Abstract
Cyclic nucleotide phosphodiesterases (PDEs) comprise a superfamily of enzymes that serve as drug targets in many human diseases. There is a continuing need to identify high-specificity inhibitors that affect individual PDE families or even subtypes within a single family. We describe a fission yeast-based high throughput screen to detect inhibitors of heterologously-expressed cAMP PDEs. The utility of this system is demonstrated by the construction and characterization of strains that express mammalian PDE2A, PDE4A, PDE4B, and PDE8A and respond appropriately to known PDE2A and PDE4 inhibitors. High throughput screens of two bioactive compound libraries for PDE inhibitors using strains expressing PDE2A, PDE4A, PDE4B, and the yeast PDE Cgs2 identified known PDE inhibitors and members of compound classes associated with PDE inhibition. We verified that the furanocoumarin imperatorin is a PDE4 inhibitor based on its ability to produce a PDE4-specific elevation of cAMP levels. This platform can be used to identify PDE activators, as well as genes encoding PDE regulators, which could serve as targets for future drug screens.
Keywords: Schizosaccharomyces pombe, cAMP, phosphodiesterase, high throughput, inhibitors
INTRODUCTION
High throughput screens allow one to test a large number of compounds for a desired property, leading to the identification of compounds for use as research tools or therapeutics. Such screens may be conducted in vitro using protein extracts or purified reagents. Alternatively, one can use cells whose growth or other behavior is altered by compounds of interest. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe are popular model eukaryotes due to the ease of genetic manipulation, through both classical and molecular genetic approaches. These yeasts carry out many biological processes in common with human cells, while allowing researchers to use simple growth phenotypes to follow the status of the process in question. These same features commend the use of these yeasts in high throughput screens to detect compounds that alter the activity of heterologously-expressed proteins that replace an endogenous protein in the host strain.
Cyclic AMP (cAMP) signaling pathways in mammals respond to the detection of hormones, odorants, and neurotransmitters, and are complicated due to the presence of multiple cAMP-producing adenylyl cyclases and cAMP-destroying cAMP phosphodiesterases (PDEs) (1, 2). There are 11 mammalian PDE families encoded by 21 genes, which produce more than 100 isoenzymes (2, 3). PDEs from the PDE4, PDE7, and PDE8 families specifically act on cAMP, PDEs from the PDE1, PDE2, PDE3, PDE10, and PDE11 families act on both cAMP and cGMP, while PDEs from the PDE5, PDE6, and PDE9 families act specifically on cGMP. The presence of multiple PDE isoenzymes in various tissues complicates efforts to determine the relative roles of specific enzymes in any given biological process. Even so, chemical inhibitors of PDEs, and in some cases chemical activators, are seen as potential therapeutic compounds for the treatment of a variety of conditions including neurological diseases such as anxiety, depression, and Alzheimer's disease; inflammatory diseases such as asthma, chronic obstructive pulmonary disease, and pulmonary hypertension; metabolic diseases such as diabetes and obesity; and other conditions such as memory loss, chronic lymphocytic leukemia, prostate cancer, and erectile dysfunction (2-9). We describe here the development of a cell-based screen for identifying both chemical inhibitors and activators of cAMP PDEs using a simple growth assay in the fission yeast S. pombe.
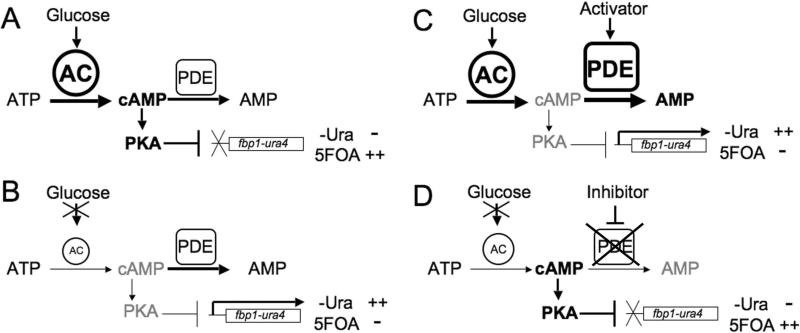
This screening platform makes use of two reporters expressed from the glucose-repressible fbp1+ promoter (10). S. pombe detects glucose via a cAMP signaling pathway to the activate the cAMP-dependent protein kinase PKA, which represses fbp1+ transcription (11, 12). Proteins required for cAMP signaling include the Git3 G protein coupled receptor (GPCR) and its cognate heterotrimeric G protein composed of the Gpa2 Gα subunit, the Git5 Gβ subunit, and the Git11 Gγ subunit. Meanwhile, glucose starvation activates the Spc1/Sty1 stress-activated MAP kinase pathway required for fbp1+ transcription (13-15). The fbp1-ura4 reporter places uracil biosynthesis under the control of the glucose/cAMP pathway, such that cells with high cAMP levels due to glucose signaling repress reporter expression, and cannot grow in medium lacking uracil (SC-ura), but grow in medium containing the pyrimidine-analog 5-fluoro-orotic acid (5FOA; Figure 1A). In contrast, cells with low cAMP levels due to defects in glucose signaling, express the reporter. This allows growth in medium lacking uracil, but not in 5FOA medium (Figure 1B). A second reporter, fbp1-lacZ, allows for easy quantitation of expression from the fbp1+ promoter.
Figure 1.
Schematic of cAMP-regulated growth phenotypes in fission yeast strains expressing the fbp1-ura4 reporter. A) Glucose signaling leads to adenylyl cyclase activation and a cAMP signal, which activates PKA to repress fbp1-ura4 transcription. These cells cannot grow in medium lacking uracil (-Ura), but do grow in medium containing 5FOA. B) Cells carrying mutations in genes required for glucose signaling have reduced adenylyl cyclase activity to lower cAMP levels. This results in low PKA activity and a failure to repress fbp1-ura4 transcription. These cells grow in medium lacking uracil (-Ura), but do not grow in medium containing 5FOA. C) A screen for PDE activators can be carried out by taking a strain such as the one in panel A and screening for compounds that enhance growth in medium lacking uracil. The compounds identified will include ones that stimulate PDE activity to lower cAMP levels. D) A screen for PDE inhibitors can be carried out by taking a strain such as the one in panel B and screening for compounds that enhance growth in 5FOA medium. The compounds identified will include ones that inhibit PDE activity to raise cAMP levels.
We show here that strains expressing the mammalian enzymes PDE2A, PDE4A, PDE4B, and PDE8A produce functional PDEs whose activities affect the expression of these fbp1-driven reporters. In addition, reporter expression in PDE4A- and PDE4B-expressing strains is repressed by the PDE4 inhibitor rolipram, while reporter expression in a PDE2A-expressing strain is repressed by the PDE2A inhibitor erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA). Results from high throughput screens for chemical inhibitors of the PDE2A, PDE4A, PDE4B, and yeast PDE Cgs2 validate the utility of this platform by identifying known PDE inhibitors and novel candidate inhibitors. Once such candidate, imperatorin, was verified to inhibit PDE4 enzymes by its ability to produce a PDE4-specific elevation in cAMP levels. We discuss additional capabilities of this screening platform to identify chemical activators of PDEs, as well as genes that encode biological activators or inhibitors of PDEs, which could serve as target proteins in future drug screens. The flexibility and versatility of this system suggest that this should be an effective way to identify both chemical and biological regulators of PDEs from a variety of organisms.
MATERIALS AND METHODS
Strains, media, and general techniques
Strains used are listed in Table 1. Methods for yeast growth and transformations have been previously described (16). Murine PDE genes were amplified by PCR using oligonucleotides containing approximately 60 nt of sequence flanking the S. pombe cgs2+ gene to direct homologous recombination to this locus. The recipient strain carries a ura4+-marked disruption of cgs2+ (17) (also referred to as pde1+) to allow for 5FOA-counterselection for candidate transformants. PCR was used to confirm the homologous integration events. Subsequent strains were constructed by standard genetic crosses and tetrad dissection to introduce the fbp1-lacZ and fbp1-ura4 reporters, as well as the pap1Δ allele.
Table 1.
S. pombe strain list.
| Strain name |
Genotype |
|---|---|
| CHP861 h+ | fbp1:: ura4+ ura4::fbp1-lacZ ade6-M210 his3-D1 leu1-32 gpa2::his3+ |
| CHP932 h+ | fbp1:: ura4+ ura4::fbp1-lacZ leu1-32 git3Δ::kan |
| CHP1085 h+ | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 his7-366 pap1Δ::ura4− cgs2::PDE4A1 gpa2::his3+ |
| CHP1098 h+ | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 pap1Δ::ura4− cgs2::PDE4A1 gpa2::his3+ |
| CHP1113 h− | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 his3D-1 pap1Δ::ura4− cgs2::PDE4B3 gpa2::his3+ |
| CHP1114 h− | bp1::ura4+ ura4::fbp1-lacZ leu1-32 ade6-M210 his3D-1 pap1Δ::ura4− cgs2::PDE4B3 |
| CHP1141 h90 | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 pap1Δ::ura4− cgs2::PDE8A |
| DDP13 h+ | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 ade6-M210 cgs2::PDE8A[LEU2+] gpa2::his3+ |
| DDP26 h− | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 cgs2::PDE4A1 gpa2::his3+ |
| DIP72 h90 | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 ade6-M210 cgs2::PDE4B3 gpa2::his3+ |
| LWP98 h+ | fbp1::ura4+ ura4::fbp1-lacZ leu1-32 ade6-M216 his3-D1 cgs2-2 gpa2::his3+ |
| LWP364 h− | fbp1:: ura4+ ura4::fbp1-lacZ leu1-32 his7-366 cgs2::PDE2A gpa2::his3+ |
| LWP367 h− | fbp1:: ura4+ ura4::fbp1-lacZ leu1-32 his7-366 pap1Δ::ura4− cgs2::PDE2A gpa2::his3+ |
| LWP369 h− | fbp1:: ura4+ ura4::fbp1-lacZ leu1-32 his7-366 cgs2::PDE2A git3Δ::kan gpa2::his3+ |
| LWP371 h− | fbp1:: ura4+ ura4::fbp1-lacZ leu1-32 his7-366 pap1Δ::ura4− cgs2::PDE2A |
β-galactosidase assays and characterization of 5FOA-sensitivity were carried out as previously described (10). cAMP assays were performed on exponential phase cells grown in EMM complete medium (3% glucose), using the Assay Designs cAMP EIA kit, according to manufacturer's instructions.
High throughput screening
High throughput screens were carried out at the Broad Institute's Chemical Biology Program screening facility. Depending upon the strain, cultures were pregrown to exponential phase in EMM complete medium containing from 0.5 to 2.5 mM cAMP to repress fbp1-ura4 transcription. Cells were collected by centrifugation, resuspended in 5FOA medium, and 25 μl were transferred to 384-well microtiter dishes (untreated, with flat clear bottoms) that had been pre-filled with 25 μl 5FOA medium and pre-pinned with 100 nl of compounds (stock solutions were generally 10mM) from a subset of the Prestwick Bioactive and the Microsource Spectrum compound libraries. Starting cell concentrations ranged from 0.5 × 105 to 4 × 10 5 cells/ml depending on the screening strain. As appropriate, control plates received either 100 nl 10mM rolipram or DMSO. Other positive control dishes contained 5mM cAMP in the 5FOA medium. Cultures were grown for 48 hours at 30°C while sealed in an airtight container with moist paper towels to prevent evaporation. Optical densities (OD600) of cultures were measured using a microplate reader. Bioinformatic analysis of the results to determine composite Z scores was performed as previously described (18, 19). The Z factor of an assay is determined by multiplying the sum of the standard deviations of the positive and negative controls by three and dividing by the absolute difference in the means of the positive and negative controls. This value is then substracted from the number one to obtain the Z factor. An assay with a Z factor of greater than 0.5 is considered to be sufficiently robust for high throughput screening. Within a screen, individual wells are assigned a Z score, which represents the number of standard deviations above or below the mean of the negative controls within that same assay plate. Duplicate Z scores for each compound are plotted onto a grid and projected perpendicularly to the diagonal that represents identity between duplicate Z scores. The composite Z score is the distance from this point on the diagonal to the origin.
RESULTS
Construction of fission yeast strains expressing mammalian PDEs
In an effort to develop yeast strains whose growth behaviors could serve as a reflection of the activity of heterologously-expressed PDEs, we used homologous recombination to replace the only S. pombe PDE gene, cgs2+, with each of four murine PDE genes, PDE2A, PDE4A, PDE4B, and PDE8A (20-22). Strains expressing these enzymes regain mating competence, which is deficient in strains lacking PDE activity (23), indicating that these PDEs are functional when expressed in S. pombe (data not shown).
We next constructed strains expressing the murine PDEs together with the fbp1-driven reporters, and carrying mutant alleles of either the git3+ glucose receptor gene or the gpa2+ Gα subunit gene, both of which are required for glucose detection, adenylyl cyclase activation, and transcriptional repression of the fbp1-ura4 and fbp1-lacZ reporters (11, 24-26). The relative level of reporter expression reflects PDE activity. β-galactosidase activity in the gpa2− mutant strains, as compared with similar strains expressing either the wild type S. pombe Cgs2+ PDE or the frame-shifted, and presumably inactive, Cgs2-2 truncated PDE (16) demonstrate that all four murine PDEs are active in S. pombe (Table 2; vehicle values). The relative level of PDE activity, as reflected by the degree to which β-galactosidase activity is elevated by the reduction in cAMP levels, is Cgs2+ > PDE4A > PDE2A ≥ PDE4B ≥ PDE8A > Cgs2-2. This order of activity is consistent with the ability of git3− and gpa2− mutations to confer 5FOA-sensitive (5FOAS) growth to strains expressing the murine PDEs (see below).
Table 2.
β-galactosidase activity in response to PDE inhibitor treatment
| β-galactosidase activity | ||||
|---|---|---|---|---|
| Strain | PDE | Vehicle | 50μM Rolipram | 100μM Rolipram |
| CHP861 | Cgs2 | 1661 ± 121 | 1807 ± 446 | 1784 ± 429 |
| DDP26 | PDE4A | 998 ± 154 | 271 ± 30 | 162 ± 17 |
| DIP72 | PDE4B | 432 ± 170 | 32 ± 12 | 21 ± 7 |
| DDP13 | PDE8A | 241 ± 61 | 253 ± 46 | 237 ± 67 |
| LWP98 | Cgs2-2 | 23 ± 10 | 19 ± 9 | 20 ± 11 |
| β-galactosidase activity | |||||
|---|---|---|---|---|---|
| Strain | PDE | Vehicle | 5μM EHNA | 20μM EHNA | 200 μM EHNA |
| LWP367 | PDE2A | 587 ± 7 | 473 ± 19 | 197 ± 51 | 45 ± 3 |
β-galactosidase activity was determined from 3 to 4 independent exponential phase cultures. The values presented are average ± SD in units of specific activity per milligram of soluble protein.
Effect of known PDE inhibitors on reporter expression and cAMP levels
We next tested the effect of known PDE inhibitors on the expression of the fbp1-lacZ fusion in murine PDE-expressing strains. As seen in Table 2, the PDE4 inhibitor rolipram reduces β-galactosidase activity in PDE4A- and PDE4B-expressing cells, but not in Cgs2- or PDE8A-expressing cells. These results support previous studies indicating that PDE8A is insensitive to rolipram. In addition, the PDE2A inhibitor EHNA reduces β-galactosidase activity expressed from a PDE2A strain (Table 2), with an ED50 of approximately 14 μM. The effect on fbp1-lacZ expression is not a direct measurement of PDE activity, therefore an IC50 cannot be determined by these data. The ED50 values of rolipram on PDE4A and PDE4B appear to be approximately 37μM and 26μM, respectively, however these values assume that the 50μM assays are in the linear range, which is not the case. A more accurate assessment of rolipram inhibition of PDE4A is presented below. No PDE8A inhibition was observed using dipyridamole (data not shown), a known PDE8A inhibitor (21). As a DNA sequence analysis of the integrated PDE8A gene showed that it encodes the wild type PDE8A protein, we presume that the failure to observe a response to dipyridamole is due to a permeability problem in fission yeast or a solubility problem with dipyridamole in the growth medium.
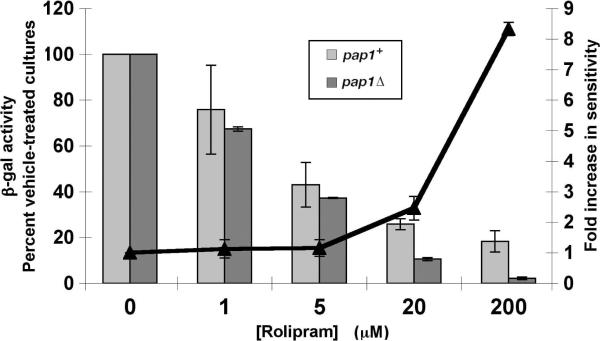
In an effort to increase the sensitivity to PDE inhibitors, we examined whether deleting pap1+, encoding a zinc finger transcriptional activator required for ABC transporter expression and whose overexpression confers staurosporine-resistance (27, 28), would enhance inhibition of PDE4A by rolipram. As shown in Figure 2, PDE4A-expressing cells lacking pap1+ (pap1Δ), are more sensitive (ED50 ≈ 3.6μM) to rolipram than pap1+ cells (ED50 ≈ 4.2μM). Moreover, pap1Δ strains that are 5FOAS due to low cAMP levels maintain the 5FOAS growth phenotype for longer periods of incubation than equivalent pap1+ strains (data not shown). This enhanced 5FOA-sensitivity should aid in the detection of PDE inhibitors that confer 5FOAR growth.
Figure 2.
Deletion of pap1+ enhances rolipram-mediated fbp1-lacZ repression. β-galactosidase activity from two independent exponential phase cultures was determined in pap1+ (light gray bars) and pap1Δ (dark gray bars) gpa2− mutant strains grown in EMM complete medium containing various concentrations of rolipram as indicated, while receiving identical volumes of DMSO (vehicle). Values are plotted as a percent of the vehicle-treated cultures that did not receive rolipram. The ratio of fold-inhibition in the pap1Δ strain versus the pap1+ strain is shown for each concentration of rolipram.
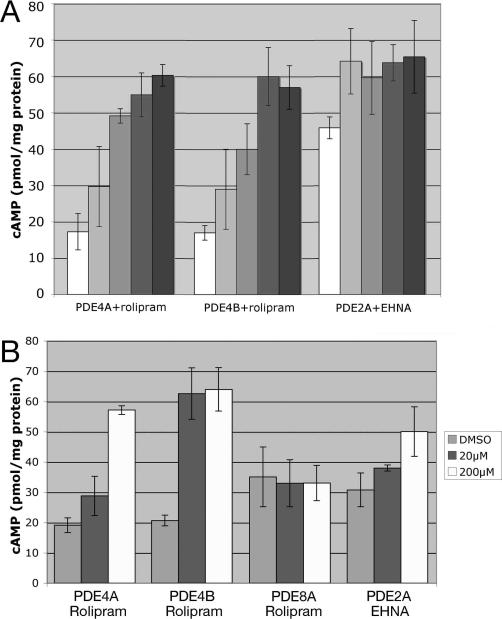
To determine if the effect of rolipram on PDE4-expressing cells and of EHNA on PDE2A-expressing cells is through inhibition of the heterologously-expressed PDEs, cAMP levels were measured before and after drug treatment. As shown in Figure 3A, cAMP levels increase within 10 minutes of exposure to 200 μM inhibitor and reach peak levels within one hour. We next examined whether varying degrees of PDE inhibition could be detected by measuring cAMP levels at the one-hour timepoint in cells exposed to lower concentrations of inhibitor. Figure 3B shows that PDE4A is only partially inhibited by 20 μM rolipram, while PDE4B is completely inhibited at this concentration, suggesting that PDE4B is more sensitive than PDE4A to rolipram in this system. Furthermore, cAMP levels in a strain expressing PDE8A are completely insensitive to rolipram treatment, consistent with previous studies of PDE8A (21), and also indicating that rolipram does not affect cAMP generation in fission yeast. Finally, PDE2A shows partial inhibition by EHNA at 20 μM as compared to 200 μM EHNA. Thus, PDE inhibition can be indirectly quantitated by measuring the effect of a compound on cAMP levels in target yeast strains.
Figure 3.
PDE inhibitors alter cAMP levels in yeast strains. A) cAMP levels were measured in exponential phase cells immediately prior to 200 μM drug addition (white bars: rolipram for strains CHP1085 (PDE4A) and CHP1114 (PDE4B), and EHNA for strain LWP371 (PDE2A)), and 10, 30, 60, and 120 minutes (from light gray to black bars) after drug addition. Values represent the average and SD of two or three independent experiments. B) cAMP levels were measured 60 minutes after addition of either vehicle (DMSO), 20 μM drug, or 200 μM drug as indicated. The strains used are as in panel A, together with strain CHP1141 (PDE8A). Values represent the average and SD of two or three independent experiments.
High throughput screening of PDE-expressing strains
While the fbp1-lacZ reporter allows for a measurement of PDE inhibition, the true power of this system is in the growth phenotype conferred by transcription of the fbp1-ura4 reporter. PDE inhibitors should restore 5FOAR growth to strains possessing low basal cAMP levels by elevating cAMP levels to repress fbp1-ura4 transcription (Figure 1D). Conversely, PDE activators should confer growth in SC-ura medium to strains possessing high cAMP levels by reducing cAMP levels to increase fbp1-ura4 transcription (Figure 1C). As mentioned above, mutations in either the git3+ or gpa2+ genes were introduced into various PDE-expressing strains. While a gpa2− mutant allele conferred 5FOA-sensitivity on PDE2A-, PDE4A-, PDE4B-, and PDE8A-expressing strains, only Cgs2- and PDE4A-expressing strains became 5FOAS when carrying a mutant allele of git3+ (data not shown). These results are consistent with previous observations that loss of Gpa2 confers a greater defect in cAMP signaling than does loss of Git3 (10, 25, 26), and that Cgs2 and PDE4A are more active than the other three PDEs in our strains (Table 2).
To determine whether the 5FOA growth phenotype could be exploited for high throughput screening, strains expressing Cgs2, PDE2A, PDE4A, PDE4B, or PDE8A were pregrown in EMM medium containing cAMP and then transferred to 5FOA medium in 384 well microtiter plates in the presence or absence of cAMP. OD600 measurements were taken after 24 and 48 hours of incubation at 30°C. After 24 hours, some cAMP-stimulated growth could be detected, however 48 hours incubation was required for cAMP-treated cells to grow to saturation (data not shown). Similar experiments in which 20 μM rolipram (final concentration) was pinned into 192 of the 384 wells, in place of cAMP addition to the medium, produced 5FOAR growth in the PDE4A and PDE4B-expressing strains. In a typical experiment with CHP1113 cells (PDE4B), the OD600 of the rolipram-treated cultures was 1.28 +/− 0.07 while the OD600 of the untreated wells was 0.18 +/− 0.02. When using CHP1098 cells (PDE4A), the OD600 of the rolipram-treated cultures was 1.15 +/− 0.06, while the OD600 of the untreated wells was 0.2 +/− 0.03. The Z factors (a statistical assessment of the quality of datasets used in high throughput screening (29)) for these screens are 0.76 and 0.72, respectively, placing them well above the 0.5 minimum Z factor indicative of a robust screen.
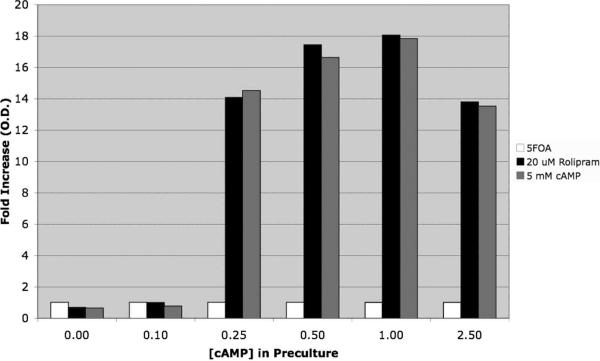
As mentioned above, cAMP was present in the growth medium prior to transferring cells to the 5FOA medium. 5FOA-sensitivity is due to the presence of the Ura4 protein, so that cultures had to be pre-grown under conditions that repress fbp1-ura4+ reporter expression. The effect of the PDE inhibitor is therefore to maintain 5FOA-resistance rather than to confer it during the assay. As shown in Figure 4, CHP1113 cells (PDE4B3 gpa2Δ) are only able to respond to exposure to either cAMP or rolipram addition to the 5FOA medium if they are pre-cultured in the presence of at least 0.25mM cAMP.
Figure 4.
Effect of cAMP in the preculture on rolipram- or cAMP-stimulated 5FOA-resistant growth. Strain CHP1113 (PDE4B) cells were precultured in EMM medium containing various concentrations of cAMP as indicated. Cells were then transferred to 5FOA medium containing DMSO (white bars, vehicle control), 20μM rolipram (black bars), or 5mM cAMP (gray bars) and incubated at 30°C for 48 hours. OD600 values are normalized to the DMSO controls. Cells precultured in less than 0.25mM cAMP fail to respond either rolipram or cAMP.
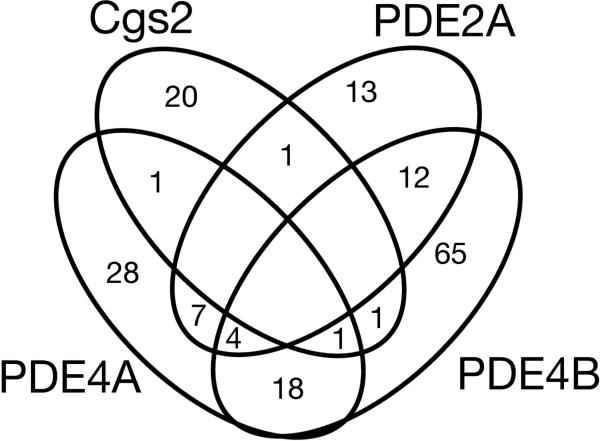
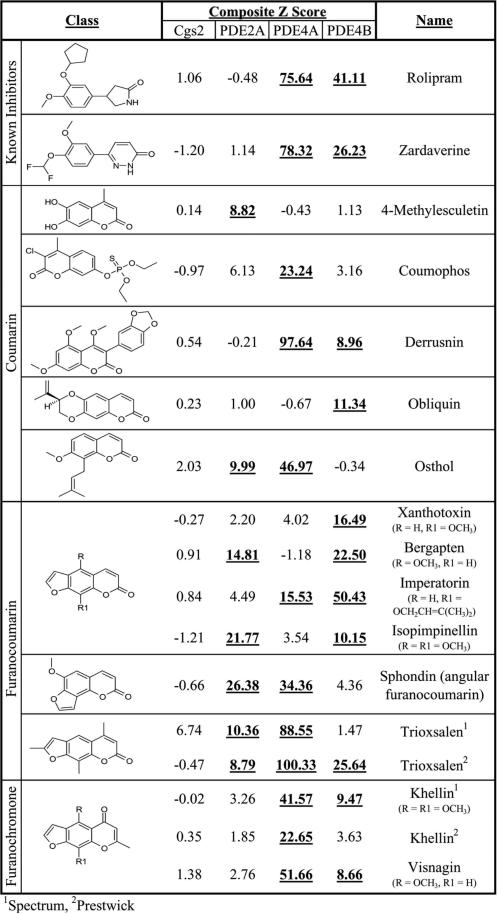
To test of the utility of this system, we screened a pair of libraries containing 3,120 bioactive compounds, including known PDE inhibitors, using 5FOAS strains expressing PDE2A, PDE4A, PDE4B, or Cgs2, for compounds that confer 5FOAR growth. Duplicate plates were screened and compounds that confer 5FOAR growth with composite Z scores of ≥8.53 (the cut-off used by the Broad Institute's Chemical Biology Program, where the screens were performed; see Materials and Methods) were identified. Figure 5 is a Venn diagram displaying the overlap of the compounds identified in these four screens. These screens identified relatively low numbers of compounds (from 0.8% to 3.2% of compounds tested per strain), which included known PDE4 inhibitors such as rolipram and zardaverine as PDE4-specific inhibitors (Figure 6). Also identified were compounds of classes that include known PDE inhibitors as discussed below.
Figure 5.
Venn diagram of compounds identified as candidate PDE inhibitors. The overlap of compounds that confer 5FOAR growth, producing a composite Z score of ≥8.53, is shown for CHP932 (Cgs2), LWP369 (PDE2A), CHP1098 (PDE4A) and CHP1113 (PDE4B) against 3,120 compounds representing a subset of the Prestwick Bioactive and the Microsource Spectrum compound libraries.
Figure 6.
Data from a subset of compounds identified in the high throughput screens. The structures of two known PDE inhibitors (rolipram and zardaverine), along with members of the coumarin, furanocoumarin, and furanochromone compound classes are presented, along with the Composite Z scores obtained from duplicate plates for each of the four target PDEs are presented. Composite Z scores that exceed the 8.53 cut-off, considered a hit in the screen, are in bold and underlined. Trioxsalen and khellin were identified from both the Microsource Spectrum and Prestwick Bioactive libraries.
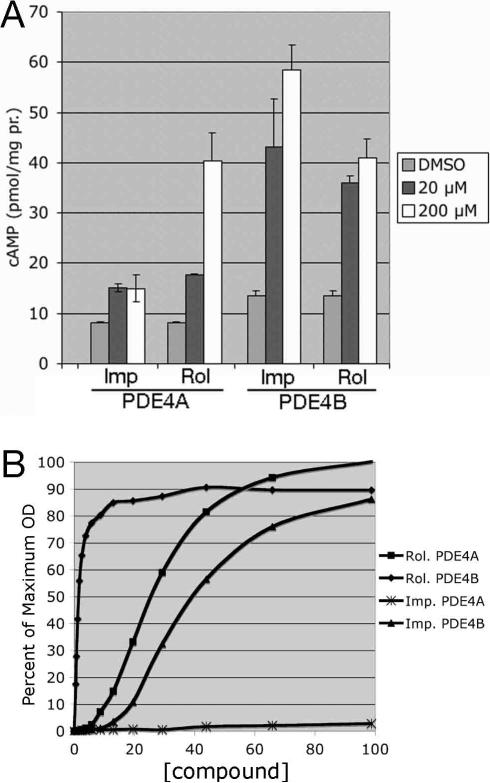
To validate the identification of one of the candidate inhibitors, we re-examined the known PDE4-specific inhibitor rolipram, together with the candidate PDE4-inhibitor imperatorin (Figure 6) to determine an ED50 (effective dose required to promote 5FOA-resistant growth to half of the cAMP-stimulated level) with respect to the 5FOA growth assay. We also examined the ability of imperatorin to elevate cAMP levels, indicative of PDE inhibition. As shown in Figure 7A, and similar to Figure 3B, rolipram elevates cAMP levels in both PDE4A- and PDE4B-expressing cells. Consistent with the composite Z scores for imperatorin in the high throughput screens (Figure 6), imperatorin significantly increases cAMP levels in a PDE4B-expressing strain (composite Z= 50.43), but only modestly increases cAMP levels in a PDE4A-expressing strain (composite Z=15.53). This is a PDE4-specific effect, as imperatorin has no significant effect on cAMP levels in a PDE2A-expressing strain (data not shown). Dose response curves for imperatorin and rolipram with respect to stimulation of 5FOA-resistant growth were also carried out (Figure 7B). Similar to the results from the cAMP assays and the high throughput screen, rolipram is highly effective on both PDE4A (ED50=25.9μM) and PDE4B (ED50=1.5μM), while imperatorin is significantly more active against PDE4B (ED50=40μM) than PDE4A (no significant growth). These data confirm that imperatorin is an effective PDE4B inhibitor, and demonstrate our ability to assess PDE inhibitors by cAMP elevation and 5FOA-resistant growth, as well as by their effect on fbp1-lacZ expression (Table 2, Figure 2).
Figure 7.
Verification of imperatorin as a PDE4 inhibitor. A) cAMP response to rolipram or imperatorin by CHP1085 (PDE4A) and CHP1114 (PDE4B) cells. cAMP levels were measured 60 minutes after addition of either vehicle (DMSO), 20 μM drug, or 200 μM drug as indicated. Values represent the average and SD of two independent experiments. B) Dose response curves for rolipram- and imperatorin-exposed CHP1098 (PDE4A) and CHP1113 (PDE4B) cells. Results from a typical experiment are given. Each datapoint represents the mean of 12 wells. Standard deviations were generally 3% to 4% of the means. OD600 values are presented as a percent of the cAMP-stimulated 5FOA-resistant growth.
DISCUSSION
We have described a novel fission yeast cell-based screening platform, amenable for high throughput screening to identify compounds that alter PDE activity. While a budding yeast system based on heat shock sensitivity of stationary phase cells has been previously reported (30), cells in that assay had to be exposed to 0.5mM to 2mM rolipram to detect an effect on PDE4B (31, 32). In contrast, we have successfully screened compound libraries at an average concentration of 20μM to detect both known and previously unidentified PDE inhibitors (Figure 6). We have gone on to determine ED50 values for rolipram against PDE4A and PDE4B of 25.9μM and 1.5μM, respectively. These values are higher than the IC50 values for rolipram against PDE4 enzymes (generally in the range of 1μM, (2)), however cell-based assays cannot be as sensitive as in vitro assays due to additional requirements of cell permeability and the presence of thousands of proteins within a cell, some of which may bind to the compound and reduce its availability to the target protein. In fact, it is even unclear how the intracellular concentration of rolipram relates to the concentation in the growth medium. In addition, we observe a significant difference in the ED50 for rolipram against PDE4A and PDE4B that is not consistent with in vitro measurements. This may reflect the fact that PDE4B appears to be less active than PDE4A in our strains (Table 2), thus less inhibition is required to produce an effect on reporter expression. This is consistent with the lower ED50 of rolipram against PDE4A for repression of β-galactosidase expression (3-4μM) than for 5FOA-resistant growth (25.9μM). For strains that express the reporter genes at a high level due to high initial PDE activity, a greater reduction in reporter expression is required to produce 5FOA-resistant growth.
Using this inexpensive assay, it will be straightforward to develop a large collection of strains expressing either mammalian cAMP-specific or dual-specificity PDEs. This platform can also be used with PDEs from pathogens, whose inhibition may either kill the target pathogen or reduce virulence. Strains representing a broad panel of PDEs could be used to identify compounds possessing desirable specificity profiles to suggest the potential of individual compounds as candidate therapeutics. Moreover, because this platform identifies compounds based on stimulation of cell growth, it will not detect compounds that, while inhibiting PDEs in vitro, are too cytotoxic or cell-impermeable for therapeutic use. The compounds identified in this screen must also remain active within the yeast for much of the 48 hour incubation period in order to promote significant cell growth. This is not the case for the majority of PDE assays, which are carried out in vitro on purified proteins or on protein extracts over a short timecourse. In addition, this cell-based screening platform should be able to detect PDE inhibitors that may not be identified by in vitro screens. For example, compounds that prevent either intermolecular or intramolecular interactions required for enzyme formation would be overlooked in an in vitro assay on purified enzymes or protein extracts, yet should be identifiable in this assay.
This screening platform is not meant to replace in vitro assays that are well-established as a means of identifying and characterizing PDE inhibitors. Clearly, there are disadvantages of not being able to identify compounds that are not soluble in the yeast growth medium or are not permeable to the yeast cells, problems not encountered in an in vitro assay. However, our screen may represent a more holistic approach to drug discovery that identifies compounds with favorable drug-like qualities beyond having a low IC50 for the target enzyme.
Where this approach may show the greatest advantage over the in vitro assays is in identifying compounds that selectively inhibit PDEs within a single family such as PDE4. While there has been significant interest in PDE4 inhibitors based on the CNS and anti-inflammatory effect of rolipram and other PDE4-specific inhibitors for the past 25 years (33, 34), no such inhibitor has received FDA approval. This is largely due to an emetic side-effect that is thought to be caused by PDE4D inhibition (35, 36). Structural approaches to PDE4 inhibition focus on the highly conserved active sites, making it unlikely to find compounds that display a significant difference in IC50 values against the PDE4 subtypes. As this cell-based assay can theoretically detect compounds that work by mechanisms other than binding to PDE active sites, there is the potential for identifying subtype-selective compounds.
The pilot high throughput screens against 3,120 bioactive compounds using strains expressing the yeast PDE Cgs2, or the murine PDEs 2A, 4A, and 4B identified a number of compounds that promote 5FOAR growth, presumably by inhibiting the target PDEs to raise cAMP levels. These included the known PDE4 inhibitors rolipram and zardaverine, which only affected the PDE4A- and PDE4B-expressing strains (Figure 6). Other compounds identified include coumarins, furanocoumarins, and furanochromones (Figure 6), as well as flavonoids and steroids (not shown). Members of these compound classes are known to have PDE inhibitory properties (37), including the furanocourmarin trioxsalen and the furanochromones khellin, and visnagin (38, 39), although most of these compounds identified had not been shown to be PDE inhibitors. Interestingly, both rolipram and imperatorin had been previously shown to inhibit HIV replication (40, 41), though the mechanism of imperatorin action was unknown. We demonstrate here that imperatorin is an effective PDE4B inhibitor, while its effect on PDE4A is very modest (Figure 7). As with imperatorin, many compounds identified in our screens display therapeutic potential, in the absence of a complete understanding of their mechanism. Therefore, data from these screens will help to partially elucidate the effect of these compounds on mammalian cells.
While, the relative overlap of the compounds identified in each screen further validates this platform, it also alerts us to some caveats. Candidates from the Cgs2 screen display the least overlap with candidates from the other three screens (Figure 5), consistent with the fact that the murine PDEs are more closely related to each other than to Cgs2. Furthermore, eighteen compounds inhibit both PDE4A and PDE4B, but not Cgs2 or PDE2A, consistent with the pharmacological grouping of PDE4A and PDE4B into the PDE4 family. On the other hand, there was unexpected amount of overlap from the PDE2A and PDE4B screens. As these PDEs appear to be less active in fission yeast than Cgs2 and PDE4A (Table 2), some of these candidates may either weakly reduce fbp1-driven transcription by a cAMP-independent manner or raise cAMP levels by stimulating adenylyl cyclase activity. This later hypothesis is consistent with the presence of diterpenoids in this group of compounds. However, we should be able to distinguish among cAMP-independent effects, adenylyl cyclase activation and PDE inhibition by measuring cAMP levels in experimental and control strains as in Figure 3B and Figure 7A. Specificially, compounds that act in a cAMP-independent manner will not affect cAMP levels in any strains, while compounds that activate adenylyl cyclase will elevate cAMP levels in control strains that lack PDE activity. Finally, the fact that a number of compounds were detected in only one of each of the four screens lends support for this platform as a tool for identifying isoenzyme-specific inhibitors, which could lead to the development of highly specific therapeutic compounds.
The ability to identify PDE inhibitors is based on the growth phenotype conferred by the cAMP-repressible fbp1-ura4 reporter. This system can also identify compounds that stimulate PDE activity to lower cAMP levels and increase fbp1-ura4 expression. PDE activators should confer Ura+ growth to strains whose high basal cAMP levels repress fbp1-ura4 expression in the absence of drug exposure (Figure 1C). Finally, as yeast are capable of maintaining autonomously-replicating plasmids, one could screen cDNA libraries for genes that encode biological inhibitors or activators of target PDEs, which could serve as novel targets for high throughput screens. Thus, this screening platform has the potential for identifying novel PDE inhibitors and activators, as well as new ways to moderate cAMP signaling pathways in an effort to improve therapeutic approaches to treating a wide array of human diseases.
Acknowledgments
This work was supported by a grant from Boston College and by NIH grant R21 GM079662 to C.S.H.. We thank Drs. James Cherry and Joe Beavo for generously supplying PDE cDNAs, and Drs. Joe Beavo, Paul Epstein, and Guillaume Cottarel for advice and encouragement, and Lynn VerPlank, Jason Burbank, and Stephanie Norton for guidance on high throughput screening. We also thank Drs. Joe Beavo, Nicola Tolliday, and Lynn VerPlank for critical reading of the manuscript. The project has been funded in whole or in part with Federal funds from the National Cancer Institute's Initiative for Chemical Genetics, National Institutes of Health, under Contract No. N01-CO-12400 and has been performed with the assistance of the Chemical Biology Platform of the Broad Institute of Harvard and MIT. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Service, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Abbreviations
- PDE
cyclic nucleotide phosphodiesterase
- cAMP
adenosine 3’,5’-cyclic monophosphate
References
- 1.Kamenetsky M, Middelhaufe S, Bank EM, Levin LR, Buck J, Steegborn C. Molecular details of cAMP generation in mammalian cells: a tale of two systems. J Mol Biol. 2006;362(4):623–39. doi: 10.1016/j.jmb.2006.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58(3):488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 3.Lerner A, Epstein PM. Cyclic nucleotide phosphodiesterases as targets for treatment of haematological malignancies. Biochem J. 2006;393(Pt 1):21–41. doi: 10.1042/BJ20051368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasta V, Shimizu-Albergine M, Beavo JA. Modulation of Leydig cell function by cyclic nucleotide phosphodiesterase 8A. Proc Natl Acad Sci U S A. 2006;103:19925–30. doi: 10.1073/pnas.0609483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyke HJ, Montana JG. Update on the therapeutic potential of PDE4 inhibitors. Expert Opin Investig Drugs. 2002;11(1):1–13. doi: 10.1517/13543784.11.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Boswell-Smith V, Spina D, Page CP. Phosphodiesterase inhibitors. Br J Pharmacol. 2006;147(Suppl 1):S252–7. doi: 10.1038/sj.bjp.0706495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4). Trends Pharmacol Sci. 2004;25(3):158–63. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol Ther. 2006;109(3):366–98. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Hebb AL, Robertson HA. Role of phosphodiesterases in neurological and psychiatric disease. Curr Opin Pharmacol. 2006 doi: 10.1016/j.coph.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman CS, Winston F. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics. 1990;124(4):807–16. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman CS. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem Soc Trans. 2005;33(Pt 1):257–60. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman CS. Except in every detail: Comparing and contrasting G protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryotic Cell. 2005;4:495–503. doi: 10.1128/EC.4.3.495-503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degols G, Shiozaki K, Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16(6):2870–7. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stettler S, Warbrick E, Prochnik S, Mackie S, Fantes P. The wis1 signal transduction pathway is required for expression of cAMP- repressed genes in fission yeast. J Cell Sci. 1996;109(Pt 7):1927–35. doi: 10.1242/jcs.109.7.1927. [DOI] [PubMed] [Google Scholar]
- 15.Stiefel J, Wang L, Kelly DA, Janoo RTK, Seitz J, Whitehall SK, Hoffman CS. Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe. Eukaryotic Cell. 2004;3(3):610–619. doi: 10.1128/EC.3.3.610-619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Griffiths K, Zhang YH, Ivey FD, Hoffman CS. Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics. 2005;171:1523–33. doi: 10.1534/genetics.105.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki N, Yamamoto M. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol Gen Genet. 1992;233(1-2):17–24. doi: 10.1007/BF00587556. [DOI] [PubMed] [Google Scholar]
- 18.Kim YK, Arai MA, Arai T, Lamenzo JO, Dean EF, 3rd, Patterson N, Clemons PA, Schreiber SL. Relationship of stereochemical and skeletal diversity of small molecules to cellular measurement space. J Am Chem Soc. 2004;126(45):14740–5. doi: 10.1021/ja048170p. [DOI] [PubMed] [Google Scholar]
- 19.Franz AK, Dreyfuss PD, Schreiber SL. Synthesis and cellular profiling of diverse organosilicon small molecules. J Am Chem Soc. 2007;129(5):1020–1. doi: 10.1021/ja067552n. [DOI] [PubMed] [Google Scholar]
- 20.Cherry JA, Thompson BE, Pho V. Diazepam and rolipram differentially inhibit cyclic AMP-specific phosphodiesterases PDE4A1 and PDE4B3 in the mouse. Biochim Biophys Acta. 2001;1518(1-2):27–35. doi: 10.1016/s0167-4781(01)00164-6. [DOI] [PubMed] [Google Scholar]
- 21.Soderling SH, Bayuga SJ, Beavo JA. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci U S A. 1998;95(15):8991–6. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu AY, Tang XB, Martinez SE, Ikeda K, Beavo JA. Molecular determinants for cyclic nucleotide binding to the regulatory domains of phosphodiesterase 2A. J Biol Chem. 2004;279(36):37928–38. doi: 10.1074/jbc.M404287200. [DOI] [PubMed] [Google Scholar]
- 23.DeVoti J, Seydoux G, Beach D, McLeod M. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. Embo J. 1991;10(12):3759–68. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivey FD, Hoffman CS. Direct activation of fission yeast adenylate cyclase by the Gpa2 Gα of the glucose signaling pathway. Proc Natl Acad Sci U S A. 2005;102(17):6108–13. doi: 10.1073/pnas.0502270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nocero M, Isshiki T, Yamamoto M, Hoffman CS. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8). Genetics. 1994;138(1):39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welton RM, Hoffman CS. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ, and the git3 putative glucose receptor. Genetics. 2000;156:513–21. doi: 10.1093/genetics/156.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998;12(10):1453–63. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5(1):60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 30.Colicelli J, Nicolette C, Birchmeier C, Rodgers L, Riggs M, Wigler M. Expression of three mammalian cDNAs that interfere with RAS function in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991;88(7):2913–7. doi: 10.1073/pnas.88.7.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillai R, Kytle K, Reyes A, Colicelli J. Use of a yeast expression system for the isolation and analysis of drug-resistant mutants of a mammalian phosphodiesterase. Proc Natl Acad Sci U S A. 1993;90(24):11970–4. doi: 10.1073/pnas.90.24.11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atienza JM, Colicelli J. Yeast model system for study of mammalian phosphodiesterases. Methods. 1998;14(1):35–42. doi: 10.1006/meth.1997.0563. [DOI] [PubMed] [Google Scholar]
- 33.Wachtel H. Characteristic behavioural alterations in rats induced by rolipram and other selective adenosine cyclic 3', 5'-monophosphate phosphodiesterase inhibitors. Psychopharmacology (Berl) 1982;77(4):309–16. doi: 10.1007/BF00432761. [DOI] [PubMed] [Google Scholar]
- 34.Wachtel H. Potential antidepressant activity of rolipram and other selective cyclic adenosine 3',5'-monophosphate phosphodiesterase inhibitors. Neuropharmacology. 1983;22(3):267–72. doi: 10.1016/0028-3908(83)90239-3. [DOI] [PubMed] [Google Scholar]
- 35.Lamontagne S, Meadows E, Luk P, Normandin D, Muise E, Boulet L, Pon DJ, Robichaud A, Robertson GS, Metters KM, et al. Localization of phosphodiesterase-4 isoforms in the medulla and nodose ganglion of the squirrel monkey. Brain Res. 2001;920(1-2):84–96. doi: 10.1016/s0006-8993(01)03023-2. [DOI] [PubMed] [Google Scholar]
- 36.Miro X, Perez-Torres S, Puigdomenech P, Palacios JM, Mengod G. Differential distribution of PDE4D splice variant mRNAs in rat brain suggests association with specific pathways and presynaptical localization. Synapse. 2002;45(4):259–69. doi: 10.1002/syn.10100. [DOI] [PubMed] [Google Scholar]
- 37.Peluso MR. Flavonoids attenuate cardiovascular disease, inhibit phosphodiesterase, and modulate lipid homeostasis in adipose tissue and liver. Exp Biol Med (Maywood) 2006;231(8):1287–99. doi: 10.1177/153537020623100802. [DOI] [PubMed] [Google Scholar]
- 38.Duarte J, Lugnier C, Torres AI, Perez-Vizcaino F, Zarzuelo A, Tamargo J. Effects of visnagin on cyclic nucleotide phosphodiesterases and their role in its inhibitory effects on vascular smooth muscle contraction. Gen Pharmacol. 1999;32(1):71–4. doi: 10.1016/s0306-3623(98)00083-4. [DOI] [PubMed] [Google Scholar]
- 39.Bovalini L, Lusini P, Simoni S, Vedaldi D, Andreassi L, Dall'Acqua F, Martelli P. Inhibition of cAMP phosphodiesterase by some phototherapeutic agents. Z Naturforsch [C] 1987;42(7-8):1009–10. doi: 10.1515/znc-1987-7-849. [DOI] [PubMed] [Google Scholar]
- 40.Sancho R, Marquez N, Gomez-Gonzalo M, Calzado MA, Bettoni G, Coiras MT, Alcami J, Lopez-Cabrera M, Appendino G, Munoz E. Imperatorin inhibits HIV-1 replication through an Sp1-dependent pathway. J Biol Chem. 2004;279(36):37349–59. doi: 10.1074/jbc.M401993200. [DOI] [PubMed] [Google Scholar]
- 41.Angel JB, Saget BM, Walsh SP, Greten TF, Dinarello CA, Skolnik PR, Endres S. Rolipram, a specific type IV phosphodiesterase inhibitor, is a potent inhibitor of HIV-1 replication. Aids. 1995;9(10):1137–44. doi: 10.1097/00002030-199510000-00004. [DOI] [PubMed] [Google Scholar]