Summary
Membrane subdomains have been implicated in T cell signaling, although their properties and mechanisms of formation remain controversial. Here, we have used single-molecule and scanning confocal imaging to characterize the behavior of GFP-tagged signaling proteins in Jurkat T cells. We show that the coreceptor CD2, the adaptor protein LAT, and tyrosine kinase Lck cocluster in discrete microdomains in the plasma membrane of signaling T cells. These microdomains require protein-protein interactions mediated through phosphorylation of LAT and are not maintained by interactions with actin or lipid rafts. Using a two color imaging approach that allows tracking of single molecules relative to the CD2/LAT/Lck clusters, we demonstrate that these microdomains exclude and limit the free diffusion of molecules in the membrane but also can trap and immobilize specific proteins. Our data suggest that diffusional trapping through protein-protein interactions creates microdomains that concentrate or exclude cell surface proteins to facilitate T cell signaling.
Introduction
In response to antigen, T cells proliferate and mount an immune response through coordinated processes of cell-cell adhesion, signal transduction, and cytoskeletal rearrangements. The discovery of the immunological synapse, a highly organized array of signaling, adhesion, and cytoskeletal proteins at the interface between a T cell and an antigen-presenting cell (APC) or a planar membrane bilayer containing APC-associated proteins (Monks et al., 1998; Grakoui et al., 1999), has highlighted the potential importance of spatial organization in T cell signaling. The molecular patterning observed at the immunological synapse has been proposed to contribute to the sensitivity of signaling initiated by the T cell receptor (TCR; Monks et al., 1998; Grakoui et al., 1999; Huppa et al., 2003). However, the precise role of the synapse and its mechanism of formation remain poorly understood (Lee et al., 2002a; Lee et al., 2003).
Studies of other model systems for T cell signaling also have suggested that membrane subdomains contribute to signal transduction. These model systems include primary and immortalized T cells (e.g., Jurkat cells) that can be activated by anti-TCR antibodies applied to glass coverslips or microspheres. The TCR and many downstream signaling molecules (e.g., tyrosine kinases such as Lck and ZAP-70 and adaptor proteins such as LAT and Grb2) redistribute to the interface between the T cell and its stimulating ligand (Bunnell et al., 2002; Ehrlich et al., 2002; Ike et al., 2003). Moreover, even within the interface between Jurkat T cells and anti-TCR-coated surfaces, small and transient clusters of signaling molecules have been observed (Bunnell et al., 2002). Several studies also have suggested that actin and myosin motor proteins are involved in the polarization and clustering of cell surface components upon TCR ligation (Wulfing and Davis, 1998; Gil et al., 2002; Jacobelli et al., 2004).
Clustering of T cell plasma membrane proteins into lipid raft microdomains has been suggested to play an important role in signal transduction. Lipid rafts are created by a partial phase separation of cholesterol, un-saturated sphingolipids, lipid-modified proteins, and proteins with longer transmembrane regions within a membrane bilayer (Rietveld and Simons, 1998). The TCR and several downstream signaling molecules have been classified as lipid raft associated (Kabouridis et al., 1997; Xavier et al., 1998; Lin et al., 1999; Yang and Reinherz, 2001). Removal of acylation motifs from either the Src family kinase Lck or the transmembrane adaptor protein LAT interferes with signaling, supporting the idea that lipid rafts serve as “platforms” for signal transduction in T cells (Yurchak and Sefton, 1995; Zhang et al., 1998b). The most commonly used criterion for classifying a protein as lipid raft associated is insolubility in cold 1% Triton X-100 (Xavier et al., 1998). However, others have questioned whether detergent insolubility gives rise to artifactual associations (Heer-klotz, 2002; Munro, 2003), thereby creating controversy over the existence of lipid rafts. In any event, this biochemical method does not provide insight into the dynamics of rafts in living cells.
Microscopy provides another avenue for investigating lipid rafts and other types of membrane micodomains. Conventional wide-field or confocal microscopies can visualize large-scale organization of molecules in the membrane but are not necessarily well suited for studying lipid rafts, which might form and dissociate on a rapid timescale and may be smaller than the resolution limit of the light microscope (Varma and Mayor, 1998; Sharma et al., 2004). The development of techniques for imaging single fluorophores in vitro and in living cells (Sako et al., 2000; Nakada et al., 2003) offers another means of probing membrane structure and the dynamics of proteins in the plasma membrane. Single-molecule tracking can yield information on diffusion coefficients and boundaries or “fences” that limit diffusion, both of which provide insight into local membrane environments and interactions that constrain mobility (Schutz et al., 1997; Dietrich et al., 2002; Fujiwara et al., 2002). These techniques also provide information on spatial and temporal heterogeneity in molecular behavior. In T cells, a single-molecule study has reported a large reduction in the diffusion coefficient of the Lckkinase upon TCR crosslinking, which the authors suggest reflects an association with lipid rafts and the actin cytoskeleton (Ike et al., 2003).
Imaging of plasma membrane proteins by confocal microscopy and single-molecule techniques provide different but potentially complementary views of membrane structure and protein dynamics, but the two approaches have not been combined in a single study on a given cell type. Moreover, published single-molecule studies have focused on single proteins, but comparisons of several proteins can potentially provide new information on factors that influence protein mobility in the plasma membrane. In this study, using Jurkat T cells, we have combined confocal imaging to examine the overall membrane distributions and total internal reflection microscopy to monitor the single-molecule behaviors of several key molecules involved in T cell signaling. Our results reveal that TCR activation induces the formation of membrane subdomains, distinct from lipid rafts and built by a network of protein-protein interactions, that can exclude or trap distinct signaling molecules.
Results
Single-Molecule Dynamics of T Cell-Surface Proteins in Unstimulated and Activated Cells
We prepared GFP fusions of two raft-associated proteins (the tyrosine kinase Lck and the adaptor protein LAT) and two nonraft-associated proteins (the costimulatory transmembrane protein CD2 and the transmembrane tyrosine phosphatase CD45), as well as selected mutant versions of these proteins. Lck, a Src family kinase, is acylated and partially partitions (∼25%) into a Triton X-100-insoluble fraction (Xavier et al., 1998). To probe lipid raft behavior without the influence of protein-protein interactions, we fused GFP to the 10 aa N terminus of Lck (Lck10), which partitions into a detergent-insoluble raft fraction by virtue of the dual lipid modification of two cysteines in this peptide (Shenoy-Scaria et al., 1994). LAT, an adaptor protein that becomes heavily tyrosine phosphorylated after TCR activation and presents docking sites for several SH2 domain-containing downstream signaling proteins (e.g., PLC-γ1 and Grb2; Hartgroves et al., 2003), also is lipid modified and fractionates with lipid rafts (Zhang et al., 1998b). We also studied a mutant of LAT in which its nine tyrosine phosphorylation sites were mutated to phenylalanine (LAT(Y-F)); this protein partitions into the detergent-insoluble fraction but does not recruit SH2-containing proteins (Lin and Weiss, 2001). Conversely, we analyzed a dual cysteine mutant of LAT (LAT(C-S)) that is not palmitoylated and does not partition into lipid rafts (Lin et al., 1999). CD2 is a transmembrane protein that interacts with CD58 on the APC. This interaction contributes to cell-cell adhesion and generates intracellular signals that enhance the TCR response (Bachmann et al., 1999). Human CD2 is not raft associated by the standard criterion of insolubility in 1% Triton X-100, although it demonstrates partial resistance to extraction at lower (0.2%) Triton X-100 concentrations (Yang and Reinherz, 2001). Finally, CD45 is a nonraft, transmembrane tyrosine phosphatase that can dephosphorylate Lck as well as other targets (Janes et al., 1999). We fused a truncated version of CD45 lacking the majority of its cytoplasmic domain to GFP to track its behavior. Data on the detergent solubility of all of the GFP-tagged proteins used in this study except CD45 and LAT(C-S), which were not present at the cell surface in high enough abundance for analysis by Western blotting, are presented in Figure S1 in the Supplemental Data available with this article online.
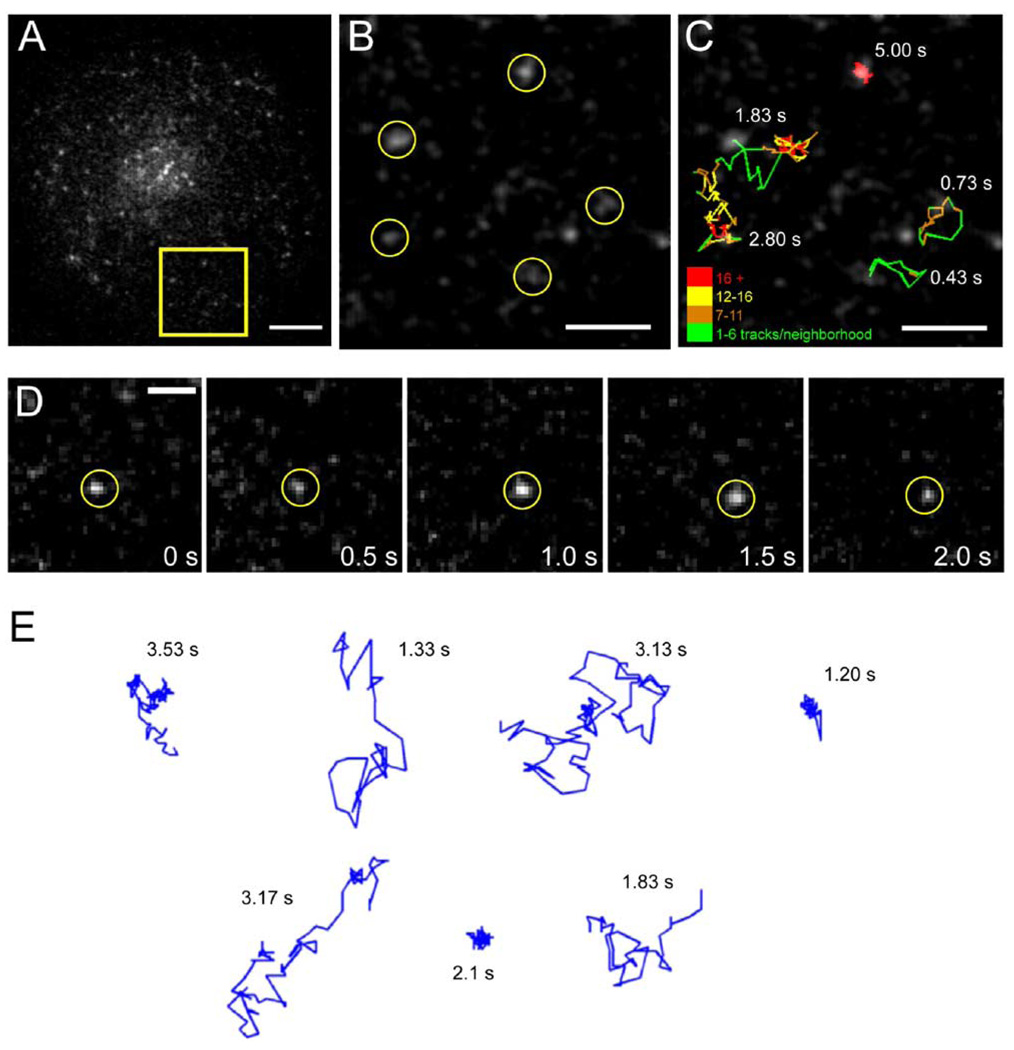
In our experimental system, Jurkat T cells were transfected with a GFP- (or mRFP)-tagged protein construct, and then the cells were plated on coverslips that were coated with either anti-TCR antibodies (stimulatory) or nonspecific IgG (nonstimulatory). The former condition triggers actin-mediated cell spreading, induction of calcium transients, and tyrosine phosphorylation (Bunnell et al., 2001; Bunnell et al., 2002), though it has been shown that a low, basal level of signaling occurs even in unstimulated Jurkat cells (Roose et al., 2003). To visualize single GFP-tagged molecules on the T cell surface at the interface with the substrate, we used total internal reflection fluorescence (TIRF) illumination (Sako et al., 2000; Axelrod, 2003) and recorded images at video rate using a cooled, intensified CCD camera (Figure 1A; Movie S1). Individual, diffraction-limited spots typically exhibited single-step photobleaching and single-exponential distributions of bleaching times and were very similar in intensity to purified GFP, indicating that they represent single molecules (Figure S2). Tracking GFP-tagged proteins for 0.5–10 s (Figure 1D) revealed multiple modes of diffusion, even for individual molecules (Figure 1B, 1C, and 1E). In some instances, single molecules were highly mobile in the plane of the membrane, but in other cases, movement was highly restricted. Similar single-molecule behavior for plasma membrane proteins has been described elsewhere (Schutz et al., 1997; Dietrich et al., 2002; Lommerse et al., 2004). Moreover, individual molecules often made abrupt transitions between “immobile” and “mobile” states (Figure 1C and 1E). For this reason, we employed a method for calculating diffusion coefficients that could measure transitions in mobility over time for individual molecules (Figure 2A and 2B; see Experimental Procedures).
Figure 1. Single-Molecule Imaging of Lck-GFP Reveals Heterogeneous Diffusion Behavior.
Jurkat T cells were transiently transfected with Lck-GFP and imaged as described in the Experimental Procedures by TIRF microscopy to observe single molecules at the ventral cell surface.
(A) A whole-cell image of spots corresponding to single Lck-GFP molecules. Scale bar, 5 µm.
(B) Subregion of (A) showing the initial locations of five single molecules. Dim objects elsewhere in the image represent intensifier noise and background fluorescence. Scale bar, 2 µm.
(C) Trajectories of the five molecules shown in (B), tracked at 30 frames/s. The total elapsed time for each trajectory is indicated in the figure. See also Movie S1. Trajectory color indicates the spatial density of single-molecule centroids within a 150 nm2 neighborhood, as a means of illustrating spatial confinement. Scale bar, 2 µm.
(D) Time-lapse series of raw images showing diffusion of a single molecule of Lck-GFP. Scale bar, 1 µm.
(E) Representative single-molecule trajectories showing highly mobile and immobile behavior, as well as transitions between the two modes. Scale bar, 2 µm.
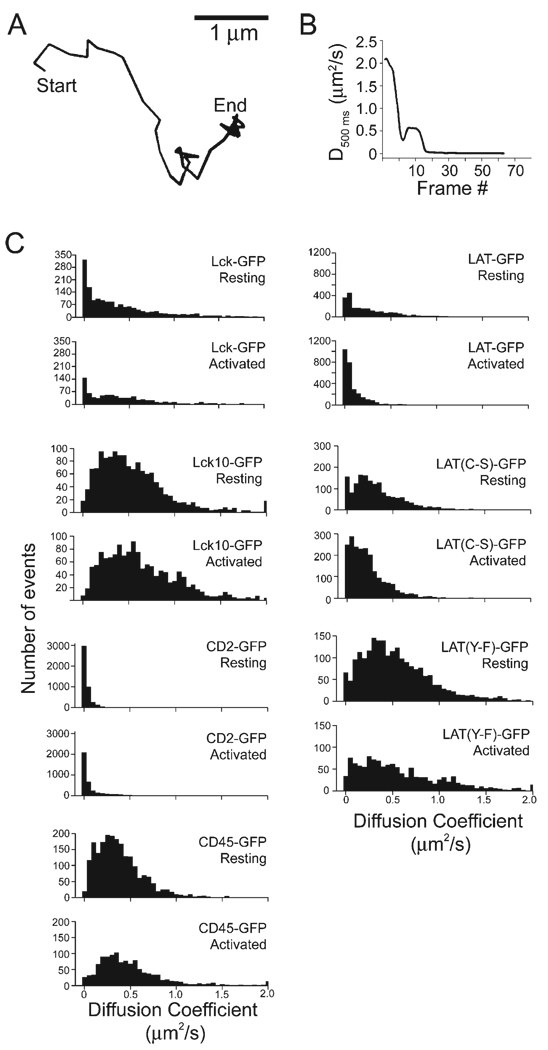
Figure 2. Diffusional Immobilization and Response to TCR Signaling for Several GFP-Tagged Signaling Molecules.
(A) and (B) illustrate the experimental determination of diffusion coefficients. In (A), a representative single-molecule trajectory is shown (duration = 2.90 s).
(B) shows the diffusion coefficients for this trajectory, by generating mean square displacement versus time plots (0.5 s) for each sequential frame (see Experimental Procedures).
(C) Histograms of diffusion coefficients are shown for unstimulated and TCR-activated cells for Lck-GFP, Lck10-GFP, LAT-GFP, LAT(C-S)-GFP, LAT(Y-F)-GFP, CD2-GFP, and CD45-GFP. Average values are reported in Table S1.
Given the potentially large difference in radius between a lipid raft and a single membrane protein, our a priori prediction was that raft-associated proteins would have lower diffusion coefficients than raft nonassociated proteins. We tested this prediction by examining the distributions of diffusion coefficients for all the above constructs. Histograms of the diffusion coefficients revealed distinct behaviors for the different GFP-tagged proteins in unstimulated T cells (Figure 2; Table S1). At one extreme, single CD2-GFP molecules were primarily immobile with only occasional episodes of rapid mobility (average diffusion coefficient of 0.04 µm2/s; Table S1). At the other extreme, the raft-targeted Lck10-GFP was primarily mobile and displayed the highest average diffusion coefficient of the proteins tested (0.73 µm2/s). In between these two extremes were Lck, LAT, and LAT(C-S). The histograms of these proteins revealed a peak of very low diffusion coefficients and then a long trailing shoulder of higher values. In contrast, the histograms for the raft-targeting motif of Lck (Lck10-GFP) and the nonphosphorylatable LAT (LAT(Y-F)-GFP) both lacked the relatively immobile population peak at the far left of the histogram. CD45 displayed a similar distribution and also had an intermediate mean value (0.45 µm2/s). In summary, the diffusional behaviors of different proteins in the T cell signaling pathway vary significantly and do not correlate with biochemical fractionation into lipid rafts. For example, CD2, which does not behave as a classical lipid raft protein, is less mobile than the lipid raft-associated proteins Lck, Lck10, LAT, and LAT(Y-F). In addition, interfering with lipid raft association of LAT through mutations did not alter its diffusion behavior. Thus, we conclude that factors other than partitioning into a detergent-insoluble raft fraction determine the diffusional properties of signaling molecules in the T cell membrane.
We also examined how activation of TCR signaling by anti-TCR antibodies affects single-molecule diffusion. TCR activation triggered a significant reduction in diffusion coefficient for LAT and the nonpalmitoylated LAT(C-S) mutant, but not for any of the other proteins (Figure 2; Table S1). This result indicates that signaling alters the local environment of LAT and does so independently of lipid rafts.
Formation of Signaling Clusters in Activated Cells
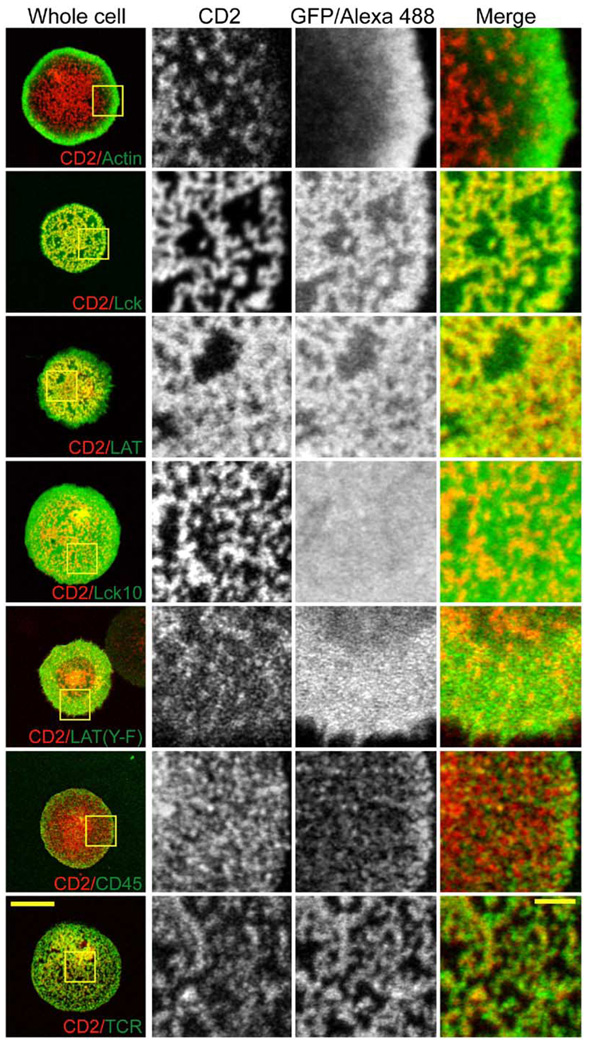
To better understand the differences in the single-molecule diffusion behavior of various signaling proteins, we examined their distributions by confocal and epifluorescence microscopy in living cells. We simultaneously imaged CD2-mRFP along with GFP fusions of a number of other proteins involved in TCR signaling, stimulating the cells for 10 min prior to imaging. CD2 localized to numerous discrete clusters at the antibody-proximal surface of the cell (Figure 3). In unstimulated cells, CD2-GFP was more diffuse, although it also displayed some weak clustering (data not shown). Although CD2 has been biochemically linked to F-actin or actin-associated proteins (Dustin et al., 1998; Badour et al., 2003), CD2 and actin did not colocalize in living, activated cells. Rather, as has been noted by others (Bunnell et al., 2001), F-actin was located predominantly at the periphery of the contact where the CD2 density was lowest (Figure 3). Surprisingly, while treating cells with Latrunculin A prior to activation prevented CD2 clustering and cell spreading, adding the drug 10 min after activation had no discernable effect on the distribution of CD2 (Figure S3. Together, these results suggest that F-actin is required for the formation, but not maintenance of CD2 clusters.
Figure 3. Formation of CD2-Enriched Signaling Domains in the Activated T Cell Surface.
Jurkat cells were transiently transfected with CD2-mRFP and the indicated GFP fusion proteins. The planes of contact between the cells and anti-TCR antibody-coated coverslips were imaged by laser scanning confocal microscopy. Yellow boxes in column 1 indicate the expanded regions shown in columns 2–4. All imaging was performed in living cells at 37°C except for the CD2 + CD45 samples, which had to be fixed and stained by immunofluorescence since expression of CD45-RO-GFP at the cell surface is very low. Primary antibodies were labeled with Zenon labeling kits (Molecular Probes) to avoid labeling of the glass-adsorbed stimulatory antibody. Scale bars, 10 µm (whole cells) and 2 µm (expanded regions).
CD45 also did not colocalize with CD2, being reduced at the center compared to the periphery as shown previously (Johnson et al., 2000; Leupin et al., 2000). The small amount of CD45 present at the center of the contact was noticeably excluded from the CD2 zones (Figure 3). In contrast, Lck and LAT, which mediate several of the earliest events in TCR signaling, coclustered with CD2 (Figure 3). Clustering of LAT and Lck in activated Jurkat cells was also observed by Bunnell et al. (2002), although CD2 was not examined in their work. This colocalization of CD2, Lck, and LAT was not a consequence of lipid raft association, since the raft marker Lck10 and the raft-associated, non-phosphorylated LAT(Y-F) showed no preferential accumulation in CD2-mRFP clusters (Figure 3). The latter result also suggests that recruitment of LAT to CD2 zones requires protein interactions mediated through its phosphorylated tyrosine residues. Consistent with the notion that these clusters are enriched in signaling activity, a phosphotyrosine antibody selectively stained the CD2 zones (Figure S4). At early times (0–2 min) after cells were applied to the anti-TCR surface, LAT and CD2 both exhibited very strong colocalization with the TCR (Figure S5). However, by 5–10 min CD2 and the TCR still overlapped but exhibited a slight offset, with TCR accumulating immediately adjacent to regions of highest CD2 concentration (Figure 3). This result also illustrates that signaling zones are reorganized as signaling progresses.
Signaling Clusters Are Largely Static in Space but Exchange Molecules
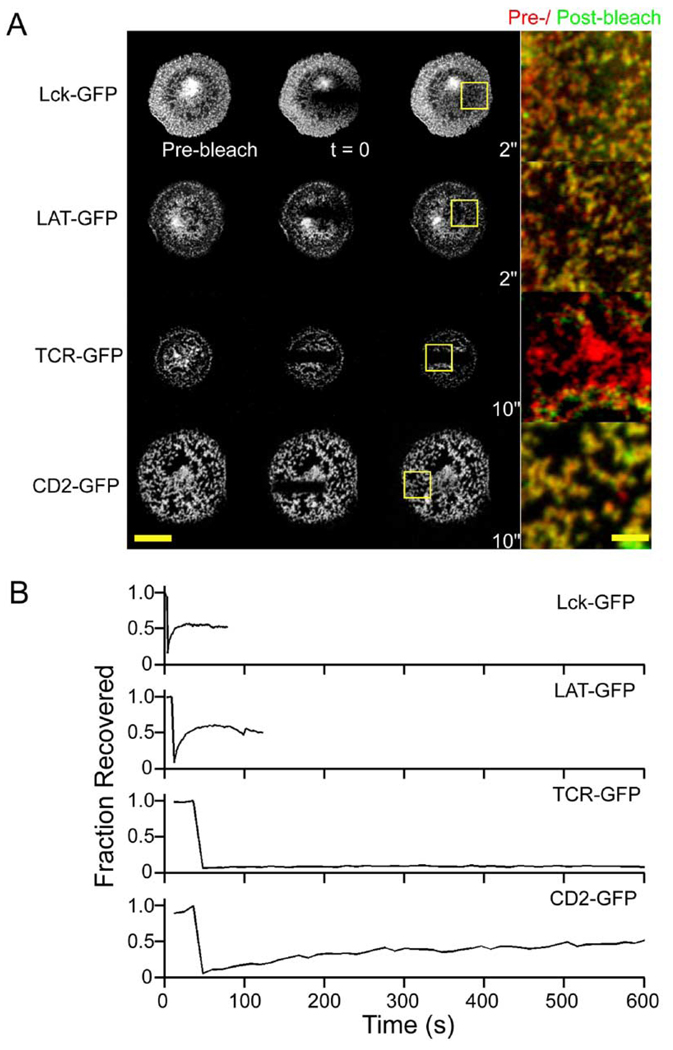
To characterize the dynamics of CD2, LAT, and Lck at the population level, we performed fluorescence recovery after photobleaching (FRAP) of these GFP-tagged molecules to measure turnover of molecules in the clusters of activated Jurkat cells (Figure 4A). Fluorescence recovery was observed for all of these proteins, but the time courses of recovery were significantly faster for LAT-GFP and Lck-GFP than for CD2-GFP (Figure 4A and 4B). We did not observe different recovery rates at the periphery versus the center of the contacts (data not shown). These results are consistent with single-molecule measurements showing that CD2 molecules are largely immobile, while LAT and Lck diffusion coefficients reveal both immobile and mobile populations of molecules. In contrast, a photobleach of TCR-GFP did not recover, as might be expected if it is bound to immobilized antibody on the glass surface (Figure 4A and 4B). Interestingly, the locations of Lck, LAT, and CD2 clusters remained unchanged between the pre-photobleaching and recovery periods, as seen when these images were overlaid (Figure 4A, merge panels). Thus, the clusters remain largely static in space, at least over a time span of several minutes, although the photobleaching and recovery reveals that there is flux of proteins into and out of these structures.
Figure 4. Signaling Clusters Are Static in Location but Exchange Molecules.
Activated cells were subjected to FRAP analyses to determine the population-level diffusion dynamics of CD2-GFP, Lck-GFP, LAT-GFP, and TCRζ-GFP. A stripe of fluorescence at the cell-antibody interface was photobleached, and fluorescence recovery in these regions was examined after the bleach.
(A) shows the raw images before, immediately after, and upon reaching maximal recovery after the bleach. Scale bar, 10 µm. The merged images show that the fluorescence in signaling clusters is replenished in their original locations after photobleaching. Scale bar, 2 µm.
(B) Representative examples of fluorescence photobleaching recovery as a function of time.
CD2 Clustering Requires Signaling Competent LAT
Since the CD2 clusters were still present in the absence of actin, we reasoned that they might be maintained by a network of protein-protein interactions in the cortical membrane. A candidate molecule that could be in-volved in such a network is LAT, a multifunctional adaptor protein that can interact with multiple SH2 domain-containing proteins (Zhang et al., 1998a; Lin and Weiss, 2001). LAT’s adaptor function requires the presence of multiple SH2 binding sites on a single protein rather than in trans (Lin and Weiss, 2001). Conflicting data exist as to the importance of LAT’s lipid raft association in regulating its signaling functions (Zhang et al., 1998b; Zhu et al., 2005). LAT previously has been shown to accumulate at sites of TCR activation in a manner that depends upon protein-protein interactions (Harder and Kuhn, 2000; Tanimura et al., 2003). Consistent with this idea and as described earlier, LAT colocalizes with clustered CD2 after TCR activation, but LAT(Y-F)-GFP does not.
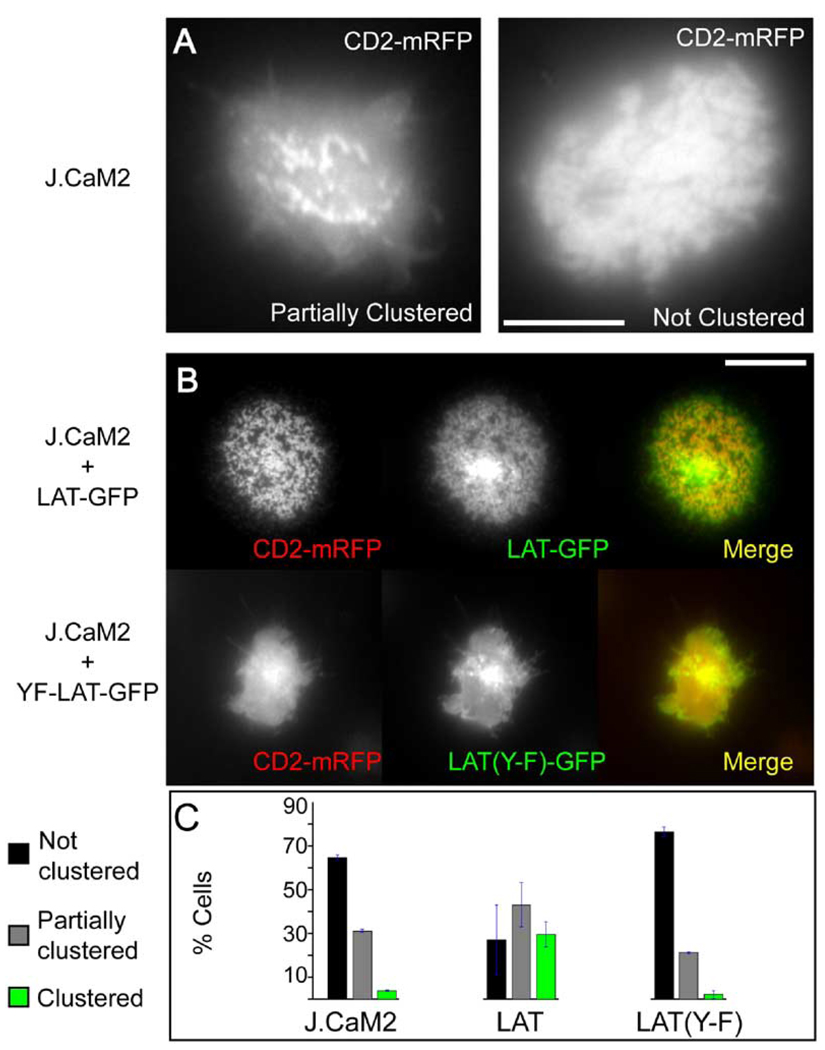
To further investigate the relationship between LAT and CD2 clustering, we examined the distribution of CD2-mRFP in the J.CaM2 cell line, a derivative of the Jurkat line in which LAT expression is severely compromised although not completely eliminated (Finco et al., 1998; Roose et al., 2003). CD2-mRFP clustering was severely impaired in J.CaM2 cells after they were plated on anti-TCR antibody surfaces. The majority of cells showed either no (63%) or a very low (31%) degree of clustering, and almost no cells displayed the tight CD2 clustering pattern observed in wild-type cells (Figure 5A and 5C). This phenotype was accompanied by reduced cell spreading, as has been reported previously for this line (Bunnell et al., 2001). Since J.CaM2 cells harbor changes in the expression of genes other than LAT (Roose et al., 2003), we wished to demonstrate that the CD2 clustering defect is specifically due to the absence of LAT by restoring LAT function in these cells. When J.CaM2 cells were transfected with LAT-GFP, TCR-induced cell spreading was restored to roughly normal levels, and CD2 clustering was observed in the majority of transfected cells (Figure 5B and 5C). In contrast, reconstitution with even low levels of LAT(Y-F)-GFP eliminated the small amount of CD2 clustering observed in the J.CaM2 cells (Figure 5B and 5C). This result can be explained if residual LAT in J.CaM2 enables weak CD2 clustering in some cells and that LAT(Y-F)-GFP produces a dominant-negative effect on this LAT activity. In conclusion, these results provide direct evidence for a requirement for LAT and tyrosine phosphorylation of LAT in the clustering of CD2.
Figure 5. Functional LAT Is Required for CD2 Clustering.
(A) shows the LAT-deficient J.CaM2 cells that were transfected with CD2-mRFP and imaged by epifluorescence microscopy at the interface with the anti-TCR antibody-coated glass. Most cells either showed no (63%) or partial (31%) clustering of CD2, and rarely (5%) showed pronounced clustering like wild-type cells. Scale bar, 10 µm.
(B) shows that CD2 clustering can be restored by transfection with LAT-GFP but not with LAT(Y-F)-GFP. Scale bar, 10 µm.
(C) shows quantitation of CD2 clustering. Cells were visually scored as either unclustered, partially clustered, or fully clustered (see examples in [A] and [B]). Data shown are the mean and SEM of three independent transfection experiments (200–300 cells scored per experiment).
Single-Molecule Tracking Relative to CD2 Clusters
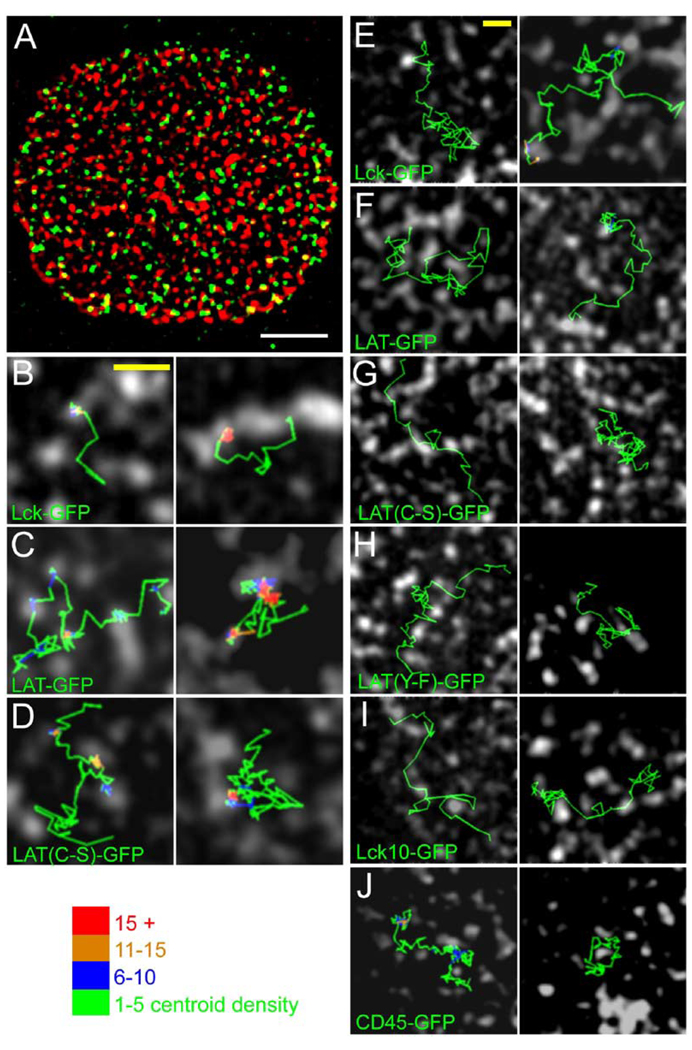
Our single-molecule tracking of Lck and LAT revealed that individual molecules undergo abrupt transitions between restricted diffusion and more mobile, free diffusion. We wished to know whether the CD2/Lck/LAT clusters in activated T cells might influence these transitions. To explore this question, we developed a strategy in which we could compare the trajectories of single GFP-tagged molecules relative to the signaling zones. We chose CD2 as a marker for these zones, not to attach any particular importance to this protein over the others but to simply provide a landmark against which to evaluate diffusion. First, a population level image of CD2-mRFP was captured using TIRF at low laser intensity and low camera gain, and then single GFP-tagged proteins were imaged in the same cell for 3–5 min. Each frame of video-rate GFP sequence was then overlaid onto the CD2-mRFP image in order to examine diffusion of single molecules relative to CD2 clusters (Figure 6A; Movies S2 and S3). This sequential imaging process was made possible by the fact that CD2’s distribution does not change appreciably over a period of several minutes (Figure 4). To quantitate diffusion relative to signaling clusters, the cell surface was segmented into CD2 clustered and nonclustered regions. The centroid positions of single molecules at each point in their trajectories were assigned to one of these two zones (see Figure 6 legend). It should be noted that several seconds of illumination and concomitant photobleaching were usually necessary to reduce the density of single GFP spots to a level that allows single-molecule tracking. As a result, the distribution of single molecules in images like the one shown in Figure 6A does not necessarily represent the equilibrium distribution because the majority of imaged molecules recently diffused into the contact zone.
Figure 6. Dual-Color Imaging of Single GFP-Tagged Molecules Relative to CD2-mRFP Clusters.
(A) A single frame from an image sequence of single molecules of Lck-GFP (green) that was superimposed upon a snapshot, bandpass-filtered image of CD2-mRFP clusters (red) (see Experimental Procedures). The movie of this cell can be found in Movie S3.
(B)–(D) show centroid trajectories of single molecules of Lck-GFP, LAT-GFP, and LAT (C-S)-GFP, illustrating that they alternate between periods of immobilization within CD2 clusters and more rapid mobility outside of the clusters (see also Movies S7–S9). (E)–(J) show examples of several single-molecule trajectories in which centroid trajectories navigate in channels between CD2 zones (see also Movies S5 and S6). In (B)– (I), the trajectory color indicates the spatial density of single-molecule centroids within a 150 nm2 neighborhood, as a means of showing spatial confinement.
Scale bars: (A) 5 µm, (B–I) 2 µm. The durations of each trace in seconds, in the left and right panels, are: (B) 1.00, 2.80 s; (C) 5.17, 6.03 s; (D) 3.77, 3.80 s; (E) 3.27, 4.57 s; (F) 2.83, 3.0 s; (G) 1.27, 3.63 s; (H) 2.63, 1.90 s; (I) 1.47, 2.10 s; (J) 4.93, 2.23 s.
When we analyzed the spatial overlap between CD2 clusters and the centroids of tracked, single molecules, we observed that centroids of CD45-GFP molecules exhibited the least overlap and Lck-GFP and LAT-GFP exhibited the greatest (Table 1), which is in general agreement with the population distributions observed by confocal microscopy (Figure 3). LAT(C-S)-GFP similarly showed a large cluster-associated fraction. In contrast, centroids of Lck10-GFP and LAT(Y-F)-GFP were approximately randomly distributed between CD2 and non-CD2 zones, as might be expected from their more uniform population distributions (Figure 3). When we inspected the paths of individual molecules outside of the CD2-mRFP-enriched zones, we observed that they often followed narrow channels and appeared to deflect off of the CD2 boundaries. Static representations of individual trajectories are shown in Figure 6E–6J, but the exclusion is best appreciated by examining the movies (Movies S5 and S6). This phenomenon was observed for all constructs and not just the excluded CD45-GFP. Thus, the membrane microdomains enriched in CD2 can, at least in some instances, act as barriers to the free diffusion of single molecules.
Table 1.
Diffusion and Single-Molecule Centroid Positions Relative to CD2-mRFP Clusters
| Construct | Din (µm2/s) | Dout (µm2/s) | Din/Dout | p value (Din/Dout) | Nin/Nout | p value (Nin/Nout) |
|---|---|---|---|---|---|---|
| LAT | 0.18 | 0.30 | 0.59 ± 0.06 | < 0.005 | 1.80 ± 0.31 | < 0.05 |
| LAT(Y-F) | 0.66 | 0.55 | 1.23 ± 0.26 | – | 0.89 ± 0.12 | – |
| LAT(C-S) | 0.14 | 0.43 | 0.32 ± 0.11 | <0.005 | 2.1 ± 0.21 | <0.01 |
| Lck | 0.26 | 0.46 | 0.55 ± 0.11 | <0.10 | 1.45 ± 0.28 | – |
| Lck10 | 1.08 | 1.14 | 0.95 ± 0.04 | – | 0.89 ± 0.11 | – |
| CD45 | N.A. | N.A. | N.A. | N.A. | 0.64 ± 0.07 | <0.01 |
The centroids of images of single GFP-tagged proteins were overlaid onto the CD2-mRFP image, as described in the Experimental Procedures. Din refers to the mean diffusion coefficients for trajectory segments overlapping a CD2 zone for five or more consecutive frames, and Dout indicates the mean diffusion coefficient for trajectory segments lying outside of CD2 zones for five or more consecutive frames. CD45 is listed as N.A. (not analyzable), since the number of trajectories that had overlapping centroids with CD2 zones for five consecutive frames was >10-fold lower than LAT-GFP, and some cells did not have any CD45 trajectories that fit this criteria. Nin/Nout refer to the ratio of centroid coordinates that are found inside or outside of CD2 clusters, normalized to the fraction of the area occupied by CD2 clusters. A value of one would be expected for a random distribution. Four to nine cells were analyzed for each construct. Error estimates represent the SEM for the different cells analyzed. p values indicate the significance of the deviation from a value of 1.
We also observed diffusional trapping of LAT-GFP (Movies S4 and S7), Lck-GFP (Movies S8 and S9), and LAT(C-S) in CD2 zones (Figure 6B–6D). Dual-color imaging revealed that regions of restricted mobility most often overlapped spatially with the CD2 clusters (red/ orange color-coded segments in Figure 6; Movies S7–S9), although immobilization sometimes also occurred outside the CD2 zones. Consistent with preferential trapping within CD2 zones, the average diffusion coefficients of Lck-GFP, LAT-GFP, and LAT(C-S)-GFP molecules were significantly reduced inside of CD2 clusters compared to outside (Table 1). In contrast, Lck10 and LAT(Y-F) did not display significantly different diffusion coefficients inside and outside of CD2 clusters (Table 1). In summary, we conclude that diffusional trapping of single molecules occurs selectively within CD2 clusters. From the results of the different constructs described above, we also propose that such diffusional trapping results from protein-protein interactions and not association with lipid rafts.
Discussion
In this study, we document the presence of signaling microdomains in the plasma membrane of Jurkat T cells. By combining observations of population distributions in the membrane by confocal microscopy with observations of single-molecule behavior by TIRF microscopy, we have been able to derive new insights into the properties and formation of these microdomains. In general, the population and single-molecule studies agree well with one another. Proteins (e.g., LAT, Lck, CD2) that showed immobilization at the single-molecule level are enriched in signaling clusters, and sites of single-molecule immobilization overlapped spatially with these microdomains. The transient nature of immobilization for Lck and LAT compared with CD2 also correlated with their faster recovery in population FRAP experiments. Our analyses of mutations that interfere either with targeting to lipid rafts or with protein interactions suggest that the observed microdomains are created by a network of protein-protein interactions, rather than by lipid rafts or an underlying actin cytoskeleton. We postulate that much of the observed single-molecule behavior can be explained by exclusion or trapping by protein binding in these microdomains, as will be discussed in more detail below and summarized in Movie S10.
Patterning of Molecules in the Plasma Membranes of Activated T Cells
Our confocal imaging reveals a spatial organization of plasma membrane proteins at the contact site between a T cell and a stimulatory surface. CD2, LAT, and Lck accumulate more centrally in the contact zone, consistent with other studies showing higher concentrations of LAT and Lck at sites of T cell contact with a stimulating surface (Harder and Kuhn, 2000; Ehrlich et al., 2002; Hartgroves et al., 2003). However, in contrast to our findings and those of Ehrlich, the study by Harder and Kuhn did not find an accumulation of Lck at contact sites.
In addition to the overall accumulation at the contact site, our studies also reveal a clustering of CD2/LAT/Lck into discrete microdomains. CD45 is excluded from these microdomains, and Lck10, a marker for lipid rafts, is neither concentrated within nor excluded from CD2/LAT/Lck clusters. Our FRAP studies show that CD2/LAT/Lck clusters are largely stationary in space but can exchange molecules on the time scale of minutes. Molecular patterning at the TCR-activated contact site of Jurkat cells was previously reported by Bunnell et al. (2002), who described a coclustering of TCR, ZAP-70, LAT, and several LAT-associated proteins but did not examine CD2, Lck, Lck10, or the LAT mutants described here. The data presented here support and extend most of the findings of that study, though LAT clustering in their system was more transient than we observe, and the patterns of LAT relative to the TCR were slightly different in the two studies (for further discussion, see Figure S5).
Formation of Clusters and Single-Molecule Immobilization Involves a Network of Protein-Protein Interactions
Several theories have been proposed to explain the segregation or immobilization of molecules at the contact between a T cell and a stimulatory surface, the most widely cited of which involve lipid rafts or the actin cytoskeleton. Our comparison of the population distributions and single-molecule behaviors of several plasma membrane proteins, the use of mutants that selectively interfere with raft association or protein-protein interactions, and the use of actin-destabilizing drugs have allowed us to probe these hypotheses.
Several studies have proposed that protein clustering and polarization are driven by association with lipid rafts, and there is evidence that raft aggregation results from (Viola et al., 1999) and facilitates (Janes et al., 1999; Yang and Reinherz, 2001) T cell signaling. However, other studies did not observe accumulation of raft markers at sites of T cell stimulation (Harder and Kuhn, 2000), and some have suggested that raft proteins do not cluster in the T cell surface, at least at the level of sensitivity afforded by FRET microscopy (Glebov and Nichols, 2004). Our work indicates that rafts are not a primary factor in the clustering and immobilization of molecules in the T cell plasma membrane. First, the degree of immobility in the membrane of living cells does not correlate well with detergent insolubility. Second, two raft-associated proteins (the lipid-modified N terminus of Lck [Lck10] and a nonphosphorylated mutant of LAT [LAT(Y-F)]) are not concentrated in CD2 clusters. Third, the average diffusion coefficients among raft-associated proteins varied widely (Table S1), indicating that they are not constitutively part of the same complex, a result that agrees with FRAP studies of other cell types (Kenworthy et al., 2004). We also have depleted cholesterol with methyl-β-cyclodextrin (a frequently used method for disrupting lipid rafts) but found that preincubation with this drug prevented cell attachment to anti-TCR-coated surfaces, most likely due to a general toxic effect (Munro, 2003). However, if 12 mM methyl-β-cyclodextrin was added for 30 min after cell attachment, we did not observe a disruption of CD2 clusters (not shown). Thus, our collective evidence suggests that lipid rafts are unlikely to be the primary determinant of diffusional immobility or the formation of CD2/Lck/LAT clusters.
Although our data do not require the existence of rafts, they also do not exclude their existence. The potentially small size of rafts (<70 nm; Varma and Mayor, 1998; Sharma et al., 2004) and their highly dynamic nature (Dietrich et al., 2002; Sharma et al., 2004) may elude our temporal or spatial detection. Thus, rafts could contribute to some aspects of signaling and spatial organization. For example, raft-mediated clustering at the nanometer scale might precede the formation of the large domains described here, or rafts might serve an organizational role for proteins within the signaling clusters.
Other studies have argued that actin is required for segregating molecules in the T cell membrane (Wulfing and Davis, 1998; Delon et al., 2001). We find that actin is essential for the formation of CD2 clusters. The initial requirement for actin may be due either to interference with signaling or to an active role of actin or actomyosin in the initial steps of clustering. In this light, it is notable that F-actin is present at the center of the contact at early time points (Bunnell et al., 2001 and data not shown). However, once clusters are formed, actin is not needed for their maintenance, as depolymerization of actin does not perturb preformed CD2 clusters (Figure S3).
Our results are most consistent with clusters being formed and maintained by a network of protein-protein interactions that involve the adaptor protein LAT. LAT-deficient cells (J.CaM2) display little or no CD2 clustering, and clustering can be rescued by transfection with LAT, but not a phosphorylation-deficient mutant of LAT (LAT(Y-F)) that cannot form interactions with SH2-domain-containing proteins. These findings are consistent with results from Harder and colleagues (Harder and Kuhn, 2000; Hartgroves et al., 2003), who showed that LAT, but not phosphorylation-deficient forms of LAT, accumulates at the interface of a T cell with an anti-TCR-coated bead, although fine spatial clustering was not described in those studies. LAT is a critical hub in the T cell signaling pathway, as it mediates downstream signaling to actin, calcium, and Ras (Finco et al., 1998; Zhang et al., 1998a). LAT’s many known binding partners include proteins that are themselves adaptor molecules that in turn mediate an additional layer of protein-protein interactions. Because of its many interactions, phosphorylated LAT could plausibly trigger the formation of a protein-protein interaction network (Harder and Kuhn, 2000; Trautmann and Valitutti, 2003). However, the details of the connections between Lck, LAT, and CD2 are not clear from the published protein-protein interactions and will require further investigation.
Dynamics of Single-Molecule Exclusion and Trapping in CD2/Lck/LAT Microdomains
Single-molecule imaging studies have shown that many plasma membrane proteins do not obey simple rules for Brownian motion but rather exhibit rapid and reversible transitions between different diffusion states. Kusumi and colleagues have performed elegant analyses at high temporal resolution demonstrating constrained diffusion of proteins and lipids in localized zones of a few hundred nanometers with abrupt transitions between zones (Fujiwara et al., 2002; Ike et al., 2003). Their evidence has suggested that this “hop diffusional” motion involves corrals established by the actin cytoskeleton (Fujiwara et al., 2002; Ike et al., 2003; Nakada et al., 2003). Other investigators also have shown transient confinement behavior at a lower temporal resolution (video rate), and such behavior appeared to be related to lipid raft association in some cases (Dietrich et al., 2002), but not in others (Lommerse et al., 2004).
In our studies of single molecules, we also have observed abrupt changes between immobility and rapid diffusion. This two state behavior was particularly obvious for LAT and Lck. The relatively stationary position of CD2 clusters has allowed us to examine single-molecule trajectories relative to these membrane microdomains, and we find that transient immobilization of single LAT or Lck molecules correlates strongly with their encounters with CD2 clusters. We believe that such single-molecule immobilization reflects a binding event between the diffusing molecule and a protein partner contained within the CD2 cluster. The trapping and dissociation of molecules from the clusters are most likely the molecular events that underlie the fluorescence recovery of photobleached clusters at a population level.
Our single-molecule observations also suggest that plasma membrane proteins have restricted access to CD2/LAT/Lck clusters, as their diffusion tends to be channeled in the space outside of the clusters. The phenomenon is particularly evident for CD45, whose single-molecule trajectories rarely pass through CD2 zones, though it is probably not mediated just by steric exclusion of CD45’s large extracellular domain. Indeed, even molecules such as Lck and LAT that lack significant extracellular portions have limited access to the clusters, as evidenced by examples of channeling of these molecules outside of CD2 zones. Moreover, when they do enter the zones they are often trapped at the cluster periphery rather than at the center (Movies S7–S9). This partial exclusion and peripheral trapping might reflect a high density and degree of order among the intracellular portions of proteins in the cluster and possibly also a saturation of available binding sites.
Implications for Signaling
High local densities of signaling molecules (e.g., within the immunological synapse or lipid rafts) have been speculated to facilitate signal transduction from the TCR (Xavier et al., 1998; Janes et al., 1999; Lin et al., 1999). The microdomains described here are enriched in Lck and LAT, two key signaling proteins, and selectively stain for phosphotyrosine, suggesting that they also are sites of active signaling. The two step process of first localizing signaling molecules at the interface and then further concentrating them into microdomains is likely to create an almost “solid-state” network of the signaling machinery. Precedence for a signal transduction mechanism proceeding through a clustered and ordered array of components can be found in bacterial chemotaxis, where the receptors, kinase, phosphatase, and adaptor proteins are clustered in an almost crystalline array at one pole of the bacterium (Weis et al., 2003). In silico modeling studies have suggested that this high degree of spatial order facilitates the gain of the signaling response (Mello et al., 2004). The vertebrate phototransduction signaling network is similarly ordered, and perturbations to the system that increase the mobility of its component proteins interfere with signaling (Calvert et al., 2001).
In addition to the stimulatory effects of concentrating signaling molecules, the exclusion of CD45 from these structures might represent a form of signal enhancement, since CD45 may inhibit TCR signaling by dephosphorylating Lck and other components of the TCR cascade (Hermiston et al., 2002). Thus, CD2/LAT/Lck clusters might create protected “sanctuaries” for signaling, and kinetic partitioning of molecules into and out of these zones might determine a net balance of phosphorylation and dephosphorylation.
Our results also suggest that CD2 might facilitate TCR signaling by contributing to the organization and perhaps nucleation of signaling microdomains. CD2’s best known function is as an adhesion protein that interacts with a partner on the APC (CD48 in rodents and CD58 in humans; Dustin et al., 1997). The inclusion of CD2 in these signaling domains is surprising, since CD2 is not thought to be a central component in T cell activation (Killeen et al., 1992). However, CD2 also has been reported to enhance the TCR signal (Bierer and Hahn, 1993; Dustin et al., 1997; Bachmann et al., 1999; Green et al., 2000), and our results raise the possibility that a membrane scaffolding role for CD2 might be responsible for this effect.
Relevance to the Immunological Synapse
An important question is how antibody-induced patterns in Jurkat cells are related to the synapses observed in cell-cell or cell-bilayer contacts. In the case of antibody-mediated crosslinking, the stimulus is immobile, whereas in the interface between two cells, the peptide-MHC complexes are freely mobile. Additional interactions also occur in T cell-APC synapses, such as those between adhesion complexes and costimulatory receptors. These differences are likely to explain why signaling proteins do not coalesce into a central super-molecular activation cluster (cSMAC) in surface antibody-stimulated Jurkat cells, as they do in cell-cell and cell-bilayer conjugates. Despite these fundamental distinctions, the signaling clusters described have similarities to immunological synapses and are likely to be relevant for understanding the spatial organization of signaling components in a more physiological context. The cSMAC in the immunological synapse is enriched in LAT, Lck, and CD2 (Dustin et al., 1998; Ehrlich et al., 2002; Bonello et al., 2004) and excludes CD45 (Johnson et al., 2000; Leupin et al., 2000), as we have described here. Smaller microclusters of Lck, CD4, and the TCR also coalesce prior to synapse formation in cell-cell contacts (Krummel et al., 2000; Ehrlich et al., 2002) and might be directly related to the structures observed here. In this way the Jurkat-antibody contact might be viewed as a frozen intermediate in T cell membrane organization during signaling. Thus, the organization and dynamics of Jurkat cell signaling microdomains is likely to provide insight into signaling in other settings in the immune system.
The mechanism of molecular patterning within the immunological synapse remains a mystery, although several hypotheses have been proposed. Recent studies have demonstrated that myosin motors actively polarize cell surface components toward the APC and also might be involved in molecular sorting for synapse formation (Wulfing and Davis, 1998; Jacobelli et al., 2004). Lipid raft-mediated superclustering of proteins also has been suggested as a potential mediator of synapse formation (Janes et al., 1999; Viola et al., 1999). Theoretical studies (Qi et al., 2001; Lee et al., 2002b) also have hypothesized that the synapse could form in the absence of active, energy-expending processes simply due to differences in the binding kinetics and ectodomain sizes of cell-surface proteins. Our data further suggest that diffusional trapping based upon protein-protein interactions should be considered as another potential driver of synapse formation. The potential of inhomogeneous diffusion to create large-scale patterns of macromolecules was formalized by Turing (Turing, 1990). Recently this model has been applied to membrane dynamics (Weiss and Nilsson, 2004), where simulations have shown that transient diffusional slowing can establish large-scale clustering and segregation in two-dimensional mixtures. A better understanding of the forces that drive synapse formation will require more detailed imaging of molecular behavior in the early stages of this process. The two-color imaging approach developed here should enable an investigation of single-molecule diffusion and behavior relative to the characteristic “bullseye” pattern of the immunological synapse, both during its formation and after it is stabilized (Grakoui et al., 1999).
Experimental Procedures
Reagents and Cell Preparation
Monoclonal antibodies against CD3ε (Hit3a) and CD2 were obtained from BD Pharmingen (San Diego, Clifornia), goat anti-mouse IgG was from Molecular Probes (Eugene, Oregon), and the monoclonal mouse anti-CD45 antibody (9.4) was the gift of Art Weiss (University of California, San Francisco). Latrunculin A was obtained from Sigma Chemical Co. (St. Louis, Missouri).
Jurkat E6.1 and J.CaM2 cells were cultured and transfected by electroporation, as described elsewhere (Lin et al., 1999). pEGFP-Lck, a gift from Drs. Lauren Richie-Ehrlich and Mark Davis (Stanford Medical School), has been described (Ehrlich et al., 2002). Lck10-GFP was generated by ligation of oligonucleotide sequences encoding the N-terminal ten amino acids of Lck into the N-terminal NcoI site of EGFP, in pCI-neo (Promega, Madison, Wisconsin). The CD45RO-GFP construct, obtained from Art Weiss and Zheng Xu (University of California, San Francisco), was generated by fusing the extracellular and transmembrane domains of murine CD45RO and a linker RVKFSRKKRGGPGSGS in frame into pEGFP-N1 (Clontech, Palo Alto, California). CD2-GFP and CD2-mRFP were generated by PCR of the human CD2 gene from pCDM8-CD2 (obtained from Mike Dustin, Skirball Institute) and ligation to the N-termi-nus of the relevant fluorescent protein in pCI-neo. pEGFP-N1-LAT and pEGFP-N1-LAT(Y-F) (all nine conserved tyrosine residues [Y36, 45, 64, 110, 127, 132, 171, 191, and 226] mutated to alanine) were gifts from Art Weiss and Joe Lin (University of California, San Francisco). For preparing a nonpalmitoylated (C26/29S) form of LAT, we found that fusion of EGFP to the C terminus resulted in the mislocalization of the protein to the cytoplasm. As a result, we prepared another construct in which we fused the signal sequence from human preprolactin in-frame with EGFP to the N-terminus of LAT(C26/29S) (Art Weiss and Joe Lin), followed by ligation of this insert into pCI-neo. The resulting fusion protein was expressed at very low levels, and so we were unable to determine the population-level distribution of this mutant by confocal microscopy, although it was well suited for single-molecule imaging. pEGFP-actin was purchased from Clontech. pEGFP-TCRζ was obtained from Max Krummel (University of California, San Francisco).
Microscopy and Analysis
Single-molecule images in living cells were obtained by objective-type TIRF on a custom-built microscope using 60 mW laser illumination at 488 nm. Glass-bottom dishes (MatTek, Ashland, Massachusetts) were cleaned by immersion in 70% sulfuric acid plus 9% hydrogen peroxide over three days during which time the cleaning solution was aspirated daily, rinsed off with ddH2O, and replaced. Surfaces were then rinsed and coated overnight at 37°C with Hit3a or a control anti-mouse IgG at 10 µg/ml in PBS. Imaging was performed 18–48 hr after transient transfection of the reporter constructs. The cells were allowed to adhere to antibody-coated glass at 37°C for 10 min prior to imaging, and images were acquired during the subsequent 10 min. A constant temperature of 37°C was maintained throughout imaging using an objective heater (Bioptechs, Butler, Pennsylvania). Single-molecule images were acquired at video rate using a cooled, intensified CCD camera (Mega10-S30Z, Stanford Photonics).
Prior to analysis, standard image processing steps were carried out using ImageJ (http://rsb.info.nih.gov/ij/). Single molecules were tracked using a set of algorithms written in the IDL language, obtained from Eric Weeks (http://www.physics.emory.edu/faculty/weeks/). All subsequent numerical analysis was carried out using custom-designed MATLAB and IDL routines. Short-range diffusion coefficients were determined as described (Klopfenstein et al., 2002). In brief, mean-squared displacement (MSD) versus time plots were generated for all possible half-second intervals throughout each trajectory. The slope of the first four time points in the MSD plots was determined, and relationship MSD = 4Dt was used to derive the diffusion coefficient.
For tracking single GFP-tagged proteins relative to CD2 clusters, still images of CD2-mRFP were acquired prior to imaging of the GFP component, and the still image was overlaid onto each frame of the video sequence. Proper registration between channels was ensured by aligning the centroids of 200 nm Tetraspeck beads (Molecular Probes), which were applied to the samples prior to imaging. Camera noise and signals from CD2-poor regions of the cell surface were minimized by applying the ImageJ fast Fourier transform bandpass filter to the CD2-mRFP image. Image brightness and contrast were then adjusted manually to maximize the intensity difference between clustered and nonclustered regions of the cell surface, and an intensity threshold was applied to binarize the image.
The bulk localization of fluorescently tagged proteins (e.g., Figure 2) was observed in living cells that were transiently cotransfected with EGFP and mRFP fusion proteins, using a Zeiss LSM 510 scanning confocal microscope with a 63x oil immersion objective at 37°C.
Supplementary Material
Acknowledgments
We thank Jeremy Ebstein for assistance with molecular biology, N. Stuurman and R. Kelso for assistance with microscopy and analysis, and A. Weiss, M. Dustin, and M. Krummel for reagents and helpful discussions. We also thank L. Richie-Ehrlich and M. Davis for experimental help in an early stage of this project. Animation in Movie S10 is by Janet Iwasa (janet@onemicron.com). This work was supported in part by a grant from the Sandler Program in Basic Sciences.
Footnotes
Supplemental Data
Supplemental Data include five figures, one table, and ten movies and can be found with this article online at http://www.cell.com/cgi/content/full/121/6/937/DC1/.
References
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Methods Enzymol. 2003;361:1–33. doi: 10.1016/s0076-6879(03)61003-7. [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Barner M, Kopf M. CD2 sets quantitative thresholds in T cell activation. J. Exp. Med. 1999;190:1383–1392. doi: 10.1084/jem.190.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badour K, Zhang J, Shi F, McGavin MK, Rampersad V, Hardy LA, Field D, Siminovitch KA. The Wiskott-Aldrich syndrome protein acts downstream of CD2 and the CD2AP and PSTPIP1 adaptors to promote formation of the immunological synapse. Immunity. 2003;18:141–154. doi: 10.1016/s1074-7613(02)00516-2. [DOI] [PubMed] [Google Scholar]
- Bierer BE, Hahn WC. T cell adhesion, avidity regulation and signaling: a molecular analysis of CD2. Semin. Immunol. 1993;5:249–261. doi: 10.1006/smim.1993.1029. [DOI] [PubMed] [Google Scholar]
- Bonello G, Blanchard N, Montoya MC, Aguado E, Langlet C, He HT, Nunez-Cruz S, Malissen M, Sanchez-Madrid F, Olive D, et al. Dynamic recruitment of the adaptor protein LAT: LAT exists in two distinct intracellular pools and controls its own recruitment. J. Cell Sci. 2004;117:1009–1016. doi: 10.1242/jcs.00968. [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Kapoor V, Trible RP, Zhang W, Samelson LE. Dynamic actin polymerization drives T cell receptor-induced spreading: A role for the signal transduction adaptor LAT. Immunity. 2001;14:315–329. doi: 10.1016/s1074-7613(01)00112-1. [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Govardovskii VI, Krasnoperova N, Anderson RE, Lem J, Makino CL. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 2001;411:90–94. doi: 10.1038/35075083. [DOI] [PubMed] [Google Scholar]
- Delon J, Kaibuchi K, Germain RN. Exclusion of CD43 from the immunological synapse is mediated by phosphorylation-regulated relocation of the cytoskeletal adaptor moesin. Immunity. 2001;15:691–701. doi: 10.1016/s1074-7613(01)00231-x. [DOI] [PubMed] [Google Scholar]
- Dietrich C, Yang B, Fujiwara T, Kusumi A, Jacobson K. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 2002;82:274–284. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Golan DE, Zhu DM, Miller JM, Meier W, Davies EA, van der Merwe PA. Low affinity interaction of human or rat T cell adhesion molecule CD2 with its ligand aligns adhering membranes to achieve high physiological affinity. J. Biol. Chem. 1997;272:30889–30898. doi: 10.1074/jbc.272.49.30889. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, Widder P, Rosenberger F, van der Merwe PA, Allen PM, Shaw AS. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T cell contacts. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- Ehrlich LI, Ebert PJ, Krummel MF, Weiss A, Davis MM. Dynamics of p56lck translocation to the T cell immunological synapse following agonist and antagonist stimulation. Immunity. 2002;17:809–822. doi: 10.1016/s1074-7613(02)00481-8. [DOI] [PubMed] [Google Scholar]
- Finco TS, Kadlecek T, Zhang W, Samelson LE, Weiss A. LAT is required for TCR-mediated activation of PLCγ1 and the Ras pathway. Immunity. 1998;9:617–626. doi: 10.1016/s1074-7613(00)80659-7. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alar-con B. Recruitment of Nck by CD3e reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- Glebov OO, Nichols BJ. Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nat. Cell Biol. 2004;6:238–243. doi: 10.1038/ncb1103. [DOI] [PubMed] [Google Scholar]
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Green JM, Karpitskiy V, Kimzey SL, Shaw AS. Coordinate regulation of T cell activation by CD2 and CD28. J. Immunol. 2000;164:3591–3595. doi: 10.4049/jimmunol.164.7.3591. [DOI] [PubMed] [Google Scholar]
- Harder T, Kuhn M. Selective accumulation of raft-associated membrane protein LAT in T cell receptor signaling assemblies. J. Cell Biol. 2000;151:199–208. doi: 10.1083/jcb.151.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartgroves LC, Lin J, Langen H, Zech T, Weiss A, Harder T. Synergistic assembly of linker for activation of T cells signaling protein complexes in T cell plasma membrane domains. J. Biol. Chem. 2003;278:20389–20394. doi: 10.1074/jbc.M301212200. [DOI] [PubMed] [Google Scholar]
- Heerklotz H. Triton promotes domain formation in lipid raft mixtures. Biophys. J. 2002;83:2693–2701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, Weiss A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J. Clin. Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat. Immunol. 2003;4:749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- Ike H, Kosugi A, Kato A, Iino R, Hirano H, Fujiwara T, Ritchie K, Kusumi A. Mechanism of Lck recruitment to the T-cell receptor cluster as studied by single-molecule-fluorescence video imaging. ChemPhysChem. 2003;4:620–626. doi: 10.1002/cphc.200300670. [DOI] [PubMed] [Google Scholar]
- Jacobelli J, Chmura SA, Buxton DB, Davis MM, Krummel MF. A single class II myosin modulates T cell motility and stopping, but not synapse formation. Nat. Immunol. 2004;5:531–538. doi: 10.1038/ni1065. [DOI] [PubMed] [Google Scholar]
- Janes PW, Ley SC, Magee AI. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Bromley SK, Dustin ML, Thomas ML. A supramolecular basis for CD45 tyrosine phosphatase regulation in sustained T cell activation. Proc. Natl. Acad. Sci. USA. 2000;97:10138–10143. doi: 10.1073/pnas.97.18.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabouridis PS, Magee AI, Ley SC. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 1997;16:4983–4998. doi: 10.1093/emboj/16.16.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen N, Stuart SG, Littman DR. Development and function of T cells in mice with a disrupted CD2 gene. EMBO J. 1992;11:4329–4336. doi: 10.1002/j.1460-2075.1992.tb05532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel MF, Sjaastad MD, Wulfing C, Davis MM. Differential clustering of CD4 and CD3zeta during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- Lee KH, Holdorf AD, Dustin ML, Chan AC, Allen PM, Shaw AS. T cell receptor signaling precedes immunological synapse formation. Science. 2002a;295:1539–1542. doi: 10.1126/science.1067710. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Hori Y, Groves JT, Dustin ML, Chakraborty AK. Correlation of a dynamic model for immunological synapse formation with effector functions: two pathways to synapse formation. Trends Immunol. 2002b;23:492–499. doi: 10.1016/s1471-4906(02)02285-8. [DOI] [PubMed] [Google Scholar]
- Lee KH, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, Sims TN, Burack WR, Wu H, Wang J, et al. The immunological synapse balances T cell receptor signaling and degradation. Science. 2003;302:1218–1222. doi: 10.1126/science.1086507. [DOI] [PubMed] [Google Scholar]
- Leupin O, Zaru R, Laroche T, Muller S, Valitutti S. Exclusion of CD45 from the T cell receptor signaling area in antigen-stimulated T lymphocytes. Curr. Biol. 2000;10:277–280. doi: 10.1016/s0960-9822(00)00362-6. [DOI] [PubMed] [Google Scholar]
- Lin J, Weiss A. Identification of the minimal tyrosine residues required for linker for activation of T cell function. J. Biol. Chem. 2001;276:29588–29595. doi: 10.1074/jbc.M102221200. [DOI] [PubMed] [Google Scholar]
- Lin J, Weiss A, Finco TS. Localization of LAT in gly-colipid-enriched microdomains is required for T cell activation. J. Biol. Chem. 1999;274:28861–28864. doi: 10.1074/jbc.274.41.28861. [DOI] [PubMed] [Google Scholar]
- Lommerse PH, Blab GA, Cognet L, Harms GS, Snaar-Jagalska BE, Spaink HP, Schmidt T. Single-molecule imaging of the H-ras membrane-anchor reveals domains in the cytoplasmic leaflet of the cell membrane. Biophys. J. 2004;86:609–616. doi: 10.1016/S0006-3495(04)74139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello BA, Shaw L, Tu Y. Effects of receptor interaction in bacterial chemotaxis. Biophys. J. 2004;87:1578–1595. doi: 10.1529/biophysj.104.042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: Elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, Kusumi A. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 2003;5:626–632. doi: 10.1038/ncb1009. [DOI] [PubMed] [Google Scholar]
- Qi SY, Groves JT, Chakraborty AK. Synaptic pattern formation during cellular recognition. Proc. Natl. Acad. Sci. USA. 2001;98:6548–6553. doi: 10.1073/pnas.111536798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld A, Simons K. The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta. 1998;1376:467–479. doi: 10.1016/s0304-4157(98)00019-7. [DOI] [PubMed] [Google Scholar]
- Roose JP, Diehn M, Tomlinson MG, Lin J, Alizadeh AA, Botstein D, Brown PO, Weiss A. T cell receptor-independent basal signaling via Erk and Abl kinases suppresses RAG gene expression. PLoS Biol. 2003;1:e53. doi: 10.1371/journal.pbio.0000053. 10.1371/journal.pbio.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- Schutz GJ, Schindler H, Schmidt T. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 1997;73:1073–1080. doi: 10.1016/S0006-3495(97)78139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Varma R, Sarasij RC, Ira X, Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- Shenoy-Scaria AM, Dietzen DJ, Kwong J, Link DC, Lublin DM. Cysteine3 of Src family protein tyrosine kinase determines palmitoylation and localization in caveolae. J. Cell Biol. 1994;126:353–363. doi: 10.1083/jcb.126.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura N, Nagafuku M, Minaki Y, Umeda Y, Hayashi F, Sakakura J, Kato A, Liddicoat DR, Ogata M, Hamaoka T, Kosugi A. Dynamic changes in the mobility of LAT in aggregated lipid rafts upon T cell activation. J. Cell Biol. 2003;160:125–135. doi: 10.1083/jcb.200207096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann A, Valitutti S. The diversity of immunological synapses. Curr. Opin. Immunol. 2003;15:249–254. doi: 10.1016/s0952-7915(03)00040-2. [DOI] [PubMed] [Google Scholar]
- Turing AM. The chemical basis of morphogenesis. 1953. Bull. Math. Biol. 1990;52:153–197. doi: 10.1007/BF02459572. [DOI] [PubMed] [Google Scholar]
- Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Viola A, Schroeder S, Sakakibara Y, Lanzavecchia A. T lymphocyte costimulation mediated by reorganization of membrane microdomains. Science. 1999;283:680–682. doi: 10.1126/science.283.5402.680. [DOI] [PubMed] [Google Scholar]
- Weis RM, Hirai T, Chalah A, Kessel M, Peters PJ, Subramaniam S. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 2003;185:3636–3643. doi: 10.1128/JB.185.12.3636-3643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Nilsson T. In a mirror dimly: tracing the movements of molecules in living cells. Trends Cell Biol. 2004;14:267–273. doi: 10.1016/j.tcb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Wulfing C, Davis MM. A receptor/cytoskeletal movement triggered by costimulation during T cell activation. Science. 1998;282:2266–2269. doi: 10.1126/science.282.5397.2266. [DOI] [PubMed] [Google Scholar]
- Xavier R, Brennan T, Li Q, McCormack C, Seed B. Membrane compartmentation is required for efficient T cell activation. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- Yang H, Reinherz EL. Dynamic recruitment of human CD2 into lipid rafts. Linkage to T cell signal transduction. J. Biol. Chem. 2001;276:18775–18785. doi: 10.1074/jbc.M009852200. [DOI] [PubMed] [Google Scholar]
- Yurchak LK, Sefton BM. Palmitoylation of either Cys-3 or Cys-5 is required for the biological activity of the Lck tyrosine protein kinase. Mol. Cell. Biol. 1995;15:6914–6922. doi: 10.1128/mcb.15.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998a;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Trible RP, Samelson LE. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998b;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- Zhu M, Shen S, Liu Y, Granillo O, Zhang W. Cutting edge: localization of linker for activation of T cells to lipid rafts is not essential in T cell activation and development. J. Immunol. 2005;174:31–35. doi: 10.4049/jimmunol.174.1.31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.