Abstract
Background
Phosphorus (P) is often a limiting mineral nutrient for plant growth. Many soils worldwide are deficient in soluble inorganic phosphate (Pi), the form of P most readily absorbed and utilized by plants. A network of elaborate developmental and biochemical adaptations has evolved in plants to enhance Pi acquisition and avoid starvation.
Scope
Controlling the deployment of adaptations used by plants to avoid Pi starvation requires a sophisticated sensing and regulatory system that can integrate external and internal information regarding Pi availability. In this review, the current knowledge of the regulatory mechanisms that control Pi starvation responses and the local and long-distance signals that may trigger Pi starvation responses are discussed. Uncharacterized mutants that have Pi-related phenotypes and their potential to give us additional insights into regulatory pathways and Pi starvation-induced signalling are also highlighted and assessed.
Conclusions
An impressive list of factors that regulate Pi starvation responses is now available, as is a good deal of knowledge regarding the local and long-distance signals that allow a plant to sense and respond to Pi availability. However, we are only beginning to understand how these factors and signals are integrated with one another in a regulatory web able to control the range of responses demonstrated by plants grown in low Pi environments. Much more knowledge is needed in this agronomically important area before real gains can be made in improving Pi acquisition in crop plants.
Keywords: Phosphate signal, phosphate regulon, transcription factor, non-coding RNAs, phosphate starvation responses
INTRODUCTION
Phosphorus (P), in the form of phosphate (Pi), is an essential macronutrient for all living organisms, including plants. It is a structural element in nucleic acids and in the phospholipids that make up biomembranes. Many phosphoesters have an indispensible role in metabolic reactions, particularly those that involve energy transfer. Pi, through protein phosphorylation and dephosphorylation, is also a key component of the numerous signal transduction cascades that establish adaptive patterns of gene expression.
A ‘phosphorus paradox’ in relation to plant nutrition has been pointed out by numerous authors (Bieleski, 1973; Marschner, 1995). Although the total P content of soil is generally high, P availability is frequently a limiting factor for plant growth and productivity. This paradox arises because the concentration of available Pi in the soil solution averages about 1 µm and seldom exceeds 10 µm (Bieleski, 1973). A number of morphological, physiological, biochemical and molecular responses have evolved in plants that allow them to grow and prosper in the presence of such low levels of available Pi. These responses include the development of lateral roots and root hairs, as well as more dramatic root structures such as proteoid and dauciform roots, the secretion from roots of phosphatases and organic acids, and the induction of high-affinity and some low-affinity Pi transporters (Lambers et al., 2006; Ai et al., 2009; Fang et al., 2009). Many plants also establish symbiotic associations with mycorrhizal fungi that aid Pi acquisition (Burleigh et al., 2002).
The application of P fertilizer can compensate for low Pi availability in cropping systems, but high Pi input can cause severe environmental problems such as eutrophication. In addition, the global source of rock P is non-renewable and is being rapidly depleted. It is predicted that easily accessed global P reserves may be depleted in 50–100 years (Steen, 1998; Cordell et al., 2009). A fuller understanding of the strategies used by plants to acquire and utilize Pi efficiently, therefore, is vitally important for the rational breeding and engineering of crop plants with greater capacity to acquire, store and recycle soil Pi. In this review, recent advances in our understanding of the molecular aspects of plant responses to Pi starvation will be discussed. The regulation of Pi transporters, while centrally important, is not covered here, but has been reviewed elsewhere (Raghothama and Karthikeyan, 2005; Bucher, 2007; Lin et al., 2009).
COMPONENTS OF THE Pi REGULON IN PLANTS
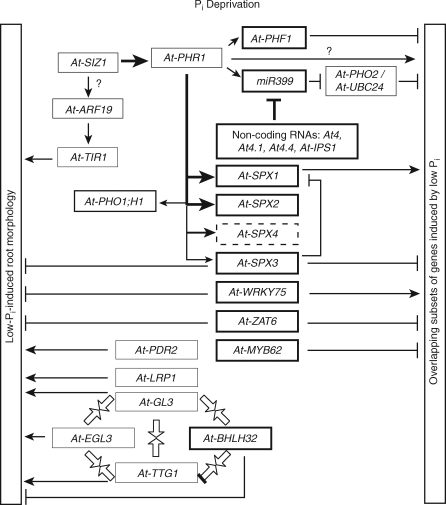
A variety of adaptive strategies have evolved in plants that alleviate or help them cope with Pi deficiency. The implementation of these strategies requires changes in the expression profiles of hundreds of genes, as demonstrated by transcriptome analyses in Arabidopsis thaliana (arabidopsis), Oryza sativa (rice), Lupinus albus (white lupin) and Phaseolus vulgaris (common bean) (Hammond et al., 2003; Uhde-Stone et al., 2003; Wasaki et al., 2003a; Wu et al., 2003; Misson et al., 2005; Hernández et al., 2007). The extent and complexity of the network of regulatory genes necessary to sense Pi status and regulate the deployment of these adaptive strategies is now being revealed. The network components identified so far include transcription factors, SPX sub-family proteins, non-coding RNAs and protein modifiers, including proteins involved in SUMOylation, phosphorylation, dephosphorylation and protein translocation (Fig. 1).
Fig. 1.
A model for the plant Pi regulon. For simplicity, the interactions of genes that are induced (heavily outlined boxes), do not respond (lightly outlined boxes) or are repressed (dashed boxes) by Pi deficiency in arabidopsis are shown. The activities of genes from other plants are discussed in the text. The target genes and functions are under either positive (arrowheads) or negative (T-junctions) regulation by the indicated factors. Genes that have been experimentally determined to be influenced by At-SIZ1 are indicated by heavy arrows. Convergent box arrows show gene products that physically interact with one another as part of a complex. Question marks indicate possible signalling pathways that are not currently supported by experimental evidence.
Transcription factors
Transcription factors bind to specific DNA sequences and regulate gene expression by altering the ability of RNA polymerase to bind to a target promoter sequence. These proteins have vital roles in many plant responses to stress conditions, including Pi deprivation. Transcription factors themselves are quite heavily regulated in response to Pi. In an experiment involving 6172 arabidopsis genes, the transcript abundance of about 30 % of the genes is changed >2-fold within 72 h of Pi deprivation (Wu et al., 2003). Among these were one-third of the 333 transcription factor genes assessed.
At-PHR1 from arabidopsis was the first transcription factor identified to be involved in the control of the Pi starvation response in a vascular plant (Rubio et al., 2001). The gene was identified in a genetic screen set up using a transgenic line harbouring an At-IPS1::GUS chimeric reporter gene. The At-IPS1 promoter is specifically responsive to Pi deprivation. Screening an ethylmethane sulfonate (EMS)-mutagenized M2 population of the reporter line identified a mutant, subsequently named phr1, that had a reduced response of the At-IPS1::GUS reporter gene to Pi deprivation. Analysis of the mutant revealed that other well-known Pi starvation responses, such as the accumulation of transcripts from a sub-set of Pi-responsive genes and the accumulation of anthocyanin, were also impaired (Rubio et al., 2001). In addition, the response of 56 of 64 Pi-responsive genes was attenuated in At-phr1 mutants (Bari et al., 2006). The latter result, together with the knowledge that the Pi responsiveness of many of those genes examined was fully reversible within 3 h of Pi resupply, led to the conclusion that At-PHR1 may be a central positive regulator of most, but not all, Pi starvation-induced genes (Fig. 1). The At-PHR1 gene was mapped to locus At4g28610 and encodes a member of the MYB transcription factor superfamily. The At-PHR1 protein sequence has high similarity to the sequence of the PHOSPHORUS STARVATION RESPONSE1 (PSR1) gene product from Chlamydomonas reinhardii (Wykoff et al., 1999). Both At-PHR1 and Cr-PSR1 have a MYB DNA-binding domain and a coiled-coil domain, indicating the potential for protein–protein interactions. At-PHR1 binds DNA as a dimer to an imperfect palindromic sequence (GNATATNC) present in the promoters of many Pi starvation-responsive genes (Rubio et al., 2001; Franco-Zorrilla et al., 2004). Thus, At-PHR1 acts downstream in the Pi starvation signalling pathway. The At-PHR1 gene is not itself particularly Pi responsive and the protein is located in the nucleus independently of the Pi status of the plant, indicating that induction of At-PHR1 activity does not require transcription (Rubio et al., 2001).
Two rice genes, Os-PHR1 and Os-PHR2, orthologous to At-PHR1 and with functions in the Pi stress signalling network similar to At-PHR1 (Fig. 1), have recently been characterized (Zhou et al., 2008). Transgenic plants with reduced expression levels of either Os-PHR1 or Os-PHR2 had lower transcript amounts for several Pi starvation response genes. These same Pi-responsive genes had increased expression in transgenic plants that over-expressed Os-PHR1 and Os-PHR2. Of the two rice PHR genes, only Os-PHR2 over-expression led to an increase of Pi in shoots under Pi-sufficient conditions, phenocopying over-expression of At-PHR1 in both wild-type and At-phr1 arabidopsis. Thus, Os-PHR2 is probably a functional homologue of At-PHR1, although it remains possible that Os-PHR1 and Os-PHR2 share the regulatory duties encompassed by At-PHR1.
At-MYB62 encodes another MYB transcription factor involved in the Pi deprivation response of arabidopsis. Unlike At-PHR1, At-MYB62 expression is induced by Pi starvation (Misson et al., 2005), but only in leaves of young seedlings (Devaiah et al., 2009). The response to low Pi is not mimicked by potassium, iron or nitrogen deficiencies. Expression in roots is apparently always low. At-MYB62 represses at least some Pi-inducible genes when over-expressed. Thus, At-MYB62, itself induced by Pi deprivation, is likely to be a negative regulator of other Pi starvation-inducible genes, and may moderate their activity during Pi starvation. The At-MYB62 protein, like At-PHR1, is localized to the nucleus irrespective of the Pi status of the plant. This observation, coupled with rapid loss of At-MYB62 transcripts upon Pi resupply and its role in the regulation of genes involved in Pi signalling, high-affinity Pi transport and mobilization, suggests a global role for At-MYB62 during Pi deficiency. Over-expression of At-MYB62 under Pi-sufficient conditions induced responses reminiscent of Pi starvation, including increased anthocyanin production, reduced primary root length and increased root acid phosphatase activity. Despite increases in Pi uptake and root Pi concentration that probably arose from changes in root architecture, the shoot Pi concentration of over-expressing plants was lower than that of the wild type (Devaiah et al., 2009). The partial rescue of the phenotype by exogenous gibberellic acid and the suppression to varying degrees of the expression of all five genes involved in gibberellic acid biosynthesis suggests that MYB62 may have a role in regulating biosynthesis of the hormone. Thus, MYB62 may regulate the Pi starvation responses through changes in gibberellic acid concentration.
The Os-PTF1 gene encodes a protein with a basic helix–loop–helix (bHLH) motif and was identified by differential screening of rice plants grown under normal and Pi starvation conditions (Yi et al., 2005). Expression of Os-PTF1 is induced in roots during Pi deprivation, while expression is constitutive in shoots. Like At-PHR1 and At-MYB62, Os-PTF1 is located in the nucleus independently of Pi status. Under Pi-deficient conditions, both in the field and in hydroponics, plants over-expressing Os-PTF1 have greater tillering, higher root and shoot biomass, and a 25 % higher P content than wild-type plants. Microarray analysis showed that the transcript abundance of 158 genes is altered by >2-fold in plants over-expressing Os-PTF1 (Fig. 1). The induced genes encode proteins such as nutrient transporters, transcription factors and ATP-binding proteins. The promoters of the induced genes generally have E-box elements (Massari and Murre, 2000), with about 20 % of the genes having at least one copy of the G-box element (Atchley et al., 1999). Further analysis of the responsive genes will enhance our understanding of the function of Os-PTF1 in the regulation of Pi response pathways.
Microarray analysis revealed that At-ZAT6 (At5g04340), encoding a cysteine-2/histidine-2 (C2H2) zinc finger transcription factor, is strongly induced by Pi starvation (Hammond et al., 2003). Green fluorescent protein (GFP) fusion experiments showed that At-ZAT6 is located in the nucleus. Suppression of At-ZAT6 expression through RNA interference (RNAi) is lethal (Devaiah et al., 2007b). Over-expression of the gene retards primary root growth independently of the Pi status of the plant and reduces Pi uptake. Over-expression of At-ZAT6 reduces the expression of a number of Pi starvation-induced genes, confirming its role in regulating Pi homeostasis (Fig. 1). Hence, At-ZAT6 appears to be a repressor of primary root growth, regulating Pi homeostasis through the control of root architecture.
At-WRKY75 is another transcription factor strongly induced by Pi deficiency in arabidopsis (Misson et al., 2005). The At-WRKY75 protein is located in the nucleus and its transcripts increase in abundance, but to various extents, in different parts of the plant during Pi deprivation. RNAi suppression of At-WRKY75 expression causes anthocyanin accumulation, indicating that the mutant plants are more susceptible to Pi stress than wild-type plants. In At-WRKY75 RNAi plants deprived of Pi, the expression of several Pi starvation-inducible genes is reduced. In contrast, lateral root length and number, as well as root hair number, significantly increase under both Pi-deficient and Pi-sufficient conditions. Thus, At-WRKY75 has a role in the Pi starvation response as well as in root development (Fig. 1) (Devaiah et al., 2007a).
Microarray experiments indicate that the expression of At-BHLH32 (At3g25710) is induced in both leaves and roots after 48 h of Pi starvation (Wu et al., 2003). In an At-bhlh32 T-DNA insertion mutant grown under Pi-sufficient conditions, there is a significant increase in the expression of the Pi starvation-inducible genes At-PPCK1 and At-PPCK2 (encoding isoforms of phosphoenolpyruvate carboxykinase), as well as increases in the accumulation of anthocyanins, the formation of root hairs and the total Pi content compared with wild-type plants (Chen et al., 2007). These results indicate that At-BHLH32 acts as a negative regulator in the Pi starvation response (Fig. 1). During Pi deprivation, mutations in At-TTG1, At-GL3 and At-EGL3, genes that take part in root hair formation (Bernhardt et al., 2005), lead to the decrease in transcript abundance of At-PPCK1 and At-PPCK2 compared with wild-type plants. These results indicate that the three genes are positive regulators in the Pi starvation response pathway (Fig. 1). Saccharomyces cerevisiae (yeast) two-hybrid experiments showed that At-BHLH32 can physically interact with At-TTG1 and At-GL3. Thus, At-BHLH32 was inferred to interfere with the function of TTG1-containing complexes (Fig. 1) and consequently influence the biochemical and morphological processes that respond to Pi status (Chen et al., 2007).
SPX domain-containing proteins
The SPX domain appears at the N-terminus of various proteins, especially those involved in signal transduction (Barabote et al., 2006). In yeast, proteins containing the SPX domain are involved in Pi transport and sensing, or the sorting of proteins to endomembranes (Wang et al., 2004). Most SPX-domain proteins with known functions in plants are involved in the regulation of either nutritional homeostasis or the response to environmental cues. The arabidopsis genome contains 20 genes encoding SPX-domain proteins. These proteins have been grouped into four sub-families based on sequence similarity.
Members of three of the four SPX protein sub-families in arabidopsis, a total of 16 proteins, possess an EXS domain in addition to the SPX domain. The At-PHO1 gene (At3g23430) encodes one such protein that is involved in the regulation of Pi homeostasis (Wang et al., 2004). The N-terminal half of At-PHO1 is mainly hydrophilic, while the C-terminal half has six potential membrane-spanning domains. The At-pho1 mutant of arabidopsis contains approx. 95 % less Pi and 50–75 % less total P in shoots than wild-type plants (Poirier et al., 1991). The At-pho1 mutant is deficient in the transfer of Pi from root epidermal and cortical cells to the xylem, but the role of At-PHO1 in Pi loading to the xylem is still unclear. The gene is expressed mainly in roots and is slightly upregulated during Pi deficiency. There are 11 At-PHO1 homologues in arabidopsis (Hamburger et al., 2002). Among these, only At-PHO1 and At-PHO1;H1 (At1g68740) are able to complement the At-pho1 mutant (Stefanovic et al., 2007). At-PHO1;H1, like At-PHO1, is involved in the Pi transport pathway; however, its response to Pi status is through a separate signal transduction pathway. The transcript abundance of At-PHO1;H1 is upregulated by the At-PHR1 transcription factor under Pi starvation conditions (Fig. 1) and suppressed by the Pi analogue phosphite, while At-PHO1 expression is independent of both At-PHR1 and phosphite (Stefanovic et al., 2007).
The fourth sub-family of SPX-domain proteins does not contain an EXS domain. There are four proteins of this type in arabidopsis, encoded by At-SPX1 (At5g20150), At-SPX2 (At2g26660), At-SPX3 (At2g45130) and At-SPX4 (At5g15330). In response to Pi starvation, At-SPX1 and At-SPX3 are strongly induced, while At-SPX2 is weakly induced and At-SPX4 is repressed in both shoots and roots (Duan et al., 2008). Of the six rice homologues in this sub-family, Os-SPX1–Os-SPX6, five are induced by Pi starvation in the roots and/or shoots, while no members are repressed (Z. Wang et al., 2009). The SPX1 and SPX2 isoforms from both arabidopsis and rice are targeted to the nucleus, while other the forms are located elsewhere in the cell (Duan et al., 2008; Z. Wang et al., 2009). The induction by Pi, the diversity of cellular locations, coupled with a variety of cell type-dependent transcription patterns suggest that that these SPX proteins may be involved in the Pi signalling networks that regulate the expression of Pi-responsive genes and that each SPX protein has a unique physiological function (Fig. 1).
The strong repression of At-SPX1 and At-SPX2 in At-siz1 (a SUMO E3 ligase gene, see below) and At-phr1 mutants indicates that both genes are positively regulated by the At-PHR1 regulon (Duan et al., 2008). At-SPX3 is also strongly repressed in the At-phr1 mutant but only weakly repressed in the At-siz1 mutant, suggesting that At-SPX3 acts downstream of At-PHR1. The point of action for At-SPX4 is unclear, because its expression is repressed about 50 % in both At-siz1 and At-phr1 mutants, which may mean that At-SPX4 is regulated by At-PHR1 in combination with other factors, or independently of At-PHR1.
Over-expression of At-SPX1 increases the transcript abundance of three Pi starvation-induced genes, more significantly under Pi-sufficient than Pi-deficient conditions, indicating that At-SPX1 is a positive regulator in the Pi signalling pathway (Duan et al., 2008). Partial RNAi repression of At-SPX3 leads to primary root growth retardation, increased Pi transportation from roots to shoots and enhanced expression of a sub-set of Pi starvation-responsive genes, including At-SPX1. These observations suggest that At-SPX3 provides negative feedback in the At-SPX1 response to Pi starvation (Duan et al., 2008). The functions of At-SPX2 and At-SPX4 require further exploration to determine the level of their involvement in the Pi starvation response.
Much more work is needed before strong inferences can be drawn about the functional equivalence, if any, among the SPX isoforms between rice and arabidopsis. Overlapping expression patterns and nuclear localization suggest that Os-SPX1 might be functionally equivalent to At-SPX1. Also, Os-SPX1 and At-SPX1 are each positively regulated by PHR1 orthologues, Os-PHR2 (C. Wang et al., 2009) and At-PHR1, respectively. However, Os-SPX1 is also functionally dissimilar to At-SPX1. Os-SPX1 and At-SPX1 are negative and positive regulators, respectively, of different sets of Pi starvation-induced genes. Unfortunately, until overlapping sets of target genes are assessed, it remains possible that the rice and arabidopsis SPX1 proteins act as both positive and negative regulators, targeting similar gene sets in similar ways. Another difference is that Os-SPX1 regulates the expression of the other five genes in this rice SPX sub-family in various ways depending on the tissue and Pi status (Z. Wang et al., 2009), a function that has not been ascribed to At-SPX1. However, At-SPX3 has been shown to be a negative regulator of At-SPX1, highlighting functional similarities between At-SPX3 and Os-SPX1. RNAi suppression of Os-SPX1 in rice also led to an increase in leaf Pi concentration (C. Wang et al., 2009), as seen in arabidopsis where At-SPX3 is suppressed by RNAi (Duan et al., 2008). The transcript abundance of certain PHT-type Pi transporters is enhanced in the roots of SPX1-suppressed lines of both rice and arabidopsis, suggesting a mechanism for the enhanced Pi abundance in the leaves. Taken together, these studies show that SPX proteins have complex, and as yet unclear, roles in the response to Pi deficiency.
SUMOylation
Small ubiquitin-related modifier (SUMO) proteins are small polypeptide tags showing greatest primary sequence similarity to ubiquitin. Like ubiquitin, SUMO proteins function through conjugation with other proteins, a post-translational modification that is involved in various cellular processes (Colby et al., 2006). While the ubiquitin system typically tags proteins for proteosome degradation, SUMO conjugation can stabilize the target proteins and alter their sub-cellular localization, as well as indirectly influence ubiquitination and protein degradation (Colby et al., 2006). At-SIZ1 functions as a SUMO E3 ligase in the arabidopsis Pi regulon (Fig. 1), as well as in other cellular processes, including abscisic acid signalling (Miura et al., 2007, 2009). The At-SIZ1 gene (At5g60410) was originally identified in a genetic screen designed to isolate genes that confer NaCl tolerance (Miura et al., 2005). An At-siz1 T-DNA insertion mutant showed retarded primary root growth compared with wild-type plants when the nutrient supply was restricted. Wild-type root growth was restored by the addition of Pi, but not other nutrients. The original At-siz1-1 mutant displayed a similar set of symptoms to wild-type plants in response to Pi deprivation, but to a more severe degree, despite similar intracellular Pi levels. Under Pi-deficient conditions, the abundance of transcripts from several Pi starvation responsive genes is similar in the At-siz1-1 mutant and wild-type plants, but under Pi-sufficient conditions these transcripts are more abundant in the At-siz1-1 mutant. Additionally, other genes are induced more slowly by Pi limitation in the At-siz1-1 mutant. At-SIZ1 was localized to the nucleus and can replace the yeast Sc-Siz2 SUMO E3 ligase in the in vitro SUMOylation of an Sc-Cdc3 substrate (Miura et al., 2005), indicating that At-SIZ1 is a SUMO E3 ligase. In vitro experiments demonstrated that At-SIZ1 can mediate the SUMOylation of At-PHR1, indicating a role for At-SIZ1 in the Pi deficiency response pathway (Fig. 1) (Miura et al., 2005). In this regard, it is noteworthy that MYB62 contains two functionally untested sites for potential SUMOylation (Devaiah et al., 2009).
At-SIZ1 may work in conjunction with At-PHO2/At-UBC24 in the SUMOylating pathway (Fujii et al., 2005). The At-UBC24 gene (At2g33770) encodes a putative E2 ubiquitin-conjugase (Aung et al., 2006; Bari et al., 2006). It is responsible for the Pi misallocation phenotype in the At-pho2 mutant. The At-pho2 mutant accumulates up to 3-fold more total P in leaves, mostly as Pi, than wild-type plants (Delhaize and Randall, 1995). Like the At-pho2 mutant, At-UBC24 T-DNA knockout mutants display increased uptake and translocation of Pi from roots to shoots and reduced Pi remobilization within leaves (Aung et al., 2006). In the At-pho2 mutant, transcripts from a number of Pi starvation-induced genes remained abundant under Pi-replete conditions, in contrast to wild-type plants where transcripts from these genes are repressed by high Pi supply (Shin et al., 2004; Bari et al., 2006). At-PHO2 and At-PHR1 evidently share a number of downstream targets. A study on the expression of 64 Pi-responsive genes in At-pho2 and At-phr1 mutants revealed 21 genes with an altered Pi response in both backgrounds (supplementary data in Bari et al., 2006). That is, the repression of these genes by Pi-replete conditions was impaired in the At-pho2 mutant, while their induction by Pi-deficient conditions was weakened in the At-phr1 mutant. This same study found that the abundance of primary transcripts from all five At-MIRNA399 genes in a At-pho2 mutant grown under Pi-replete conditions was the same as in wild-type plants, but they were not fully induced in the At-phr1 background. These results and the location of putative At-PHR1-binding sites 160–270 nucleotides (nt) upstream of several At-MIRNA399 genes place At-PHO2/At-UBC24 downstream of At-PHR1 in the plant Pi signalling pathway (Fig. 1) (Bari et al., 2006). While the interactions of At-PHO2/At-UBC24 with downstream elements of the Pi signalling pathway remain unclear, the At-SPX genes are likely candidates, as Os-SPX1 functions downstream of Os-PHO2 (C. Wang et al., 2009).
Protein phosphorylation and dephosphorylation
Protein phosphorylation by kinases and dephosphorylation by phosphatases is a potent binary switch involved in the regulation of most cellular activities and processes (Luan, 2003). Microarray analysis revealed differential regulation of several protein phosphatases at the onset of Pi deprivation in arabidopsis (Wu et al., 2003). However, Le-PS2 from Lycopersicon esculentum (tomato) is the only protein phosphatase induced by Pi starvation that has been studied in any detail (Baldwin et al., 2001, 2008). The ability of recombinant Le-PS2 expressed in bacteria to dephosphorylate a phosphopeptide substrate, an activity that is suppressed by okadaic acid, demonstrates that Le-PS2 is a phosphoprotein Ser/Thr phosphatase (Baldwin et al., 2008). However, its target proteins are not known. Given the central importance of reversible protein phosphorylation in other cellular processes and signal transduction pathways, it is surprising that its involvement in the Pi deprivation response has received so little attention.
Protein translocation
One crucial determinant for the activity of Pi transporters is their delivery through the secretory pathway to the plasma membrane. At-PHF1, the first Pi starvation-responsive component of the trafficking pathway to be isolated, enables the high-affinity Pi transporters to exit from the endoplasmic reticulum (ER). The At-phf1 mutant was isolated (González et al., 2005) using a reporter gene screen similar to that used to isolate the At-phr1 mutant (Martín et al., 2000; Rubio et al., 2001) and described above. The At-phf1 mutation causes the retention of the At-PHT1;1 Pi transporter in the ER and leads to a decrease in the concentration of Pi in whole plants. The lesion in the At-phf1 mutant was positionally cloned to At3g52190, which encodes a SEC12-related protein specific to plants. The At-PHF1 gene is expressed widely in the plant, but mainly in roots, flowers and senescing leaves. At-PHF1 has a promoter motif that conforms to the core binding sequence of At-PHR1 (Rubio et al., 2001). The decrease in At-PHF1 transcript abundance in the At-phr1 mutant suggests that At-PHF1 is indeed under direct transcriptional control of At-PHR1 (Fig. 1). The mechanism by which At-PHF1 regulates the intracellular transport of proteins and determines sub-cellular localization is still unclear. The possibility of a separate trafficking apparatus for an individual protein, or a group of proteins involved in a single process, is intriguing, and further investigations may provide new ideas on how plant cells deliver proteins to specific intracellular locations, especially during acclimation to physiological change.
Non-coding RNA
Non-coding RNA genes produce functional RNA molecules rather than proteins. They play key roles in chromosomal silencing, transcriptional regulation, translational repression, developmental control and responses to stress (Axtell et al., 2007). Non-coding RNAs are derived predominantly from introns and intergenic regions, but also from the opposite strand of protein-coding genes (Storz, 2002). MicroRNAs (miRNAs) are non-coding RNAs 20–24 nt in length. They are derived from the stem of hairpin-like precursors of about 75 nt that are in turn derived from longer pri-miRNAs (Zhu, 2008). miRNAs silence genes with complementary or partly complementary sequences by aiding mRNA cleavage or translational repression (Carrington et al., 2003; Bartel, 2004; He and Hannon, 2004). A screen of miRNAs regulated by Pi deprivation led to the identification of miR399 (Sunkar and Zhu, 2004). There are six MIRNA loci in arabidopsis that encode miR399 species a–f. In arabidopsis, At-PHO2/At-UBC24 was confirmed to be a target gene for At-miR399 (Fig. 1); it has five miR399 target sites in the 5′-untranslated region (UTR) of its transcripts (Sunkar and Zhu, 2004; Allen et al., 2005). At-miR399 accumulation is induced by Pi deprivation, a condition that suppresses the At-PHO2/At-UBC24 target gene (Fujii et al., 2005). Experiments with transgenic plants that produce artificial At-PHO2/At-UBC24 mRNAs with or without the 5′UTR indicated that miR399 downregulates At-PHO2/At-UBC24 mRNA accumulation by targeting the 5′UTR (Fujii et al., 2005).
The degradation of At-PHO2/At-UBC24 transcripts by miR399 in the Pi regulon is itself regulated by non-coding RNAs from the Mt4/TPSI1 gene family (Fig. 1). These non-coding RNAs originally came to prominence because of their strong induction in Pi-starved plants (Burleigh and Harrison, 1997, 1998; Liu et al., 1997). Members of the Mt4/TPSI1 gene family include Le-TPSI1 in tomato (Liu et al., 1997), Mt4 in Medicago truncatula (Burleigh and Harrison, 1997, 1998), an Mt4-like gene in Glycine max (soybean) (Burleigh and Harrison, 1999), At4, At4.1, At4.2 and At-IPS1 in arabidopsis (Burleigh and Harrison, 1999; Martín et al., 2000; Shin et al., 2006) and Os-PI1 in rice (Wasaki et al., 2003b). The transcripts of Mt4/TPSI1 genes typically contain multiple short open reading frames that are not conserved among the family members and are unlikely to encode proteins. The lack of conserved open reading frames originally suggested that the transcripts from Mt4/TPSI1 were the active gene products. The overall sequence identity across the family is quite low, except for a fairly well conserved 23 nt motif in the central region of each transcript.
The Mt4/TPSI1 genes have a dramatic impact on Pi distribution. For example, an At4 T-DNA insertion mutant does not redistribute shoot Pi to the roots. Instead, Pi accumulates in the shoots, causing an increase in the shoot to root Pi ratio compared with that in wild-type plants (Shin et al., 2006). The first clue to the mechanism mediating the action of the Mt4/TPSI1 RNAs came from the observation that the 23 nt sequence in the middle of At4 that is well conserved among all Mt4/TPSI1 family members hybridizes to an approx. 22 nt RNA expressed during Pi starvation (Shin et al., 2006). It was also noted that the 23 nt conserved sequence had extensive homology to miR399. However, there are critical mismatches, including a bulge opposite positions 10–11 of miR399, disrupting the base pairing that is required for miR399-guided cleavage of mRNA targets (Jones-Rhoades et al., 2006; Franco-Zorrilla et al., 2007). Indeed, At-IPS1 RNA is not cleaved in an At-miR399-dependent manner, but instead sequesters At-miR399 (Franco-Zorrilla et al., 2007). Moreover, At-IPS1 over-expression results in the accumulation of At-PHO2/At-UBC24 mRNA, an miR399 target, demonstrating that At-IPS1 represses miR399-dependent transcript cleavage. Therefore, Mt4/TPSI1 RNAs may function as non-cleavable miR399 substrate competitors, sequestering miR399 in a state where it cannot act upon target gene transcripts (Franco-Zorrilla et al., 2007). Interestingly, At-miR399b and At-miR399c are less efficient in suppressing At-PHO2/At-UBC24 than At-miR399f. This differential efficiency may be due in part to differences in complementary between the various At-miR399 isoforms and At4/At-IPS1 transcripts, where higher complementarity may lead to more efficient sequestration by the At4/At-IPS1 transcript (Doerner et al., 2008). The differential efficiency of its members suggests that the At-miR399 family is part of a fine-tuning mechanism that allows the cell to respond subtly to the dynamics of Pi availability.
Revealing the details of regulatory circuits such as the miR399, Mt4/TPSI1 and PHO2/UBC24 system is only the first step in understanding what is likely to be a complex involvement for non-coding RNAs within the Pi starvation regulatory network. Deep sequencing and other global RNA analysis tools are revealing a rapidly growing number of small RNAs whose abundance responds to Pi status (Hsieh et al., 2009; Pant et al., 2009). Several of these small RNAs are miRNAs and are likely to be involved in regulating processes such as Pi uptake and translocation (i.e. At-miR399), anthocyanin biosynthesis, oxidative stress reduction, sulfate translocation and nutrient recycling. While the expression of some of the miRNAs seems to respond only to Pi, others respond not only to Pi status, but also to the availability of other mineral nutrients, such as N, K, S, Cu and Fe. Among the miRNAs in this latter group, the regulatory response can be either in the same or the opposite direction as the response to Pi status. Another group of Pi-responsive small RNAs are miRNA star strands, such as At-miR399*, indicating that these molecules may have important biological functions. The role of still other Pi-responsive small RNAs in the Pi regulon is entirely unknown. Together, these molecules provide the prospect of many exciting discoveries in the future.
Pi SENSING IN HIGHER PLANTS
Local Pi sensing
The local availability of Pi controls responses such as root hair number and length, and primary root length. Pre-existing root hairs of plants grown under Pi-replete conditions are shorter than new root hairs that grow after plants are transferred to Pi-deficient conditions (Bates and Lynch, 1996). Reciprocally, root hair growth is suppressed in plants transferred from Pi-deficient to Pi-replete conditions. Primary root growth also slows dramatically when the root tip reaches patches of growth medium that contain little Pi (Linkohr et al., 2002). This Pi deficiency-dependent arrest is not a nutritional response but a response to a local Pi signal because primary roots that detached from the growth medium and entered the air phase did not stop growing despite the lack of Pi uptake by the growing tip (Svistoonoff et al., 2007). Moreover, growth of these roots stopped immediately when the tips encountered medium containing low concentrations of Pi (Svistoonoff et al., 2007). Using a reporter gene approach to identify dividing cells at the G2/M transition, it was shown that the root tips growing through air divided normally, but ceased to divide when the root tips encountered growth medium containing low Pi (Svistoonoff et al., 2007).
Mutant analysis is revealing the mechanism of local Pi sensing. The EMS-induced At-pdr2 mutant of arabidopsis showed reduced primary root growth compared with wild-type plants when DNA was the only source of Pi (Chen et al., 2000). The mutant had reduced rates of root cell division and elongation when grown in Pi-deficient conditions. The mutant was not affected in the P concentration within the root tip or in Pi uptake rates, excluding a defect in high-affinity Pi acquisition. These results helped lead to the conclusion that the At-pdr2 phenotype is caused by a defect in the sensing of the local external Pi concentration (Ticconi et al., 2004). At-PDR2 was recently identified by map-based cloning (Ticconi et al., 2009) and found to encode the single P5-type ATPase (At5g23630). At-PDR2 was localized to the ER and found to regulate stem cell differentiation and meristem activity through a pair of GRAS family transcription factors, SCR and SHR (Ticconi et al., 2009) that are key regulators of radial root patterning (Di Laurenzio et al., 1996; Gallagher and Benfey, 2009).
At-PDR2 was found to interact genetically with At-LPR1 in an ER-resident pathway (Ticconi et al., 2009). At-LPR1, a quantitative trait locus (QTL) responsible for determining root responses to Pi starvation in arabidopsis, was mapped to the At1g23010 locus, which encodes a multicopper oxidase required for low Pi-dependent growth arrest (Svistoonoff et al., 2007). At-LPR1, like At-PRD2, is expressed in the root tip, including the meristem and root cap. At-LPR1 may be involved in the switching of primary root growth from indeterminate to determinate, moderating the activity and/or distribution of a hormone-like compound (Svistoonoff et al., 2007). At-LPR1 and At-PDR2 are thought to function together to adjust meristem activity in response to external Pi status (Ticconi et al., 2009). Elucidating the nature of this interaction and uncovering other components of the ER-resident control pathway will provide new insights into the adjustment of root characteristics to local Pi signals.
Pi itself can function as a local signal. Both Pi and phosphite, a supposedly metabolically inert analogue of Pi, could rescue the root meristematic activity of the At-pdr2 mutant under Pi-limiting conditions (Ticconi et al., 2004). The appearance within 1 h of Pi limitation of transcripts from genes that are Pi starvation inducible and the decrease in abundance of these transcripts within 30 min of Pi resupply, while shoot Pi and carbohydrate status did not change, has also been taken as evidence that local Pi concentration acts as a signal (Wang et al., 2002; Muller et al., 2004).
Long-distance signals
Plants acquire mineral nutrients from the rhizosphere through the roots and transport them to shoots. The balance between shoot demand and root supply for any particular mineral nutrient is determined by long-distance signals that report on nutrient status in the various tissues (Lin et al., 2008). The existence of long-distance signals that report on Pi status in the tissues has been demonstrated in split-root experiments, where the roots of Pi-starved plants were divided, with one part exposed to Pi-replete medium and the other part exposed to Pi-deficient medium. In tomato, genes that were normally induced in response to Pi starvation were systemically repressed in the portion of the root system exposed to Pi-deficient medium when the remaining roots were exposed to Pi-sufficient medium (Liu et al., 1998; Baldwin et al., 2001). Similarly, lateral root elongation is also controlled by shoot Pi status (Linkohr et al., 2002; Shane and Lambers, 2006; Shane et al., 2008). Long-distance signals are also important for the regulation of resource allocation during leaf development, flowering and pathogen defence. These long-distance signals move through the phloem, which contains proteins, sugars, organic acids, amino acids, phytohormones, miRNAs and other types of small RNAs, all of which are potential signal molecules (Lough and Lucas, 2006).
Sugars have been suggested to be a systemic signal for the Pi starvation response in plants, in addition to their more established roles as signals in other plant adaptive responses (Koch et al., 2000; Grigston et al., 2008), substrates for the biosynthesis of complex carbohydrates and in energy metabolism, and in driving phloem transport and delivery of C skeletons to sink tissues for growth and development. Sugars are required for Pi starvation responses, including the stimulation of lateral root growth and the induction of Pi starvation-responsive genes (Karthikeyan et al., 2007). Sucrose induces the accumulation of At-Pht1;1 and At4 transcripts in plants grown under Pi-sufficient conditions, and its absence under Pi-deficient conditions represses transcript accumulation from these genes. Since plants grown under Pi sufficiency contain significantly more Pi than those grown under Pi deficiency, the effect of sucrose on the expression of Pi starvation-responsive genes is not merely the result of sequestration of Pi in sugar–phosphates. Instead, it appears that sucrose acts as a signal in the Pi starvation response pathway (Karthikeyan et al., 2007; Hammond and White, 2008).
The mapping of the At-pho3 mutation to At-SUC2, which encodes a sucrose–proton symporter that is important for phloem loading of sucrose, provides further evidence that sugar acts as a signal during Pi starvation (Lloyd and Zakhleniuk, 2004). The At-pho3 mutant has reduced root acid phosphatase activity when grown under Pi-deficient conditions (Zakhleniuk et al., 2001). When grown under Pi-sufficient conditions, the total P content is much lower in the shoots and roots of 11-d-old At-pho3 mutant seedlings than in wild-type seedlings. The At-pho3 mutant also accumulates much more anthocyanin and starch than wild-type seedlings, clear indicators of a Pi starvation response (Zakhleniuk et al., 2001). While suggesting that sugar acts as a systemic signal in the plant Pi starvation response, these experiments cannot exclude the possibility that sugar is only being used as a substrate. More research needs to be done to understand fully the role of sugars as components of the Pi starvation response pathway.
Small non-coding RNAs are likely to be another important class of long-distance signalling molecules. miR399 has been detected in the phloem sap of Cucurbita maxima (Yoo et al., 2004) and Brassica napus (Buhtz et al., 2008). The high Pi deficiency-dependent abundance of miR399 in the roots was dwarfed by the abundance of miR399 in phloem sap (Pant et al., 2008). This led to the demonstration through grafting experiments that miR399 is transported from the shoots to the roots through the phloem in both arabidopsis and Nicotiana benthamiana (Lin et al., 2008; Pant et al., 2008). When shoots over-expressing miR399 due to the presence of a miR399 transgene were grafted onto the wild-type rootstocks lacking miR399, the roots accumulated mature miR399 to very high levels, while the corresponding primary transcripts were virtually absent. The grafted plants also accumulated 5-fold more Pi in the shoots than the wild-type plants did. Thus, the phloem transport of miR399 from the shoots to the roots apparently systemically controls the maintenance of plant Pi homeostasis (Lin et al., 2008; Pant et al., 2008).
The At-pho1 mutant may give important insights into how miR399 exerts this systemic control. Under Pi-sufficient conditions, At-pho1 plants have decreased amounts of Pi in the shoots, despite wild-type amounts of Pi in the roots. The Pi deficiency in the At-pho1 shoots results in the accumulation in this tissue of both primary and mature miR399 transcripts. The amount of primary miR399 in roots is also increased. It is not yet clear whether the increased abundance of miR399 in roots is the result of increased expression of miR399 in the roots or the translocation of miR399 from the shoots (Lin et al., 2008). Distinguishing between these two possibilities will give us a greater appreciation of the mechanism through which miR399 controls Pi homeostasis.
Phloem sap contains a large variety of RNA molecules, ranging in size from miRNAs to mRNAs (summarized in Zhang et al., 2009), and including miRNA star strand sequences (Buhtz et al., 2008; Pant et al., 2009). Among these RNAs are a group of small RNAs with sequences related to nuclear or organelle genes encoding rRNA and tRNA, as well as nuclear genes for small nucleosomal RNA, processing-related small RNA and signal recognition particle RNA (Zhang et al., 2009). The recent finding that several RNA fragments falling into these classes, as well as several miRNA star sequences, accumulate specifically in Pi-starved plants (Buhtz et al., 2008; Hsieh et al., 2009; Pant et al., 2009) suggests that these classes of RNA may have a biological function. Moreover, their presence in phloem sap (Buhtz et al., 2008; Pant et al., 2009; Zhang et al., 2009) suggests that some may have systemic regulatory functions. This possibility adds a potentially exciting new dimension to the RNA-based regulatory components of the Pi starvation response.
Global integration of Pi signalling
Many of the factors described above are components of a complex regulatory web that dynamically assesses and responds to the availability of Pi at the cellular or whole-plant level. The establishment and maintenance of a mineral nutrient balance that allows optimal growth under the prevailing environmental conditions and within the specific developmental stage of the plant requires the integration of these Pi-specific responses with other signalling networks operating at the whole-plant level. Such signals include information on the availability of other mineral nutrients, such as iron and zinc, and long-distance and local signals embedded within molecules such as phytohormones. A thorough understanding of the integration of the relevant signals and responses is central to obtaining a holistic view of plant nutrient acquisition. Aspects of this integration have been discussed in detail elsewhere (Ward et al., 2008; Rubio et al., 2009; Santner et al., 2009).
The interaction of P with iron is a long-known example where the availability of one nutrient affects the activity of another. The chemical interaction of P and iron in the growth medium, at the root surface or within the plant itself can lead to the formation of complexes that lower the bioavailability of both nutrients. The inhibition of primary root elongation that is commonly observed in arabidopsis upon Pi deprivation is not typical of all plants and was recently linked to iron toxicity (Ward et al., 2008). When plants were grown in media containing decreased iron, the removal of Pi had no effect on primary root elongation. This observation goes some way to explain why a number of iron-responsive genes were found among the genes that respond to Pi deficiency in tomato and arabidopsis (Wang et al., 2002; Misson et al., 2005; Zheng et al., 2009). These genes were likely to be responding to an increase in available iron under low Pi conditions. Also in arabidopsis, increases in plant iron concentration and modifications to iron storage under low Pi conditions are related to enhanced iron availability in the plant and the external medium (Hirsch et al., 2006).
It has also been known for a long time that zinc deficiency can cause high levels of Pi to accumulate in plant tissues, which can lead to toxicity when Pi is readily available (Cakmak and Marschner, 1986). In barley, the expression of genes encoding the Hv-PT1 and Hv-PT2 high-affinity Pi transporters was induced by zinc deficiency, independently of Pi availability (Huang et al., 2000). The role of zinc in the regulation of the high-affinity Pi transporter genes is specific, and cannot be mimicked by manganese, another divalent cationic micronutrient. From these results, it was suggested that zinc may have a specific role in the signal transduction pathway regulating the high-affinity Pi transporter genes (Huang et al., 2000).
It is becoming clearer that phytohormones are involved in the responses of plants to the availability of mineral nutrients, including Pi (Rubio et al., 2009). However, this role is often secondary to the initial response to low Pi and may only affect specific components or segments of the Pi regulon. Plant hormones are structurally unrelated small molecules that have profound effects as growth regulators (Santner et al., 2009). Generally acting at low concentrations, they mediate plant responses to both biotic and abiotic stresses by acting at a distance from the site of synthesis, and/or by acting locally, at or near the site of synthesis. Hormone signalling generally leads to major changes in transcript profiles. Non-genomic hormonal responses are also likely to occur, but these are not as well characterized. Recent work has made it clear that regulated protein degradation mediated by ubiquitin is a common theme in hormone signalling. Several hormone receptors have now been identified as enzymes in the ubiquitin–protein conjugation pathway, and the abundance of key downstream signalling proteins has also been found to be regulated by ubiquitin-dependent degradation (Santner et al., 2009).
Cytokinins are negative regulators of Pi starvation responses in arabidopsis. The expression of Pi starvation-induced genes, such as At-PHT1;1, At-ACP5 and members of the Mt4/TPSI1 gene family, was repressed by the application of exogenous cytokinin (Martín et al., 2000; Shin et al., 2006). Mutation of the cytokinin receptor genes At-CRE1/WOL/AHK4 and At-AHK3 can attenuate the repression in the presence of sugar (Franco-Zorrilla et al., 2002, 2005). Moreover, Pi deprivation repressed the accumulation of cytokinin and the expression of At-CRE1, conditions that would be expected to favour activation of the Pi starvation response (Franco-Zorrilla et al., 2002). Evidence indicating that cytokinins, generally acting through At-CRE1, also repress a number of genes that respond to nitrogen, sulfur or iron deficiency (Rubio et al., 2009) is a measure of the complexity of the regulatory network necessary to integrate the nutrient-dependent signals within plants.
Several studies in arabidopsis have examined the role of auxin as a mediator for the reduction in primary root growth and the proliferation of lateral roots induced by Pi starvation. Auxin-resistant mutants show largely wild-type primary root growth responses under both Pi sufficiency and deficiency, suggesting an auxin-independent mechanism for this response (Williamson et al., 2001; López-Bucio et al., 2002; Al-Ghazi et al., 2003). On the other hand, Pi starvation enhances the responsiveness of the root system to the induction of lateral root proliferation caused by the application of exogenous auxin (López-Bucio et al., 2002). The accumulation of auxin at the primary root apex, in pre-initiated lateral root primordia and in young lateral roots suggested that auxin transport or biosynthesis was important (Nacry et al., 2005). However, plants defective either in auxin sensing or in polar transport due to mutation or inhibitors are still able to produce increased lateral roots in response to Pi starvation (Williamson et al., 2001; López-Bucio et al., 2002; Jain et al., 2007; Pérez-Torres et al., 2008). Together, these results point to increased auxin sensitivity, not increased auxin transport or synthesis, as the mediator of proliferative lateral root growth.
The low Pi-induced increase in auxin sensitivity leading to lateral root formation is likely to be mediated by the At-TIR1 auxin receptor (Pérez-Torres et al., 2008). Many auxin-responsive genes are held in a repressed state by the binding of an Aux/IAA protein to an auxin response factor occupying the promoter (Santner et al., 2009). At-TIR1 is induced in response to low Pi and encodes an F-box protein that binds directly with Aux/IAA, an interaction that is enhanced by auxin. The combination of auxin, At-TIR1 and Aux/IAA induces the ubiquitin-dependent degradation of Aux/IAA by the 26S proteosome, allowing the auxin response factor to proceed with transcription. The arabidopsis transcription factor AUXIN RESPONSE FACTOR 19 (At-ARF19) has been implicated in the increase in lateral root growth in response to low Pi, and is a candidate participant in the TIR1-mediated induction of lateral root formation in response to low Pi (Pérez-Torres et al., 2008).
An involvement of gibberellic acid in the Pi starvation response is beginning to emerge. Gibberellic acid at least partially represses low Pi-induced changes to arabidopsis root and shoot growth and architecture, including the inhibition in primary root growth, the increase in lateral root density and the increase in root-to-shoot ratio (Jiang et al., 2007). Moreover, over-expression of At-MYB62 led to symptoms of gibberellic acid deficiency, which could be partially rescued by the application of exogenous gibberellic acid (Devaiah et al., 2009). The gibberellic acid deficiency arose from a decrease in bioactive hormone brought about by associated changes in the transcript abundance for genes involved in gibberellic acid metabolism.
At least some of the molecular events leading to low Pi-induced changes in plant architecture and anthocyanin accumulation are dependent on the gibberellic acid–DELLA signalling pathway, which involves ubiquitin-dependent protein degradation (Jiang et al., 2007). Gibberellic acid induces plant growth by promoting the destruction of the growth-restraining DELLA proteins (there are five DELLA proteins in arabidopsis). Similarly to the auxin signalling pathway, the binding of gibberellic acid to a specific receptor (GID1a, b or c) promotes binding of the hormone–receptor complex to the DELLA proteins (Santner et al., 2009). DELLA proteins are typically bound to, and inactivate, various transcription factors. The binding of the gibberellic acid–receptor complex to DELLA initiates the ubiquitin-dependent destruction of DELLA by the 26S proteosome, releasing the transcription factors to activate the gibberillin response (Santner et al., 2009).
Other hormones seem to have only a limited role in regulating the Pi starvation response. Mutants with impaired abscisic acid sensitivity (aba2-1) or biosynthesis (aba1) have a reduced Pi starvation response, including reduced expression of some Pi-responsive genes and accumulation of anthocyanin (Trull et al., 1997; Ciereszkoa and Kleczkowsk, 2002). However, the mutants were not impaired in the production of Pi starvation-induced phosphatases or in the ability to modulate biomass allocation between roots and shoots. Application of the hormone to the roots of Pi-deprived wild-type plants decreased the transcript abundance in the roots for several members of the Mt4/TPSI1 gene family (Shin et al., 2006). It is interesting to note in this regard that At-miR399 homologues were identified in a population of arabidopsis plants subjected separately to a number of stresses and abscisic acid before pooling (Sunkar and Zhu, 2004). At-RAB18, which is induced by Pi deprivation and sugar (Ciereszkoa and Kleczkowsk, 2002), is also induced by abscisic acid. The induction is through an At-ABI5-dependent signalling pathway, a process attenuated by the At-SIZ1-dependent SUMOylation of At-ABI5, a transcription factor containing a basic leucine-zipper domain (Miura et al., 2009).
Ethylene was found to inhibit root elongation in Pi-sufficient plants, but stimulated it in Pi-deficient plants (Borch et al., 1999; Ma et al., 2003). Conversely, the inhibition of ethylene production or action inhibited root elongation in Pi-deficient plants while stimulating it in Pi-sufficient plants. However, the mechanism mediating the ethylene-dependent response of root elongation to Pi deficiency remains unclear.
It is likely that there is a complex interplay among Pi, hormones and sugar signalling. Mutation of the cytokinin receptor genes At-CRE1/WOL/AHK4 and At-AHK3 can attenuate the repression in the presence of sugar (Franco-Zorrilla et al., 2002, 2005). The abscisic acid and Pi starvation-inducible At-RAB18 gene is also induced by sugar (Ciereszkoa and Kleczkowsk, 2002).
UNIDENTIFIED MUTANT GENES RELEVANT TO Pi NUTRITION IN HIGHER PLANTS
Many of the genes described above that are central to the Pi starvation response network were identified by mutant analysis, clearly reiterating the power of using mutants to determine gene function and dissecting the genetic pathways. There are a number of interesting mutants available that are relevant to Pi nutrition where identification of the mutated gene and functional analysis of the gene product will give insights into the details of the Pi starvation response network.
The At-lpi mutants
The reduction in primary root elongation is a conspicuous root developmental change that occurs during Pi starvation (Williamson et al., 2001). The At-lpi mutants, representing four different genetic loci, were isolated from an EMS-mutagenized population due to their ability to maintain primary root growth during Pi starvation (Sánchez-Calderón et al., 2006). The abundance of transcripts from a sub-set of Pi starvation-induced genes is reduced during Pi deprivation in the At-lpi mutant compared with wild-type plants. The At-lpi phenotype is indicative of a function that may be central to the cross-talk between low Pi status and the activation of Pi deficiency-responsive genes that control root development. Isolation of the affected genes from the At-lpi mutants may, therefore, identify major control points in the Pi starvation-induced signalling network (Sánchez-Calderón et al., 2006).
The At-psr1 mutant
Arabidopsis can grow on a medium containing DNA as the main Pi supply (Chen et al., 2000). The At-psr1 mutant was isolated from an EMS-mutagenized arabidopsis population due to its inability to use exogenous DNA when Pi is limited. The mutant grows well when Pi is supplied. Biochemical analysis showed that RNase and acid phosphatase activities in the At-psr1 mutant are generally reduced compared with those in wild-type plants. Genetic analysis indicated that the mutant phenotype is caused by a single recessive allele, implying that At-PSR1 influences the expression of a sub-set of genes encoding enzymes that degrade exogenous organophosphate substrates, increasing the ability of the plant to scavenge Pi (Chen et al., 2000). Identifying the corresponding gene will give added insight into how plants regulate the scavenging of Pi from the rhizosphere.
The At-pup1 mutant
The At-pup1 mutant was isolated from a T-DNA-mutagenized arabidopsis population due to reduced root staining for phosphatase activity when grown on a Pi-deficient medium (Trull and Deikman, 1998). Analysis of the phosphatases produced by the mutant showed that a 160 kDa acid phosphatase isoform is missing. The response of the At-pup1 mutant to Pi deprivation, such as the accumulation of anthocyanin and the altered partitioning of P between root and shoot, were the same as in wild-type plants, while the root to shoot ratio was lower in the mutant under Pi-sufficient conditions (Trull and Deikman, 1998). Identifying At-PUP1 within the 5 cM interval on chromosome 2 to which it has been mapped (Trull and Deikman, 1998) will give us another gene involved in scavenging Pi from the rhizosphere, providing further opportunities to dissect the regulatory pathways used to control the deployment of these strategies.
CONCLUSIONS AND FUTURE PERSPECTIVES
In the past few years, there has been rapid progress made in understanding the ways that plants respond and acclimate to Pi-deficient growth conditions. The PHR1 transcription factors At-PHR1 and Os-PHR2 have emerged as central regulators in the deployment of the adaptive strategies required to cope with Pi deficiency in arabidopsis and rice, respectively. Their constitutive presence in the nucleus suggests they are poised for action, awaiting the signal that Pi concentration is insufficient. But what is the nature of that signal? In future, it will be interesting to see if there are other central regulators, and to determine the conservation of functions across evolutionary distances. For example, why does rice require a second PHR-type transcription factor, Os-PHR1, to adapt to Pi deficiency? Does a transcription factor in arabidopsis have a role analogous to that of Os-PHR1? Are the differences seen between rice and arabidopsis simply an indication of species diversity, or do they hint at something larger, the divide between monocots and eudicots?
The PHR factors are supported by a host of other regulatory functions, including the transcription factors BHLH32, WRKY75, ZAT6 and MYB62. Some of these factors are positive regulators acting to induce the expression of genes necessary for adaptation, while others are negative regulators. The involvement of both positive and negative effectors illustrates the dynamic process underlying the deployment and adjustment of the Pi starvation response to maintain a balance between nutrient availability and acquisition to satisfy the nutritional demands imposed by the prevailing developmental programme and growth requirements. A fuller understanding of the interplay among factors such as ZAT6 and MYB62 that modulate the intensity of the low Pi response and positive regulators such as PHR1 will reveal the true complexity of plant adaptive responses to limiting Pi.
Details are now rapidly emerging about the interactions of the Pi regulon with the availability of other nutrients and the regulatory networks deployed to adapt to deficiencies in these nutrients. In this regard, hormonal responses and small RNA molecules have central roles that are just being documented. Both hormonal responses and pathways regulated by small RNAs, such as miR399, highlight the importance of post-translational modifications, such as ubiquitination and SUMOylation, in the plant response to Pi availability. Future discoveries on the targets of post-translational modification and the regulation of the modification process in response to Pi availability are areas needing much further work.
The recent finding of numerous small RNA molecules whose abundance is responsive to Pi status (Hsieh et al., 2009; Pant et al., 2009) is both exciting and sobering. The involvement of miRNAs and other small RNAs in regulating plant responses to Pi is clearly very poorly understood, despite the enlightening work done on revealing the regulation of PHO2/UBC24 by miR399. It will take much effort to reveal the roles of small RNAs in the plant response to Pi. However, the academic and practical rewards for this effort are likely to be large. Especially intriguing is the role of small RNAs, as well as hormones and sugars, as long-distance signals in the Pi starvation response pathway. In the case of RNAs, what is the mechanism of their loading into and unloading from the phloem? What cells are responsible for their synthesis and what cells receive and act upon the signals received?
There are many other intriguing questions yet to be answered in regard to the Pi starvation response in plants. Perhaps top among these is the nature and identity of the sensor that sets the whole Pi starvation response in motion. The finding that local Pi is sensed by an ER-localized pathway involving PDR2 and LPR1 is an excellent step forward, but what is the mechanism of action of these proteins and how does this pathway interlink with the systemic signalling pathways? Might it be something as simple as a protein phosphorylation that is inhibited by the lack of Pi as a substrate or will it be something much more complex? Answering these and many other questions will ultimately reveal the entire Pi starvation regulatory network and provide the understanding needed to develop new molecular genetic strategies for establishing crop plants with improved Pi acquisition and Pi use efficiency.
ACKNOWLEDGEMENTS
We thank Hans Lambers, Ricarda Jost and Weihua Chen for their excellent comments on draft versions of the manuscript. This work was supported by a China Postgraduate Scholarship from the China Scholarship Council (to X.J.Y.) and a grant from the Australian Research Council (to P.M.F.).
LITERATURE CITED
- Ai P, Sun S, Zhao J, et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. The Plant Journal. 2009;57:798–809. doi: 10.1111/j.1365-313X.2008.03726.x. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Al-Ghazi Y, Muller B, Pinloche S, et al. Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signaling. Plant, Cell and Environment. 2003;26:1053–1066. [Google Scholar]
- Atchley WR, Therhalle W, Dress A. Positional dependence, cliques and predictive motifs in the bHLH protein domain. Journal of Molecular Evolution. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology. 2006;141:1000–1011. doi: 10.1104/pp.106.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. The Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG. LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiology. 2001;125:728–737. doi: 10.1104/pp.125.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Cao AQ, Raghothama KG. Biochemical and molecular analysis of the LePS2;1: a phosphate starvation induced protein phosphatase gene from tomato. Planta. 2008;228:273–280. doi: 10.1007/s00425-008-0736-y. [DOI] [PubMed] [Google Scholar]
- Barabote RD, Tamang DG, Abeywardena SN, et al. Extra domains in secondary transport carriers and channel proteins. Biochimica et Biophysica Acta. 2006;1758:1557–1579. doi: 10.1016/j.bbamem.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Bari R, Pant BD, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:218–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant, Cell and Environment. 1996;19:529–538. [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- Bieleski RL. Phosphate pools, phosphate transport, and phosphate availability. Annual Review of Plant Physiology. 1973;24:225–252. [Google Scholar]
- Borch K, Bouma TJ, Lynch JP, Brown KM. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant, Cell and Environment. 1999;22:425–431. [Google Scholar]
- Bucher M. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytologist. 2007;173:11–26. doi: 10.1111/j.1469-8137.2006.01935.x. [DOI] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chapell L, Baulcombe D, Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. The Plant Journal. 2008;53:739–749. doi: 10.1111/j.1365-313X.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Molecular Biology. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. Characterization of the Mt4 gene from Medicago truncatula. Gene. 1998;216:47–53. doi: 10.1016/s0378-1119(98)00326-6. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to shoots. Plant Physiology. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Cavagnaro T, Jakobsen I. Functional diversity of arbuscular mycorrhizas extends to the expression of plant genes involved in P nutrition. Journal of Experimental Botany. 2002;53:1591–1601. doi: 10.1093/jxb/erf013. [DOI] [PubMed] [Google Scholar]
- Cakmak I, Marschner H. Mechanism of phosphorus induced zinc deficiency in cotton: I. Zinc-deficiency enhanced uptake rate of phosphorus. Physiologia Plantarum. 1986;68:483–490. [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta. 2000;211:13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Nimmo GA, Jenkins GI, Nimmo HG. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochemical Journal. 2007;405:191–198. doi: 10.1042/BJ20070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszkoa I, Kleczkowsk LA. Effects of phosphate deficiency and sugars on expression of rab18 in Arabidopsis: hexokinase-dependent and okadaic acid-sensitive transduction of the sugar signal. Biochimica et Biophysica Acta. 2002;1579:43–49. doi: 10.1016/s0167-4781(02)00502-x. [DOI] [PubMed] [Google Scholar]
- Colby T, Matthal A, Boeckelmann A, et al. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiology. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. The story of phosphorus: global food security and food for thought. Global Environmental Change. 2009;19:292–305. [Google Scholar]
- Delhaize E, Randall PJ. Characterization of a phosphate accumulator mutant of Arabidopsis thaliana. Plant Physiology. 1995;107:207–113. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology. 2007a;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajna VK, Raghothama KG. Phosphate homeostasis and root development in Arabidopsis are synchronized by the zing finger transcription factor ZAT6. Plant Physiology. 2007b;145:147–159. doi: 10.1104/pp.107.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Molecular Plant. 2009;2:43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Doerner P. Phosphate starvation signaling: a threesome controls systemic Pi homeostasis. Current Opinion in Plant Biology. 2008;11:536–540. doi: 10.1016/j.pbi.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. The Plant Journal. 2008;54:965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- Fang ZY, Shao C, Meng YJ, Wu P, Chen M. Phosphate signaling in Arabidopsis and Oryza sativa. Plant Science. 2009;176:170–180. [Google Scholar]
- Franco-Zorrilla JM, Martín AC, Solano R, Rubio V, Leyva A, Paz-Ares J. Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. The Plant Journal. 2002;32:353–360. doi: 10.1046/j.1365-313x.2002.01431.x. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. The transcriptional control of plant responses to phosphate limitation. Journal of Experimental Botany. 2004;55:285–293. doi: 10.1093/jxb/erh009. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martín AC, Leyva A, Paz-Ares J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiology. 2005;138:847–857. doi: 10.1104/pp.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT–ROOT movement. The Plant Journal. 2009;57:785–797. doi: 10.1111/j.1365-313X.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J. Phosphate transporter traffic facilitator1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. The Plant Cell. 2005;17:3500–3512. doi: 10.1105/tpc.105.036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigston JC, Osuna D, Scheible WR, Liu C, Stitt M, Jones AM. d-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Letters. 2008;582:3577–3584. doi: 10.1016/j.febslet.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, Perétot JMC, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. The Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, White PJ. Sucrose transport in the phloem: integrating root responses to phosphorus starvation. Journal of Experimental Botany. 2008;59:93–109. doi: 10.1093/jxb/erm221. [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, et al. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Reviews Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, Valdés-López O, et al. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiology. 2007;144:752–767. doi: 10.1104/pp.107.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, et al. Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie. 2006;88:1767–1771. doi: 10.1016/j.biochi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Huang C, Barker SJ, Langridge P, Smith FW, Graham RD. Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiology. 2000;124:415–422. doi: 10.1104/pp.124.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih ACC, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, et al. Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiology. 2007;144:232–247. doi: 10.1104/pp.106.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin–DELLA signaling pathway in Arabidopsis. Plant Physiology. 2007;145:1460–1467. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta. 2007;225:907–918. doi: 10.1007/s00425-006-0408-8. [DOI] [PubMed] [Google Scholar]
- Koch KE, Ying Z, Wu Y, Avigne WT. Multiple paths of sugar-sensing and a sugar/oxygen overlap for genes of sucrose and ethanol metabolism. Journal of Experimental Botany. 2000;51:417–427. doi: 10.1093/jexbot/51.suppl_1.417. [DOI] [PubMed] [Google Scholar]
- Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ. Root structure and functioning for efficient acquisition of phosphorus: matching morphological and physiological traits. Annals of Botany. 2006;98:693–713. doi: 10.1093/aob/mcl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. The Plant Journal. 2002;29:751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, et al. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiology. 2008;147:732–746. doi: 10.1104/pp.108.116269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Lin SL, Chiou TJ. Molecular regulators of phosphate homeostasis in plants. Journal of Experimental Botany. 2009;60:1427–1438. doi: 10.1093/jxb/ern303. [DOI] [PubMed] [Google Scholar]
- Liu CM, Muchhal US, Raghothama KG. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Molecular Biology. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Liu CM, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiology. 1998;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. Journal of Experimental Botany. 2004;55:1221–1230. doi: 10.1093/jxb/erh143. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiology. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annual Review of Plant Biology. 2006;57:203–232. doi: 10.1146/annurev.arplant.56.032604.144145. [DOI] [PubMed] [Google Scholar]
- Luan S. Protein phosphatases in plant. Annual Review of Plant Biology. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiology. 2003;131:1381–1390. doi: 10.1104/pp.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, et al. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. The Plant Journal. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix–-loop–helix proteins: regulators of transcription in eucaryotic organisms. Molecular and Cellular Biology. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences, USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences, USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Current Opinion in Plant Biology. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Nilsson L, Krintel C, Nielsen TH. Gene expression during recovery from phosphate starvation in roots and shoots of Arabidopsis thaliana. Physiologia Plantarum. 2004;122:233–243. [Google Scholar]
- Nacry P, Canivenc G, Muller B, et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiology. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. The Plant Journal. 2008;53:731–773. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiology. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. The Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiology. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG, Karthikeyan AS. Phosphate acquisition. Plant and Soil. 2005;274:37–49. [Google Scholar]
- Rubio V, Linhares F, Solano R, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes and Development. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen MA, Cardona-López X, Rojas-Triana M, Paz-Ares J. Plant hormones and nutrient signaling. Plant Molecular Biology. 2009;69:361–373. doi: 10.1007/s11103-008-9380-y. [DOI] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiology. 2006;140:879–889. doi: 10.1104/pp.105.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos LIA, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nature Chemical Biology. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- Shane MW, Lambers H. Systemic suppression of cluster root formation and net P-uptake rates in Grevillea crithmifolia at elevated P supply: a proteacean with resistance for developing symptoms of ‘P toxicity. Journal of Experimental Botany. 2006;57:413–423. doi: 10.1093/jxb/erj004. [DOI] [PubMed] [Google Scholar]
- Shane MW, Lambers H, Cawthray GR. Impact of phosphorus mineral source (Al–P or Fe–P) and pH on cluster-root formation and carboxylate exudation in Lupinus albus L. Plant and Soil. 2008;304:169–178. [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. The Plant Journal. 2004;39:629–642. doi: 10.1111/j.1365-313X.2004.02161.x. [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Chen RJ, Harrison MJ. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. The Plant Journal. 2006;45:712–726. doi: 10.1111/j.1365-313X.2005.02629.x. [DOI] [PubMed] [Google Scholar]
- Steen I. Phosphorus availability in the 21st century: management of a non-renewable resource. Phosphorus and Potassium. 1998;217:25–31. [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, et al. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to shoot, and are regulated by phosphate deficiency via distinct pathways. The Plant Journal. 2007;50:982–994. doi: 10.1111/j.1365-313X.2007.03108.x. [DOI] [PubMed] [Google Scholar]
- Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nature Genetics. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. Short on phosphate: plant surveillance and countermeasures. Trends in Plant Science. 2004;9:548–555. doi: 10.1016/j.tplants.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt D, Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. The Plant Journal. 2004;37:801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, et al. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proceedings of the National Academy of Sciences, USA. 2009;106:14174–14179. doi: 10.1073/pnas.0901778106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trull MC, Deikman J. An Arabidopsis mutant missing one acid phosphatase isoform. Planta. 1998;206:544–550. doi: 10.1007/s004250050431. [DOI] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J. The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant, Cell and Environment. 1997;20:85–92. [Google Scholar]
- Uhde-Stone C, Zinn KE, Ramirez-Yáñez M, Li A, Vance CP, Allan DL. Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorus deficiency. Plant Physiology. 2003;131:1064–1079. doi: 10.1104/pp.102.016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang HJ, Li K, Wu P, Shou HX. Involvement of OsSPX1 in phosphate homeostasis in rice. The Plant Journal. 2009;57:895–904. doi: 10.1111/j.1365-313X.2008.03734.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiology. 2004;135:400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Garvin DF, Kochian LV. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiology. 2002;130:1361–1370. doi: 10.1104/pp.008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. Journal of Integrative Plant Biology. 2009;51:663–674. doi: 10.1111/j.1744-7909.2009.00834.x. [DOI] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiology. 2008;147:1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, et al. Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant, Cell and Environment. 2003a;26:1515–1523. [Google Scholar]
- Wasaki J, Yonetani R, Shinano T, Kai M, Osaki M. Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytologist. 2003b;158:239–248. [Google Scholar]
- Williamson LC, Ribrioux S, Fitter AH, Leyser HMO. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiology. 2001;126:875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, et al. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proceedings of the National Academy of Sciences, USA. 1999;96:15336–15341. doi: 10.1073/pnas.96.26.15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, et al. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiology. 2005;138:2087–2096. doi: 10.1104/pp.105.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, et al. A systemic small RNA signaling system in plants. The Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhleniuk OV, Raines CA, Lloyd JC. pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta. 2001;212:529–534. doi: 10.1007/s004250000450. [DOI] [PubMed] [Google Scholar]
- Zhang S, Sun L, Kragler F. The phloem delivered RNA pool contains small non-coding RNAs and interferes with translation. Plant Physiology. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Huang F, Narsai R, et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiology. 2009;151:262–274. doi: 10.1104/pp.109.141051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao FC, Wu ZC, et al. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology. 2008;146:1673–1686. doi: 10.1104/pp.107.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Reconstituting plant miRNA biogenesis. Proceedings of the National Academy of Sciences, USA. 2008;105:9851–9852. doi: 10.1073/pnas.0805207105. [DOI] [PMC free article] [PubMed] [Google Scholar]