Abstract
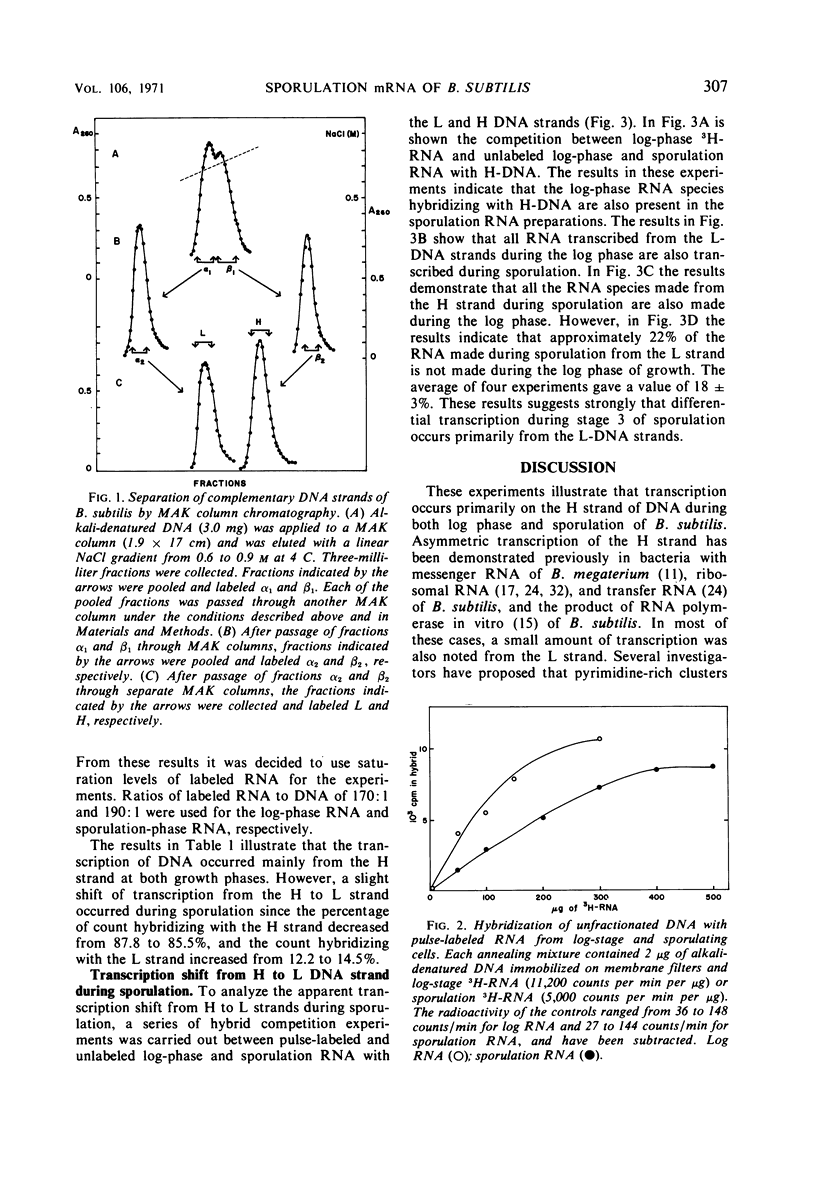
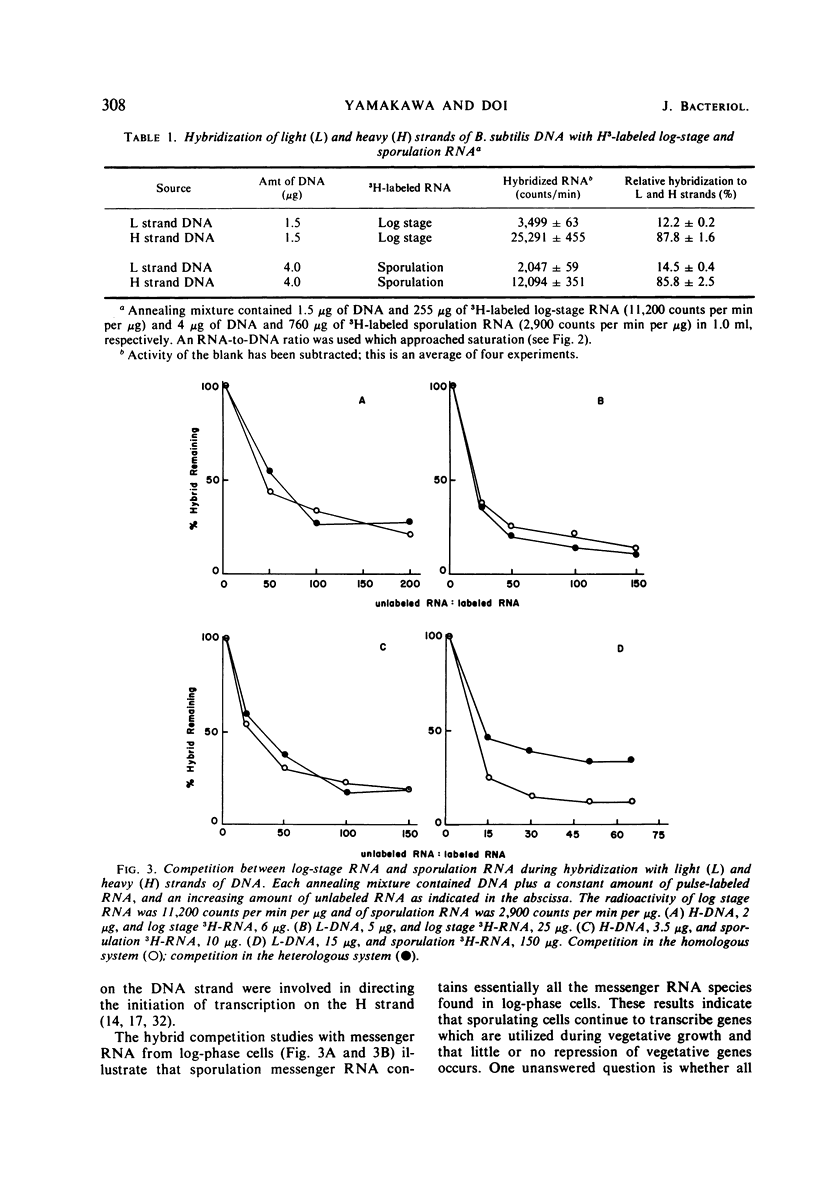
Messenger ribonucleic acid (RNA) from log and sporulation stages of growth were transcribed mainly from the heavy strand of the complementary strands of Bacillus subtilis deoxyribonucleic acid (DNA). During sporulation a slight transcription shift from heavy to light DNA strands was observed. RNA-DNA hybrid competition experiments revealed that this shift was due to sporulation-specific transcription from light-DNA strands.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARONSON A. I. CHARACTERIZATION OF MESSENGER RNA IN SPORULATING BACILLUS CEREUS. J Mol Biol. 1965 Mar;11:576–588. doi: 10.1016/s0022-2836(65)80012-2. [DOI] [PubMed] [Google Scholar]
- Borst P., Aaij C. Identification of the heavy strand of rat-liver mitochondrial DNA as the messenger strand. Biochem Biophys Res Commun. 1969 Feb 7;34(3):358–364. doi: 10.1016/0006-291x(69)90841-9. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- DOI R. H., IGARASHI R. T. GENETIC TRANSCRIPTION DURING MORPHOGENESIS. Proc Natl Acad Sci U S A. 1964 Sep;52:755–762. doi: 10.1073/pnas.52.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R. H., Igarashi R. T. Heterogeneity of the conserved ribosomal ribonucleic acid sequences of Bacillus subtilis. J Bacteriol. 1966 Jul;92(1):88–96. doi: 10.1128/jb.92.1.88-96.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigner J., Doty P. The native, denatured and renatured states of deoxyribonucleic acid. J Mol Biol. 1965 Jul;12(3):549–580. doi: 10.1016/s0022-2836(65)80312-6. [DOI] [PubMed] [Google Scholar]
- GIERER A., SCHRAMM G. Infectivity of ribonucleic acid from tobacco mosaic virus. Nature. 1956 Apr 14;177(4511):702–703. doi: 10.1038/177702a0. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Guha A., Szybalski W. Fractionation of the complementary strands of coliphage T4 DNA based on the asymmetric distribution of the poly U and poly U,G binding sites. Virology. 1968 Apr;34(4):608–616. doi: 10.1016/0042-6822(68)90082-2. [DOI] [PubMed] [Google Scholar]
- HAYASHI M., HAYASHI M. N., SPIEGELMAN S. RESTRICTION OF IN VIVO GENETIC TRANSCRIPTION TO ONE OF THE COMPLEMENTARY STRANDS OF DNA. Proc Natl Acad Sci U S A. 1963 Oct;50:664–672. doi: 10.1073/pnas.50.4.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habich A., Weissmann C., Libonati M., Warner R. C. Isolation of a fraction of Bacillus megaterium DNA enriched in "minus" sequences. J Mol Biol. 1966 Nov 14;21(2):255–264. doi: 10.1016/0022-2836(66)90096-9. [DOI] [PubMed] [Google Scholar]
- Halvorson H. O., Vary J. C., Steinberg W. Developmental changes during the formation and breaking of the dormant state in bacteria. Annu Rev Microbiol. 1966;20:169–188. doi: 10.1146/annurev.mi.20.100166.001125. [DOI] [PubMed] [Google Scholar]
- Karkas J. D., Rudner R., Chargaff E. Seapration of B. subtilis DNA into complementary strands. II. Template functions and composition as determined by transcription with RNA polymerase. Proc Natl Acad Sci U S A. 1968 Jul;60(3):915–920. doi: 10.1073/pnas.60.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., Spudich J. A., Nelson D. L., Deutscher M. P. Origin of proteins in sporulation. Annu Rev Biochem. 1968;37:51–78. doi: 10.1146/annurev.bi.37.070168.000411. [DOI] [PubMed] [Google Scholar]
- Kubinski H., Opara-Kubinska Z., Szybalski W. Patterns of interaction between polyribonucleotides and individual DNA strands derived from several vertebrates, bacteria and bacteriophages. J Mol Biol. 1966 Sep;20(2):313–329. doi: 10.1016/0022-2836(66)90067-2. [DOI] [PubMed] [Google Scholar]
- Kumar S., Szybalski W. Orientation of transcription of the lac operon and its repressor gene in Escherichia coli. J Mol Biol. 1969 Feb 28;40(1):145–151. doi: 10.1016/0022-2836(69)90303-9. [DOI] [PubMed] [Google Scholar]
- Losick R., Sonenshein A. L. Change in the template specificity of RNA polymerase during sporulation of Bacillus subtilis. Nature. 1969 Oct 4;224(5214):35–37. doi: 10.1038/224035a0. [DOI] [PubMed] [Google Scholar]
- Lozeron H. A., Szybalski W., Landy A., Abelson J., Smith J. D. Orientation of transcription for the amber suppressor gene su 3 as determined by hybridization between tyrosine tRNA and the separated DNA strands of transducing coliphage phi80d su3. J Mol Biol. 1969 Jan 14;39(1):239–243. doi: 10.1016/0022-2836(69)90345-3. [DOI] [PubMed] [Google Scholar]
- OPARA-KUBINSKA Z., KUBINSKI H., SZYBALSKI W. INTERACTION BETWEEN DENATURED DNA, POLYRIBONUCLEOTIDES, AND RIBOSOMAL RNA: ATTEMPTS AT PREPARATIVE SEPARATION OF THE COMPLEMENTARY DNA STRANDS. Proc Natl Acad Sci U S A. 1964 Oct;52:923–930. doi: 10.1073/pnas.52.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi M. The transcribing strands of bacillus subtilis DNA for ribosomal and transfer RNA. Proc Natl Acad Sci U S A. 1969 Jan;62(1):256–262. doi: 10.1073/pnas.62.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pène J. J. Modification of transcription following interaction of template specific monomers of Bacillus subtilis RNA polymerase. Nature. 1969 Aug 16;223(5207):705–707. doi: 10.1038/223705a0. [DOI] [PubMed] [Google Scholar]
- Riva S. C. Asymmetric transcription of B. subtilis phage SPP1 DNA in vitro. Biochem Biophys Res Commun. 1969 Mar 31;34(6):824–830. doi: 10.1016/0006-291x(69)90254-x. [DOI] [PubMed] [Google Scholar]
- Roger M. Chromatographic resolution of complementary strands of denatured Pneumococcal DNA. Proc Natl Acad Sci U S A. 1968 Jan;59(1):200–207. doi: 10.1073/pnas.59.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner R., Karkas J. D., Chargaff E. Separation of B. subtilis DNA into complementary strands, I. Biological properties. Proc Natl Acad Sci U S A. 1968 Jun;60(2):630–635. doi: 10.1073/pnas.60.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. A., Kornberg A. Biochemical studies of bacterial sporulation and germaination. VII. Protein turnover during sporulation of Bacillus subtilis. J Biol Chem. 1968 Sep 10;243(17):4600–4605. [PubMed] [Google Scholar]
- Szybalski W., Kubinski H., Sheldrick P. Pyrimidine clusters on the transcribing strand of DNA and their possible role in the initiation of RNA synthesis. Cold Spring Harb Symp Quant Biol. 1966;31:123–127. doi: 10.1101/sqb.1966.031.01.019. [DOI] [PubMed] [Google Scholar]
- Taylor K., Hradecna Z., Szybalski W. Asymmetric distribution of the transcribing regions on the complementary strands of coliphage lambda DNA. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1618–1625. doi: 10.1073/pnas.57.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]


