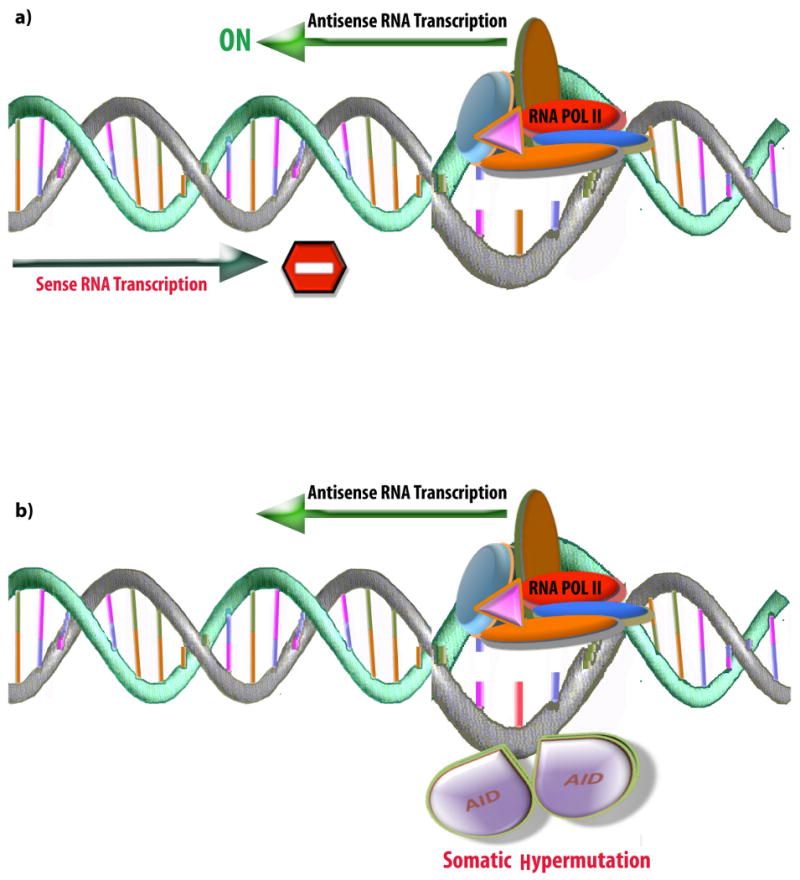
(a) Transcription collision model in which RNA synthesis from one DNA strand might clash with transcription from the other strand. According to this model, transcription only occurs in one direction at any given time, and active antisense transcription would suppress sense RNA transcription. Alternatively, supercoiling of DNA strand due to transcription of converging genes is suggested to affect transcription in both directions. (b) Genomic DNA rearrangement in hypervariable regions of B and T lymphocytes. Natural antisense transcription from the opposite strand will open the transcription bubble and expose the single-stranded DNA (ssDNA). With the ssDNA exposed, the activation-induced cytidine deaminase (AID) enzyme can bind to DNA and convert deoxycytidine to deoxyuridine, which underlies hypermutation processes. Considering the small size of the transcription bubble (17 +/- 1 melted bases), which is entirely covered by the elongating RNA polymerase II (RNAPII), it is not clear how AID fits into this region to modify the non-template strand of DNA. Additionally, the selectivity of the somatic hypermutation phenomenon to particular DNA regions suggests that more complex machinery must be involved, in addition to antisense RNA transcription per se. Nevertheless, in this proposed model the act of transcription in antisense direction, but not the antisense RNA molecule by itself, would be important for genomic rearrangements.
Transcriptional collision
The transcriptional collision model is based on the assumption that RNA polymerases bind to the promoters of convergent genes on opposing strands of DNA and then proceed towards the 3′ end of sense and antisense genes. RNA polymerase complexes collide in the overlapping region, blocking further transcription15 (Figure 1a). Collision of RNA polymerases has been observed in Escherichia coli, using atomic force microscopy16. Transcriptional collision has also been observed in Saccharomyces cerevisiae17, where transcription from the antisense direction halts transcription of the sense mRNA. A bioinformatics study found that, in humans and mice, the length of overlapping regions between sense and antisense transcripts is inversely correlated to the expression levels of antisense transcript. This finding may suggest a possible clash of RNA polymerases, pertaining to the point that the likelihood of collision increases as the length of overlapping region increases18. In this model, the antisense RNA per se is not involved in interfering with transcription of the sense RNA: instead, it is transcription in the antisense direction that decreases synthesis of sense RNA.
Transcriptional collision may affect a subset of mammalian cis-encoded natural antisense transcripts, but it is apparently not the predominant route of antisense-mediated gene regulation. Transcription of the sense and antisense RNAs may occur at different times at the same chromosome locus, or sense and antisense RNA may be transcribed simultaneously from paternal and maternal chromosomes. Allelic-specific transcription may explain why the X chromosome shows a significantly lower degree of antisense transcription compared with other chromosomes1.
Genomic rearrangements
Immunoglobulin production in B lymphocytes and receptor selection by T lymphocytes depend on transcription from hypervariable regions. To generate variability, T and B lymphocytes need a recombination process that occurs through hypermutation in the variable regions of immunoglobulin and T cell receptor genes. Activation-induced cytidine deaminase (AID), which deaminates deoxycytidine to deoxyuridine in single-stranded DNA (ssDNA), is required for both somatic hypermutation (SHM) and class switch recombination (CSR), two processes that are involved in secondary immune response. DNA must locally uncoil or melt, and become single-stranded for DNA recombination, replication and transcription to occur. The transcription complex consists of an RNA polymerase molecule enclosing approximately 17 +/- 1 melted bases (the transcription bubble) of the template DNA19; however, the size of transcription bubble may vary in different instances. The AID enzyme optimally deaminates a small ssDNA bubble, in vitro, consisting of ∼5 nucleotides with a ssDNA motif, called WRC20. It has been hypothesized that antisense transcription in the variable region could make the ssDNA accessible for AID21,22 (Figure 1b).
Antisense transcription is frequently observed in the variable23,24, but not in the constant, regions and it may be initiated in preparation for SHM and CSR (reviewed in Ref. 25). According to this model, antisense transcription is proposed to open the transcription bubble and expose the ssDNA, which is a substrate for AID23,24, or antisense transcription may be involved in remodelling of the chromatin structure to make the DNA sequence accessible for recombination22-24. In both of these scenarios, transcription from the antisense direction, but not the antisense RNA molecule, is important for genomic rearrangement.
RNA–DNA interactions
Natural antisense transcripts may also be involved in epigenetic regulation of transcription, through DNA methylation26, chromatin modifications27 and monoallelic expression (for example, genomic imprinting28, X-chromosome inactivation29 and random monoallelic exclusion of the autosomal loci). This gene regulatory model is based on direct or indirect RNA–DNA and RNA–chromatin interactions30. Antisense transcripts may bind to the corresponding DNA strand, resulting in either DNA methylation (Figure-2a) or providing a scaffold for histone-modifying enzyme (HME) recruitment and subsequent alterations of the chromatin status (Figure-2b).
Figure 2. Induction of chromatin and DNA epigenetic changes by natural antisense transcripts.
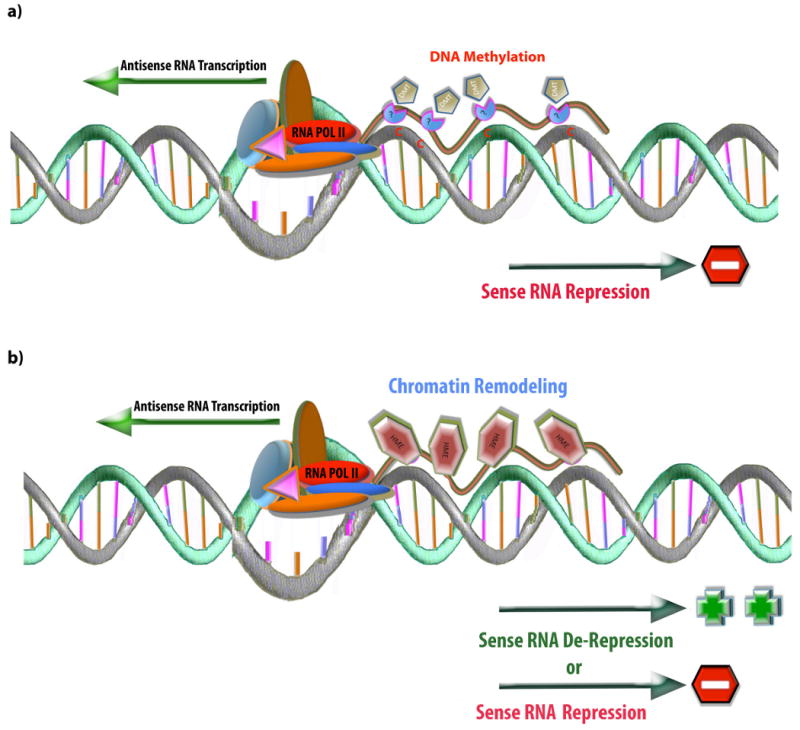
(a) RNA–DNA interaction model in which newly formed antisense RNA transcript can interact, directly or indirectly, with DNA methyltransferases (DMT) and guide methylation of DNA, resulting in repression of sense RNA transcription (b) Alternatively, newly formed antisense RNA transcript can recruit histone-modifying enzymes (HMEs) and modify chromatin, thereby modulating chromatin architecture and epigenetic memory. Activation of sense RNA transcription, by the means of removal of repressive effects of promoter-directed RNA transcripts, has been observed for several gene loci34,35,62. This de-repression effect may be particularly prominent when an antisense RNA is located in or around the transcription start site of the sense gene. Argonaute2 (AGO2) protein is thought to be involved in this largely uncharacterized process (not shown). The resultant chromatin remodelling would repress or de-repress sense RNA transcription. Similar models have been proposed for imprinted genes as well as for mammalian X-chromosome inactivation.
Some of the antisense-mediated epigenetic changes are reported to be independent of Dicer, arguing against a role for small RNA mediators27. Therefore, antisense RNA processing to small RNA, which may be mediated by Dicer in the cytoplasm, is not necessarily required for epigenetic modifications. Instead, antisense transcripts accumulate locally and trigger DNA or chromatin modifications31. These modifications then expand to neighbouring regions, even though these adjacent regions do not exhibit complementarities to the original antisense transcript32. The secondary expansion of the modifications may be restricted to the promoter or enhancer of a single gene (random monoallelic exclusion), or may include a cluster of genes, such as in the case of genomic imprinting of the Kcnq1 imprinted locus28. Finally, this expansion process occasionally involves an entire chromosome, such as X chromosome inactivation in females.
Alteration of DNA and chromatin
Natural antisense transcripts have been proposed to cause DNA methylation26, DNA demethylation33 and chromatin modifications of non-imprinted autosomal loci27,34. Suppression of transcription is usually caused by DNA and chromatin modification at the promoter region of the sense strand (Supplementary information S2 (box)). For example, an antisense RNA for the α-globulin 2 gene (HBA2) can induce DNA methylation, leading to silencing of the α-globulin 2 gene26.
Antisense-mediated transcriptional silencing also affects the p1527, p2134 and progesterone receptor (PR)35 genes, through DNA methylation and heterochromatin formation. Suppression of sense transcription is often induced by trimethylation of histone H3 lysine 27 (H3K27me3) at the sense promoter region. For example, an antisense transcript of the tumour suppressor gene p21 recruits a regulatory complex that induces H3K27me3 and suppression of the sense promoter region34. Considering the presence of RNA-binding motifs in some chromatin-modifying enzymes30, one could postulate that RNA transcripts are local modulators of chromatin structure. This proposed mechanism might also explain functionality even if the abundance of antisense RNA molecule is low9,10,34. Unlike RNAs that can be present in cells in many copies, there are two copies of DNA for any given gene in cells; therefore, only two molecules of antisense RNA per cell are sufficient to bind to the corresponding DNA strand and to exert a regulatory function. These proposed local effects of antisense transcripts might sometimes extend to the neighbouring regions, such as in the case of the imprinted locus28 (see below). These examples suggest the possibility of local and extended antisense RNA-induced DNA and chromatin modifications.
Chromatin alteration of active promoters
Overlapping transcription of small (<50 nucleotides) RNAs 36,37 as well as promoter-associated small RNAs (PASRs)36,37, termini-associated small RNAs (TASRs)36,37 and promoter upstream transcripts (PROMPTs)10 have been documented using strand-specific genomic tilling arrays in the ENCODE region of the human genome36,37. Transcription start site-associated RNAs (TSSa-RNAs) (20-90 nucleotides) have also been identified using deep sequencing techniques8,9. The enrichment of antisense transcripts in both promoter and terminal regions has been confirmed by the unbiased ASSAGE technique7. TSSa-RNAs are flanking active promoters in both the sense and antisense direction with regard to downstream protein-coding genes8,9. Divergent promoter activity, which is believed to produce this small RNA group, is documented for more than half of mouse and human genes8,9. These well-documented promoter activities, which generate promoter-directed sense and antisense transcripts, challenge the canonical ‘gene’ definition and how genes and their regulatory elements are arranged in mammalian genomes. Mammalian transcriptomes appear to contain an abundance of nested and overlapping gene structures, giving rise to both coding and noncoding transcripts. In addition, it seems that the RNA regulatory elements that control the expression of a gene are frequently distributed within or beyond other genes in both directions. Collectively, pervasive divergent promoter activity suggests a lack of definite 5′ and 3′ boundaries for the transcribed genes.
Interestingly, these small antisense transcripts do not correspond to annotated natural antisense transcripts36,37. There is also no evidence for double-stranded RNA (dsRNA) or hairpin RNA precursors that might represent intermediates in the biogenesis of such small natural antisense transcripts36,37. Generation of TSSa-RNA is also shown to be independent of Dicer, suggesting that the possible regulatory function of these transcripts is not mediated through common RNA interference (RNAi), such as RISC and RITS pathways. Additionally, the expression of promoter-directed antisense transcripts may be associated with the presence of abortive RNA transcripts38.
The transcribed regions of PROMPTs and TSSa-RNAs overlap with paused RNA polymerase II (RNAPII) and active chromatin marks, such as trimethylation of histone H3 lysine 4 (H3K4me3), suggesting a role for local RNA accumulation in maintaining the dynamic chromatin state required for promoter activity9,10. The abundance of short RNA transcripts at the transcription initiation and termination sites8,9 as well as transcription of unstable transcripts upstream of genes10, which are usually co-localized with particular chromatin marks, imply involvement of these local and transient RNA transcripts in the regulation of the sense gene expression and possible participation in chromatin remodelling. We hypothesize that the frequent presence of these RNA classes in or around the promoter region of actively transcribed genes, in a very low concentration, suggests the possibility of local RNA-induced chromatin modifications.
Genomic imprinting
Imprinted genes are genes for which only one allele, maternal or paternal, is actively transcribed. Natural antisense transcripts are often associated with imprinted genes, with a frequency up to 81% in one study1 (Supplementary information S3 (box)). More than 160 imprinted genes have been identified so far in humans and mice, most of which are organized into clusters. Some imprinted genes, including insulin-like growth-factor type-2 receptor (IGFER)39 and potassium voltage-gated channel, KQT-like (Kcnq1) imprinting control region31, exhibit guided chromatin and DNA modification by antisense RNA that expands to include neighbouring genes. These effects are not mediated through the RNAi pathway40. Instead, antisense RNA appears to recruit repressor complexes that modify chromatin into an inactive state. Suppressive chromatin modifications spread in both directions to neighbouring genes, similar to X chromosome inactivation (below) but with limited penetrance31.
X chromosome inactivation
X chromosome inactivation is required for balanced expression of the genes on the X chromosome in female mammals. This dosage compensation occurs through heterochromatin formation along the X chromosome to be inactivated. Two long non-protein-coding genes are transcribed from the X chromosome inactivation centre, XIST (X-inactive specific transcript) and TSIX (X [inactive]-specific transcript, antisense). These two sense and antisense transcripts control the silencing of the X chromosome29. The X chromosome inactivation centre is necessary and sufficient for X chromosome inactivation. Elements of the mechanism have been demonstrated by showing that Tsix silences Xist through modification of the chromatin structure in the Xist promoter region. Premature termination of Tsix transcription abolishes the repressive chromatin configuration at the Xist promoter29.
Nuclear RNA duplex formation
The third type of mechanism for antisense transcript-mediated gene regulation is based on the predicted nuclear sense–antisense RNA duplex formation (Figure 3Aa). The duplex RNA between sense and antisense transcripts may result in several outcomes, all of which modulate sense mRNA expression. Nuclear RNA duplex may produce mRNA transcripts with alternative splicing. Additionally, hybrid nuclear RNA is a substrate for editing enzymes that can alter the localization, transport and stability of the sense mRNA transcript.
Figure-3. Nuclear and cytoplasmic sense–antisense RNA pairing.
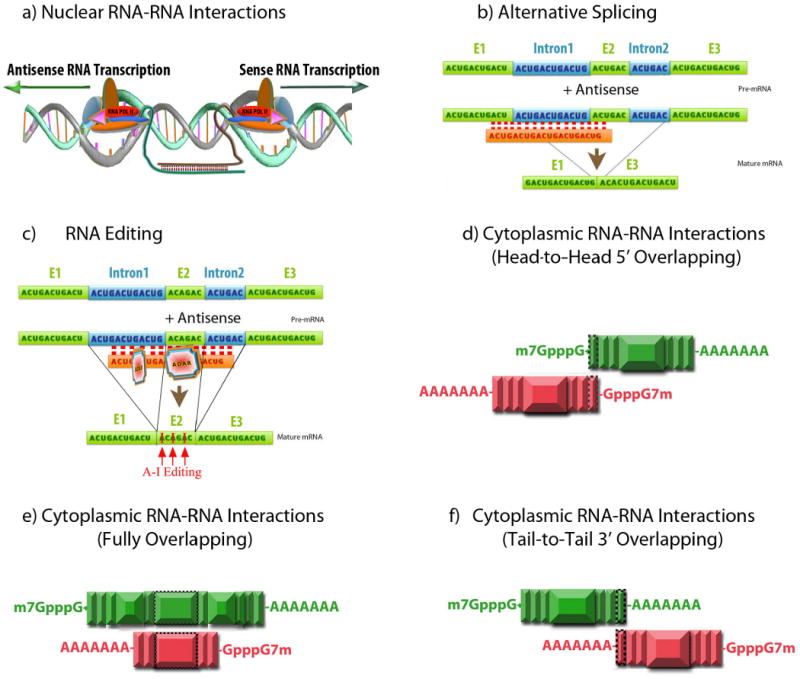
(Aa) Nuclear RNA duplex formation might occur locally after transcription, thereby inhibiting sense RNA processing. The RNA processing includes capping, polyadenylation, nuclear localization and transport, which all might be affected by nuclear sense-antisense RNA duplex formation. (Ab) Natural antisense transcripts can cover donor and acceptor splice sites in the sense pre-mRNA transcript to change alternative splicing patterns. (Ac) RNA editing is another possible consequence of nuclear RNA duplex formation, which causes A-to-I editing by the ADAR (adenosine deaminases that act on RNA) enzyme. RNA editing can induce nuclear retention and degradation of hyper-edited transcript by inosine-specific nucleases or alternatively, editing can result in alteration of the amino acid sequence. (B) Cytoplasmic sense–antisense duplex formation can occur in three different formats; (Ba) head-to-head 5′ overlapping, (Bb) fully overlapping in which antisense is embedded in the sense transcript or (Bc) tail-to-tail 3′ overlapping configuration. Exons are depicted as vertical boxes and the overlapping region is shown with a border. Cytoplasmic sense–antisense RNA duplex formation can possibly exert its own effects on sense mRNA. Hiding or exposing AU-rich elements in sense transcript can affect RNA stability. Changes in the RNA secondary structure, after binding to antisense RNA, can alter translation, subcellular localization and accessibility of the RNA degradation machinery. According to this model, natural antisense transcripts can potentially ‘mask’ miRNA-binding sites and release the miRNA-induced block of translation. Finally, Dicer can also process sense–antisense RNA duplexes to endogenous siRNAs.
Alternative splicing and termination
Antisense RNA may also bind to the sense RNA, masking the splice sites and thereby changing the balances between splice variants (Figure 3Ab). Thyroid hormone receptor alpha gene (TRα) is an example where the antisense transcript RevErbAα influences splicing of TRα1 and TRα2 mRNAs41. Using a similar arrangement, antisense transcripts can potentially cause alternative polyadenylation and termination of transcription.
Transport, nuclear retention and editing
Natural antisense transcripts may modulate mRNA nuclear transport by a mechanism that involves duplex formation between sense and antisense RNAs. Nuclear retention of the antisense RNA is commonly observed and may account for some antisense RNA-mediated regulation. Some cellular stressors, such as hypoxia, serum starvation and hydrogen peroxide, can change the nuclear retention pattern of antisense transcripts, thereby altering the levels of their sense partners42. Direct interactions with nuclear proteins or other nuclear RNAs likely cause nuclear retention of antisense transcripts, although the precise mechanism for each case has not yet been discovered.
Natural antisense transcripts have also been linked to mRNA editing (Figure 3Ac). Interaction between the Drosophila melanogaster 4f-rnp gene and its antisense transcript, sas-10, is reported to induce adenosine-to-inosine (A-to-I) editing in the overlapping region of 4f-rnp mRNA43. A-to-I RNA editing is induced by dsRNA formation, resulting in the recruitment of the enzyme ADAR (adenosine deaminases that act on RNA), which deaminates the targeted adenosine to inosine44.
Cytoplasmic RNA duplex formation
In the fourth proposed mechanism of sense–antisense interference, a duplex forms between sense and antisense RNA in the cytoplasm (Figure 3Ba–c). Cytoplasmic RNA hairpins may affect sense mRNA stability or translation. The sense–antisense RNA duplex may also, in theory, cover microRNA (miRNA)-binding sites or serve as a hairpin template for generating endogenous small interfering RNA (siRNA).
Changes in mRNA stability and translation
Cytoplasmic sense–antisense duplex formation can alter sense mRNA stability and translation efficiency. The overlapping region may affect mRNA stability by reducing mRNA decay, whereby mRNA undergoes endo- or exonucleolytic degradation by various RNases. Indeed, we have recently demonstrated that an antisense BACE1 transcript (BACE1-AS) increases the stability of BACE1 mRNA through a mechanism involving RNA duplex formation. We hypothesize that transient RNA duplex formation may alter the secondary or tertiary structure of BACE1 and thereby increase its stability42.
Antisense transcripts for inducible nitric oxide synthase (iNOS), an important gene in inflammatory diseases, increase the stability of iNOS mRNA45. Enhancement of iNOS mRNA stability is mediated through interactions of antisense RNA molecule with the AU-rich element (ARE)-binding HuR protein. The HuR protein, in turn, may suppress RNA degradation by inhibiting deadenylase or exonuclease enzymes45. Alterations in the secondary structure of HIF-1α sense transcript by an antisense RNA may expose the ARE and reduce the stability of the transcript by making this RNA prone to degradation46.
Translational inhibition is yet another proposed function for some naturally occurring antisense, as in the case of B cell maturation antigen (BCMA) transcript, where over-expression of the antisense RNA has been reported to reduce sense protein, but not sense mRNA, levels47. Another well-documented case of translational inhibition is the antisense for PU.1 mRNA. PU.1 mRNA translation is inhibited by a noncoding antisense transcript48, which is a polyadenylated RNA with a lower concentration but a longer half-life time than the sense PU.1 transcript. Processed antisense RNA in the cytoplasm may bind to the sense transcript and stall translation between initiation and elongation steps48. It also seems likely that formation of duplexes may alter ribosome entry, and consequently the protein output of the target.
Masking miRNA-binding sites
We propose that many natural antisense transcripts may have the ability to cover miRNA-binding sites following cytosolic RNA duplex formation. This appears to be the case for an antisense BACE1 transcript (BACE1-AS) in addition to its ability to increase the stability of BACE1 mRNA42. Thus, one of the regulatory functions of antisense transcripts may be to ‘mask’ the miRNA-binding site on the sense mRNA. The 3′UTR mRNAs are frequently shown to contain target sites for miRNAs, and at least 34% of natural antisense transcripts in the FANTOM3 dataset showed tail-to-tail format with a 3′UTR overlapping region.
Formation of endogenous siRNA
The presence of endogenous processing machinery for exogenous siRNA, which mediate sequence-specific knockdown of targeted genes, implies that endogenous siRNA should exist. Endogenous siRNAs derived from a natural antisense transcripts were observed in Arabidopsis thaliana, where they regulate salt tolerance49. Plant endogenous siRNA are derived from sense–antisense RNA duplex formation of several genes: for example, the Sho gene antisense transcript50, SRLK/AtRAP antisense transcript51 and 64% of protein-coding natural antisense transcripts in A. thaliana are reported to generate endogenous siRNAs52.
Endogenous siRNAs derived from transposable elements and pseudogenes have also been identified in mouse oocytes and cultured human cells53-57. Endogenous siRNAs originating from mRNAs and their corresponding antisense were also recently identified in mouse oocytes54 and human HepG2 liver carcinoma cells55. Both Piwi-interacting RNAs (piRNAs)58,59 and siRNAs originating from mRNAs were found in mouse oocytes54. Therefore, formation of endogenous siRNA from naturally occurring antisense transcripts appears to occur in mammalian cells.
Transposable elements, inverted repeat structures, sense–antisense genes (cis-natural antisense transcript) and antisense transcripts from remote loci (trans-natural antisense transcripts) have also been recognized as sources of dsRNAs and subsequent, Dicer-dependent, endogenous siRNA production54,55. One example includes the overlapping transcripts in the kinesin family member 4A (KIF4A) and PDZ domain-containing 11 (Pdzd11) locus that generates endogenous siRNA derived from its cis-antisense transcript. Importantly, almost all of the endogenous siRNAs (117 unique sequences) are derived from the overlapping region of the sense and antisense RNA, suggesting that these endogenous siRNAs are produced from an intermolecular dsRNA formed between the oppositely oriented transcripts. In Dicer mutants, levels of the siRNAs derived from the Pdzd11/Kif4 locus are decreased, and Pdzd11 and Kif4 mRNA levels are increased, suggesting that endogenous siRNA production is cytoplasmic and Dicer dependent, and that expression of both sense and antisense RNAs is regulated by the RNAi pathway54. Therefore, endogenous siRNAs may regulate both sense and antisense transcript levels.
Co-expression of natural antisense transcripts with their sense counterparts, as well as frequently observed concordant regulation of sense and antisense RNAs in many tissues and cell lines, argues against endogenous siRNA being the sole mechanism of antisense-mediated regulation of gene expression. Additionally, many co-expressed sense-antisense transcripts in D. melanogaster S2 cells do not generate endogenous siRNAs60,61. It is unclear how the majority of co-expressed sense and antisense RNAs escape the endogenous siRNA formation pathway 63. It is also not clear whether there is active selection for entry into the RNAi pathway and endogenous siRNA formation.
Concluding remarks
We have presented examples of functional natural antisense transcripts in order to show the multilayered involvement of these molecules in regulating gene expression. It can be concluded that antisense transcripts are not a uniform group of regulatory elements, but that they display some common features, including sequence complementarities to conventional sense genes. We have summarized the proposed mechanisms of antisense transcript actions into four models, each corresponding to a putative regulatory mechanism for certain antisense transcripts.
Although we have provided examples for each model, it is important to note that not all four models have equally strong experimental support. For instance, the frequently observed convergent promoter activity and engagement of multiple RNAPII complexes in the opposite orientation of active promoters would argue against the transcriptional collision model as a widely used mode of action. On the other hand, nuclear and cytoplasmic RNA–RNA interactions with various outcomes, such as RNA editing, splicing and dicing, have been observed for several functionally validated antisense transcripts, but it is not a predominant basis of natural antisense regulation. Furthermore, antisense RNA-induced alteration in sense promoter DNA methylation has only a few documented examples and, considering the static nature of DNA methylation, we postulate that it might be operational only in certain early developmental stages. Conversely, antisense RNA-induced chromatin remodelling seems to be a more feasible and dynamic mode of action for many low-copy natural antisense transcripts. In the latter case, antisense RNA might predominantly act locally to maintain or modify chromatin structure and ultimately to activate or suppress sense gene expression. Natural antisense transcripts are involved in different gene regulatory pathways, but it is still not clear which intrinsic properties of natural antisense RNA molecules, or extrinsic features such as protein interactions, cellular and developmental context, are decisive for any given pathway.
It is becoming clear that the expression of natural antisense transcripts and other non-protein-coding RNAs are pervasive in the human genome, although the mechanisms of their production and preferential sites of action are poorly understood. Particularly, the exact molecular machineries behind the antisense RNA-induced chromatin modifications are not currently known. It will be important to learn which histone-modifying enzymes might be involved, how they interact with antisense RNAs and which histone modifications they induce over time. It will also be of interest to define the exact contexts in which perturbation of the same antisense transcript35 results in either activation or repression of sense RNA expression. It is remarkable that these fundamental and powerful regulatory mechanisms relating to natural antisense transcription remain to be elucidated in large part. It can be safely assumed that examination of natural antisense phenomena will remain a rich field of investigation for the foreseeable future.
Supplementary Material
Acknowledgments
We are grateful to M. P. van der Brug, B. H. Miller, J. Silva and S. Brothers for insightful comments and careful reading of the manuscript. Discussions with Y. Hayashizaki, G. St-Laurent and other colleagues within the FANTOM Transcriptomics consortium have also been highly valuable to us.
Glossary box
- Transcription unit
A group of expressed sequence tags or mRNAs, usually with alternative splice patterns, in which they share exonic overlap of at least one nucleotide and are in the same chromosomal orientation
- Dicer
An RNase III family endonuclease that processes double-stranded RNA and pre-microRNAs into small interfering RNAs and microRNAs, respectively
- ENCODE
The ENCyclopedia Of DNA Elements is a publicly founded project that aims to find functional elements in human genome (http//www.genome.gov/10005107#1)
- MicroRNA
A small (20–25 nucleotides long) single-stranded RNA that is thought to regulate the expression of other genes, either through inhibiting protein translation or through degrading a target mRNA transcript, by a process that is similar to RNA interference
- Short interfering RNA
A short (21–23 nucleotides long) RNA molecule that is processed from a long double-stranded RNA. siRNAs are functional components of the RNAi-induced silencing complex, (RISC) and typically target mRNAs by binding perfectly complementary sequences in the mRNA and causing their degradation
- RISC
RNA-induced silencing complex is a multi-protein complex that incorporates one strand of siRNA and use them to recognize the target mRNA for degradation
- RITS
RNA-induced transcriptional silencing complex is a multi-protein complex, e.g. in fission yeast, that incorporates short RNA molecules, such as siRNA and trigger the down-regulation of transcription of a particular gene or genomic region. This is usually accomplished by modification of histone tails, which target the genomic region for heterochromatin formation
- AU-rich element
A region in an RNA transcript with frequent A and U nucleotides, such as AUUUA, that targets the RNA for degradation
- Piwi-interacting RNA
A small (25–35 nucleotides long) RNA species that is processed from single-stranded precursor RNA, independent of Dicer, and forms a complex with the Piwi protein. piRNAs are probably involved in transposon silencing and stem-cell function
References
- 1.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–75. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosok O, Sioud M. Systematic identification of sense-antisense transcripts in mammalian cells. Nat Biotechnol. 2004;22:104–8. doi: 10.1038/nbt925. [DOI] [PubMed] [Google Scholar]
- 5.Ge X, Wu Q, Jung YC, Chen J, Wang SM. A large quantity of novel human antisense transcripts detected by LongSAGE. Bioinformatics. 2006;22:2475–9. doi: 10.1093/bioinformatics/btl429. [DOI] [PubMed] [Google Scholar]
- 6.Ge X, Rubinstein WS, Jung YC, Wu Q. Genome-wide analysis of antisense transcription with Affymetrix exon array. BMC Genomics. 2008;9:27. doi: 10.1186/1471-2164-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The Antisense Transcriptomes of Human Cells. Science. 2008 doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 11.Finocchiaro G, et al. Localizing hotspots of antisense transcription. Nucleic Acids Res. 2007;35:1488–500. doi: 10.1093/nar/gkm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun M, Hurst LD, Carmichael GG, Chen J. Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 2005;33:5533–43. doi: 10.1093/nar/gki852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada Y, et al. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum Mol Genet. 2008;17:1631–40. doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- 14.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–8. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–45. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–25. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci U S A. 2002;99:8796–801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sastry SS, Hoffman PL. The influence of RNA and DNA template structures during transcript elongation by RNA polymerases. Biochem Biophys Res Commun. 1995;211:106–14. doi: 10.1006/bbrc.1995.1784. [DOI] [PubMed] [Google Scholar]
- 20.Larijani M, Martin A. Single-stranded DNA structure and positional context of the target cytidine determine the enzymatic efficiency of AID. Mol Cell Biol. 2007;27:8038–48. doi: 10.1128/MCB.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci U S A. 2008;105:3843–8. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronai D, et al. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–90. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolland DJ, et al. Antisense intergenic transcription in V(D)J recombination. Nat Immunol. 2004;5:630–7. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 24.Bolland DJ, et al. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol. 2007;27:5523–33. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roa S, Kuang FL, Scharff MD. Does antisense make sense of AID targeting? Proc Natl Acad Sci U S A. 2008;105:3661–2. doi: 10.1073/pnas.0800935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tufarelli C, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–65. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 27.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanduri C. Functional insights into long antisense noncoding RNA Kcnq1ot1 mediated bidirectional silencing. RNA Biol. 2008;5 doi: 10.4161/rna.7113. [DOI] [PubMed] [Google Scholar]
- 29.Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–35. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 31.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 32.Mohammad F, et al. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–28. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imamura T, et al. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 34.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz JC, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borel C, et al. Mapping of small RNAs in the human ENCODE regions. Am J Hum Genet. 2008;82:971–81. doi: 10.1016/j.ajhg.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 38.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–8. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 40.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–29. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 41.Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–300. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters NT, Rohrbach JA, Zalewski BA, Byrkett CM, Vaughn JC. RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts. Rna. 2003;9:698–710. doi: 10.1261/rna.2120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohman M. A-to-I editing challenger or ally to the microRNA process. Biochimie. 2007;89:1171–6. doi: 10.1016/j.biochi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Matsui K, et al. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–97. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 46.Uchida T, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–8. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 47.Hatzoglou A, et al. Natural antisense RNA inhibits the expression of BCMA, a tumour necrosis factor receptor homologue. BMC Mol Biol. 2002;3:4. doi: 10.1186/1471-2199-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebralidze AK, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008;22:2085–92. doi: 10.1101/gad.1654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zubko E, Meyer P. A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an antisense mechanism in cytokinin regulation. Plant J. 2007;52:1131–9. doi: 10.1111/j.1365-313X.2007.03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–34. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 55.Kawaji H, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–43. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 59.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 60.Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- 61.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 63.Faghihi MA, Wahlestedt C. RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals. Genome Biol. 2006;7:R38. doi: 10.1186/gb-2006-7-5-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


