Abstract
Ovarian cancer is a disease characterised by complex genomic rearrangements but the majority of the genes that are the target of these alterations remain unidentified. Cataloguing these target genes will provide useful insights into the disease etiology and may provide an opportunity to develop novel diagnostic and therapeutic interventions. High resolution genome wide copy number and matching expression data from 68 primary epithelial ovarian carcinomas of various histotypes was integrated to identify genes in regions of most frequent amplification with the strongest correlation with expression and copy number. Regions on chromosomes 3, 7, 8, and 20 were most frequently increased in copy number (>40% of samples). Within these regions, 703/1370 (51%) unique gene expression probesets were differentially expressed when samples with gain were compared to samples without gain. 30% of these differentially expressed probesets also showed a strong positive correlation (r≥0.6) between expression and copy number. We also identified 21 regions of high amplitude copy number gain, in which 32 known protein coding genes showed a strong positive correlation between expression and copy number. Overall, our data validates previously known ovarian cancer genes, such as ERBB2, and also identified novel potential drivers such as MYNN, PUF60 and TPX2.
Introduction
While progress has been made in elucidating the molecular events that underlie the development of ovarian cancer, the identity of the majority of genes which drive the development of this disease remain elusive. Numerous gene expression studies have identified lists of genes with significantly altered expression, but disappointingly there is little consensus between studies [1]. While gene expression studies are useful in identifying broad categories of pathways altered in cancer and clinically important subtypes [2], on their own they may not be able to distinguish the genetically altered key driver genes. An alterative strategy used to identify driver genes has been annotation of recurrent chromosomal aberrations. Early studies were hampered because the technologies for genome-wide genomic analysis lacked the resolution to adequately refine cancer associated loci [3]. The problem of resolution has been overcome with the development of ultra-high resolution aCGH and SNP arrays. Recently, our group has used these latest-generation SNP arrays to annotate even small regions (as small as 25 kb) of genomic alteration [4]. This data also demonstrated that the genetic events occurring in ovarian cancers are more numerous and complex than previously suspected. While some potential driver genes could be rapidly identified from this data due to their location on focal alterations, the majority of recurrent alterations are large and encompass numerous genes.
To expedite identification of ovarian cancer growth promoting genes we have integrated matching DNA copy number and gene expression data from a cohort of 68 primary epithelial ovarian cancers. We have particularly focused on genes in regions of copy number gain, with the expectation that expression of a driver gene within an amplicon will be more tightly correlated with gene copy number than co-amplified genes whose expression is agnostic to tumorigenesis. Integration of copy number and expression has provided a list of candidate dominantly acting driver genes, which can be used to underpin functional analysis that will be necessary to validate their contribution to ovarian tumorigenesis. In addition, the amplified and over expressed genes have the potential to serve as useful therapeutic or diagnostic markers for ovarian cancer.
Results
Frequency of copy number alterations (CNA) in ovarian cancer
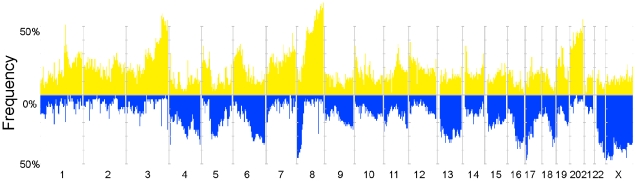
Assessment of CNA in 72 epithelial ovarian tumours (Table 1, Table S1) yielded a total of 36,534 segments comprising 20,570 CN gains and 15,964 CN losses. The median number of regions with CN gain per tumour was 208, accounting for an average of 13.6% of the genome per sample (Table S2). The median number of regions with CN loss was 194 representing 12.2% of the genome. These CNAs occurred across the genome but there were some very frequent recurrent regions of CNA among the 72 tumours (Figure 1) including gains located on 1q, 3q, 6, 7q, 8q, 19, and 20 and losses on chromosomes 4, 6, 8, 13, 16, 17, 18, 22q and X. Within epithelial ovarian cancer histotypes we noted that mucinous and to a lesser extent clear cell cases appeared to have fewer CNAs and a smaller proportion of the genome was involved compared to the other subtypes (Figure S1). However, the numbers of samples in the minor subtypes were small, making it difficult to draw statistically valid conclusions about subtype specific changes. Most of the samples were of the serous or related high grade endometrioid subtype and many of the regions of gain and loss are primarily driven by these subtypes.
Table 1. Summary of samples analysed by SNP and expression array.
| Grade | FIGO Stage | |||||||
| Subtype | 1 | 2 | 3 | NK | 1 | 2 | 3 | NK |
| Clear Cell (9) | 2 | 2 | 3 | 2 | 3 | 1 | 2 | 3 |
| Endometrioid (14) | 2 | 4 | 8 | 0 | 8 | 2 | 4 | 0 |
| Mucinous (7) | 5 | 2 | 0 | 0 | 4 | 0 | 2 | 1 |
| Serous (37) | 3 | 11 | 20 | 3 | 3 | 10 | 17 | 7 |
| Undifferentiated (1) | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
NK, grade or stage not known. Information for 68 tumours that had both high quality expression and copy number data is listed here. Four more samples that were used in the copy number analyses alone are detailed in Table S1.
Figure 1. Overview of genomic aberrations in the ovarian cancer dataset (N = 72).
Frequency of occurrence of genomic gains (yellow) and losses (blue) across the genome, depicted in chromosome order from 1p to Xq.
Integration of mRNA expression in regions of frequent copy number gain
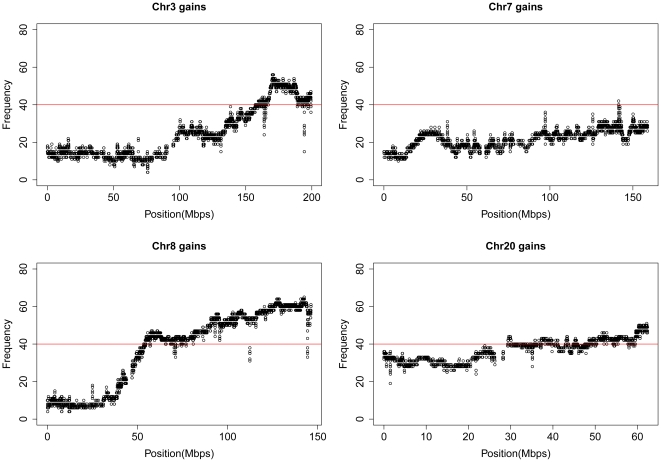
A common mechanism of activation of gene function in cancer development is through over expression as a consequence of gene amplification. While many genes may be located within a particular amplicon, the targeted gene(s) would be expected to consistently show elevated expression compared with adjacent bystander genes [5]. We have previously conducted an integrated expression analysis of candidate tumour suppressor genes within regions of loss of heterozygosity on an overlapping tumour cohort [6], thus for this study we chose to focus on the identification of candidate genes located within amplicons. An arbitrary frequency threshold of at least 40% was chosen as a filter for selecting key regions, resulting in the demarcation of multiple chromosomal regions on 3q, 7q, 8q and 20q (Figure 2). Each segment of frequent CN gain was labelled by the cytoband it belonged to; following which regions with the same cytoband tag were collapsed into one larger region (Figure S2-A). Those regions overlapping with germline copy number polymorphism (CNPs, Table S3) were excluded as described in Figure S2-B. The final 106 amplicons ranged in size from 11 kb to 7 Mb (Table S4) and 90 of these regions in total contained 1370 gene expression probesets on the Affymetrix Gene 1.0ST array corresponding to 938 known protein coding genes. The other 16 amplicons were not represented by probesets on the Gene 1.0ST arrays.
Figure 2. Detailed view of chromosomes showing frequent gains.
Frequent gains occur on chromosomes 3, 7, 8 and 20, with each point indicating the frequency of gain of a CN segment. The red line in all panels indicates the 40% frequency threshold.
Expression analyses were carried out for probesets within each of the 90 regions (Tables 2, 3, 4, Table S5). For each region groups of samples that showed copy number gain (3 or more copies) were tested for differential expression against groups of samples that showed normal copy number (∼2 copies). Across all regions, there were 703 (51%) differentially expressed probesets corresponding to 629 genes with unique identifiers such as an HGNC gene symbol or Ensembl ID (Table S5). Only one gene, hCG_16001, showed a negative log fold change (−0.34, Figure S3). On average (in regions with at least 5 probesets), 50% of the probesets were found to be differentially expressed suggesting a generalised increase in expression of genes within CN gains. Interestingly, we observed that MYC, an oncogene characterised by copy number gain in a wide variety of tumour types, was not significantly differentially expressed between amplified and unamplified groups of samples. One possibility is that MYC is expressed at a high level across all tumours irrespective of the copy number status and hence is not different between groups of tumours that show a gain and those that do not. To test this possibility we compared expression of MYC in amplified ovarian cancer samples to expression in normal fallopian tube epithelium. We did not find any increase in MYC expression when comparing tumours to these samples (p = 0.41, Welch corrected unpaired t-test, Figure S4).
Table 2. Genes with increased expression on chromosomes 3 and 7.
| Region ID | Chr | Start1 | End | Samples “G”2 | Samples “N”2 | DE Probesets (%)3 | Most significant DE Genes4 |
| 3_1 | 3 | 157.223 | 157.972 | 30 | 37 | 3 (60) | SSR3; TIPARP; KCNAB1 |
| 3_2 | 3 | 158.260 | 159.895 | 31 | 36 | 8 (62) | MLF1; GFM1; RSRC1; CCNL1; PTX3; VEPH1; LXN; SHOX2 |
| 3_3 | 3 | 159.895 | 159.959 | 30 | 37 | 2 (100) | RARRES1 |
| 3_4 | 3 | 159.959 | 161.006 | 32 | 35 | 2 (50) | MFSD1; SCHIP1 |
| 3_5 | 3 | 161.006 | 161.392 | 30 | 37 | 3 (75) | SCHIP1;IL12A |
| 3_7 | 3 | 161.392 | 168.660 | 33 | 35 | 8 (24) | KPNA4; SMC4; B3GALNT1; NMD3; TRIM59; hCG_16001; IFT80 |
| 3_8 | 3 | 168.697 | 168.916 | 37 | 31 | 1 (50) | PDCD10 |
| 3_9 | 3 | 168.916 | 169.209 | 38 | 30 | 2 (100) | PDCD10; SERPINI1 |
| 3_10 | 3 | 169.209 | 172.478 | 41 | 27 | 12 (40) | MYNN; PHC3; SKIL; MDS1; ARPM1;TLOC1; PRKCI; EVI1; EIF5A2; SLC7A14 |
| 3_12 | 3 | 172.586 | 177.095 | 39 | 29 | 2 (8) | ECT2; AADACL1 |
| 3_14 | 3 | 177.366 | 180.518 | 39 | 29 | 4 (27) | TBL1XR1; PIK3CA |
| 3_15 | 3 | 180.518 | 180.608 | 35 | 33 | 3 (100) | ZNF639; MFN1;GNB4 |
| 3_17 | 3 | 180.608 | 181.970 | 36 | 32 | 6 (43) | ACTL6A; MRPL47; NDUFB5; GNB4; LOC442098; TTC14 |
| 3_18 | 3 | 181.971 | 184.153 | 34 | 34 | 4 (57) | FXR1; DNAJC19; DCUN1D1; ATP11B |
| 3_19 | 3 | 184.153 | 184.291 | 35 | 33 | 2 (100) | DCUN1D1;MCCC1 |
| 3_20 | 3 | 184.291 | 185.996 | 34 | 34 | 18 (50) | ABCF3*; PSMD2; AP2M1; EIF4G1; PARL; ALG3; KLHL24; POLR2H; EIF2B5*; DVL3*; YEATS2; MAGEF1; MCCC1; LAMP3; ABCC5 |
| 3_22 | 3 | 186.007 | 187.399 | 36 | 32 | 5 (31) | SENP2; TMEM41A*; SFRS10; VPS8;EHHADH |
| 3_24 | 3 | 187.519 | 189.379 | 35 | 33 | 10 (42) | RFC4*; RPL39L*; DNAJB11; EIF4A2; TBCCD1; SNORA4; ST6GAL1; BCL6; RTP4 |
| 3_25 | 3 | 189.379 | 189.430 | 38 | 30 | 2 (100) | LPP; FLJ42393 |
| 3_27 | 3 | 193.766 | 193.936 | 35 | 33 | 1 (50) | FGF12 |
| 3_28 | 3 | 193.936 | 199.337 | 31 | 37 | 36 (46) | WDR53*; FBXO45*; NCBP2; LSG1; PIGX; RNF168; SENP5; OPA1; FYTTD1; CENTB2; UBXD7; PCYT1A; ATP13A3; KIAA0226*; DLG1 |
| 7_1 | 7 | 141.416 | 141.431 | 34 | 33 | 1 (100) | MGAM |
1. Start and end position of regions in Mbp based on hg18 (March 2006 release).
2. Number of samples with copy number gains (G) and normal copy number (N) used in the expression analysis. Samples with copy number loss were not included.
3. Number of differentially expressed (DE) probesets by expression microarray. Regions with no DE named genes are not shown here but are listed in Table S4.
4. Only coding genes with a symbol (from Affymetrix array annotation) are listed here and hence can differ from the number quoted in brackets. For a full list see Table S5. Genes for each region are listed in decreasing order of significance, with only the top 15 most significant listed.
*indicates genes with a correlation coefficient of >0.7.
Table 3. Genes with increased expression on chromosome 8.
| Region ID | Chr | Start1 | End | Samples “G”2 | Samples “N”2 | DE Probesets (%)3 | Most significant DE Genes4 |
| 8_1 | 8 | 53.390 | 55.545 | 29 | 39 | 11 (73) | ATP6V1H*; MRPL15; TCEA1; LYPLA1; RB1CC1; RGS20; NPBWR1; SOX17; UNQ9433 |
| 8_5 | 8 | 60.387 | 61.696 | 35 | 32 | 6 (60) | XKR4; TGS1*; TMEM68*; RP1 |
| 8_7 | 8 | 61.696 | 61.817 | 35 | 32 | 2 (100) | RAB2A; CHD7 |
| 8_13 | 8 | 62.549 | 65.928 | 33 | 33 | 3 (21) | RLBP1L1*; YTHDF3 |
| 8_15 | 8 | 66.237 | 68.051 | 33 | 33 | 14 (67) | ARMC1; VCPIP1; C8orf44; RRS1; SGK3; C8orf45; MYBL1; ADHFE1; MTFR1; C8orf46 |
| 8_16 | 8 | 68.051 | 68.292 | 29 | 36 | 6 (100) | COPS5; ARFGEF1; CSPP1 |
| 8_19 | 8 | 70.815 | 73.993 | 32 | 35 | 9 (41) | KCNB2; NCOA2; TRPA1; TRAM1; MSC |
| 8_21 | 8 | 74.016 | 78.270 | 34 | 33 | 8 (32) | TMEM70; STAU2; PXMP3; TERF1; UBE2W; TCEB1 |
| 8_25 | 8 | 80.419 | 84.683 | 36 | 31 | 10 (40) | CHMP4C; ZNF704; ZBTB10; SNX16; ZFAND1 |
| 8_27 | 8 | 85.122 | 87.055 | 34 | 33 | 16 (67) | C8orf59; REXO1L2P; REXO1L1; E2F5 |
| 8_30 | 8 | 87.250 | 89.422 | 34 | 33 | 5 (50) | WWP1; FAM82B; CPNE3; WDR21C; CNGB3 |
| 8_31 | 8 | 89.426 | 93.278 | 37 | 29 | 7 (35) | OTUD6B; NBN; TMEM55A; SLC26A7; RUNX1T1; TMEM64 |
| 8_33 | 8 | 93.587 | 98.637 | 38 | 28 | 17 (53) | UQCRB; TP53INP1; C8orf38; MTERFD1; PLEKHF2; PTDSS1; KIAA1429; RBM35A; INTS8; TSPYL5 |
| 8_34 | 8 | 98.637 | 99.159 | 35 | 32 | 7 (88) | MTDH; LAPTM4B; MATN2; RPL30 |
| 8_36 | 8 | 99.159 | 100.102 | 37 | 30 | 9 (82) | POP1*; NPAL2; STK3; VPS13B; HRSP12; OSR2; KCNS2 |
| 8_37 | 8 | 100.112 | 101.579 | 37 | 30 | 6 (55) | COX6C; RNF19A; POLR2K; VPS13B; FBXO43 |
| 8_38 | 8 | 101.579 | 101.675 | 36 | 31 | 2 (100) | ANKRD46; MGC39715 |
| 8_39 | 8 | 101.675 | 105.906 | 39 | 28 | 25 (50) | YWHAZ; WDSOF1; FLJ45248; ATP6V1C1; ZNF706; UBR5; FZD6; PABPC1; AZIN1; MGC39715 |
| 8_43 | 8 | 107.681 | 110.578 | 39 | 28 | 4 (31) | ENY2; TTC35; NUDCD1; OXR1 |
| 8_45 | 8 | 110.578 | 110.700 | 38 | 29 | 2 (67) | EBAG9; GOLSYN |
| 8_52 | 8 | 113.663 | 117.487 | 40 | 27 | 1 (25) | TRPS1 |
| 8_54 | 8 | 117.713 | 119.186 | 42 | 25 | 8 (89) | RAD21; C8orf53; MED30; EXT1; EIF3H; SLC30A8 |
| 8_56 | 8 | 119.298 | 121.983 | 44 | 23 | 6 (35) | MTBP;DCC1; TAF2; MRPL13; SAMD12; MAL2 |
| 8_59 | 8 | 122.661 | 122.935 | 40 | 27 | 1 (100) | HAS2 |
| 8_60 | 8 | 122.935 | 127.209 | 44 | 23 | 24 (60) | C8orf76*; RNF139; DERL1; ATAD2; TRMT12; NDUFB9; ZNF572; TMEM65; C8orf32; SQLE |
| 8_63 | 8 | 127.320 | 129.639 | 46 | 21 | 2 (25) | FAM84B |
| 8_66 | 8 | 129.735 | 131.499 | 47 | 20 | 4 (50) | FAM49B; MLZE; DDEF1 |
| 8_69 | 8 | 131.596 | 135.232 | 44 | 23 | 9 (45) | TG; OC90; KCNQ3; NDRG1; KIAA0143; PHF20L1; WISP1; SLA |
| 8_72 | 8 | 135.435 | 136.466 | 43 | 24 | 2 (67) | ZFAT1* |
| 8_76 | 8 | 137.616 | 139.944 | 42 | 25 | 2 (100) | COL22A1; FAM135B |
| 8_78 | 8 | 140.056 | 146.269 | 43 | 24 | 89 (74) | ZC3H3*; PUF60; GPR172A; CYHR1; SCRIB; HSF1 ; ZNF7*; MAF1; SHARPIN; BOP1 |
1.-4. Please see legend to Table 2, except that only the top 10 genes are listed and genes present in more than one region are only shown in one of these.
Table 4. Genes with increased expression on chromosome 20.
| Region ID | Chr | Start1 | End | Samples “G”2 | Samples “N”2 | DE Probesets (%)3 | Most significant DE Genes4 |
| 20_1 | 20 | 29.299 | 31.465 | 34 | 34 | 34 (62) | POFUT1; PDRG1; PLAGL2 ;ASXL1; TM9SF4; TPX2; CDK5RAP1; MAPRE1; COMMD7; KIF3B; C20orf112; RP11-49G10.8; DEFB118; DUSP15; DNMT3B |
| 20_2 | 20 | 31.466 | 31.648 | 29 | 39 | 2 (100) | CBFA2T2; SNTA1 |
| 20_4 | 20 | 31.649 | 33.694 | 31 | 37 | 34 (74) | PIGU; DYNLRB1; GGTL3; RBM12; RALY; NCOA6*;CEP250*; APBA2BP; TRPC4AP; EIF6; EDEM2; GSS; UQCC; PXMP4; EIF2S2 |
| 20_5 | 20 | 33.696 | 33.760 | 36 | 32 | 5 (100) | RBM12; NFS1; RBM39; C20orf52 |
| 20_6 | 20 | 33.958 | 37.049 | 29 | 39 | 38 (64) | CTNNBL1*; LOC388796; KIAA0406P; DHX35*; C20orf77; ACTR5; MANBAL; FAM83D; DSN1; RBL1; C20orf198; RPN2; SCAND1; C20orf117; C20orf24 |
| 20_7 | 20 | 37.107 | 41.095 | 29 | 37 | 11 (65) | PLCG1; CHD6; LPIN3; TOP1; PTPRT; LOC149692; ZHX3; EMILIN3; MAFB |
| 20_8 | 20 | 41.095 | 41.113 | 30 | 36 | 1 (100) | PTPRT |
| 20_9 | 20 | 41.124 | 41.226 | 28 | 38 | 2 (100) | PTPRT |
| 20_10 | 20 | 42.962 | 45.772 | 30 | 37 | 45 (62) | PIGT; UBE2C; ZSWIM1; TOMM34; DNTTIP1*; NCOA5; SLC35C2; ACOT8; NEURL2; KCNS1; C20orf67; SNX21; ELMO2; ZMYND8; TP53RK; |
| 20_12 | 20 | 45.850 | 49.180 | 29 | 38 | 23 (61) | TMEM189*; MOCS3*; DPM1; STAU1; DDX27; CSE1L; ARFGEF2; ADNP; SPATA2*; PTPN1; SLC9A8*; C20orf199; PARD6B; ZNF313; KCNG1 |
| 20_14 | 20 | 49.222 | 54.379 | 30 | 37 | 10 (42) | ZFP64*; AURKA; PFDN4; ATP9A; MC3R; TSHZ2; SUMO1P1 |
| 20_15 | 20 | 54.379 | 54.417 | 27 | 41 | 2 (100) | CSTF1; AURKA |
| 20_16 | 20 | 54.417 | 55.828 | 31 | 36 | 8 (32) | C20orf43; RAE1; BMP7; RBM38; GCNT7 |
| 20_18 | 20 | 55.991 | 57.887 | 33 | 34 | 12 (57) | VAPB; TUBB1; RAB22A; TH1L; SLMO2; STX16; ATP5E; GNAS; SYCP2; PPP4R1L; NPEPL1 |
| 20_20 | 20 | 57.901 | 62.427 | 33 | 34 | 38 (45) | LSM14B*; YTHDF1; SS18L1; DIDO1; GTPBP5; PSMA7; TAF4; C20orf11; C20orf20; TCFL5; C20orf177; MYT1; PCMTD2; DNAJC5; TPD52L2; |
1.-4. Please see legend to Table 2.
To further refine this list of 703 copy number driven, differentially expressed probesets, we reasoned that those genes showing the strongest correlation of copy number and expression may be the most likely genes targeted by the CN gain. Thus, we calculated the correlation co-efficient for all differentially expressed genes with copy number probeset coverage in the candidate amplicons (Table S5). Of the 692 probesets tested (11 did not contain copy number probes), 219 (corresponding to 206 protein-coding genes) showed a strong positive correlation (r≥0.6) between expression and copy number.
Genes targeted by high CN amplification
Our main approach to identify cancer-related genes was to filter for the most frequent aberrations but we noted that well characterised cancer driver genes, such as CCNE1 and ERBB2 [7], were not identified since they were amplified in less than 40% of tumours. Rather than using a lower cut-off which would risk including many regions altered due to generalised genomic instability (for example ∼67% of the genome would be considered as candidate regions if a cut-off of >10% was used), we instead filtered for genes showing a high amplitude CN gain. Here, we looked at all segments that had a copy number greater than or equal to 5 and were present in at least 5 samples, which identified 21 regions over 27.2 Mb (Table 5). These regions corresponded to 181 gene expression probesets on our Affymetrix Gene 1.0ST arrays, of which 39 (22%) had a strong positive correlation between CN and gene expression (r>0.6). These probesets corresponded to 32 known protein coding genes including well known cancer driver genes such as ERBB2 (Table S6).
Table 5. Highly amplified genes.
| Chr | Start (Mb) | End (Mb) | Length (bp) | No. samples | Genes1 |
| 3 | 170.040 | 170.248 | 208141 | 6 | None |
| 3 | 178.305 | 178.589 | 283690 | 5 | TBL1XR1 |
| 3 | 180.121 | 180.410 | 288435 | 5 | PIK3CA; ZMAT3 |
| 8 | 55.208 | 55.528 | 319922 | 5 | MRPL15* |
| 8 | 62.495 | 63.491 | 995369 | 6 | RLBP1L1*;NKAIN3;ASPH |
| 8 | 102.003 | 102.062 | 58823 | 5 | YWHAZ |
| 8 | 123.144 | 123.746 | 601615 | 6 | None |
| 8 | 123.856 | 124.369 | 513120 | 6 | DERL1; ZHX2; WDR67*; ZHX1*; C8orf76*; FAM83A |
| 8 | 124.369 | 125.825 | 1455953 | 6 | ATAD2; C8orf32*; FBXO32; ANXA13; KLHL38; FAM91A1; FER1L6; MTSS1; NDUFB9; RNF139; TATDN1; TMEM65; TRMT12* |
| 8 | 125.828 | 127.764 | 1936500 | 7 | KIAA0196; NSMCE2; SQLE; ZNF572*; TRIB1; FAM84B* |
| 8 | 127.764 | 128.973 | 1208920 | 7 | MYC; POU5F1P1 |
| 8 | 128.973 | 130.166 | 1193146 | 8 | PVT1†, TMEM75* |
| 8 | 130.166 | 138.988 | 8821634 | 7 | ADCY8; DDEF1; EFR3A; FAM49B; KCNQ3; MLZE; OC90; LRRC6; NDRG1; PHF20L1; SLA; TG*; TMEM71; WISP1; ST3GAL1; ZFAT*; KHDRBS3; CCDC26 |
| 8 | 138.988 | 144.000 | 5382420 | 7 | FAM135B; COL22A1; KCNK9; NIBP*; CHRAC1*; EIF2C2; PTK2; DENND3*; SLC45A4; FLJ43860; GPR20; PTP4A3; ARC; BAI1; C8orf55; CYP11B1; CYP11B2; GML; JRK; LY6D; LY6K; LYNX1; LYPD2; PSCA; SLURP1; TSNARE1 |
| 17 | 35.104 | 35.105 | 529 | 5 | ERBB2* |
| 19 | 34.125 | 34.639 | 513414 | 5 | UQCRFS1* |
| 19 | 34.639 | 35.610 | 971542 | 6 | C19orf12*; PLEKHF1; POP4*; CCNE1; C19orf2; ZNF536* |
| 19 | 35.968 | 36.703 | 734619 | 6 | TSHZ3* |
| 19 | 37.459 | 38.011 | 552023 | 5 | ANKRD27*; PDCD5*; RGS9BP; ECAT8; DPY19L3*; ZNF507* |
| 19 | 38.372 | 39.140 | 767924 | 5 | CEBPA; LRP3; SLC7A10; CHST8; KCTD15; CEBPG*; PEPD*; FLJ12355 |
| 20 | 29.427 | 29.849 | 421241 | 5 | BCL2L1; COX4I2; DEFB119; DEFB121; DEFB123; DEFB124; HM13; ID1; REM1; TPX2 |
1. Derived from Refseq annotation (September 2009). Genes in italics are known oncogenes (based on Cancer Gene Census [38]), *Genes that show a strong (r>0.6) positive correlation of copy number with expression, †Not on expression microarray. Note that some regions encompass multiple smaller amplicons, only genes within regions (+/−10 kb) defined by >5 samples are shown.
Prioritising candidate driver genes
In order to prioritise the most promising candidates from the previous analyses, we built a gene list using the following criteria. Firstly, we selected those known genes with a high frequency of gain (>40%), that were differentially expressed (n = 629). From this list we selected the genes most strongly over expressed by the level of log fold change (>0.7) between samples with CN gain and samples that were neutral at the locus (n = 59). As a different measure of how gene expression was affected by copy number, we also selected genes that showed a strong correlation (>0.7) of copy number and expression (n = 58). The union of these criteria produced a list of 110 genes. From this list, we identified genes on each chromosome that were the most frequently affected by copy number change; for chr8, this included genes with a frequency of ≥60%, for chr3, ≥50% and for chr20 ≥42%. This list comprised 37 genes (Table 6).
Table 6. Candidate oncogenes and current literature.
| Gene | Chr | Start | End | Total gain (%) | Comments | Other genes in region |
| PDCD10 | 3 | 168.884 | 168.935 | 43 | Angiogenesis disorder [39], ERK pathway [40] | |
| PRKCI | 3 | 171.423 | 171.506 | 51 | Oncogene in ovarian and other cancers [41], [42] | SKIL, PHC3, MYNN |
| ECT2 | 3 | 173.955 | 174.022 | 50 | Cytokinesis [43]. Transforming protein [44]. Interacts with PRKCI [45] | |
| TBL1XR1* | 3 | 178.221 | 178.398 | 50 | Oncogene in breast cancer [46], transcriptional repressor [47] | |
| PIK3CA* | 3 | 180.349 | 180.435 | 50 | Known oncogene | MRPL47, NDUFB5 |
| SENP2 | 3 | 186.787 | 186.832 | 51 | SUMO1 deconjugating peptidase. Possible role in degradation of beta-catenin [48]. | TMEM41A |
| MRPL15* | 8 | 55.210 | 55.224 | 42 | Mitochondrial ribosomal protein [49] | |
| RLBP1L1* | 8 | 62.363 | 62.577 | 46 | Clavesin 1 (CLVS1), regulates endosome morphology [50], upregulated in liver cancer [51] | |
| YWHAZ* | 8 | 102.000 | 102.035 | 53 | 14-3-3 isoform zeta, oncogenic functions in inhibiting apoptosis and adhesion [52] | |
| DERL1* | 8 | 124.095 | 124.124 | 60 | Endoplasmic reticulum protein [53] with role in stress response. Elevated expression in cancer [54], [55] | WDR67*, C8orf76* |
| ATAD2* | 8 | 124.401 | 124.478 | 60 | ATPase. E2F target, binds MYC, expression correlates with poor outcome in breast cancer [56]. Interacts with ER and AR and is required for target gene expression [57] | WDYHV1/C8ORF32*, FBXO32*, FAM91A1* |
| RNF139* | 8 | 125.556 | 125.570 | 60 | Translocation causes hereditary renal cancer. Interacts with VHL [58] | NDUFB9*, TRMT12*, TMEM65*, SQLE* |
| FAM84B* | 8 | 127.634 | 127.640 | 61 | – | |
| FAM49B* | 8 | 130.923 | 131.021 | 61 | – | |
| NDRG1* | 8 | 134.319 | 134.379 | 60 | Diverse role in stress response including hypoxia [59]. Fusions with ERG in prostate cancer [60]. | |
| ZFAT* | 8 | 135.559 | 135.794 | 60 | Zinc finger and AT hook protein, anti-apoptotic role [61] | |
| PTK2* | 8 | 141.738 | 142.081 | 60 | Focal adhesion kinase. Involved in signal transduction for proliferation[62] | CHRAC1*, NIBP/TRAPPC9*, SLC45A4* |
| PTP4A3* | 8 | 142.501 | 142.511 | 60 | Protein tyrosine phosphatase. Increases proliferation and metastasis [63] | JRK*, TSTA3, ZC3H3, LY6E |
| PUF60 | 8 | 144.971 | 144.984 | 60 | mRNA splicing factor [25] | CYC1, ZNF623, ZNF7, CYHR1 |
| ERBB2* | 17 | 35.098 | 35.138 | Known oncogene in breast cancer | ||
| TPX2* | 20 | 29.791 | 29.853 | 42 | Activator of Aurora-A and involved in spindle assembly [30]. Interacts with BRCA1/BARD1 [64] | |
| UBE2C | 20 | 43.875 | 43.879 | 42 | Ubiquitin-conjugating enzyme E2C, degradation of mitotic cyclins and cell cycle progression [65] | PIGT |
| ZFP64 | 20 | 50.134 | 50.242 | 43 | Zinc finger protein, Notch signalling [66] | |
| AURKA | 20 | 54.378 | 54.401 | 43 | Aurora kinase, cell cycle regulation, chromosome segregation, microtubule/spindle function [67] | CSTF1, RAE1, C20orf43 |
| SS18L1 | 20 | 60.152 | 60.191 | 46 | Synovial sarcoma translocation fusion gene [68]); calcium-responsive transactivator [67] | GTPBP5, LSM14B, TAF4 |
Genes were selected as follows: Gain in >40% and differentially expressed, with fold change expression in gain vs. neutral of >0.7 or correlation coefficient (r) of >0.7. Of these genes (n = 121), the most frequently gained in each chromosome were selected: Chr 3 n≥50, Chr8 n≥60, Chr 20 n≥42. 2. High level amplification in at least 5 samples (*), and differentially expressed, with fold change expression in gain vs. neutral of >0.6 or correlation coefficient (r) of >0.6. Chr19 genes (n = 12) are not shown here.
Secondly, we also wished to include genes that were highly amplified. From our list of highly amplified genes in at least 5 samples we selected those that had a strong positive correlation between copy number and expression (r>0.6, n = 32). Some of the genes that were highly amplified were also differentially expressed based on the expression analysis of frequently gained regions, so we also included genes with a log fold change greater than 0.6 (n = 17). Taking genes satisfying one or the other of these criteria, we added 41 genes to our high priority list (Table 6).
When we combined these two gene lists, the first based on “high frequency” and the second on “high amplitude” but both with increased expression, the final number of unique genes was 70 (Table 6).
Discussion
Gene expression analysis has been widely used to identify key pathways and clinically important subgroups in ovarian cancer but identification of specific driver genes using this methodology alone has been hampered by the fact that expression is rather plastic and there has been little consensus in the genes identified between such studies [1], [8]. One reason for this lack of consistency is that most studies have analysed RNA from whole tumour samples without verification of the percentage cancer epithelium and/or have used diverse control tissues such as whole ground ovary [9]. In contrast to gene expression, genomic alterations may be a more stable and reliable predictor of the location of driver genes. Ovarian cancer has long been suspected to be cytogenetically complex [10] and recent advances in genomics technology has confirmed the profound genomic aberrations that characterise most ovarian cancers [4], [11], [12], [13]. Despite this complexity, published copy number profiles of ovarian cancers are highly comparable at a global level [3] and many studies have identified very similar regions of frequent copy number alteration. However, progress at identifying key driver genes has been slow, with different studies often identifying different candidates in the same genomic region. For example, the chromosome 20 amplicon driver has variously been suggested to be ADRM1 [14], EYA2 [15], AURKA and ZNF217 [16], among several others. Early studies integrating expression and copy number data have either used cancer cell lines to identify over expressed genes [17], [18] and/or microarray platforms with limited resolution and genome coverage [19], [20]. To date few studies have exploited a truly genome-wide integrated copy number and expression analysis on matched samples for the unbiased identification of candidate genes [21], [22], [23] and there has only been one previous study of a smaller cohort of ovarian tumours [12]. In this study we have therefore attempted to circumvent some of the issues of examining expression or copy number in isolation by integrating two data sets obtained from microdissected tumour epithelial cells.
As a first pass of the data we focussed on gains occurring in a very high proportion of cases which included regions of chromosomes 3, 7, 8 and 20. Identification of differentially expressed genes reduced our list of candidate cancer genes in these regions by approximately half (range 6–89% for regions with at least 5 probesets). We have validated several of the genes identified in Haverty et al., for example, on 3q26.2 we confirmed increased expression in 7/8 of their genes. However, we have also identified a number of additional amplified and over expressed genes (Tables 2, 3, 4), most likely due to differences in our method and larger sample size. The proportion of differentially expressed genes in our study is consistent with previous studies of other cancer types [24] supporting the concept that copy number can have a strong influence on gene expression. Consequently, for many regions we were not able to identify one particular driver gene. It is possible that there may truly be many driver genes within each amplicon and although each may individually contribute little to cancer progression, coordinate over expression of these genes in amplified regions may have an additive or synergistic oncogenic effect. Alternatively, many of the differentially expressed genes may be passengers whose over expression endows no selective advantage or disadvantage to the tumour. Discriminating between passengers and drivers within a genomic region may therefore only be achieved through large-scale functional analyses and combinatorial approaches examining many genes in concert.
Despite the relatively large number of amplified and differentially expressed genes identified in this study, we still hypothesise that those genes showing the strongest over expression, and also those genes with the highest amplitude copy number gains, may be more likely to be drivers of tumorigenesis than weakly over expressed genes. Hence, we prioritised our gene list using stringent expression criteria. For example, one of the genes most frequently targeted by copy number that is strongly over expressed is PUF60 (poly-U binding splicing factor 60 kDa). This gene encodes for a pre-mRNA splicing factor thought to be involved in the recognition of 3′ splice sites [25]. It may also inhibit transcription by interacting with the TFIIH helicase, the key factor mutated in the cancer-prone syndrome xeroderma pigmentosum, and this interaction is implicated in the correct regulation of MYC transcription [26], [27].
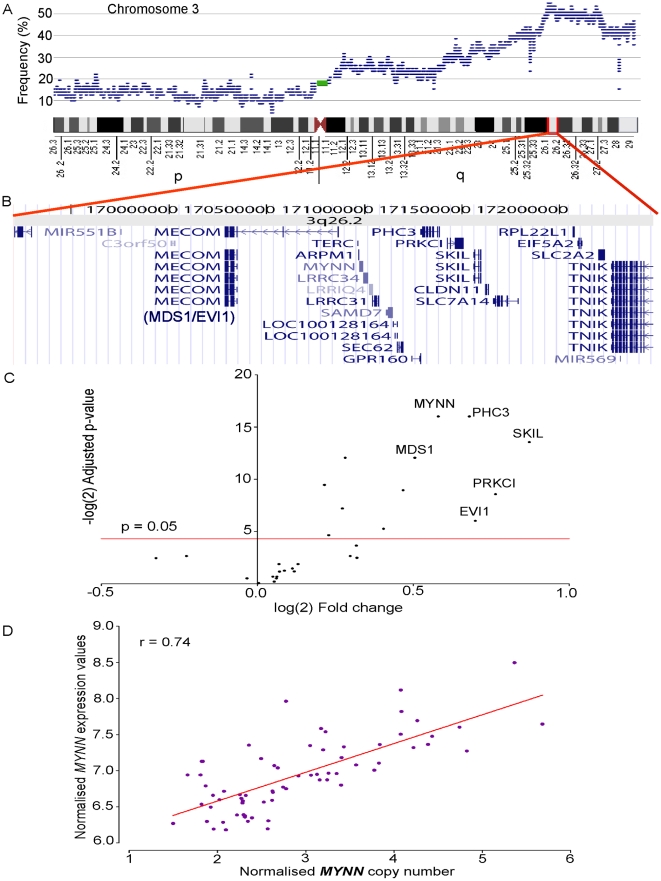
Myoneurin or MYNN is a gene that is located in a region of frequent (60%) copy number gain on 3q26.2. It is differentially expressed (adjusted p = 1.51E-05) between amplified and unamplified groups, and shows the strongest correlation between copy number and expression (r = 0.74, Figure 3) amongst all genes in this region. This gene was identified as a member of the Broad complex, Tramtrack, Bric a' brac (BTB) or poxvirus and zinc finger (POZ)-ZF i.e BTB/POZ-ZF family of transcription factors [28]. First discovered in Drosophila, this family consists of about 60 human proteins including several cancer related proteins such as leukaemia related factor (LRF/ZBTB7) and B-cell lymphoma 6 (BCL6). While the role of MYNN in cancer is yet to be characterised, other members of this family are similarly overexpressed in tumors [29].
Figure 3. Correlation between copy number and expression for a frequently gained region on cytoband 3q26.2.
A. Frequency of copy number gain on chromosome 3 from p-ter at left to q-ter at right as indicated by the ideogram. B. Genes on Chr3: 169.209–172.478 Mbp, a region gained in 60% (41/68) of all samples, including genes previously associated with ovarian cancer (PRKCI, MECOM or MDS1/EVI1) and potentially novel oncogenes (MYNN). C. A volcano plot presenting the results of expression analyses between amplified and unamplified samples in this region. The genes in the top right corner are significantly overexpressed in samples with copy number gain (p<0.05; above the red line at –logP 4.32) compared to samples without copy number change (selected genes are labelled). For full list of differentially expressed genes see Table S5. D. Plot comparing copy number and expression in all samples for the gene MYNN that showed the highest correlation (r = 0.74, Pearson's test) between copy number and expression for this region on 3q26.2.
As well as identifying high frequency, differentially expressed genes, including known cancer genes such as PIK3CA and AURKA, we also used high amplitude regions to locate additional known (e.g. ERBB2 and CCNE1) and potential oncogenes. For example, on chromosome 20, the high-amplitude approach identified a small minimal region that was not evident from the low-amplitude analysis. This 421 kb interval at 20q11.21 encompasses 10 genes, of which TPX2 showed the strongest correlation with copy number (r = 0.53). This gene was also differentially expressed between samples with any TPX2 gain and those with normal TPX2 copy number, and had the strongest fold change of any gene on chromosome 20 (log2 fold change of 1.03). The protein encoded by this gene functions as an activator of Aurora-A with a role in spindle assembly [30]. Interestingly for ovarian cancer, it has been shown to interact with the BRCA1/BARD1 complex (15). Recently, it has been identified as a potential oncogene in pancreatic cancer [31].
In summary, our study shows that combining the high frequency and high amplitude analyses and targeting the most strongly over expressed genes reduced the candidate list to just 70 genes out of the many thousands targeted by copy number change alone. We have identified many promising candidate genes not previously noted in ovarian cancer, particularly genes such as MYNN, TPX2 and PUF60. It should be noted, however, that our method of analysis is one of many that can be employed in the identification of novel cancer genes, and is unlikely to have identified all possible candidates. The example of MYC, not strongly expressed in our data but previously shown to have a functional effect in ovarian cancer cell lines [32], clearly indicates that our approach should be considered complementary to others such as functional screens and deep sequencing of primary cancer samples. Nevertheless our data provides an important platform from which to rationally pursue the validation of these potential dominant drivers of ovarian tumorigenesis. In addition, this list may include genes that are valid candidates for diagnostic or therapeutic purposes.
Materials and Methods
Ethics Statement
All samples were collected with the donor's written informed consent. This study was approved by the Peter MacCallum Cancer Centre Human Research Ethics Committee (Protocol number 01/38).
Sample collection
Tumour biopsies were obtained from 72 patients who were undergoing surgery for primary ovarian cancers (a) at hospitals in the Wessex region of Southeast England, UK and (b) in hospitals in Victoria, Australia (accessed through the Peter MacCallum Cancer Centre Tissue Bank). Blood was collected from the same patients for matching lymphocytes. Fallopian tube samples were collected through the tissue bank from BRCA1 or BRCA2 mutation carriers undergoing prophylactic bilateral salpingo-oophorectomy in hospitals around Melbourne. The accrual and use of patient samples related to this project were approved by the relevant institutional ethics committees. Clinical and histopathological information about the samples are provided in Table 1 and Table S1.
DNA and RNA extraction
Fresh-frozen tissue was embedded in Optimal Cutting Temperature Compound (OCT, Sakura Finetek, Torrance, CA) and cut into 10 µm sections. Tumour DNA and tumour and fallopian tube RNA were extracted from identical regions after needle micro-dissection of >80% tumour epithelial cells. Sections for RNA were stained using Cresyl violet and RNA was extracted using Ambion mirVana total RNA extraction protocol (Applied Biosystems/Ambion, Austin, TX). Tissue sections used for DNA extraction were stained with haematoxylin and eosin and DNA was extracted using the Qiagen Blood and Tissue Kit (Qiagen, Valencia, CA, USA). DNA from matching normal lymphocytes for samples from the Peter MacCallum Cancer Centre Tissue Bank were extracted using the same kit. DNA from matching normal lymphocytes for samples from Southampton were extracted as described previously [33].
Microarray data generation and quality control
500 ng of DNA from each tumour sample was analysed using the Affymetrix Genome-wide Human SNP Array 6.0 (SNP6.0) following the manufacturer's instructions (Affymetrix, Santa Clara, CA). Where available (57 cases) DNA from matching peripheral blood lymphocytes was analysed on the same platform and in the same batch. For mRNA expression, 300 ng of total RNA from the same tumour samples were analysed using the Affymetrix Human Gene1.0 ST Array. Analysis of array performance for SNP6.0 arrays was performed using genotyping call rates (>90% call rate required) and also visual inspection of copy number traces to remove noisy samples. 72 samples passed quality control measures and were used in the copy number analysis. For expression arrays, the profiles of hybridisation controls, spike-in controls and positive-versus-negative area under the curve (AUC) were assessed using Affymetrix Expression Console. Additionally, the quality of the arrays was assessed based on Relative Log-Likelihood (RLE) and Normalised Unscaled Standard Errors (NUSE) criteria generated using the “affyPLM” package in the R open-source software. Expression arrays that were flagged as dubious by 2 out of 3 measures (AUC, RLE, NUSE) were excluded from expression analyses. 68 tumour samples (57 with normal DNA) passed for both expression and copy number and were retained in the integrated expression analyses. The final sample set in the integrated analysis included the four most commonly seen histological subtypes of ovarian cancer – serous (n = 37), endometrioid (n = 14), mucinous (n = 7) and clear cell (n = 9). One sample in the study was of unknown histotype (Table 1). Both gene expression and copy number data are MIAME compliant and have been submitted to the National Centre for Biotechnology Information's (NCBI) Gene Expression Omnibus (GEO) website, series accession number GSE19539.
Copy number analysis
Copy number generation and analyses were performed using Partek® Genomics Suite™ version 6.03 (Partek Inc., St. Louis, Missouri) and Bioconductor packages in the R-open source software framework [34], [35]. SNP 6.0 CEL files were imported into Partek using default settings for background correction and summarisation. Human Genome Build 36.1 (hg18, March 2006) was used for base pair locations. Probeset copy number ratios were calculated by comparing each tumour with its matching normal when available (n = 57). For samples that did not have matching normal data (n = 15), a pooled normal baseline from all the other normal samples was used. Circular binary segmentation [36] was performed using the R-based package “DNAcopy” to segment the data into distinct regions of change using default package settings. This analysis produced a list of regions per sample that was then filtered for those regions that showed gain (copy number ratio >2.5) or loss (copy number ratio <1.5) across ≥40% (n≥29) of all samples. These regions were collapsed into cytobands for easier data manipulation (Figure S2 for more detail). It is important to note that since these regions have undergone filtering steps defined above, they do not include the entire cytoband by which they are represented and hence the high resolution of the data is not compromised.
To identify potential germline copy number polymorphisms (CNP) that could interfere with accurate identification of somatic changes, copy number data for 57 normal samples was generated relative to a pooled baseline of all normal samples. Regions showing gain or loss in >5% of all samples were called as CNPs (Table S3). Regions of interest from the tumour data were scanned for these CNPs and matches were removed from downstream analyses (Figure S2-B). CNP-removed, cytoband-collapsed regions were queried against the entire copy-number dataset to generate accurate, region-wise values of copy number.
Copy number was extracted on a gene-by-gene basis to perform Pearson correlation analysis with expression. Since some genes were so small that there were no copy number probesets mapping to them, an additional 10 kb was added to all gene start and stop positions before extracting their copy number.
Expression microarray analysis
For each candidate region, samples were divided into two groups, G – consisting of all samples that showed gain (>3 copies) on the SNP6.0 platform; and N – consisting of all samples that showed normal copy number (1.5–2.5 copies). A test for differential expression was performed between these two groups using the “limma” package available on the R-open source software platform [34]. Histological subtype was included as a factor in the analysis. Genes were considered to be significantly differentially expressed with a p-value of <0.05 after multiple testing correction [37]. A Pearson's correlation analysis between copy number and expression was also performed. Separate analyses were performed on a gene-by-gene basis for all genes within (a) most frequently amplified regions (CN≥3; Freq≥40%) and (b) most highly amplified regions (CN≥5; Freq≥7%).
Supporting Information
Sample details. Clinicopathological features and assay information for each sample. 57 out of 72 tumours had matching lymphocytic DNA available for copy number microarray analysis.
(0.06 MB PDF)
Proportion of genome-wide gain and loss by sample. In all of these samples, the aberrant genome adds up to 95.4% on average. The missing 4.6% can be attributed to regions on chromosome Y, Mitochondrial DNA and repetitive sequences around centromeric regions that are either removed from the segmentation analysis or not covered by the Affymetrix SNP6.0 array.
(0.06 MB PDF)
Germline copy number polymorphisms on Chr 3, 7, 8, 20. The regions/segments of copy number gain that contained one or more of these CNPs were removed or altered as displayed in Figure S1-B. The type of CNP is also displayed in the far right column.
(0.05 MB PDF)
Regions of gain present in >40% of samples. This table contains genomic information for the 90 regions included in the expression analyses, i.e., all those regions that mapped to 1 or more probesets on the Human GeneST1.0 microarrays. On this microarray platform, most probesets map uniquely to a protein-coding gene. The region IDs correspond to those in Tables 2, 3, 4 and S5.
(0.13 MB PDF)
All differentially expressed probesets in frequent regions of gain. Every probeset tested for differential expression is listed and tagged by the region it belongs to. These region IDs are consistent across all tables in the paper and are derived as shown in Figure S1-A. Column 5 displays the Pearson's correlation between copy number and expression for the listed probeset. Columns 6–11 are derived from differential expression analyses performed using the “limma” package in R.
(0.06 MB PDF)
Correlation for all genes highly amplified (CN>5) in at least 5 samples. This table displays Pearson's correlation between copy number and gene expression for all 181 probesets in regions of high CN gain across the genome. The p-value displayed is a raw p-value obtained while testing for correlation. * Genes highly amplified in 4 samples but that were within 10 kb of a copy number breakpoint of 5 amplified samples.
(0.03 MB PDF)
Subtype breakdown of genome wide CN changes. (A) Overall copy number landscape for the cohort of ovarian cancer samples. This is similar to Figure 1 with the exception that the y-axis ranges from 0–100% of samples as opposed to 0–50%. Below are the distribution of copy number changes for (B) 37 serous ovarian cancers, (C) 14 endometrioid ovarian cancers, (D) 7 mucinous ovarian cancers and (E) 9 clear cell ovarian cancers. A, B and C jointly show that the major contributors for the high frequency changes are serous and endometrioid tumours. Data for the single tumor classified as undifferentiated is not shown here.
(0.43 MB TIF)
‘Cytoband collapsing’ and the exclusion of CNPs. (A) Shows the steps taken towards obtaining the copy number regions. The starting data (far left) contains genomic position and copy number information for segmental overlaps. All segments at this step of analysis occur with >40% frequency and have 3 or more copies. Letters a, b, r, s, t, u, v and w refer to genomic start/stop sites in basepairs. Regions are sorted by chromosome, then by genomic start and finally by genomic stop positions. Following this they are annotated with their cytobands and the newly defined “collapsed” region is bounded by the lowest start (a) and highest stop (b) positions and annotated with the cytoband of origin. The ‘a’ and ‘b’ from here carry through to part B of the figure. Regions that span two cytobands are listed as a separate group as shown in Table S4. (B) Shows the rules used to eliminate CNPs from the cytoband regions. Regions such as “Amp 4” are split into two, resulting in more regions after CNP elimination than before. (C) Regions of CNP across the genome and their position in relation to regions of copy number gain relevant to our study. (i) Global changes in normal (n = 57, green = gain and red = loss) and tumour (n = 72, yellow = gain and blue = loss) samples. We define a CNP as a change that occurs in at least 5% of normal samples. CNPs often show both genomic gain and loss at the same locus in normal samples. (ii) All changes on Chromosome 3 and in particular a CNP on 3q26.1 between 168.66 and 168.69 Mbp highlighted by the black oval, observed in >15% of all normal samples. (iii) The 3q26.1 CNP occurs in the middle of a region of copy number gain that we investigate further. This CNP region was removed from the data in accordance with S2-B.
(0.55 MB PDF)
Expression of all genes in regions of frequent copy number gain. This figure displays all genes in 90 regions of copy number change in terms of their average expression and t-statistic, resulting from the test for differential expression for each of these regions between amplified and unamplified samples. Genes showing a significant differential expression are represented by red dots and non-significant genes are represented by purple dots. Only one gene hCG_16001 showed a significant reduction in expression under the influence of copy number gain. This is a ribosomal protein L23a pseudogene 42 (RPL23A42) where RPL23A encodes a ribosomal protein that is a component of the 60S subunit and may be one of the target molecules involved in mediating growth inhibition by interferon.
(0.09 MB TIF)
Expression of MYC across various sample groups. RMA normalised expression of MYC based on Gene 1.0 ST array data. No significant differences were found between groups of samples that showed copy number gain in the region and those that did not.
(0.15 MB TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: MR is supported by the Cancer Council Victoria Postgraduate Scholarship. This work is funded by a grant from the National Health and Medical Research Council (NHMRC) of Australia (ID: 566603). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gyorffy B, Dietel M, Fekete T, Lage H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int J Gynecol Cancer. 2008;18:1215–1233. doi: 10.1111/j.1525-1438.2007.01169.x. [DOI] [PubMed] [Google Scholar]
- 2.Tothill RW, Tinker AV, George J, Brown R, Fox SB, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 3.Gorringe KL, Campbell IG. Large-scale genomic analysis of ovarian carcinomas. Mol Oncol. 2009;3:157–164. doi: 10.1016/j.molonc.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorringe KL, Jacobs S, Thompson ER, Sridhar A, Qiu W, et al. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13:4731–4739. doi: 10.1158/1078-0432.CCR-07-0502. [DOI] [PubMed] [Google Scholar]
- 5.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 6.Gorringe KL, Ramakrishna M, Williams LH, Sridhar A, Boyle SE, et al. Are there any more ovarian tumour suppressor genes? A new perspective using ultra high-resolution copy number and loss of heterozygosity analysis. Genes Chromosomes Cancer. 2009;48:931–942. doi: 10.1002/gcc.20694. [DOI] [PubMed] [Google Scholar]
- 7.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 8.Israeli O, Goldring-Aviram A, Rienstein S, Ben-Baruch G, Korach J, et al. In silico chromosomal clustering of genes displaying altered expression patterns in ovarian cancer. Cancer Genet Cytogenet. 2005;160:35–42. doi: 10.1016/j.cancergencyto.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Zorn KK, Jazaeri AA, Awtrey CS, Gardner GJ, Mok SC, et al. Choice of normal ovarian control influences determination of differentially expressed genes in ovarian cancer expression profiling studies. Clin Cancer Res. 2003;9:4811–4818. [PubMed] [Google Scholar]
- 10.Taetle R, Aickin M, Yang JM, Panda L, Emerson J, et al. Chromosome abnormalities in ovarian adenocarcinoma: I. Nonrandom chromosome abnormalities from 244 cases. Genes Chromosomes Cancer. 1999;25:290–300. [PubMed] [Google Scholar]
- 11.Gray JW, Suzuki S, Kuo WL, Polikoff D, Deavers M, et al. Specific keynote: genome copy number abnormalities in ovarian cancer. Gynecol Oncol. 2003;88:S16–21; discussion S22-14. doi: 10.1006/gyno.2002.6677. [DOI] [PubMed] [Google Scholar]
- 12.Haverty PM, Hon LS, Kaminker JS, Chant J, Zhang Z. High-resolution analysis of copy number alterations and associated expression changes in ovarian tumors. BMC Med Genomics. 2009;2:21. doi: 10.1186/1755-8794-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 14.Fejzo MS, Dering J, Ginther C, Anderson L, Ramos L, et al. Comprehensive analysis of 20q13 genes in ovarian cancer identifies ADRM1 as amplification target. Genes Chromosomes Cancer. 2008;47:873–883. doi: 10.1002/gcc.20592. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Yang N, Huang J, Buckanovich RJ, Liang S, et al. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumour growth. Cancer Res. 2005;65:925–932. [PubMed] [Google Scholar]
- 16.Watanabe T, Imoto I, Katahira T, Hirasawa A, Ishiwata I, et al. Differentially regulated genes as putative targets of amplifications at 20q in ovarian cancers. Jpn J Cancer Res. 2002;93:1114–1122. doi: 10.1111/j.1349-7006.2002.tb01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidenblad M, Lindgren D, Veltman JA, Jonson T, Mahlamaki EH, et al. Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic amplifications. Oncogene. 2005;24:1794–1801. doi: 10.1038/sj.onc.1208383. [DOI] [PubMed] [Google Scholar]
- 18.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 19.Gorringe KL, Boussioutas A, Bowtell DD. Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2005;42:247–259. doi: 10.1002/gcc.20136. [DOI] [PubMed] [Google Scholar]
- 20.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 21.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 23.Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 24.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS One. 2007;2:e538. doi: 10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, et al. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, He L, Collins I, Ge H, Libutti D, et al. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5:331–341. doi: 10.1016/s1097-2765(00)80428-1. [DOI] [PubMed] [Google Scholar]
- 28.Alliel PM, Seddiqi N, Goudou D, Cifuentes-Diaz C, Romero N, et al. Myoneurin, a novel member of the BTB/POZ-zinc finger family highly expressed in human muscle. Biochem Biophys Res Commun. 2000;273:385–391. doi: 10.1006/bbrc.2000.2862. [DOI] [PubMed] [Google Scholar]
- 29.Kelly KF, Daniel JM. POZ for effect–POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warner SL, Stephens BJ, Nwokenkwo S, Hostetter G, Sugeng A, et al. Validation of TPX2 as a potential therapeutic target in pancreatic cancer cells. Clin Cancer Res. 2009;15:6519–6528. doi: 10.1158/1078-0432.CCR-09-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 33.Mullenbach R, Lagoda PJ, Welter C. An efficient salt-chloroform extraction of DNA from blood and tissues. Trends Genetics. 1989;5:391. [PubMed] [Google Scholar]
- 34.R-Development-Core-Team. Vienna, Austria: R Foundation for Statistical Computing; 2008. R: A Language and Environment for Statistical Computing. 2.7.2 ed. [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - A Practical and Powerful Approach to Multiple Testing. JOURNAL OF THE ROYAL STATISTICAL SOCIETY SERIES B-METHODOLOGICAL. 1995;57:289–300. [Google Scholar]
- 38.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Zhao H, Shan J, Long F, Chen Y, et al. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18:1965–1978. doi: 10.1091/mbc.E06-07-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fields AP, Regala RP. Protein kinase C iota: human oncogene, prognostic marker and therapeutic target. Pharmacol Res. 2007;55:487–497. doi: 10.1016/j.phrs.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Huang J, Yang N, Liang S, Barchetti A, et al. Integrative genomic analysis of protein kinase C (PKC) family identifies PKCiota as a biomarker and potential oncogene in ovarian carcinoma. Cancer Res. 2006;66:4627–4635. doi: 10.1158/0008-5472.CAN-05-4527. [DOI] [PubMed] [Google Scholar]
- 43.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 45.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28:3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, et al. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res. 2009;69:7357–7365. doi: 10.1158/0008-5472.CAN-09-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. Embo J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishida T, Kaneko F, Kitagawa M, Yasuda H. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J Biol Chem. 2001;276:39060–39066. doi: 10.1074/jbc.M103955200. [DOI] [PubMed] [Google Scholar]
- 49.O'Brien TW, Fiesler SE, Denslow ND, Thiede B, Wittmann-Liebold B, et al. Mammalian mitochondrial ribosomal proteins (2). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J Biol Chem. 1999;274:36043–36051. doi: 10.1074/jbc.274.51.36043. [DOI] [PubMed] [Google Scholar]
- 50.Katoh Y, Ritter B, Gaffry T, Blondeau F, Honing S, et al. The clavesin family, neuron-specific lipid- and clathrin-binding Sec14 proteins regulating lysosomal morphology. J Biol Chem. 2009;284:27646–27654. doi: 10.1074/jbc.M109.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao S, Xu C, Qian H, Lv L, Ji C, et al. Cellular retinaldehyde-binding protein-like (CRALBPL), a novel human Sec14p-like gene that is upregulated in human hepatocellular carcinomas, may be used as a marker for human hepatocellular carcinomas. DNA Cell Biol. 2008;27:159–163. doi: 10.1089/dna.2007.0634. [DOI] [PubMed] [Google Scholar]
- 52.Niemantsverdriet M, Wagner K, Visser M, Backendorf C. Cellular functions of 14-3-3 zeta in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene. 2008;27:1315–1319. doi: 10.1038/sj.onc.1210742. [DOI] [PubMed] [Google Scholar]
- 53.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 54.Ran Y, Hu H, Hu D, Zhou Z, Sun Y, et al. Derlin-1 is overexpressed on the tumour cell surface and enables antibody-mediated tumour targeting therapy. Clin Cancer Res. 2008;14:6538–6545. doi: 10.1158/1078-0432.CCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Hua H, Ran Y, Zhang H, Liu W, et al. Derlin-1 is overexpressed in human breast carcinoma and protects cancer cells from endoplasmic reticulum stress-induced apoptosis. Breast Cancer Res. 2008;10:R7. doi: 10.1186/bcr1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciro M, Prosperini E, Quarto M, Grazini U, Walfridsson J, et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69:8491–8498. doi: 10.1158/0008-5472.CAN-09-2131. [DOI] [PubMed] [Google Scholar]
- 57.Zou JX, Guo L, Revenko AS, Tepper CG, Gemo AT, et al. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69:3339–3346. doi: 10.1158/0008-5472.CAN-08-3440. [DOI] [PubMed] [Google Scholar]
- 58.Gemmill RM, Bemis LT, Lee JP, Sozen MA, Baron A, et al. The TRC8 hereditary kidney cancer gene suppresses growth and functions with VHL in a common pathway. Oncogene. 2002;21:3507–3516. doi: 10.1038/sj.onc.1205437. [DOI] [PubMed] [Google Scholar]
- 59.Ellen TP, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29:2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- 60.Pflueger D, Rickman DS, Sboner A, Perner S, LaFargue CJ, et al. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11:804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujimoto T, Doi K, Koyanagi M, Tsunoda T, Takashima Y, et al. ZFAT is an antiapoptotic molecule and critical for cell survival in MOLT-4 cells. FEBS Lett. 2009;583:568–572. doi: 10.1016/j.febslet.2008.12.063. [DOI] [PubMed] [Google Scholar]
- 62.McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, et al. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer. 2005;5:505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 63.Bessette DC, Qiu D, Pallen CJ. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev. 2008;27:231–252. doi: 10.1007/s10555-008-9121-3. [DOI] [PubMed] [Google Scholar]
- 64.Joukov V, Groen AC, Prokhorova T, Gerson R, White E, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 65.Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci U S A. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakamoto K, Tamamura Y, Katsube K, Yamaguchi A. Zfp64 participates in Notch signaling and regulates differentiation in mesenchymal cells. J Cell Sci. 2008;121:1613–1623. doi: 10.1242/jcs.023119. [DOI] [PubMed] [Google Scholar]
- 67.Lukasiewicz KB, Lingle WL. Aurora A, centrosome structure, and the centrosome cycle. Environ Mol Mutagen. 2009;50:602–619. doi: 10.1002/em.20533. [DOI] [PubMed] [Google Scholar]
- 68.Storlazzi CT, Mertens F, Mandahl N, Gisselsson D, Isaksson M, et al. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer. 2003;37:195–200. doi: 10.1002/gcc.10210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample details. Clinicopathological features and assay information for each sample. 57 out of 72 tumours had matching lymphocytic DNA available for copy number microarray analysis.
(0.06 MB PDF)
Proportion of genome-wide gain and loss by sample. In all of these samples, the aberrant genome adds up to 95.4% on average. The missing 4.6% can be attributed to regions on chromosome Y, Mitochondrial DNA and repetitive sequences around centromeric regions that are either removed from the segmentation analysis or not covered by the Affymetrix SNP6.0 array.
(0.06 MB PDF)
Germline copy number polymorphisms on Chr 3, 7, 8, 20. The regions/segments of copy number gain that contained one or more of these CNPs were removed or altered as displayed in Figure S1-B. The type of CNP is also displayed in the far right column.
(0.05 MB PDF)
Regions of gain present in >40% of samples. This table contains genomic information for the 90 regions included in the expression analyses, i.e., all those regions that mapped to 1 or more probesets on the Human GeneST1.0 microarrays. On this microarray platform, most probesets map uniquely to a protein-coding gene. The region IDs correspond to those in Tables 2, 3, 4 and S5.
(0.13 MB PDF)
All differentially expressed probesets in frequent regions of gain. Every probeset tested for differential expression is listed and tagged by the region it belongs to. These region IDs are consistent across all tables in the paper and are derived as shown in Figure S1-A. Column 5 displays the Pearson's correlation between copy number and expression for the listed probeset. Columns 6–11 are derived from differential expression analyses performed using the “limma” package in R.
(0.06 MB PDF)
Correlation for all genes highly amplified (CN>5) in at least 5 samples. This table displays Pearson's correlation between copy number and gene expression for all 181 probesets in regions of high CN gain across the genome. The p-value displayed is a raw p-value obtained while testing for correlation. * Genes highly amplified in 4 samples but that were within 10 kb of a copy number breakpoint of 5 amplified samples.
(0.03 MB PDF)
Subtype breakdown of genome wide CN changes. (A) Overall copy number landscape for the cohort of ovarian cancer samples. This is similar to Figure 1 with the exception that the y-axis ranges from 0–100% of samples as opposed to 0–50%. Below are the distribution of copy number changes for (B) 37 serous ovarian cancers, (C) 14 endometrioid ovarian cancers, (D) 7 mucinous ovarian cancers and (E) 9 clear cell ovarian cancers. A, B and C jointly show that the major contributors for the high frequency changes are serous and endometrioid tumours. Data for the single tumor classified as undifferentiated is not shown here.
(0.43 MB TIF)
‘Cytoband collapsing’ and the exclusion of CNPs. (A) Shows the steps taken towards obtaining the copy number regions. The starting data (far left) contains genomic position and copy number information for segmental overlaps. All segments at this step of analysis occur with >40% frequency and have 3 or more copies. Letters a, b, r, s, t, u, v and w refer to genomic start/stop sites in basepairs. Regions are sorted by chromosome, then by genomic start and finally by genomic stop positions. Following this they are annotated with their cytobands and the newly defined “collapsed” region is bounded by the lowest start (a) and highest stop (b) positions and annotated with the cytoband of origin. The ‘a’ and ‘b’ from here carry through to part B of the figure. Regions that span two cytobands are listed as a separate group as shown in Table S4. (B) Shows the rules used to eliminate CNPs from the cytoband regions. Regions such as “Amp 4” are split into two, resulting in more regions after CNP elimination than before. (C) Regions of CNP across the genome and their position in relation to regions of copy number gain relevant to our study. (i) Global changes in normal (n = 57, green = gain and red = loss) and tumour (n = 72, yellow = gain and blue = loss) samples. We define a CNP as a change that occurs in at least 5% of normal samples. CNPs often show both genomic gain and loss at the same locus in normal samples. (ii) All changes on Chromosome 3 and in particular a CNP on 3q26.1 between 168.66 and 168.69 Mbp highlighted by the black oval, observed in >15% of all normal samples. (iii) The 3q26.1 CNP occurs in the middle of a region of copy number gain that we investigate further. This CNP region was removed from the data in accordance with S2-B.
(0.55 MB PDF)
Expression of all genes in regions of frequent copy number gain. This figure displays all genes in 90 regions of copy number change in terms of their average expression and t-statistic, resulting from the test for differential expression for each of these regions between amplified and unamplified samples. Genes showing a significant differential expression are represented by red dots and non-significant genes are represented by purple dots. Only one gene hCG_16001 showed a significant reduction in expression under the influence of copy number gain. This is a ribosomal protein L23a pseudogene 42 (RPL23A42) where RPL23A encodes a ribosomal protein that is a component of the 60S subunit and may be one of the target molecules involved in mediating growth inhibition by interferon.
(0.09 MB TIF)
Expression of MYC across various sample groups. RMA normalised expression of MYC based on Gene 1.0 ST array data. No significant differences were found between groups of samples that showed copy number gain in the region and those that did not.
(0.15 MB TIF)